Abstract
Cytoplasmic face-mediated integrin inside-out activation remains a paradigm in transmembrane signal transduction. Emerging evidence suggests that this process involves dissociation of the complex between the integrin cytoplasmic tails; however, a dynamic image of how it occurs on the membrane surface remains elusive. We show here that, whereas membrane-proximal helices of integrin α/β cytoplasmic tails associate in cytoplasm-like aqueous medium, they become partially embedded into membranemimetic micelles when unclasped. Membrane embedding induces substantial structural changes of the cytoplasmic tails as compared to their aqueous conformations and suggests there may be an upward movement of the membrane-proximal helices into the membrane during their separation. We further demonstrate that the β3 tail exhibits additional membrane binding site at its C terminus containing the NPLY motif. Talin, a key intracellular integrin activator, recognizes this site as well as the membrane-proximal helix, thereby promoting cytoplasmic tail separation along the membrane surface. These data provide a structural basis of membrane-mediated changes at the cytoplasmic face in regulating integrin activation and signaling.
Keywords: NMR, micelles, transmembrane signaling, cell adhesion, receptor
Integrins are a major family of α/β heterodimeric transmembrane receptors and are essential for the development and survival of multicellular organisms (1, 2). They function by interacting with matrix proteins through their large extracellular domain and with intracellular proteins by means of their small cytoplasmic domain composed of the C-terminal ≈20–50 residues of each subunit. In this manner, integrins link the exterior and interior of the cell to regulate a variety of cellular processes including adhesion, spreading, and migration. Typical of other cell surface receptors, integrins can transduce signals from outside the cell into the cytoplasm on binding of extracellular ligands (outside-in signaling). Unique to integrins is their capacity to also transduce inside-out signaling; i.e., integrins are normally expressed on the cell surface in a low-affinity state, but signals received from other cell surface receptors, such as G protein-coupled receptors, are transmitted from the integrin cytoplasmic face to the extracellular domain, thereby increasing the affinity/avidity of the receptors for ligands (integrin activation) (2). The molecular mechanism by which the cytoplasmic face mediates such conformation-based inside-out signaling has been under intensive investigation over the past decade (3). These studies have led to a model where the α and β cytoplasmic tails associate to form a clasp in maintaining the receptor at a low-affinity state, whereas cellular activators such as talin induce the dissociation of the clasp during inside-out activation (4–7). This unclasping hypothesis is supported by NMR data (7), biochemical data (8, 9), and a most recent fluorescence energy transfer experiment in vivo (10). However, although the detailed structural basis of integrin cytoplasmic clasp in regulating integrin activation has been elucidated on integrin αIIbβ3 (7), the extension of the activation mechanism to other integrins and, more importantly, the structural events that occur subsequent to unclasping to sustain the active conformer remain largely unclear.
To determine whether the unclasping hypothesis is a general mechanism for integrin activation, we have performed NMR experiments on two representative integrins: α5β1, a member of the largest integrin subfamily (the β1 subfamily), and αvβ3, an integrin implicated in diverse biological responses including angiogenesis, apoptosis, and tumor growth (1, 2), both of which can be activated in a talin-dependent manner, as recently demonstrated by using small interference RNA technology (11). We show that the α/β cytoplasmic tails of these integrins also associate in aqueous solution and dissociate on binding to talin, thus establishing that cytoplasmic unclasping is a general mechanism for initiating integrin inside-out activation. Furthermore, to gain a detailed molecular understanding of how cytoplasmic tail unclasping occurs at the membrane surface, we have characterized in detail the membrane-anchoring and structural properties of integrin αIIb/β3 cytoplasmic tails in membrane-mimetic micelles. We show that, although membrane-proximal helices of αIIb/β3 tails associate in a cytoplasm-like aqueous medium, they become partially embedded into membrane in the unclasped state, thereby inducing substantial conformational change on each tail. This provides new structural insight into the integrin activation where the membrane-proximal helices may move upward into the membrane on separation. We further show that the β3 tail contains an unanticipated additional membrane-anchoring site in the vicinity of a major docking site for several integrin-binding proteins, including talin. We demonstrate that talin binds to this site as well as to the membrane-proximal helix. These data allow us to extend the unclasping hypothesis to incorporate mechanisms for how talin facilitates cytoplasmic tail separation and movement along the membrane surface and how separation is sustained to maintain the activated state of the receptor.
Methods
Protein and Peptide Preparation. Peptides corresponding to the cytoplasmic tails of αIIb, αv, α5, β1-N, and β3-N (C-terminal truncations of the β3 and β1 tails) were synthesized by the LRI Biotechnology Core (Cleveland; see Fig. 1A for sequences). Large quantities of full-length β3 tail and β3 tail mutants were synthesized by using the procedure described (12). All peptides were HPLC purified and their molecular weights were confirmed by MS. Recombinant and/or isotope-labeled αIIb tail, β3 tail, maltose-binding protein (MBP)-β1 tail, MBP-β3 tail, talin head domain (1–429), and talin F2-F3 (206–429) were all prepared by using described procedures (7).
