Compartmentalization is a key principle for the functional organization of living cells (1). Transport across compartmental boundaries and, in particular, across the cell wall is controlled by membrane proteins that act as selective channels and transporters. Puncturing the boundaries leads to pathologies, e.g., in the case of toxins, but is also an opportunity for treatment, e.g., in the case of antimicrobial peptides. The ability of membrane channels to sort single molecules is of great interest in bioengineering and, as a result, membrane channels like α-hemolysin have been adopted for in vitro devices (2–5) or used as an inspiration for manufacturing artificial channels (6, 7).
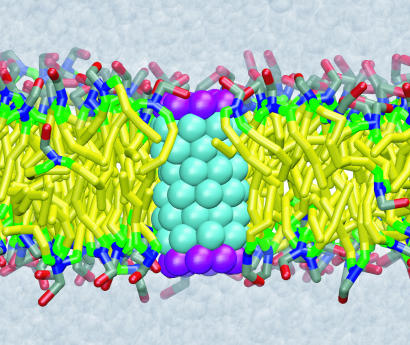
For deployment of proteins as channels, they need to be inserted into compartmental walls made of lipid bilayers. The majority of membrane proteins, socalled constitutive membrane proteins (8), are inserted into a lipid bilayer at the same time as they are synthesized. A special membrane protein, translocon (9), binds to the ribosome synthesizing the membrane protein and threads the protein into a lipid bilayer through an internal channel (10, 11). In contrast, the nonconstitutive membrane proteins, such as toxins (12) and antimicrobial peptides (13), insert themselves into a lipid bilayer without assistance. The insertion mechanism of such proteins, which is an indispensable part of their function, is poorly understood. The work of Lopez et al. (14) in this issue of PNAS demonstrates how a direct insertion of a typical nonconstitutive protein into a lipid membrane could take place. Modeling a membrane protein as a hydrophobic tube with hydrophilic sites at the tube's ends, they observed, in an unprecedented molecular dynamics (MD) simulation, a spontaneous insertion of a generic nanosyringe into a lipid bilayer. The simulation revealed microscopic details of the insertion kinetics, showing the mechanism by which lipids assist one of the ends of the nanosyringe to cross the hydrophobic core of the membrane. The simulation monitored also the facility of the nanosyringe to conduct water molecules. The inserted nanosyringe is shown in Fig. 1.
Fig. 1.
Final stage of the nanopore insertion into a lipid bilayer. The colors correspond to the atom types of the coarse-grained model that has been used to simulate the nanopore insertion (14).
The time scale of the protein insertion process is beyond that covered by conventional microscopic simulations. Even when biased by an external electrical field, insertion of a small protein such as a single α-helix could not be observed in all-atom MD simulations (15), because the lipid molecules forming the bilayer do not adjust their conformations quickly enough. Steered MD (16), which is a proven method to accelerate events along the path connecting the initial and final states of a process, cannot be deployed for studying a spontaneous insertion of a transmembrane protein, because the insertion path is not known. MD simulations using implicit solvent/membrane models (17), as well as Monte Carlo simulations (18–20), cannot reveal the true microscopic kinetics of the insertion process, because the description of the lipid bilayer in these approaches is rather rudimentary.
A hydrophobic nanotube with hydrophilic ends could be spontaneously inserted into the lipid bilayer.
In contrast, coarse-grained MD (21–23) preserves essential features of the microscopic system, at the same time expanding the time scale of the simulation. In this method, small groups of atoms are represented by sites interacting with each other by bounded and nonbounded forces, like atoms in conventional MD. The potential energy governing dynamics of the coarsegrained system is much simpler than a conventional molecular force field and is limited to describing interactions of two and three bounded atoms and van der Waals and electrostatic forces (24). Despite such simplicity, the coarse-grained method used by Lopez et al. (14) can reproduce approximately the density profile of an aqueous dimyristoylphosphatidylcholine bilayer (21). Coarse graining atomic interactions not only reduces the number of atoms in the system by a factor of 10 and thereby the number of computations per time step, but it also accelerates the overall internal dynamics, because the free-energy profile of the coarse-grained system is less bumpy. This leads to an acceleration that permits studying such rare processes as a spontaneous insertion of a protein into a lipid bilayer; however, it is also a drawback of the approach, because it hides the “real-time” interval covered by the coarse-grained MD.
