Abstract
Francisella tularensis is able to survive and grow within macrophages, a trait that contributes to pathogenesis. Several genes have been identified that are important for intramacrophage survival, including mglA and iglC. F. tularensis is also able to survive within amoebae. It is shown here that F. tularensis mglA and iglC mutant strains are not only defective for survival and replication within the macrophage-like cell line J774, but also within Acanthamoebae castellanii. Moreover, these strains are highly attenuated for virulence in mice, suggesting that a common mechanism underlies intramacrophage and intraamoebae survival and virulence. A 2D gel analysis of cell extracts of wild-type and mglA mutant strains revealed that at least seven prominent proteins were at low levels in the mglA mutant, and one MglA-regulated protein was identified as the IglC protein. RT-PCR analysis demonstrated reduced transcription of iglC and several other known and suspected virulence genes in the mglA mutant. Thus, MglA regulates the transcription of virulence factors of F. tularensis that contribute to intramacrophage and intraamoebae survival.
Francisella tularensis causes the life-threatening disease tularemia in humans. It can be acquired through skin abrasions, ingestion of contaminated food or water, by a vector-borne route (e.g., ticks), or, most seriously, by aerosols. Aerosol administration can lead to the pneumonic form of the disease, which has a high mortality rate. Because of this danger, F. tularensis was developed as a bioweapon in several national weapons programs. No approved vaccine is currently available in the United States or Europe for protection against this disease (for review, see ref. 1).
F. tularensis subsp. tularensis (type A) is the most virulent form of F. tularensis because it is associated with the highest morbidity and mortality in humans. An additional subspecies, novicida, is essentially avirulent for humans, but it causes a disease in mice that is similar to that caused by subsp. tularensis. All the subspecies of F. tularensis are closely related at the genomic level, and the reasons for the differences in tropism and virulence are yet to be determined (2).
Relatively little is known about the molecular mechanisms of Francisella pathogenesis. F. tularensis can survive and replicate within macrophages both in vitro and in vivo, and this ability to replicate appears to be its primary mechanism of pathogenesis (3, 4). The Francisella-containing vacuole has been reported to evade phagosome–lysosome fusion (5), and Francisella infection can cause macrophage apoptosis (6) and apparently also abrogate Toll-like receptor signaling (7). Several genes necessary for intramacrophage survival have been identified, including mglA (8) and iglC (9, 10), but the exact functions of these gene products have not been elucidated. MglA shares homology with SspA of Escherichia coli, which regulates stationary-phase gene transcription by interacting with RNA polymerase (11, 12). IglC has only been found in Francisella spp., and evidence has suggested a role for this protein in the disruption of TLR4-mediated signal transduction in infected macrophages (7).
It has recently been demonstrated that F. tularensis can survive and replicate within the amoebae Acanthamoebae castellanii (13), suggesting that protozoa may serve as an environmental reservoir. In the present study, we demonstrate that MglA regulates the transcription of multiple genes, including iglC, and that IglC is necessary for both intraamoebae and intramacrophage survival and also virulence in mice. Thus, MglA is a global regulator of factors necessary for virulence and presumably also environmental persistence.
Materials and Methods
Strains and Media. Construction of F. tularensis subsp. novicida strains KKF34 (ΔmglA::ermC) and KKF24 (ΔiglC::ermC) has been described (14); these strains are isogenic with the wild-type strain U112 (ATCC 15482). Strains were grown on tryptic soy broth/agar (Difco) supplemented with 0.1% cysteine (TSAcys) or on cysteine heart agar (Difco) supplemented with 5% horse blood (CHA).
Intracellular Survival Assay. The intramacrophage survival assay within J774 cells has been described (14). For intraamoebae survival assays, the protocol of Abd et al. was followed (13) by using A. castellanii strain ATCC 30234 grown in ATCC medium 712 at 30°C in microtiter wells and infected at a multiplicity of infection of ≈10:1 (≈106 F. tularensis subsp. novicida/≈105 A. castellanii). Bacterial inocula were determined by plate count. After 1 h, wells were treated with 50 μg/ml gentamicin for 1 h and then washed three times with PBS and replenished with 712 medium. Twelve days after infection, amoebae were treated with 50 μg/ml gentamicin for 1 h, then washed three times in PBS, and lysed with 0.2% deoxycholate; intracellular bacteria were determined by plate count. Control experiments determined that all strains were equally susceptible to gentamicin, and all extracellular bacteria were killed during the 1-h gentamicin treatment.
