Abstract
Genetic recombination is believed to assist HIV-1 diversification and escape from host immunity and antiviral therapies, yet this process remains largely unexamined within the natural target-cell populations. We developed a method for measuring HIV-1 recombination directly that employs reporter viruses bearing functional enhanced yellow fluorescent protein (YFP) and enhanced cyan fluorescent protein (CFP) genes in which recombination produces a modified GFP gene and GFP fluorescence in the infected cells. These reporter viruses allow simultaneous quantification of the dynamics of HIV-1 infection, coinfection, and recombination in cell culture and in animal models by flow-cytometric analysis. Multiround infection assays revealed that productive cellular coinfection was subject to little functional inhibition. As a result, generation of recombinants proceeded according to the square of the infection rate during HIV-1 replication in T lymphocytes and within human thymic grafts in severe combined immunodeficient (SCID)-hu (Thy/Liv) mice. These results suggest that increases in viral load may confer a compounding risk of virus escape by means of recombinational diversification. A single round of replication in T lymphocytes in culture generated an average of nine recombination events per virus, and infection of macrophages led to ≈30 crossover events, making HIV-1 up to an order of magnitude more recombinogenic than recognized previously and demonstrating that the infected cell exerts a profound influence on the frequency of recombination.
The continual generation of genetic variation allows HIV-1 to persist in the face of host adaptive immunity and multidrug antiviral therapy. HIV-1 variants are created by means of viral nucleic acid polymerization errors (mutations) and recombination (1–3). Recombination is a shared feature among retroviruses and occurs when reverse transcriptase (RT) transfers between the two RNA templates in the diploid virion, generating a daughter DNA provirus that is a mosaic of the parental genomes (see ref. 4 for review). Coinfection of cells with divergent strains allows copackaging of nonidentical genomes in the next generation of viruses, and these heterozygous virions provide the substrate for recombinational diversification.
To date, studies examining the cellular–biochemical aspects of recombinational strand transfer events (RSTE) have depended on reporter viruses bearing antibiotic drug-resistance genes. Such studies have been restricted to single rounds of replication in adherent fibroblastic cell lines, in which the multiplicity of infection (MOI) can be assessed by enumerating the productively infected drug-resistant cells (see ref. 4 for review). However, as for many other retroviruses, HIV-1 infects immune cells, including T lymphocytes and macrophages, in which the process of reverse transcription might be influenced by lineage-specific activation, differentiation, and proliferation processes. Although abundant coinfection has been documented recently within the spleens of HIV-1-infected individuals, indicating an environment supportive of a high level of recombination (5), the kinetics of coinfection and recombination remain undefined. As a result, recombination has been so far excluded from most quantitative models of HIV-1 dynamics and evolution (1, 6–9). Here, we examine the dynamics of HIV-1 coinfection and recombination in real time in biologically relevant cell types, T lymphocytes, and macrophages, as well as an in vivo animal model.
Materials and Methods
Viruses and Molecular Clones. Reporter viruses employ enhanced yellow fluorescent protein genes (Clontech) and enhanced cyan fluorescent protein genes (Clontech). HIV-1 pNL4–3 (10) and NL4–3 hemigenomic plasmids p83–10 and p83–2 (11) were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. Hemigenomic HIV-1 89.6 plasmids were obtained from Ronald Collman. Replication competent NL4–3-derived enhanced yellow fluorescent protein (YFP) and enhanced cyan fluorescent protein (CFP) viruses are designated NLENY1-internal ribosome entry sequence (IRES) and NLENC1-IRES. For single-round infection assays in Fig. 4, envelope-defective viruses, designated NLENY1-ES-IRES and NLENC1-ES-IRES, were complemented with CXCR4-tropic NL4–3 or CCR5-tropic YU2 envelopes. To generate recombinants between NL4–3 and 89.6 strains in Fig. 3, hemigenomic plasmids NL.C and 89.Y were constructed similarly to the construction of NLENG1 described in ref. 12, except that 200 bp of the nef gene were removed.
Fig. 4.
Measurement of HIV-1 recombination frequency in various cell types. (A) HIV-1 recombination frequencies based on the appearance of the GFP* form in a single-round infection. Values along the y axis represent the percentage of the infected cells in each sample that were GFP+. Infection of two or more cell types was performed in parallel for all data collection. Data are given as the mean of multiple samples from at least three independent experiments for each cell type. (B) Influence of myeloid differentiation on recombination rates. One of three representative experiments is shown.
Fig. 3.