Aside from acceleration due to smoothing the time scale, the advantage of the coarse-grained method is at best equal to the saving due to a reduced particle count, i.e., 10-fold in the present case (14). In fact, full-atom simulations of similar-size systems today routinely cover 10 ns, with total simulation times of >100 ns, and include complete Coulomb forces (25). However, the practical benefit of coarse graining stems from the smoothing effect that permits simulation of very slow processes.
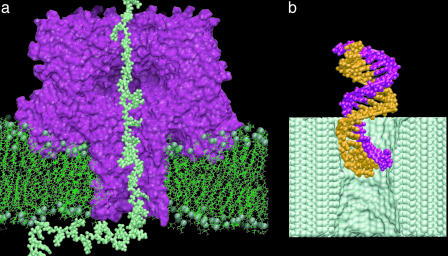
Nanopore-forming proteins of living cells are attractive for technological applications. One of the best-studied systems is the α-hemolysin toxin of Staphylococcus aureus (26), shown in Fig. 2a, which, in its monomeric form, inserts into a lipid bilayer and thereupon assembles into a heptameric transmembrane pore of ≈1.5-nm diameter. By applying a voltage bias across the lipid membrane, DNA or RNA strands can be driven inside the transmembrane pore, where they partially block the ionic current (2). Because the level of the reduced ionic current and the duration of the current blockades are indicative of the nucleotide composition, measured blockage current can discriminate strands of pyrimidine (C, T, and U) and purine (A and G) nucleotides from one another (3, 4), as well as the segments of purine and pyrimidine nucleotides within the same RNA strand (3). A single-nucleotide resolution has been demonstrated for individual Watson–Crick base pairs at the termini of single DNA hairpins (5), raising the prospect of creating a nanopore sensor capable of reading the nucleotide sequence directly from a DNA or RNA strand. Such a device can also be built, in principle, around a nanometer-size pore in a thin (2- to 5-nm) synthetic membrane. Such pores, like the one shown in Fig. 2b, have already been manufactured in silicon nitride and silicon oxide membranes with a subnanometer precision using highly focused ion and electron beams (6, 7). Not affected by aging, these pores can withstand high voltages, can operate in a broad range of temperatures, and are relatively easy to produce using the existing tools of nanofabrication technology.
Fig. 2.
Sequencing DNA with a nanopore sensor. An external electrical field drives DNA through the nanopore, transiently blocking the ionic current. By analysis of these current blockades, the DNA sequence can be discerned, in principle. (a) A strand of DNA (lime) threading the transmembrane pore of the α-hemolysin channel (maroon) shown in the cross section normal to the phospholipid membrane (green). The spheres represent the phosphate atoms; water and ions are not shown. The DNA strand has been pulled into the pore by all-atom steered MD (18). (b) Driven by an external electrical field, double-stranded DNA (orange and mauve) enters the artificial nanopore in a silicon nitride membrane (lime). The image is a snapshot from an all-atom MD simulation; water and ions are not shown.
Acknowledgments
We thank M. Klein for providing coordinates for the structure presented in Fig. 1 and Greg Timp for a collaboration on silicon nanopores. We also acknowledge support through National Science Foundation Grant CCR 02-10843.
See companion article on page 4431.