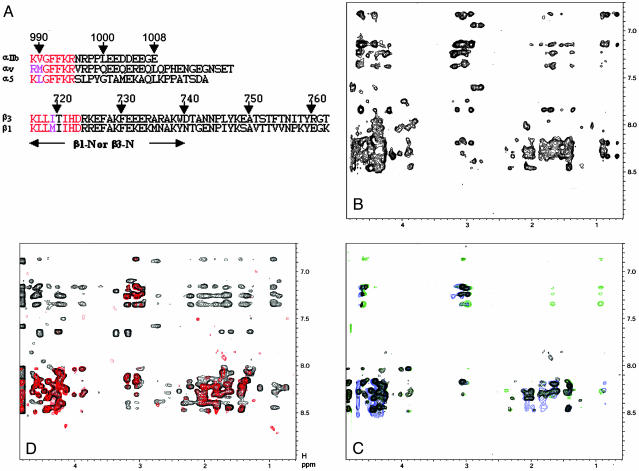
Fig. 1.
Association and dissociation of integrin αv/β3 and α5/β1 tails. (A) Primary sequences of the αIIb, αv, α5, β1, and β3 tails. Highly conserved residues involved in the membrane-proximal interface are indicated by red (identical) and pink (similar). The lengths of β1-N and β3-N with C-terminal truncations are indicated by the arrow. (B) The 2D NOESY spectrum of α5 tail in the presence of MBP-β1(α5:MBP-β1 = 1 mM:0.1 mM), showing substantial transferred NOEs due to α5/β1 tail association. (C) The 2D NOESY spectra of 1 mM α5 tail in free form (black) and in the presence of 0.1 mM MBP-β1/0.2 mM talin-H (blue) or 0.1 mM MBP-β1/0.2 mM talin F2-F3 (green). Clearly, talin-H or talin F2-F3 abolished the transferred NOEs. (D) Mathematical addition of 2D NOESY spectra of 1 mM α5 and β1-N (red, random-coiled pattern) vs. 2D NOESY spectrum of α5/β1-N mixture in 1 mM:1 mM ratio (black, highly structured with substantial number of NOEs). Note that no fusions were attached to these peptides. Mixing time is 400 ms for all experiments, which were performed at 25°C in 20 mM phosphate buffer, 5 mM Ca2+/pH 6.3.
NMR Sample Preparation. To examine α5/β1 tail association using a transferred nuclear Overhauser effect (NOE) experiment, a solution of 1 mM unlabeled α5 tail was prepared in the absence or presence of 0.1 mM MBP-β1 in 20 mM phosphate buffer, 5 mM Ca2+, pH 6.3. To examine how talin-H perturbs the α5/β1 tail interaction using the transferred NOE method, a solution of 1 mM unlabeled α5 tail was prepared in the presence of 0.1 mM MBP-β1 and 0.2 mM talin-H in 20 mM phosphate buffer/5 mM Ca2+, pH 6.3. To examine α5/β1-N tail association, a solution of 1 mM α5 or β1-N was prepared in the absence and presence of the corresponding 1 mM β1-N or α5, respectively, in 20 mM phosphate buffer/5 mM Ca2+, pH 6.3. The same procedure was used to study αv/β3 association and dissociation. The sample conditions for examining heteronuclear single quantum correlation of β3 tail/talin-F2–F3 interaction was: unlabeled talin-F2-F3:15N labeled β3 = 0.4 mM:0.2 mM = 2:1 in 20 mM phosphate buffer, 1 mM CaCl2/50 mM NaCl, pH 6.3.
To characterize the structures and membrane-binding properties of αIIb and β3 tails, 1 mM 15N- and/or 13C-labeled αIIb or β3 tail was dissolved in 50, 100, 150, 300 mM, 500 mM deuterated dodecylphosphocholine (DPC) in 20 mM phosphate buffer/5 mM Ca2+, pH 6.3. The 300 mM DPC was chosen as the optimal condition for binding (spectra no longer change when adding more DPC). To detect the intermolecular NOEs between 15N/13C-labeled αIIb/β3 tails and DPC, nondeuterated DPC was used.
NMR Spectroscopy. All heteronuclear NMR experiments were performed as described in refs. 13 and 14. All NMR experiments were performed on Varian Inova 500- or 600-MHz spectrometers equipped with triple resonance probe and shielded z-gradient unit. Pulsed-field gradients were incorporated in all experiments for solvent suppression as WATERGATE. All parameters for transferred NOE and heteronuclear single quantum correlation experiments were described previously (7). The 2D NOESY of α5/β1-N and αv/β3-N had a mixing time of 300 ms. The resonance assignments and NOE analyses (τm = 150 ms) of 15N/13C-labeled αIIb/β3 tails in DPC were made by using standard triple resonance experiments at 40°C. Intermolecular NOEs between DPC and 15N/13C-labeled β3 were obtained by using 3D 15N/13C-filtered (F1) NOESY with a mixing time of τm = 100 msec. All of the spectra were processed with nmrpipe (15) and visualized with pipp (16). Structure calculations of αIIb/β3 tails in DPC were performed as described (7, 12).