Mouse Virulence Assays. Groups of five female 4- to 6-week-old BALB/cAnNHsd mice (Harlan Sprague) were inoculated at a given dose of F. tularensis subsp. novicida delivered in 20 μl of PBS intranasally or 100 μl i.p. Actual bacterial counts delivered were determined by plate count from the inocula, and an approximate LD50 was calculated based on survival of mice within each inoculated group. Mice were monitored for 30 days after infection.
2D Gel Electrophoresis and Matrix-Assisted Laser Desorption Ionization–Time-of-Flight (MALDI-TOF) Analysis. 2D gel and in-gel trypsin digestion were carried out with whole-cell lysates from U112, KKF34, and KKF24 as described (15). Lysates were prepared from stationary-phase cultures grown in TSAcys at 37°C. MALDI-TOF mass spectrometry was used to obtain peptide mass fingerprinting profiles. The mass-to-charge ratio of each fragment generated after trypsin digestion was queried from a public database (http://prospector.ucsf.edu/ucsfhtml4.0/msfit.htm) or from the F. tularensis strain Schu 4 genome-sequencing consortium (http://artedi.ebc.uu.se/Projects/Francisella), and the protein with highest statistical certainty was chosen for identification. The locations of the genes encoding the two additional putative MglA-regulated proteins are (i) “lipoprotein,” contig 33, nucleotides 70780–71460 and (ii) “amidase,” contig 33, nucleotides 136404–136907.
Semiquantitative RT-PCR. Total RNA was isolated from midlog cultures (OD600 0.3–0.4) of U112, KKF34, and KKF24 by TRIzol reagent (GIBCO/Life Technologies, Grand Island, NY). Total RNA was treated with DNase I (Ambion, Austin, TX) and reverse-transcribed by using random hexamers to produce cDNA (Invitrogen). PCR was performed on cDNA samples matched based on total RNA. Results shown were determined to fall within a linear response curve (i.e., changing cDNA concentrations by 2-fold up or down resulted in 2-fold differences in product) by preliminary experiments that altered the number of cycles and amount of cDNA added to reaction, and control experiments demonstrated a lack of contaminating genomic DNA in the isolated RNA.
PCR primer pairs were designed based on internal sequences of the individual genes. Sequence of the subsp. novicida pdpDiglABCD locus and the pdpA gene was already available (GenBank accession no. AY293579). PCR of the tul4 locus has been reported (14).
Results
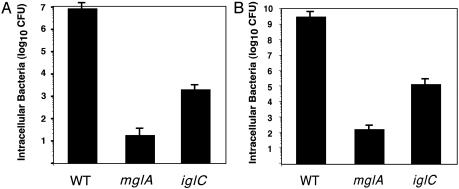
MglA and IglC Are Necessary for Intraamoebae and Intramacrophage Growth. MglA and IglC have both been shown to be necessary for F. tularensis intramacrophage survival and growth (8–10). We have recently developed a method to construct targeted F. tularensis mutants with PCR products (14) and have confirmed the defect of mglA and iglC mutants in intracellular survival and growth within the J774 macrophage cell line (Fig. 1A). The mglA strain is recovered at >105-fold lower levels than the wild-type strain, and the iglC strain is recovered at >103-fold lower levels. The lower levels of intracellular bacteria at 24 h are not caused by differences in initial uptake; similar levels of intracellular bacteria were detected at 1 h after gentamicin pulse (wild type, 1.5 × 102; mglA, 2.2 × 102; and iglC, 1.3 × 102). Similar results have been obtained in four separate experiments (not shown). Because the mglA strain is recovered at reduced levels in comparison with the iglC strain suggests it is more highly attenuated for intramacrophage survival and growth.
Fig. 1.