Measurement of HIV-1 recombination frequency in T cells by sequence analysis. (A) DNA sequence analysis of GFP* proviruses generated by a singe round of viral replication in Jurkat T cells. RSTE occurred at the junctions of yellow and blue bars, which represent regions derived from 89.6 and NL4–3, respectively. Below the bar chart is a to-scale schematic representation of the HIV-1 genome, providing a reference for the locations of the RSTE. (B) The mean and median number of crossovers in the 13 proviruses are normalized to a wild-type 9,700-bp HIV-1 genome. (C) Locations of the GFP*-generating RSTE. The mutations that generate YFP and CFP are superimposed for reference.
Cell Culture. We obtained 293T cells from the American Type Culture Collection. HeLa-CD4, Jurkat (clone EL-6), U937, and THP-1 were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. Primary human peripheral blood mononuclear cells from HIV-1 negative donors were isolated by Ficoll/Hypaque centrifugation. CD8+ cells were depleted by anti-CD8 magnetic bead separation (Dynal, Great Neck, NY). Primary monocytes were isolated by adherence of peripheral blood mononuclear cells to plastic, and macrophages were generated by culture for 4–6 days in DMEM plus 20% normal human AB serum. These cells were ≥95% CD14+/CD13+. CD4+ T cells were stimulated in RPMI-1640 with 2 μg/ml PHA-L (Roche), 50 units/ml rhIL-2 (Roche), 10% FCS (HyClone), 10% AB serum and penicillin/streptomycin (pen-strep) for 4 days before infection. We maintained 293T and HeLa-CD4 in DMEM; Jurkat, THP-1, and U937 were maintained in RPMI-1640 with 10% FBS plus pen-strep. Nucleosides (20–40 μM each of 2′-deoxycytidine, 2′-deoxyadenosine, 2′-deoxyguanosine, and thymidine) (Sigma) were added at the time of infection.
Quantitative Recombination Assays. We transfected 293T cells by using Effectene (Qiagen, Valencia, CA). In YFP and CFP cotransfected cells, the fluorescent cells were ≥90% YFP/ CFP+/+. At 24 h, the transfection medium was replaced with fresh medium. At 36–48 h, virus-containing medium was collected and then centrifuged at 1,800 × g for 20 min to remove residual cells and debris.
Flow cytometry was performed on a FACStar Plus (Becton Dickenson). GFP and YFP were excited by using an argon laser at 488 nm. GFP and YFP were detected by using a 510 ± 10-nm bandpass filter and a 550 ± 15-nm bandpass filter, respectively, and a 510-nM dichroic short-pass splitter, with electronic compensation during data collection. CFP was excited by using a spatially separated krypton laser at 405 nm and detected by using a 470 ± 10-nm bandpass filter.
Time-course assay for infection, coinfection, and recombination was performed as follows. We infected 1 × 106 PHA-L/IL-2-activated CD4+ T lymphocytes with various amounts of an equal mixture of replication-competent NLENY1-IRES and NLENC1-IRES at 1 μg/ml p24. On the days indicated in Fig. 2, one third of all cells (≈1 × 106) were analyzed by flow cytometry for YFP, CFP, and GFP expression.
Fig. 2.
Kinetics of HIV-1 replication, coinfection, and recombination in T cells. Activated CD4+ T cells were infected on day 0 with the following amounts of replication-competent homozygous YFP and CFP HIV-1 reporter viruses: 2,000 ng (red); 500 ng (blue); 200 ng (green); and 50 ng (black) p24 gag protein input dose. IL-2 was added on day 4 and on each subsequent day. A–C show one representative experiment of three independent experiments using CD4+ T lymphocytes from separate donors. D–F show results from all three experiments. (A) Percentage of all cells that were fluorescent for YFP, CFP, or GFP. (B) Coinfection rate measured as percentage of YFP/CFP+/+ cells [%YFP/CFP (+/+)]. Proportion of the infected cells that were YFP/CFP+/+. Inset shows percentage of all cells that were YFP/CFP+/+. At infection rates <10%, the percentage of cells that are YFP/CFP+/+ is very close to one-half of all coinfected cells. (C) Appearance of GFP+ cells infected with GFP* recombinant viruses. Percentage of the infected cells that were GFP+. Inset shows percentage of all cells that were GFP+.(D) The coinfection rate is proportional to nearly the square of the infection rate. Data at all time points from three independent experiments are graphed, both before and after peak infection. Theoretical square relationship is indicated by a black line. (E) The abundance of cells infected with recombinant GFP* viruses is directly proportional to the coinfection rate. Data from three independent experiments are shown. Three data sets were restricted to the days in each sample until peak infection and before target-cell depletion. The fourth data set (green triangles), represents cells from Donor 3 on days after peak infection. Black line shows theoretical linear relationship, and green line shows curve fit. (F) The emergence of recombinant viruses is proportional to the square of the infection rate. Data sets are the same as in E.(G–I) Relationships between the rates of infection, coinfection, and recombination in vivo in the SCID-hu (Thy/Liv) mouse model. We infected 12 thymic implants with YFP and CFP replication-competent reporter viruses; they were then assayed at 2–3 weeks after infection. Data are presented as in D–F. Black lines indicate theoretical square or linear functions, as indicated.