Surface Plasmon Resonance Spectroscopy (SPR). All SPR-based biosensor technology experiments were carried out on a Biacore-1000 instrument (LRI Biotechnology Core). Peptides were coupled to carboxymethylated CM5 chips by means of the SH2 function group of N-terminal cysteines following standard manufacturer's procedures {1387}{1389}{1390}. Data analysis was performed by using biaevaluation software (Ver. 3.0) provided by Biacore.
Results and Discussion
We have previously used multiple NMR approaches to demonstrate the αIIb/β3 cytoplasmic tail interaction (7). To investigate whether such interaction occurs with α5/β1 and αv/β3 tails, respectively (see sequences in Fig. 1 A), we first used the most sensitive transferred NOE method, where we examine the NOE pattern of an α cytoplasmic tail in the presence and absence of β partner fused to large MBP (7). Fig. 1B shows the 2D NOESY spectrum of the α5 tail in an aqueous medium in the presence of the β1 tail fused to MBP (α5:MBP-β1 = 10:1). Substantial transferred NOEs (Fig. 1B) are clearly induced by α5/β1 interaction as compared to the random-coiled NOESY pattern for free α5 tail (Fig. 1C). The transferred NOEs were also observed for the αv/MBP–β3 tail complex (Fig. 6, which is published as supporting information on the PNAS web site). Note that the fusion protein MBP was previously shown not to be involved in the α/β tail interaction because the MBP alone did not induce any transferred NOEs to αIIb (7). To confirm the interaction between the tails, we performed another independent NMR study: the NOESY spectrum of a mixture of the α5 tail and a C-terminally truncated β1 tail (β1-N) (no fusion attached to either α5 or β1-N) was compared to the NOESY spectrum obtained by mathematically adding the individual α5 and β1-N spectra together. This approach has been effectively used to detect αIIb/β3 tail interactions (17). Fig. 1D shows that the mathematical addition of individual α5 and β1-N NOESY spectra is significantly different from the NOESY of the α5/β1-N mixture. The actual NOESY spectrum of the mixture exhibits substantially more NOEs (Fig. 1D), reflecting the α5/β1–N tail interaction. Of note, the C-terminal β tail is clearly not involved in the complex formation, consistent with the structure of the αIIbβ3 cytoplasmic complex where the complex interface is restricted to the highly conserved membrane-proximal regions of the α/β tails (7).
The talin head domain, composed of a four-point-one, ezrin, radixin, and moesin domain containing three subdomains, F1, F2, and F3, or a smaller fragment of talin head domain, talin F2-F3, activates integrin αIIbβ3 by binding to the β cytoplasmic tails (7, 18, 19). To investigate how talin acts on integrin α5/β1 and αv/β3 tail complexes, we examined the transferred NOE effect by adding the talin head domain or talin F2-F3 to α5/MBP-β1 or αv/MBP-β3 mixtures. Fig. 1C demonstrates that either the talin head domain or talin F2-F3 completely quenches the transferred NOEs for α5 to its β1 tail partner. The same effect was observed with the αv/β3 cytoplasmic complex (Fig. 6) and the αIIb/β3 cytoplasmic complex (7). Hence, cytoplasmic clasping and talin-mediated unclasping occur with multiple integrins.
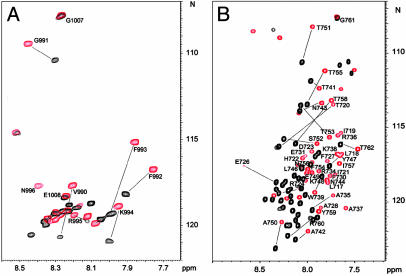
Although the structure of the αIIb/β3 tail complex in aqueous solution provided insight into the resting cytoplasmic face of the receptor (7), the structure of the active cytoplasmic face in the unclasped state is unknown, which limits our understanding of integrin activation at a molecular level. With the above additional data indicating that the unclasped state occurs as an important step in the activation of multiple integrins, it becomes essential to determine the structure of the free α and β tails and their relationship to the membrane. To investigate this, we dissolved αIIb and β3 tail peptides in DPC detergent micelles. DPC has the same zwitterionic phosphocholine head group as the dominant phospholipids of most eukaryotic membranes and has been used extensively as a membrane mimetic for NMR studies (20, 21). We have previously shown that the N-terminal myristoylated αIIb tail can be incorporated into DPC micelles and acquires structure as contrasted to the random coil that is observed in aqueous solution (12). Surprisingly, DPC induced substantial spectral changes for both nonmyristoylated αIIb and β3 tails (Fig. 2), demonstrating that each of the tail peptides bound to DPC. Resonance assignments of αIIb showed that only its membrane-proximal K989-R997 region is substantially perturbed on binding to DPC (Fig. 2 A), indicating that this region is involved in interacting with membrane. On the other hand, essentially all of the resonances in β3 are significantly perturbed on binding to DPC (Fig. 2B), suggesting that β3 underwent a more complex conformational rearrangement than αIIb. To understand how exactly DPC binds to the peptides, we performed 15N/13C-filtered NOESY experiments on αIIb and β3, each dissolved in protonated DPC. Detailed analyses showed that side chains of αIIb K989-F993 (Fig. 7, which is published as supporting information on the PNAS web site) and β3 K716-I721 exhibit extensive NOE contacts with hydrophobic methylene groups of DPC (Fig. 3), indicating that these regions are fully inserted into the membrane. The insertion regions are consistent with those predicted previously by glycosylation analysis on the individual α/β cytoplasmic peptides to map the boundaries of their membrane insertion (22) and contrast with the boundaries maintained when the α/β cytoplasmic peptides are in complex representing the integrin in its nonactivated state (7).