(A) Survival of F. tularensis strains within the J774 macrophage-like cell line. Strains U112 (WT), KKF34 (mglA), and KKF24 (iglC) were inoculated at a multiplicity of infection of ≈1:1 to J774 cells, and intracellular bacteria were enumerated at 24 h. The assay was performed in triplicate (see Materials and Methods). Bacterial inocula ranged from 5.1 to 6.5 × 105 cfu. (B) Survival of F. tularensis strains within A. castellanii. Strains U112 (WT), KKF34 (mglA), and KKF24 (iglC) were inoculated at an multiplicity of infection of ≈10:1 to A. castellanii, and intracellular bacteria were enumerated at 10 days. The assay was performed in triplicate (see Materials and Methods). Bacterial inocula ranged from 1.3 to 2.1 × 106 cfu.
A recent report demonstrated the ability of F. tularensis to survive and grow within amoebae (13). To determine whether the mechanisms of intramacrophage and intraamoebae survival and growth are related, we tested the ability of mglA and iglC mutants to survive and grow within A. castellanii (Fig. 1B). The relative amounts of intracellular bacteria were remarkably similar to those found within J774 macrophages. Compared with the wild-type U112 strain, the mglA and iglC strains were attenuated for survival and growth within A. castellanii. The mglA strain was recovered at ≈107-fold lower levels than the wild-type strain, and the iglC strain was recovered at ≈104-fold lower levels, suggesting that, as with Legionella spp. (16), the mechanism(s) of Francisella intraamoebae and intramacrophage survival may be linked. Similar results have been obtained in three separate experiments (not shown). The mglA strain was recovered at reduced levels in comparison with the iglC strain, suggesting that the mglA strain is more highly attenuated for intraamoebae survival and growth, as also seen for intramacrophage survival and growth.
mglA and iglC F. tularensis Mutants Are Attenuated for Virulence. The mglA and iglC mutants were analyzed for virulence in BALB/c mice by two different routes of inoculation. The wild-type U112 strain has a low LD50, ≈10 colony-forming units (cfus), when administered by either the i.p. or intranasal routes (ref. 17 and Table 1). In contrast, the mglA and iglC strains are highly attenuated for virulence by both routes of inoculation. No death of any animal occurred at the highest doses of the mglA and iglC strains tested, indicating >105-fold increases in the intranasal LD50 and >103 increases in the i.p. LD50.
Table 1. Virulence of F. tularensis strains in BALB/c mice.
| Strain | LD50 (intranasal) | LD50 (i.p.) |
|---|---|---|
| Wild-type U112 | ≈10* | 3† |
| mglA | >3.1 × 106 | >2.3 × 104 |
| iglC | >9.4 × 107 | >1.7 × 106 |
Approximate LD50: two of five mice survived at an inoculum of 30 cfu, and one of five mice survived at an inoculum of 300 cfu
From ref. 17
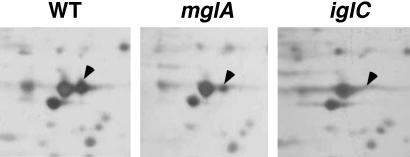
MglA Regulates the Expression of Multiple Proteins, Including IglC. MglA is predicted to be a regulatory protein (8), so to determine the proteins regulated by MglA, 2D gel electrophoretic separation was performed on wild-type, mglA, and iglC strain whole-cell lysates. At least seven individual protein spots were found at lower levels in the mglA lysate, when compared with the wild-type lysate (not shown). Only one protein spot was noticeably absent in the iglC lysate, and this protein spot corresponded to one of the seven apparent MglA-regulated proteins (Fig. 2). This protein spot was excised from the wild-type sample and identified by MALDI-TOF analysis as IglC. Two additional proteins that appear to be positively regulated by MglA were also identified by the same analysis; these proteins share highest similarity with (i) a Coxiella burnetii lipoprotein (AE016960, locus tag CBU0048; 43% similarity) and (ii) an amidase from Nostoc punctiforme (ZP_00106217; 50% similarity). The genes encoding these proteins are not located in the region of the genome near iglC and have not been studied further.
Fig. 2.
2D analysis of protein expression in F. tularensis strains. Whole-cell lysates of strains U112 (WT), KKF34 (mglA), and KKF24 (iglC) were separated by 2D gel electrophoresis. The putative MglA-regulated spot identified by MALDI-TOF from the wild-type sample is indicated by arrows and corresponds to IglC.