Single-round recombination assay (see Fig. 4) was performed as follows. Heterozygous, pseudotyped NLENY1-ES-IRES/NLENC1-ES-IRES viruses were generated by cotransfection with YFP and CFP genomes plus envelope expression plasmids and then applied to 1–2 × 106 target cells in 3-fold serial dilutions at levels shown to infect 0.1–10% of cells. At 48 or 96 h after infection for macrophages (recombination rates were identical at the two time points) and 48 h after infection for all other cells, up to 2 × 106 cells were analyzed. Multiple samples at the lowest MOI that generated sufficient GFP+ cells were averaged. All comparisons of recombination rates among cell types and viruses were made at equivalent infection rates. Macrophage differentiation was performed as follows. Jurkat and THP-1 cells were treated with a single dose (20 ng/ml) of the phorbol ester phorbol 12-myristate 13-acetate. U937 required two to three exposures to phorbol 12-myristate 13-acetate for >5 days to develop the full differentiated phenotype. Viruses were pseudotyped with VSV-G in this experiment to bypass the phorbol 12-myristate 13-acetate-induced CD4 down-modulation in THP-1 and U937. Plasmid DNA recombination within transfected cells used to generate stocks of heterozygous viruses resulted in ≈0.33% of the producer cells exhibiting GFP fluorescence, a rate too low to contribute significantly to the appearance of GFP+ infected cells.
Calculation of the frequency of recombinational strand transfer was performed as follows. Three factors determine the relationship between the percentage of cells that are GFP+ and the RSTE frequency. First, the distance between the critical YFP and CFP residues is 408 of 9,700 bp in a wild-type HIV-1 genome, or 4% of the total length of the virus. Second, one-half of the RSTE within this region will begin on the CFP gene and end on YFP, thus removing the two critical residues and generating a modified GFP gene (which we designate “GFP*”) sequence. Third, approximately one-half of all HIV-1 virions produced from coexpressing cells will be heterozygous, whereas the other half will be homozygous (13). Thus, 0.04 × 0.5 × 0.5 = 0.01, or 1% of all RSTE within the HIV-1 genome, will produce GFP*.
Generation and Sequence Analysis of Recombinant GFP* Genomes. Viral stocks of NL4–3-CFP and 89.6-YFP viruses from hemigenomic plasmids were generated as described (12). Heterozygous virions were produced by coinfecting CEMx174 cells with 1 μg of p24 gag of each of these viruses. At 24 h after infection, culture medium was collected and used to infect Jurkat T cells at low MOI (<0.5%). The protease inhibitor indinavir was added at 0, 24, and 48 h after injection to limit infection to a single round. At 48 h after infection, single cells that were positive for GFP fluorescence and negative for YFP and CFP were cloned by using the automatic cell-deposition unit (ACDU) of the FACStar Plus into 96-well plates, expanded to >106 cells in the presence of indinavir, and then verified as GFP+YFP-CFP- by flow cytometry. Genomic DNA from 2–5 × 106 GFP+ cells was isolated by using the Genomic DNA isolation kit (Qiagen). We amplified 8,870 bp of the HIV-1 proviruses between the LTRs by PCR using the High-Fidelity Long PCR kit (Roche), and we then cloned them into pCR-XL-TOPO (Invitrogen) by using Stbl2 competent cells (Life Technologies, Grand Island, NY) at 25°C. Most of the primers used for sequencing have been described (14), as well as additional primers specific for the GFP* region.
Severe Combined Immunodeficient (SCID)-hu (Thy/Liv) Mice. SCID-hu mice were prepared by implantation of human fetal liver and thymus under the left renal capsule as described (15). At 4–8 months after implantation, 50–200 ng of an equal mixture of NLENY1-IRES and NLENC1-IRES were injected into the thymic grafts. At 2–3 weeks after infection, single-cell suspensions of thymocytes were analyzed by flow cytometry for YFP, CFP, and GFP expression.