References
- 1.Koshland, D. E. (2002) Science 295, 2215–2216. [DOI] [PubMed] [Google Scholar]
- 2.Kasianowicz, J. J., Brandib, E., Branton, D. & Deamer, W. (1996) Proc. Natl. Acad. Sci. USA 93, 13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akenson, M., Branton, D., Kasianowicz, J. J., Brandin, E. & Deamer, D. W. (1999) Biophys. J. 77, 3227–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meller, A., Nivon, L., Brandin, E., Golovchenko, J. & Branton, D. (2000) Proc. Natl. Acad. Sci. USA 97, 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vercoutere, W. A., Winters-Hilt, S., DeGuzman, V. S., Deamer, D., Ridino, S. E., Rodgers, J. T., Olsen, H. E., Marziali, A. & Akeson, M. (2003) Nucleic Acids Res. 31, 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, J., Stein, D., McMullan, C., Branton, D., Aziz, M. J. & Golovchenko, J. A. (2001) Nature 412, 166–169. [DOI] [PubMed] [Google Scholar]
- 7.Storm, A. J., Chen, J. H., Ling, X. S., Zandberger, H. W. & Dekker, C. (2003) Nat. Mat. 2, 611–615. [DOI] [PubMed] [Google Scholar]
- 8.White, S. H. & Wimley, W. C. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 319–365. [DOI] [PubMed] [Google Scholar]
- 9.Andrews, D. W. & Johnson, A. E. (1996) Trends Biochem. Sci. 21, 365–369. [PubMed] [Google Scholar]
- 10.Borel, A. C. & Simon, S. M. (1996) Cell 85, 379–389. [DOI] [PubMed] [Google Scholar]
- 11.Do, H., Falcone, D., Lin, J. L., Andrews, D. W. & Johnson, A. E. (1996) Cell 85, 369–378. [DOI] [PubMed] [Google Scholar]
- 12.Gouaux, E. (1997) Curr. Opin. Struct. Biol. 7, 566–573. [DOI] [PubMed] [Google Scholar]
- 13.Zasloff, M. (2002) Nature 415, 389–395. [DOI] [PubMed] [Google Scholar]
- 14.Lopez, C. F., Nielsen, S. O., Moore, P. B. & Klein, M. L. (2004) Proc. Natl. Acad. Sci. USA 101, 4431–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tielemann, D. P., Berendsen, H. J. C. & Sansom, M. S. P. (2001) Biophys. J. 80, 331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isralewitz, B., Gao, M. & Schulten, K. (2001) Curr. Opin. Struct. Biol. 11, 224–230. [DOI] [PubMed] [Google Scholar]
- 17.Lazaridis, T. (2003) Proteins Struct. Funct. Genet. 52, 176–192. [DOI] [PubMed] [Google Scholar]
- 18.Maddox, M. W. & Longo, M. L. (2002) Biophys. J. 82, 244–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kessel, A., Shental-Bechor, D., Haliloglu, T. & Ben-Tal, N. (2003) Biophys. J. 85, 3431–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efremov, R. G., Volynsky, P. E., Nolde, D. E., van Dalen, A., de Kruijff, B. & Arseniev, A. S. (2002) FEBS Lett. 526, 97–100. [DOI] [PubMed] [Google Scholar]
- 21.Shelley, J., Shelley, M., Reeder, R., Bandyopadhyay, S., Moore, P. & Klein, M. (2001) J. Phys. Chem. B 105, 9785–9792. [Google Scholar]
- 22.Marrink, S. J. & Mark, A. E. (2003) J. Am. Chem. Soc. 125, 15233–15242. [DOI] [PubMed] [Google Scholar]
- 23.Stevens, M. J. (2003) Phys. Rev. Lett. 91, 188102–188104. [DOI] [PubMed] [Google Scholar]
- 24.Lopez, C., Moore, P., Shelley, J., Shelley, M. & Klein, M. (2002) Comput. Phys. Commun. 147, 1–6. [Google Scholar]
- 25.Gullingsrud, J. & Schulten, K. (2004) Biophys. J., in press.
- 26.Song, L., Hobaugh, M. R., Shustak, C., Cheley, S., Bayley, H. & Gouaux, J. E. (1996) Science 274, 1859–1866. [DOI] [PubMed] [Google Scholar]