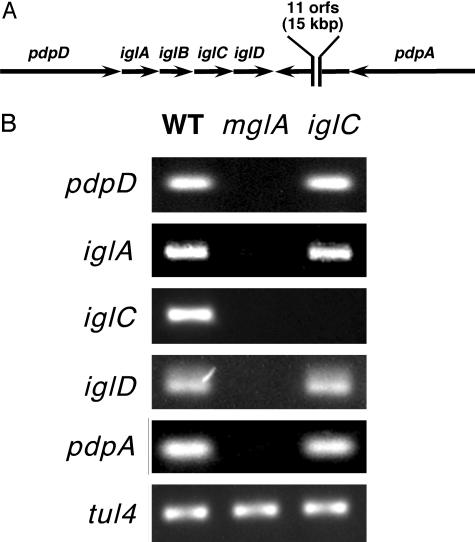
MglA Is a Positive Transcriptional Regulator of the iglC Operon. The iglC gene is found within a region of the genome containing additional virulence genes that has been proposed to be a pathogenicity island (F.E.N., unpublished data). iglC lies within a putative operon consisting of pdpDiglABCD (Fig. 3A), based on the close or overlapping stop and start codons and lack of any apparent transcriptional terminators. A mutation within iglA also leads to decreased macrophage survival and virulence (9). The pdpD gene encodes a large protein (140.7 kDa) with no significant similarity to any known proteins; although its role in virulence is unknown, several other very large genes found downstream of iglC, including pdpA, are necessary for intramacrophage survival and virulence and show some weak homology to Plasmodium rhoptry proteins (F.E.N., unpublished data). The pdpA gene is located 15 kbp downstream of iglD and is transcribed convergently.
Fig. 3.
RT-PCR analysis of MglA-regulated genes. (A) Diagram of the F. tularensis iglC locus. Genes surrounding iglC are shown as arrows (not drawn to scale) oriented in the direction of transcription; the region between iglD and pdpA contains 11 additional ORFs and is not shown. (B) RT-PCR analysis of the iglC locus. RT-PCR analysis (see Materials and Methods) was performed on total RNA prepared from F. tularensis strains U112 (WT), KKF34 (mglA), and KKF24 (iglC). PCR primers are specific to internal coding sequences of pdpD, iglA, iglC, iglD, pdpA, and tul4.
Semiquantitative RT-PCR analysis was performed with total RNA from the wild-type, mglA, and iglC strains (Fig. 3B). Lower levels of pdpD, iglA, iglC, iglD, and pdpA transcripts were detected in the mglA strain in comparison with the wild-type strain. These results demonstrate that MglA regulates multiple promoters of virulence genes. The data also suggest that pdpDiglABCD may be arranged in a single MglA-dependent transcriptional operon. The iglC strain demonstrated an absence of iglC transcript, as expected, but it had levels of pdpA, pdpD, iglA, and iglD transcripts similar to the wild-type strain, indicating that IglC has little effect on the transcription of these genes and, moreover, that the ΔiglC::ermC mutation has no polar effect on iglD. RT-PCR analysis was performed on midlog cultures, whereas 2D analysis (Fig. 2) was performed with stationaryphase cultures, which may account for the apparent greater levels of IglC protein vs. iglC transcripts in the mglA strain.
Transcription of the tul4 gene was used as an MglA-independent control. We have shown equivalent amounts of Tul4 protein in wild-type, mglA, and iglC strains (14), and the amount of tul4 transcript was similar in all strains (Fig. 3). Our results demonstrate that MglA positively regulates the transcription of multiple virulence genes that contribute to intraamoebae and intramacrophage survival.
Discussion
F. tularensis is a human pathogen and potential bioterrorist agent that can cause significant morbidity and mortality, and yet very little is known about its mechanism(s) of pathogenesis. However, it is clear that the ability of Francisella spp. to survive and replicate within macrophages is an important aspect of virulence. Consistent with this ability, mutations that inhibit intramacrophage survival, such as mglA and iglC mutations, dramatically attenuate virulence, as shown here.