Results
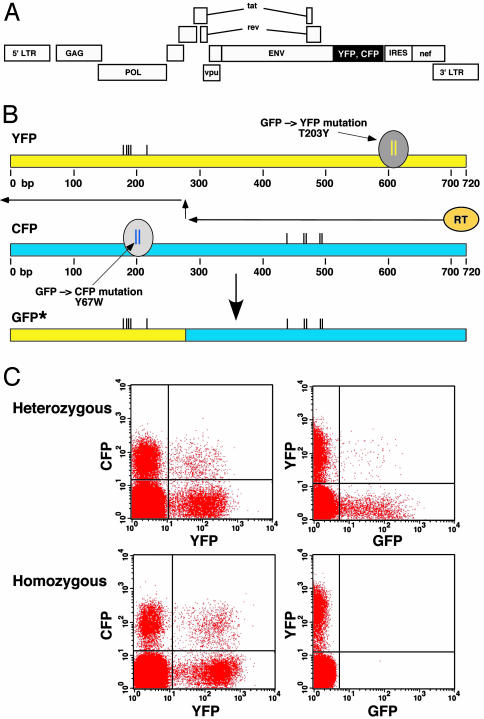
Full-length HIV-1 reporter viruses containing the genes for YFP and CFP (Fig. 1A) allow quantitation by flow cytometry of the infection frequency as the sum of all fluorescent cells, with the coinfection frequency being proportional to the percentage of cells that are YFP/CFP+/+. YFP and CFP are mutational derivatives of GFP, in which single amino acid changes 408-bp apart on the GFP backbone confer the spectral characteristics of the respective proteins (Fig. 1B) (16). When YFP and CFP heterozygous virions are generated by cotransfection of producer cells with YFP and CFP genomes, recombination between the YFP and CFP genes during reverse transcription can generate a modified gene, termed GFP*, which confers GFP fluorescence to the infected cells (Fig. 1C Upper Right). Efficient recombination only occurs between copackaged genomes because YFP and CFP viruses that have been generated independently generate very few GFP+ cells (Fig. 1C Lower Right).
Fig. 1.
System to measure HIV-1 infection and recombination. (A) Schematic representation of YFP and CFP reporter viruses. A replication-competent virus for multiround infection assays is shown. For single-round infection assays two-stop codons were introduced into the envelope reading frame after the vpu gene. YFP or CFP were inserted between the env and nef ORFs, and an IRES directs wild-type level of nef expression. (B) Generation of GFP* from YFP and CFP by means of recombination. Diagram depicts recombinational strand transfer during minus-strand DNA synthesis. The spectrum-determining mutations within YFP and CFP are indicated. The recombinant origin of GFP* genes is demonstrated by means of DNA sequence analysis by the presence of several mutations that are unique to the parental YFP and CFP genes. These mutations are indicated by the black vertical lines. (C) Three-color flow-cytometric analysis of infection, coinfection, and recombination. (Upper) Infection with heterozygous HIV-1 reporter viruses containing copackaged YFP and CFP genomes results in coinfection (Left) and the appearance of GFP+ cells by means of recombination (Right). (Lower) Infection with an equal mixture of homozygous YFP and CFP viruses generated by independent transfection of producer cells produces identical coinfection as heterozygous viruses (Left) but few, if any, GFP+ cells (Right).
To examine the dynamics of HIV-1 coinfection and recombination, we infected activated CD4+ T lymphocytes with an equal mixture of replication-competent YFP and CFP reporter viruses at four different MOIs (Fig. 2 A–F). As expected, the infection frequency rose in a time- and MOI-dependent manner and then declined as a result of target-cell depletion and loss of cell activation and permissiveness to infection (Fig. 2 A). The appearance of coinfected YFP/CFP+/+ cells tracked with the infection rate, whether the coinfected cells are counted as a percentage of all cells (Fig. 2B Inset) or as a percentage of only the infected cells (Fig. 2B). At all time points, including after peak infection, the percentage of all cells that were YFP/CFP+/+ was nearly proportional to the square of the overall infection rate (Fig. 2D). Importantly, these relationships were completely independent of the input MOI, the number of rounds of replication, or the time since cell activation, and identical results were obtained in independent experiments by using CD4+ T lymphocytes from different donors (Fig. 2D).
When equal amounts of YFP and CFP viruses are used to infect cells simultaneously, the chance of a cell becoming infected with both a YFP and a CFP virus should be the product of the probability of its being infected with either virus. In other words, in the first round of infection, it is understandable that the coinfection frequency is proportional to the square of the overall infection rate. However, during later rounds of replication, coinfection does not need to be simultaneous and may be sequential. In this situation, the opportunity for reinfection could increase the coinfection rate, whereas viral interference to reinfection could result in a lowering of the coinfection rate (17, 18). The relationship between the infection and coinfection rates showed only a slight decline from this theoretical square function at later time points. This result indicates that functionally there was very little inhibition to multiple infection events during HIV-1 replication.
Recombination was evident by day 3, based on the appearance of GFP+ cells, and was ongoing at all subsequent stages of infection (Fig. 2C) even when the percentage of all cells that were GFP+ dropped in correspondence to the overall decline in infection rates (Fig. 2C Inset). During virus expansion, i.e., before peak virus infection, the abundance of recombinant GFP* viruses (measured as the abundance of GFP+ cells) was directly proportional to the coinfection frequency (Fig. 2E) and proportional to the square of the infection frequency (Fig. 2F). Identical results were observed by using multiple cell donors in independent experiments (Fig. 2F).