Fig. 2.
Spectral perturbation for αIIb/DPC and β3/DPC interactions. (A) Heteronuclear single quantum correlation (HSQC) spectra of 15N-labeled αIIb in the absence (black) and presence (red) of 300 mM DPC showing dramatic perturbation in the membrane-proximal region K989-R997. (B) HSQC spectra of 15N-labeled β3 in the absence (black) and presence (red) of 300 mM DPC. A global spectral perturbation occurs indicating a complex structural rearrangement as compared to αIIb in A.
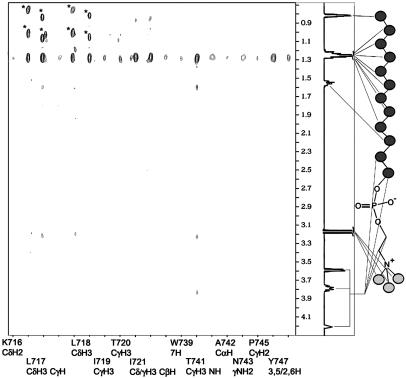
Fig. 3.
Selective strip plot of intermolecular NOEs between β3 and protonated DPC. The structure and chemical-shift assignment of DPC are shown on the right. Circles in DPC represent CH2 or CH3 groups. Signals marked by * are diagonal peaks.
Surprisingly, the filtered NOESY experiments also revealed an additional DPC-binding site on the β3 tail at its NPLY region (Fig. 3). A β3 C-terminal peptide containing the NPLY region (with no membrane-proximal region) also binds to DPC, thus providing additional evidence for membrane anchoring involving the NPLY motif (Fig. 8, which is published as supporting information on the PNAS web site). In comparison with the membrane-proximal regions, this region does not have extensive contacts with DPC (Fig. 3). The major contacts at this site appear to involve the side chains of T741 and Y747 (strong NOEs) (Fig. 3). These two side chains interact with each other in the β3 tail structure (see below) to stabilize the NPLY turn, indicating structurally how this region might orient and anchor onto the membrane surface. The membrane anchoring of the NPLY region has important functional implications: because the cytoplasmic tails are separated during integrin activation (7, 8, 10), the membrane anchoring of this region may restrict the movement of the β3 tail during tails separation. However, because this region overlaps with the talin-binding site (7, 23), talin binding could relieve this additional membrane-anchoring constraint, which then facilitates the movement of the β3 tail during tail separation. Consistently, although β3 D723R mutation would induce cytoplasmic unclasping for constitutive inside-out integrin activation (7), talin knock-down suppressed integrin activation (11), suggesting that the unclasping-induced lateral movement of β3 tail is restricted due to the membrane anchoring of the NPLY region. The membrane detachment of this region by talin may also allow it to bind to other proteins such as kinases for phosphorylation as well as to adaptors involved in outside-in signaling (24).
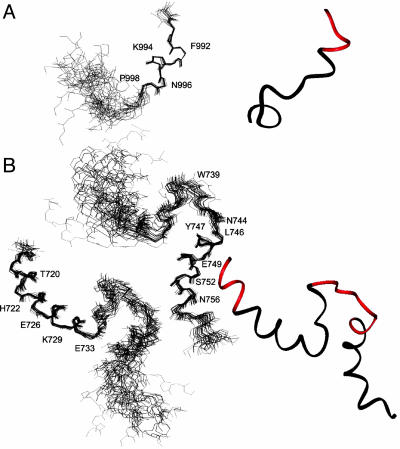
To gain more detailed structural insight into the membrane-anchoring topology of cytoplasmic tails, we determined the NMR structures of αIIb/β3 tails in DPC (see Table 1 for structural statistics). As expected, αIIb K899-N996 adopts a helical structure. However, in contrast to the “closed/inactive” αIIb structure obtained for the myristoylated form (12), which was also observed for this tail in the αIIb/β3 complex in aqueous solution (7), the C-terminal part of nonmyristoylated αIIb tail does not fold back to interact with the membrane-proximal region (Fig. 4A). The DPC embedding of K989-F993 in the αIIb tail clearly precludes interaction between F993 in the membrane proximal region and P1000 and the methylene group of D1004 in the C-terminal region, as observed previously (7, 12). It is very likely that the long and hydrophobic myristoyl group previously used to anchor the αIIb tail onto the DPC allowed for little or only partial membrane embedding of αIIb F993, which permitted its interaction with the C-terminal loop, thus forming the “closed” conformation (12) seen in the tail complex (7).
