Abstract
Taxonomic and phylogenetic analyses of great apes and humans have identified two potential areas of conflict between molecular and morphological data: phylogenetic relationships among living species and differentiation of great ape subspecies. Here we address these problems by using morphometric data. Three-dimensional landmark data from the hominoid temporal bone effectively quantify the shape of a complex element of the skull. Phylogenetic analysis using distance-based methods corroborates the molecular consensus on African ape and human phylogeny, strongly supporting a Pan–Homo clade. Phenetic differentiation of great ape subspecies is pronounced, as suggested previously by mitochondrial DNA and some morphological studies. These results show that the hominoid temporal bone contains a strong phylogenetic signal and reveal the potential for geometric morphometric analysis to shed light on phylogenetic relationships.
Since the advent of genetic investigations of hominoid diversity and relationships, most evidence has favored the view that African great apes and humans form a clade and that Homo and Pan are sister taxa (1, 2). Although this conclusion has been supported with morphological data (3–7), linear measurements and qualitative cranial characters failed to replicate the molecular phylogeny in the most comprehensive tests (8, 9). Moreover, the conflicts between morphological data sets in studies of the hominoid fossil record have led some to suggest that the utility of skeletal characters must be confirmed by comparison with molecular phylogenies of living groups before these characters can be used for phylogenetic reconstruction of fossil taxa (10).
Below the species level, analysis of great ape mitochondrial DNA has shown that genetic differentiation among subspecies of Pan troglodytes, Gorilla gorilla, and Pongo pygmaeus is pronounced. This level of differentiation may be greater than the degree of morphological differentiation among subspecies (11–14). For example, Gagneux et al. (ref. 14, p. 5081) concluded that “The extensive genetic differentiation in chimpanzees has not been accompanied by concordant morphological and behavioral differentiation in the different clades. The three geographically defined subspecies of chimpanzees are morphologically indistinguishable except at the level of minor dental/craniometric features.”
However, some morphological studies are concordant with the patterns of differentiation indicated by genetic evidence. For example, populations of gorillas and orangutans are divergent in hard and soft tissue anatomy (3, 15–17). Also, although traditional craniometric analysis has not identified major differences between chimpanzee subspecies (18), detailed studies of skeletal traits have revealed differences that are generally consistent with the molecular relationships (16, 17, 19).
Thus, controversy exists regarding morphological support for molecular relationships and differentiation at two levels in hominoids. In determining whether morphological data sets contain an accurate phylogenetic signal, it is important also to ask whether the methods used to study morphology are adequate for detecting that signal. Therefore, in this study we not only take advantage of an underutilized source of skeletal evidence, but we also evaluate geometric morphometric methods of studying shape to determine whether they improve on the qualitative and quantitative data that have failed to obtain the hominoid molecular tree (8). Improvement would be expected for several reasons. Landmark data are repeatable to a greater degree than qualitative character assessment, and 3D relationships among landmarks offer greater resolution of shape differences than do chord measurements and angles.
We chose to focus on great ape and human temporal bones because this anatomical region (i) presents a complex surface with numerous replicable landmarks and (ii) reflects the influence of brain size and cognition, mastication, hearing, posture, and other functional systems that have been important in higher primate evolution. Because of these factors, the temporal bone is a natural target for phylogenetic study. Its functional complexity should minimize the possibility that a single behavioral shift in unrelated taxa could lead to homoplastic similarity across a suite of features. In addition, the temporal bone is a common element in hominin fossil assemblages, and therefore evaluation of its phylogenetic signal is strongly relevant to studies of fossil hominin taxa.
Materials and Methods
Our hominoid sample represents eight great ape subspecies (wild-shot individuals) and one cadaver-based human sample, for a total of 405 adult specimens. Approximately equal numbers of males and females are represented. Sample size for each group ranges from 10 (P. pygmaeus abelii) to 78 (P. troglodytes troglodytes) (Table 1). G. gorilla graueri, the eastern lowland gorilla, was not digitized for this study and is not included. Females and males were analyzed separately, because shape dimorphism occurs in orangutans and gorillas.