Theoretically, an equilibrium state should be reached in which the rate of creation of the GFP* form and its reversion back to YFP and CFP are balanced. We were able to achieve this equilibrium by replenishing the donor 3 target-cell population with activated uninfected cells every 2 days, allowing virus replication to continue for ≥2 weeks (data not shown). Although the relationships among infection, coinfection, and recombination were identical to those of previous experiments until the time of maximal infection (Fig. 2 D–F), replenishment of the target-cell population resulted in a plateauing of the GFP* form, as predicted (Fig. 2 E and F, green triangles).
In vivo lymphoid tissue provides a more natural context for studies of HIV-1 dynamics, and therefore we continued this study in the SCID-hu (Thy/Liv) mouse model (15). Infection of 12 human thymic grafts, representing four different tissue donors, with replication-competent YFP and CFP HIV-1 resulted in one log variation in infection rates after 2–3 weeks of virus replication. Nonetheless, the relationships among infection rates, coinfection rates, and the abundance of recombinant viruses was nearly identical to that observed during the expansion phase of infection in CD4+ T lymphocytes in culture (Fig. 2 G–I). It is noteworthy that, despite down-modulation of CD4 expression to undetectable levels on >90% of infected cells (data not shown), the constant relationship between infection and coinfection rates paralleled the results in tissue culture, suggesting that viral interference fails to lower the coinfection/infection ratio to any significant degree. It is likely that the kinetics of infection and target-cell depletion result in most infected cells being infected and reinfected simultaneously (or nearly simultaneously). The close correspondence between cell culture and the in vivo model suggests that generally applicable relationships are described by these systems.
In addition to the frequency of coinfection, the frequency with which RT transfers between the two available genomic templates during reverse transcription is an important factor in establishing the dynamics of recombinational diversification. Prior studies quantifying HIV-1 RT-strand transfer frequencies across the HIV-1 genome have been conducted only during infection of HeLa-CD4 fibroblastic cells and have demonstrated a minimal average of three to four RSTE per virus per round of replication (19–21). Because the intracellular environments of diverse cell types vary, we sought to determine the frequency of RT-strand transfer directly during HIV-1 infection of a natural target of HIV-1 infection. Therefore, we performed DNA sequencing of recombinant GFP* proviruses generated during infection of Jurkat T cells. First, we copackaged YFP and CFP genomes by using strains of HIV-1 that are 6% divergent in DNA sequence, infected Jurkat T lymphocytes with these heterozygous virions in a single round of infection, and then cloned the resulting GFP+ cells and sequenced the integrated proviruses. Fig. 3A shows the pattern of recombination events within the 13 GFP* recombinant viruses. A total of 90 RSTE were detected, with one in each provirus being the YFP- and CFP-strand transfer that generated the GFP* form. In agreement with studies (20), RSTE were evident throughout the genomes, indicating that all regions of HIV-1 are subject to diversification by means of recombination. We could identify 3–13 RSTE in each provirus, yielding an average of seven and a half RSTE per wild-type genome (Fig. 3B), or approximately twice the rate reported during infection of HeLa-CD4 cells (19–21).
None of the RSTE was associated with an alteration of the primary sequence, including point mutations, deletion, or duplication, consistent with the previously described (19, 21) high fidelity of the recombination process. Regions of the HIV-1 genome containing RNA secondary structures (e.g., the rev responsive element) or repeats did not appear to have a greater frequency of recombination than other regions, suggesting that during infection of cells, recombination is not enhanced in such structures. Recombination between YFP and CFP genes was identified in three discrete regions (Fig. 3C), indicating that, as for the HIV-1 genome proper, all regions are subject to recombination.
The recombination frequency in Jurkat T cells suggested that HIV-1 RT-strand-transfer rates may be influenced by the infected cell type; therefore, we performed a single-cycle infection analysis of several cell types under identical experimental conditions. As shown in Fig. 4A, infection of HeLa-CD4 with pseudotyped YFP/CFP heterozygous viruses resulted in the lowest frequency of appearance of GFP recombinants of any examined cell type. Because 1% of all RSTE between YFP and CFP viruses generate the GFP* form (see Materials and Methods), we calculate five and a half RSTE per virus in HeLa-CD4, which, being slightly higher than previous minimal estimates, is in substantial agreement with them. Importantly, we calculate eight RSTE in Jurkat, which corresponds well with the seven and a half RSTE detected by sequence analysis. We calculate nine RSTE per virus in primary CD4+ T lymphocytes. The agreement between the sequence analysis, which provides RSTE frequency in individual viruses known a priori to be recombinants, and the flow-cytometric method, which yields an average RSTE frequency across the entire population, suggests that most or all viruses are undergoing recombination, consistent with two recent reports (13, 22). If recombination were occurring within only a subset of viruses (23), then the set of viruses identified as recombinants by their GFP expression would display a greater than average number of RSTE, which is not the case. The route of viral entry had little effect on recombination rates because pseudotyping of HIV-1 with CCR5-tropic and CXCR4-tropic HIV-1 envelopes, amphotropic murine leukemia virus envelope, or VSV-G protein yielded rates of recombination that varied by no more than 20% (data not shown).