Table 1. Structural statistics for αIIb and β3 tails.
| αIIb | β3 | ||
|---|---|---|---|
| NOE distance constraints | |||
| All | 198 | 926 | |
| Sequential (|i - j| = 1) | 53 | 306 | |
| Medium (1 <|i - j| ≤ 5) | 28 | 218 | |
| Long range (|i - j| > 5) | 0 | 13 | |
| Intraresidue | 117 | 399 | |
| Largest rms deviation from experimental distance restraint, Å | 0.109 ± 0.008 | 0.077 ± 0.007 | |
| rms deviation from idealized covalent geometry | |||
| Bonds, Å | 0.010 ± 0.004 | 0.009 ± 0.001 | |
| Angles, ° | 1.406 ± 0.048 | 1.102 ± 0.013 | |
| Impropers, ° | 0.953 ± 0.090 | 2.082 ± 0.010 | |
| EL-J (kcal·mol-1)* | -26.4 ± 7.5 | -30.1 ± 4.68 | |
| procheck (Ramachandran plot) | |||
| Residues in allowed regions, % | 100 | 100 | |
| Most favored regions, % | 57.9 | 87.2 | |
| Additionally allowed regions, % | 41.4 | 12.0 | |
| Generously allowed regions, % | 0.7 | 0.0 | |
| Disallowed regions, % | 0.0 | 0.0 | |
| Average rmsd to the mean structure | Residues 990-997 | Residues 717-733 | Residues 744-756 |
| Backbone, Å | 0.23 ± 0.13 | 0.35 ± 0.11 | 0.31 ± 0.14 |
| Heavy atoms, Å | 1.31 ± 0.23 | 1.11 ± 0.18 | 0.80 ± 0.17 |
Statistics were derived from the ensemble of 20 final simulated annealing structures.
EL-J is the Lennard-Jones van der Waals energy value calculated with the charmm (Accelrys, San Diego) empirical energy function and is not included in the target function for simulated annealing or restrained minimization.
Fig. 4.
Structures of αIIb and β3 cytoplasmic tails in DPC. (A) Backbone superposition of the 20 best structures and ribbon diagram of the αIIb tail. The membrane-anchoring site is highlighted in the ribbon diagram (red). (B) The 20 best structures of β3 superimposed on either N-terminal helix K716-R734 or C-terminal N744-T755. The relative orientation between the two helices is less defined due to the lack of interhelical NOEs. The membrane-anchoring sites for both tails are highlighted in the ribbon diagram (red).
The membrane-bound structure of the β3 tail is shown in Fig. 4B. The prominent features of this structure are a long membrane-proximal α-helix (K716-R734) followed by a flexible loop and another short helix at Y747-T755. One interesting, but not so surprising, feature of the membrane-proximal helix in our structure is a kink at residue D723, which allows this helix to bend slightly and brings the flexible loop into possible contact with the membrane surface (see below). An irregular structural feature in the D723 region was also suggested in a secondary structural analysis of the β3 cytoplasmic-transmembrane domain performed by Li et al. (25). However, instead of the slightly bent structure that we determined, Li et al. (25) proposed a hinge that breaks the membrane proximal helix into two parts based on the J-coupling constant and some dynamics data. Another notable difference between our study and that of Li et al. (25) is that we precisely defined membrane-anchoring sites using filtered NOESY experiments. The connecting loop between the two helices in our structure is less defined and flexible, as indicated by the absence of medium and long-range NOEs. Immediately next to the loop is the N744PLY747 motif that forms an inverse turn, which is the start point for the second helix. The turn is stabilized by long-range interaction between the T741 methyl group and the Y747 ring. The formation of the C-terminal helix is surprising, because it was absent in the structure of the αIIb/β3 tail complex in aqueous solution. Because no long-range NOEs were observed between the N- and C-terminal helices, the formation of the C-terminal helix is likely induced and stabilized by the docking of the NPLY turn onto DPC. The N756ITY759 motif after the helix exhibits a helical turn feature and is followed by the unstructured C terminus of the peptide.