F. tularensis can also survive and replicate within amoebae, and it has been suggested that protists may provide an environmental niche for Francisella outside of mammalian hosts (13). We demonstrate that the mglA and iglC mutations, which diminish intramacrophage survival and growth, also diminish intraamoebae survival and growth. Thus, the mechanism(s) necessary for intraamoebae survival and growth may be similar to those necessary for intramacrophage survival and growth. Legionella pneumophila is likewise able to survive and grow inside both amoebae and macrophages, and overlap occurs in the genes necessary for intraamoebae and intramacrophage survival, such as the genes encoding the Dot/Icm type IV secretion apparatus (18, 19). F. tularensis, like L. pneumophila, may have initially been a protozoan parasite that evolved the ability to survive within macrophages because of the similarity in resistance mechanisms necessary to survive in both cell types (16).
The mechanism(s) that F. tularensis uses to evade intramacrophage (and intraamoebae) killing are unknown; no type III or type IV secretion systems that other intracellular pathogens use are apparent within the partially completed F. tularensis genome sequence. Rather, F. tularensis contains several large ORFs with weak homology to rhoptry proteins from Plasmodium spp., and insertions in several of these pdp genes, including pdpA, result in a decrease in virulence and intramacrophage survival (F.E.N, unpublished data). It has been suggested that F. tularensis utilizes these factors to modify the intracellular macrophage environment to survive. The pdp genes are localized to the region of the genome that also contains the iglC gene, and, as shown here, pdpD and pdpA are also regulated by MglA. The function of IglC is unknown, but it likely contributes to the modification of the Francisella-containing intramacrophage environment, which may include a down-regulation of TLR4-dependent signaling (7).
MglA is also necessary for intramacrophage and intraamoebae survival and virulence. MglA has homology with SspA, a “stringent starvation” transcriptional regulatory protein of E. coli that interacts with RNA polymerase and generally regulates stationary-phase genes. We have found that MglA appears to positively regulate the expression of at least seven different proteins in F. tularensis, as determined by 2D analysis, including IglC. RT-PCR analysis demonstrated that MglA positively regulates the transcription of the pdpA, pdpD, iglA, iglC, and iglD genes, of which three encode proven virulence factors (IglA, IglC, and PdpA). Thus, MglA appears to be a global transcriptional regulator of F. tularensis virulence, similar to PhoP of Salmonella typhimurium (20, 21) and ToxT of Vibrio cholerae (22). Given the pleiotropic nature of a mglA mutation, this finding likely explains the greater attenuation in intracellular growth seen in the mglA mutant than in the iglC mutant (Fig. 1).
SspA homologues have been demonstrated to play roles in the virulence of other bacterial pathogens: in pilin expression in Neisseria gonorrhea (23), invasin expression in Yersinia enterocolitica (24), and multiple drug resistance in Providencia stuartii (25). Because of the homology of MglA with SspA, it seems likely that MglA activates transcription in a similar manner. SspA is necessary for activation of bacteriophage P1 late genes in E. coli, and a recent study showed that SspA appears to interact directly with RNA polymerase to facilitate closed-complex formation but requires an additional phage-encoded protein (Lpa) to specifically activate P1 late promoters (12). Transcription of mglA is stimulated within the intramacrophage environment (26), but it remains to be determined whether MglA activity is also modulated by the intracellular environment and whether additional factors are necessary for MglA-dependent transcription activation.
Acknowledgments
We thank Rob Edwards for assistance with the F. tularensis genome sequence and Yousef Abu Kwaik for assistance with intraamoebae survival assays. Preliminary sequence data were obtained from the F. tularensis strain Schu 4 genome-sequencing consortium. This study was supported by the University of Texas Health Science Center at San Antonio Presidential Research Excellence Fund and National Institutes of Health Grant AI50564 (to K.E.K.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: MALDI-TOF, matrix-assisted laser desorption ionization–time-of-flight; cfu, colony-forming unit.