Infection of macrophages resulted in an average of ≈30 RSTE per virus (Fig. 4A), or about one order of magnitude greater than previously documented in fibroblastic cell lines. The only cellular cofactors necessary for successful reverse transcription are nucleotide precursors; thus, the most likely means to increase HIV-1-strand-transfer rates would be by altered intracellular nucleotide (dNTP) availability. Low dNTP availability reduces RT activity and can increase RT template switching (24–26). Low dNTP availability in differentiated macrophages lowers reverse transcription rates, whereas the addition of exogenous nucleosides allows completion of reverse transcription and restores permissiveness to infection (27, 28). In our study, addition of exogenous nucleosides to macrophages at the time of infection enhanced productive infection up to 3-fold but failed to lower the frequency of recombination (see Supporting Text and Table 1, which are published as supporting information on the PNAS web site). Differentiation of two monocytic cell lines enhanced RSTE rates ≈3-fold (Fig. 4B), suggesting that myeloid differentiation results in the expression or modulation of cellular factors that influence the reverse transcription process and recombination.
Discussion
Despite a mutation rate that is only of average magnitude among RNA viruses (2, 29), the high rate of HIV-1 replication in vivo ensures rapid diversification (1, 9). Recombination represents a complementary mechanism that has the potential to accelerate HIV-1 diversification and evolution greatly (30, 31), although some models challenge this view (32, 33). By introducing multiple simultaneous alterations in the genome, recombination can generate constellations of alleles that would be otherwise unlikely or unable to assemble by means of mutation alone (31, 34). Recombination breaks linkage among alleles, increasing the effective viral population size, and in doing so it can prevent degradation of population fitness from the effects of evolutionary drift and allow selection to be dominant over stochastic events (6, 31). A better understanding of the parameters of HIV-1 recombination is vital to the development of more complete models of HIV-1 evolution and escape from adaptive immunity and antiviral therapies. We propose that mathematical models of HIV-1 dynamics may reasonably incorporate recombination based on the parameters defined in this study.
Our results indicate that the opportunity for recombination is favored rather than inhibited. For example, coinfection occurred with little inhibition, indicating that rapid or simultaneous infection and reinfection of cells provides ample opportunity for recombination. The close correspondence in results from multiple cell donors and between the cell culture and SCID-hu systems suggests that the relationships described may be generally applied to the in vivo situation. Likewise, by examining the rate of RT-strand transfer in the proper cellular contexts, CD4+ T cells and macrophages, we find that HIV-1 is substantially more recombinogenic than it was previously believed to be.
Recent studies (13, 22) and the unpublished work of D.N.L. and G.M.S. have shown that HIV-1 is about one order of magnitude more recombinogenic than several other retroviruses, including Maloney murine leukemia virus and human T cell leukemia virus type 1 (HTLV-1), during infection of identical cell types. HIV-1 infection of macrophages results in recombination frequencies nearly two logs higher than reported for infection of murine leukemia virus and spleen necrosis virus in fibroblast cell lines (13, 22). It is possible that HIV-1 may have evolved higher recombination rates to foster more rapid diversification and promote its survival. Because our results, as well as the results of other studies (19), indicate that recombination occurs in a manner that preserves the integrity of ORFs and cis elements, there may be little risk of virus inactivation from the high rates of recombinational strand transfer that we report here during infection of immune cells. Such a scenario would stand in contrast to mutation, in which population fitness will decline if the error rate exceeds a defined level (the “error threshold”), which results in a loss of population fitness (35, 36). As a result, there may be a rationale for enhanced recombination rates as a means of fostering efficient diversification that does not exist for mutation.