The structures of the αIIb and β3 tails and their detailed intermolecular NOEs with DPC allowed us to construct a membrane-anchoring topology of the unclasped (active) cytoplasmic face (Fig. 5). For αIIb, the membrane-proximal K989-F993 insert into the membrane with the rest of the tail being below the membrane surface. Because of the insertion, the C-terminal acidic loop of αIIb moves away, which otherwise interacts with the insertion region (7, 12). This results in an “open” conformation of the αIIb tail, which we know to be an active form of the receptor because mutations that induce this open form activate αIIbβ3 (12). For β3, the N-terminal K716-I721 is embedded into the membrane. The C-terminal part of β3 tail, which is otherwise anchored onto the membrane surface through the NPLY region in the inactive integrin, is detached from membrane surface by talin (Fig. 5). Thus, talin likely plays a dual role in integrin inside-out signaling: it detaches the NPLY region from the membrane, and it dissociates the membrane-proximal clasp. Although crystallographic study has confirmed the direct interaction between talin F2-F3 and the NPLY motif (23), the exact talin-binding site in the membrane-proximal region is not unambiguously defined at this time, but two lines of evidence suggest that it may contain H722-R724: (i) when titrated into 15N-labeled β3 tail, talin F2-F3 noticeably perturbs only the T720-D723 region and the middle segment containing the NPLY turn (Fig. 5B); and (ii) point mutations of H722D723 or D723R724 to AA substantially reduce talin binding to β3 (Fig. 5C), independently implicating HDR in binding to talin. Because D723 forms a salt bridge with αIIb R995 (7), these data also indicate that talin disrupts the salt bridge and initiates cytoplasmic tail separation.
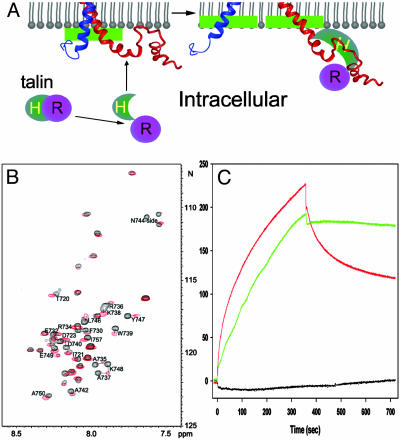
Fig. 5.
Structural illustration of integrin activation by talin. (A) A model for membrane-mediated change of cytoplasmic face during integrin activation. Agonist stimulation induces a conformational change in talin that exposes its head domain (Talin-H). Talin-H binds to the β3 tail at both the NPLY-containing region and the membrane-proximal helix. The binding to the membrane-proximal region displaces the αIIb tail from its complex with the β3 tail, leading to an unclasping, and the binding in the NPLY region releases a membrane-anchoring constraint on β3, which further facilitates the unclasping movement along the membrane surface. Notice the shifted membrane interface for both membrane-proximal helices before and after unclasping (green bars), which suggests a “fanning-out” unclasping process because the transmembrane domains may also undergo separation or open-scissor motion. The unclasping initiates the opening of the integrin C-terminal stalks, which is necessary for the rearrangement of the extracellular headpiece for high-affinity ligand binding. (B) HSQC spectra of the 15N-labeled β3 tail in the absence (black) and presence (red) of unlabeled talin F2-F3 at 35°C. Residues with significant chemical shift changes were labeled, which primarily involve membrane-proximal T720-D723 and C-terminal A735–A750, containing the N744-Y747 turn. (C) Surface plasmon resonance data. One hundred nanomol of talin-H (1–429) was passed over CM5 sensor chips coated with β3 (716–762) (red), a β3 membrane-proximal mutant (H722A/D723A, black), or a β3 single mutant (F730A, green), with association and dissociation phases of 360 sec. The former mutant had diminished binding to talin, but the latter has about the same as the binding capacity to talin, indicating that H722D723 is critical for talin binding. D723A/R724A mutations also had the same effect as H722A/D723A (data not shown). Talin-H made no detectable interaction with αIIb(989–1008) when this peptide was coupled to a CM5 chip.
The insertion of αIIb/β3 membrane-proximal regions in the dissociated state suggests an upward movement of these regions into the membrane during cytoplasmic tail separation (Fig. 5A). This process would result in more elongated transmembrane domains (Fig. 5A), which is supported by the previously proposed C-terminal separation model (7, 8, 10) where transmembrane domains are expected to be more elongated from an inactive parallel coiled-coil state (26) to an active v-shaped (7, 8, 10) or open-scissor state (2, 6). Note that the proposed cytoplasmic location of the clasped membrane-proximal regions (Fig. 5A) is not merely based on the detection of the tail complex in the cytoplasm-like aqueous solution (7) but also is supported by the following observations: (i) Although the membrane-proximal αIIb K989-R997 interacts with the C-terminal part of the αIIb tail in the αIIb/β3 tail complex in aqueous solution (7), partial membrane embedding of the former results in an “open” conformation by precluding such interaction (Fig. 4A). We know the open conformation represents the active form of αIIb, because it was observed in the activating αIIb P999A/P1000A mutant (12). Thus, αIIb adopts a “closed/inactive” conformation in cytoplasm but an “open/active” conformation when embedded into the membrane. (ii) A cryoelectron microscopy structure of inactive αIIbβ3 determined in a membrane-mimetic environment revealed that the αIIb/β3 tails are associated (26). This indicates that the membrane-proximal clasp is preserved in the intact receptor and is in the cytoplasm. The clasp would be disrupted if it were in the membrane due to the membrane embedding of the membrane-proximal interface; as shown by NMR, i.e., αIIb/β3 tail peptides weakly associate in aqueous solution but not in DPC (7). (iii) A number of integrin-activating proteins, such as talin (18), CIB (27, 28), and β3-endonexin (29), bind to the membrane-proximal regions of either αIIb or β3, suggesting that these regions are in the cytoplasm for the low-affinity state of the integrin to be accessible to these intracellular activators.