References
- 1.Ellis, J., Oyston, P. C. F., Green, M. & Titball, R. (2002) Clin. Microbiol. Rev. 15, 631-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broekhuijsen, M., Larsson, P., Johansson, A., Bystrom, M., Eriksson, U., Larsson, E., Prior, R. G., Sjostedt, A., Titball, R. & Forsman, M. (2003) J. Clin. Microbiol. 41, 2924-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony, L. S. D., Burke, R. D. & Nano, F. E. (1991) Infect. Immun. 59, 3291-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tarnvik, A. (1989) Rev. Infect. Dis. 11, 440-451. [PubMed] [Google Scholar]
- 5.Fortier, A. H., Green, S. J., Polsinelli, T., Jones, T. R., Crawford, R. M., Leiby, D. A., Elkins, K. L., Meltzer, M. S. & Nacy, C. A. (1994) Immunol. Ser. 60, 349-361. [PubMed] [Google Scholar]
- 6.Lai, X. H., Golovliov, I. & Sjostedt, A. (2001) Infect. Immun. 69, 4691-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telepnev, M., Golovliov, I., Grundstrom, T., Tarnvik, A. & Sjostedt, A. (2003) Cell. Microbiol. 5, 41-51. [DOI] [PubMed] [Google Scholar]
- 8.Baron, G. S. & Nano, F. E. (1998) Mol. Microbiol. 29, 247-259. [DOI] [PubMed] [Google Scholar]
- 9.Gray, C. G., Cowley, S. C., Cheung, K. K. & Nano, F. E. (2002) FEMS Microbiol. Lett. 215, 53-56. [DOI] [PubMed] [Google Scholar]
- 10.Golovliov, I., Sjostedt, A., Mokrievich, A. N. & Pavlov, V. M. (2003) FEMS Microbiol. Lett. 222, 273-280. [DOI] [PubMed] [Google Scholar]
- 11.Williams, M. D., Ouyang, T. X. & Flickinger, M. C. (1994) Biochem. Biophys. Res. Commun. 201, 123-127. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, A. M., Lehnherr, H., Wang, X., Mobley, V. & Jin, D. J. (2003) Mol. Microbiol. 48, 1621-1631. [DOI] [PubMed] [Google Scholar]
- 13.Abd, H., Johansson, T., Golovliov, I., Sandstrom, G. & Forsman, M. (2003) Appl. Environ. Microbiol. 69, 600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauriano, C. M., Barker, J. R., Nano, F. E., Arulanandam, B. P. & Klose, K. E. (2003) FEMS Microbiol. Lett. 229, 195-202. [DOI] [PubMed] [Google Scholar]
- 15.Yoon, S. S., Hennigan, R. F., Hilliard, G. M., Ochsner, U. A., Parvatiyar, K., Kamani, M. C., Allen, H. L., DeKievit, T. R., Gardner, P. R., et al. (2002) Dev. Cell 3, 593-603. [DOI] [PubMed] [Google Scholar]
- 16.Harb, O. S., Gao, L. Y. & Abu Kwaik, Y. (2000) Environ. Microbiol. 2, 251-265. [DOI] [PubMed] [Google Scholar]
- 17.Kieffer, T. L., Cowley, S. C., Nano, F. E. & Elkins, K. L. (2003) Microb. Infect. 5, 397-403. [DOI] [PubMed] [Google Scholar]
- 18.Gao, L. Y., Harb, O. S. & Abu Kwaik, Y. (1997) Infect. Immun. 65, 4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel, J. P., Andrews, H. L., Wong, S. K. & Isberg, R. R. (1998) Science 279, 873-876. [DOI] [PubMed] [Google Scholar]
- 20.Miller, S. I., Kukral, A. M. & Mekalanos, J. J. (1989) Proc. Natl. Acad. Sci. USA 86, 5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groisman, E. A., Chiao, E., Lipps, C. J. & Heffron, F. (1989) Proc. Natl. Acad. Sci. USA 86, 7077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DiRita, V. J., Parsot, C., Jander, G. & Mekalanos, J. J. (1991) Proc. Natl. Acad. Sci. USA 88, 5403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Reuse, H. & Taha, M. K. (1997) Res. Microbiol. 148, 289-303. [DOI] [PubMed] [Google Scholar]
- 24.Badger, J. L. & Miller, V. L. (1998) J. Bacteriol. 180, 793-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding, X., Baca-DeLancey, R. R. & Rather, P. N. (2001) FEMS Microbiol. Lett. 196, 25-29. [DOI] [PubMed] [Google Scholar]
- 26.Baron, G. S. & Nano, F. E. (1999) Gene 229, 59-65. [DOI] [PubMed] [Google Scholar]