How HIV-1 RSTE rates are enhanced in different cell types is not clear, but we were able to exclude the most likely explanation, namely, low nucleotide levels frequently present in these cells. We propose that there is an uncharacterized and potentially important virus–cell interaction within the HIV-1 reverse transcription process that influences recombination. The first mechanistic model, to our knowledge, of retroviral recombination envisioned strand breaks within the RNA molecule that forced the RT to switch to the alternate template to continue polymerization (37). Subsequently, it has been shown that RSTE can occur independently of template breaks (4), but if the higher levels of nucleases present in macrophages result in increased RNA strand breakage, enhanced RSTE frequency might result. An implication of this model is the possibility that retroviral diploidy and recombination originally may have emerged primarily as a means of repairing genetic damage. However, the enhanced recombination rates observed in T cells versus HeLa-CD4 cells, in addition to our recent observation that HIV-1 recombination frequencies during infection of B lymphocytes are double those in T cells (D.N.L. and G.M.S., unpublished data), suggest that cell factor(s) in diverse cell types can affect recombination rates. As an alternative mechanistic model, there is evidence that the ratio of the RT polymerization rate to the rate of digestion of the trailing template by the RT RNase H activity can drive strand transfer (38). Thus, if the RNase H activity exceeds the polymerization rate, strand transfer is induced. It is possible that the cell influences the relative activities of these independent RT functions, thus altering the strand-transfer rate. This putative mechanism would be consistent with the observation that RT polymerization kinetics are lower in macrophages than they are in T cells (27, 28).
It is clearly desirable to maintain maximal viral suppression in patients when possible, but even highly active antiretroviral therapy fails to halt HIV-1 replication and evolution completely in some cases (39–41). Recent mathematical analyses predict that the emergence of new drug-resistant strains is directly proportional to the residual amount of virus replication (42, 43). Our finding that recombinational diversification proceeds according to the square of the infection frequency may suggest the higher risk of viral evolution and escape that are inherent in the failure to maintain suppression of viral replication and provide a further incentive to sustain highly suppressive therapy.
Supplementary Material
Acknowledgments
We thank Eric Hunter for useful discussions, Truman Grayson for animal work, and Marion Spell for cell sorting. This work was supported by a University of Alabama Center for AIDS Research Developmental Award and an Elizabeth Glaser Pediatric AIDS Foundation Scholar Award (to D.N.L.). Animal work was supported by National Institute of Allergy and Infectious Diseases Grant R01 AI41924 (to G.M.A.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CFP, cyan fluorescent protein; YFP, yellow fluorescent protein; RT, reverse transcriptase; RSTE, recombinational strand transfer events; SCID, severe combined immunodeficient; MOI, multiplicity of infection; IRES, internal ribosome entry sequence.
References
- 1.Coffin, J. M. (1995) Science 267, 483-489. [DOI] [PubMed] [Google Scholar]
- 2.Bonhoeffer, S. & Sniegowski, P. (2002) Nature 420, 367-369. [DOI] [PubMed] [Google Scholar]
- 3.Malim, M. H. & Emerman, M. (2001) Cell 104, 469-472. [DOI] [PubMed] [Google Scholar]
- 4.Negroni, M. & Buc, H. (2001) Annu. Rev. Genet. 35, 275-302. [DOI] [PubMed] [Google Scholar]
- 5.Jung, A., Maier, R., Vartanian, J. P., Bocharov, G., Jung, V., Fischer, U., Meese, E., Wain-Hobson, S. & Meyerhans, A. (2002) Nature 418, 144. [DOI] [PubMed] [Google Scholar]
- 6.Rouzine, I. M., Wakeley, J. & Coffin, J. M. (2003) Proc. Natl. Acad. Sci. USA 100, 587-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leigh-Brown, A. J. (1997) Proc. Natl. Acad. Sci. USA 94, 1862-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colgrove, R. & Japour, A. (1999) Antiviral Res. 41, 45-56. [DOI] [PubMed] [Google Scholar]
- 9.Rouzine, I. M., Rodrigo, A. & Coffin, J. M. (2001) Microbiol. Mol. Biol. Rev. 65, 151-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adachi, A., Gendelman, H. E., Koenig, S., Folks, T., Willey, R., Rabson, A. & Martin, M. A. (1986) J. Virol. 59, 284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs, J. S., Regier, D. A. & Desrosiers, R. C. (1994) AIDS Res. Hum. Retroviruses 10, 343-350. [DOI] [PubMed] [Google Scholar]