Conclusion
The data presented allow us to propose how cytoplasmic unclasping occurs along the membrane inner surface during integrin activation and inside-out signaling (Fig. 5A). On agonist stimulation, a series of intracellular signaling events are initiated that propagate to the αIIb/β3 cytoplasmic face. One of these signaling events involves induction of a conformational change in an integrin activator. In the case of talin, the change exposes its head domain such that it can bind to the β3 tail at both the NPLY-containing region and the membrane-proximal helix, as definitively shown here (Fig. 5 B and C) and suggested previously (7, 23, 30). The binding displaces the αIIb tail from its complex with the β3 tail, leading to an upward movement of the membrane-proximal regions into the membrane. Such movement may occur as a “fanning-out” unclasping process, because the transmembrane domains may also undergo separation or open-scissor motion (Fig. 5A). The binding also releases the C-terminal membrane-anchoring constraint of β3, which further facilitates the unclasping movement along the membrane surface. The unclasping or reordering of the transmembrane segments initiates the opening up of the integrin C-terminal stalks in the extracellular domain, which is necessary for the structural rearrangement of the integrin headpiece for high-affinity ligand binding (31). This model of unclasping and upward movement is consistent with the recent fluorescence energy transfer data of Kim et al. (10), suggesting the occurrence of marked separation of the tails on integrin activation in intact cells. It is also consistent with the data of Tadokoro et al. (11), indicating a prominent role of talin in the activation of integrins. Such C-terminal opening may also facilitate integrin clustering and further enhanced ligand affinity through oligomerization of the transmembrane domains, as suggested by Li et al. (32). On the other hand, it is known that ligand binding induces additional conformational change (1, 2), which is transmitted to the cytoplasmic face (outside-in signaling). Although it remains to be determined how this conformational change occurs, one possibility is that ligand binding induces a piston motion that moves the membrane-embedded regions partially or fully back out of the membrane. There are several elegant examples for such ligand-induced piston motion (33, 34). A number of proteins such as calreticulin, FAK, and Rac1 (35) have been suggested to bind to these regions in ligated integrins, and the accessibility of their binding sites would be restricted to the ligated integrin by this mechanism, thus mediating outside-in signaling.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants (to J.Q. and E.F.P.) and an American Heart Association Scientist Development Grant (to O.V.).
Abbreviations: NOE, nuclear Overhauser effect; MBP, maltose-binding protein; DPC, dodecylphosphocholine.
Data deposition: The atomic coordinates in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1S4W and 1S4X).
References
- 1.Giancotti, F. G. & Ruoslahti, E. (1999) Science 285, 1028–1032. [DOI] [PubMed] [Google Scholar]
- 2.Hynes, R. O. (2002) Cell 110, 673–687. [DOI] [PubMed] [Google Scholar]
- 3.Liddington, R. C. & Ginsberg, M. H. (2002) J. Cell Biol. 158, 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Toole, T. E., Mandelman, D., Forsyth, J., Shattil, S. J. & Plow, E. F. (1991) Science 254, 845–847. [DOI] [PubMed] [Google Scholar]
- 5.O'Toole, T. E., Katagiri, Y., Faull, R. J., Peter, K., Tamura, R., Quaranta, V., Loftus, J. C., Shattil, S. J. & Ginsberg, M. H. (1994) J. Cell. Biol. 124, 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes, P. E., Diaz-Gonzalez, F., Leong, L., Wu, C., McDonald, J. A., Shattil, S. J. & Ginsberg, M. H. (1996) J. Biol. Chem. 271, 6571–6574. [DOI] [PubMed] [Google Scholar]
- 7.Vinogradova, O., Velyvis, A., Velyviene, A., Hu, B., Haas, T., Plow, E. F. & Qin, J. (2002) Cell, 110, 587–597. [DOI] [PubMed] [Google Scholar]
- 8.Takagi, J., Erickson, H. P. & Springer, T. A. (2001) Nat. Struct. Biol. 8, 412–416. [DOI] [PubMed] [Google Scholar]
- 9.Lu, C., Takagi, J. & Springer, T. A. (2001) J. Biol. Chem. 276, 14642–14648. [DOI] [PubMed] [Google Scholar]
- 10.Kim, M., Carman, C. V. & Springer, T. A. (2003) Science 301, 1720–1725. [DOI] [PubMed] [Google Scholar]
- 11.Tadokoro, S., Shattil, S. J., Eto, K., Tai, V., Liddington, R. C., de Pereda, J. M., Ginsberg, M. H. & Calderwood, D. A. (2003) Science 302, 103–106. [DOI] [PubMed] [Google Scholar]
- 12.Vinogradova, O., Haas, T., Plow, E. F. & Qin, J. (2000) Proc. Natl. Acad. Sci. USA 97, 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clore, G. M. & Gronenborn, A. M. (1998) Trends Biotechnol. 16, 22–34. [DOI] [PubMed] [Google Scholar]
- 14.Ferentz, A. E. & Wagner, G. (2000) Q. Rev. Biophys. 33, 29–65. [DOI] [PubMed] [Google Scholar]
- 15.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 16.Garrett, D. S., Powers, R., Gronenborn, A. M. & Clore, G. M. (1991) J. Magn. Reson. 95, 214–220. [DOI] [PubMed] [Google Scholar]
- 17.Weljie, A. M., Hwang, P. M. & Vogel, H. J. (2002) Proc. Natl. Acad. Sci. USA 99, 5878–5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calderwood, D. A, Zent, R., Grant, R., Rees, D. J., Hynes, R. O. & Ginsberg, M. H. (1999) J. Biol. Chem. 274, 28071–28074. [DOI] [PubMed] [Google Scholar]
- 19.Calderwood, D. A., Yan, B., de Pereda, J. M., Alvarez, B. G., Fujioka, Y., Liddington, R. C. & Ginsberg, M. H. (2002) J. Biol. Chem. 277, 21749–21758. [DOI] [PubMed] [Google Scholar]
- 20.Henry, G. D. & Sykes, B. D. (1994) Methods Enzymol. 239, 515–535. [DOI] [PubMed] [Google Scholar]
- 21.MacKenzie, K. R., Prestegard, J. H. & Engleman, D. M. (1997) Science 276, 131–133. [DOI] [PubMed] [Google Scholar]
- 22.Armulik, A., Nilsson, I., von Heijne G. & Johansson, S. (1999) J. Biol. Chem. 274, 37030–37034. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Alvarez, B., de Pereda, J. M., Calderwood, D. A., Ulmer, T. S., Critchley, D., Campbell, I. D., Ginsberg, M. H. & Liddington, R. C. (2003) Mol. Cell 11, 49–58. [DOI] [PubMed] [Google Scholar]
- 24.Phillips, D. R., Prasad, K. S., Manganello, J., Bao, M. & Nannizzi-Alaimo, L. (2001) Curr. Opin. Cell Biol. 13, 546–554. [DOI] [PubMed] [Google Scholar]
- 25.Li, R., Babu, C. R., Valentine, K., Lear, J. D., Wand, A. J., Bennett, J. S. & DeGrado, W. F. (2002) Biochemistry 41, 15618–15624. [DOI] [PubMed] [Google Scholar]
- 26.Adair, B. D. & Yeager, M. (2002) Proc. Natl. Acad. Sci. USA 99, 14059–14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsuboi, S. (2002) J. Biol. Chem. 277, 1919–1923. [DOI] [PubMed] [Google Scholar]
- 28.Barry, W. T., Boudignon-Proudhon, C., Shock, D. D., McFadden, A., Weiss, J. M., Sondek, J. & Parise, L. V. (2002) J. Biol. Chem. 277, 28877–28883. [DOI] [PubMed] [Google Scholar]
- 29.Eigenthaler, M., Hofferer, L., Shattil, S. J. & Ginsberg, M. H. (1997) J. Biol. Chem. 272, 7693–7698. [DOI] [PubMed] [Google Scholar]
- 30.Ulmer, T. S., Calderwood, D. A., Ginsberg, M. H. & Campbell, I. D. (2003) Biochemistry 42, 8307–8312. [DOI] [PubMed] [Google Scholar]
- 31.Takagi, J., Petre, B. M., Walz, T. & Springer, T. A. (2002) Cell 110, 598–611. [DOI] [PubMed] [Google Scholar]
- 32.Li, R., Mitra, N., Gratkowski, H., Vilaire, G., Litvinov, R., Nagasami, C., Weisel, J. W., Lear, J. D., DeGrado, W. F. & Bennett, J. S. (2003) Science 300, 795–798. [DOI] [PubMed] [Google Scholar]
- 33.Ottemann, K. M., Xiao, W., Shin, Y.-K. & Koshland, D. E., Jr. (1999) Science 285, 1751–1754. [DOI] [PubMed] [Google Scholar]
- 34.Falke, J. J. & Hazelbauer, G. L. (2001) Trends Biochem. Sci. 26, 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, S., Calderwood, D. A. & Ginsberg, M. H. (2000) J. Cell Sci. 113, 3563–3571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.