- 12.Kutsch, O., Benveniste, E. N., Shaw, G. M. & Levy, D. N. (2002) J. Virol. 76, 8776-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onafuwa, A., An, W., Robson, N. D. & Telesnitsky, A. (2003) J. Virol. 77, 4577-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, F., Robertson, D. L., Carruthers, C. D., Morrison, S. G., Jian, B., Chen, Y., Barre-Sinoussi, F., Girard, M., Srinivasan, A., Abimiku, A. G., et al. (1998) J. Virol. 72, 5680-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aldrovandi, G. M., Feuer, G., Gao, L., Jamieson, B., Kristeva, M., Chen, I. S. & Zack, J. A. (1993) Nature 363, 732-736. [DOI] [PubMed] [Google Scholar]
- 16.Lippincott-Schwartz, J. & Patterson, G. H. (2003) Science 300, 87-91. [DOI] [PubMed] [Google Scholar]
- 17.Le Guern, M. & Levy, J. A. (1992) Proc. Natl. Acad. Sci. USA 89, 363-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volsky, D. J., Simm, M., Shahabuddin, M., Li, G., Chao, W. & Potash, M. J. (1996) J. Virol. 70, 3823-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuang, J., Jetzt, A. E., Sun, G., Yu, H., Klarmann, G., Ron, Y., Preston, B. D. & Dougherty, J. P. (2002) J. Virol. 76, 11273-11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jetzt, A. E., Yu, H., Klarmann, G. J., Ron, Y., Preston, B. D. & Dougherty, J. P. (2000) J. Virol. 74, 1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu, H., Jetzt, A. E., Ron, Y., Preston, B. D. & Dougherty, J. P. (1998) J. Biol. Chem. 273, 28384-28391. [DOI] [PubMed] [Google Scholar]
- 22.Rhodes, T., Wargo, H. & Hu, W. S. (2003) J. Virol. 77, 11193-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, W. S., Bowman, E. H., Delviks, K. A. & Pathak, V. K. (1997) J. Virol. 71, 6028-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeiffer, J. K., Topping, R. S., Shin, N. H. & Telesnitsky, A. (1999) J. Virol. 73, 8441-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svarovskaia, E. S., Delviks, K. A., Hwang, C. K. & Pathak, V. K. (2000) J. Virol. 74, 7171-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeStefano, J. J., Mallaber, L. M., Rodriguez-Rodriguez, L., Fay, P. J. & Bambara, R. A. (1992) J. Virol. 66, 6370-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neil, S., Martin, F., Ikeda, Y. & Collins, M. (2001) J. Virol. 75, 5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Brien, W. A., Namazi, A., Kalhor, H., Mao, S. H., Zack, J. A. & Chen, I. S. (1994) J. Virol. 68, 1258-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preston, B. D. & Dougherty, J. P. (1996) Trends Microbiol. 4, 16-21. [DOI] [PubMed] [Google Scholar]
- 30.Temin, H. M. (1986) Cell Biophys. 9, 9-16. [DOI] [PubMed] [Google Scholar]
- 31.Temin, H. M. (1991) Trends Genet. 7, 71-74. [DOI] [PubMed] [Google Scholar]
- 32.Otto, S. P. & Lenormand, T. (2002) Nat. Rev. Genet. 3, 252-261. [DOI] [PubMed] [Google Scholar]
- 33.Bretscher, M. T., Althaus, C. L., Müller, V. & Bonhoeffer, S. (2004) Bioessays 26, 180-188. [DOI] [PubMed] [Google Scholar]
- 34.Katz, R. A. & Skalka, A. M. (1990) Annu. Rev. Genet. 24, 409-445. [DOI] [PubMed] [Google Scholar]
- 35.Eigen, M. (2002) Proc. Natl. Acad. Sci. USA 99, 13374-13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domingo, E. (2000) Virology 270, 251-253. [DOI] [PubMed] [Google Scholar]
- 37.Coffin, J. M. (1979) J. Gen. Virol. 42, 1-26. [DOI] [PubMed] [Google Scholar]
- 38.Hwang, C. K., Svarovskaia, E. S. & Pathak, V. K. (2001) Proc. Natl. Acad. Sci. USA 98, 12209-12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furtado, M. R., Callaway, D. S., Phair, J. P., Kunstman, K. J., Stanton, J. L., Macken, C. A., Perelson, A. S. & Wolinsky, S. M. (1999) N. Engl. J. Med. 340, 1614-1622. [DOI] [PubMed] [Google Scholar]
- 40.Sharkey, M. E., Teo, I., Greenough, T., Sharova, N., Luzuriaga, K., Sullivan, J. L., Bucy, R. P., Kostrikis, L. G., Haase, A., Veryard, C., et al. (2000) Nat. Med. 6, 76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, L., Ramratnam, B., Tenner-Racz, K., He, Y., Vesanen, M., Lewin, S., Talal, A., Racz, P., Perelson, A. S., Korber, B. T., et al. (1999) N. Engl. J. Med. 340, 1605-1613. [DOI] [PubMed] [Google Scholar]
- 42.Ribeiro, R. M. & Bonhoeffer, S. (2000) Proc. Natl. Acad. Sci. USA 97, 7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraser, C., Ferguson, N. M. & Anderson, R. M. (2001) Proc. Natl. Acad. Sci. USA 98, 15167-15172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.