Table 1. Centroid size for hominoid taxa.
| Mean female | n | Mean male | n | Average of sex means | |
|---|---|---|---|---|---|
| P. troglodytes verus | 88.1 | 24 | 93.1 | 24 | 90.6 |
| P. troglodytes schweinfurthii | 91.2 | 20 | 97.2 | 20 | 94.2 |
| P. troglodytes troglodytes | 91.7 | 39 | 96.5 | 39 | 94.1 |
| P. paniscus | 81.5 | 23 | 82.3 | 19 | 81.9 |
| H. sapiens | 83.1 | 29 | 86.6 | 32 | 84.9 |
| G. gorilla gorilla | 109.9 | 36 | 132.1 | 36 | 121.0 |
| G. gorilla beringei | 114.0 | 6 | 133.2 | 11 | 123.6 |
| P. pygmaeus abelii | 94.8 | 5 | 103.6 | 5 | 99.2 |
| P. pygmaeus pygmaeus | 89.5 | 20 | 108.4 | 17 | 98.9 |
Centroid size is the square root of the sum of squared distances from each landmark to the centroid of each specimen.
Twenty-two 3D ectocranial landmarks were used in the analysis, representing the glenoid, petrous, tympanic, and mastoid parts of the temporal bone (see Fig. 3 and Table 3, which are published as supporting information on the PNAS web site, and ref. 20). These landmarks were chosen based on repeatability and comprehensiveness and were collected by using a Micro-Scribe (Immersion, San Jose, CA) 3DX digitizer.
After obtaining raw 3D data, all individuals were subjected to generalized Procrustes analysis in morphologika (21). The generalized Procrustes analysis is a least-squares (LS) superimposition method that translates, rotates, and scales the landmarks for each individual (22, 23). This method adjusts for isometric effects of size, which are expressed as centroid size. The new coordinate data are referred to informally as Procrustes coordinates. Although Procrustes coordinates represent shape data, they still are subject to potential allometric effects.
In this article we focus on distance-based methods to avoid making assumptions about which components of the continuous variation are discrete “characters.” Euclidean distances among taxa were calculated based on mean Procrustes coordinates for each subspecies in the overall generalized Procrustes analysis. Although not identical to true Procrustes distances, which exist in curved space, these Euclidean distances are a close approximation [r = 0.999975, determined by using the program tpssmall (24)].
Two phylogenetic methods were used: neighbor joining (NJ) (25) and ordinary LS (26) with global rearrangement and forcing nonnegative branch lengths. Although NJ and LS usually provide similar results, they diverge slightly in our analyses, as discussed below. All NJ and LS results are based on analysis in phylip 3.57c (27). Minimum evolution algorithms (28) and weighted LS algorithms (29) also were evaluated in the course of the study. These algorithms provided similar results to those of ordinary LS analyses and therefore will be discussed only briefly.
The NJ and LS methods share some assumptions. Principally, we assume that shape distances among taxa provide phylogenetic information, given knowledge of an outgroup. It should be noted that these methods are not phenetic as used in this study (30). The distances themselves are based on overall similarity, but the use of an outgroup provides an axis of polarity and a measure of derived similarity.
Similar to other phylogenetic methods, NJ and LS favor results that minimize the frequency of convergence and reversal. Neither method assumes that morphological evolution is continuous or at a constant rate (i.e., both allow branch lengths to vary). All landmarks were given equal weight in distance calculations, which implies that, if a set of landmarks covary, they will contribute more information to the analysis. Discrete characters were not specified, although the morphology represented by shape differences among taxa was explored by principal components analysis (20).
The methods differ in how trees are determined. NJ starts with an unresolved tree and progressively identifies clusters that result in the greatest decrease in total branch length. NJ thus provides a single result and does not compare alternative trees. On the other hand, LS methods are optimization methods that compare trees and choose the one that minimizes the difference between observed shape distances and patristic shape distances (estimated from the tree).
Both NJ and LS methods produce unrooted trees. To root the trees, P. pygmaeus was made the outgroup to all other taxa. The phylogenetic relationship of Pongo to African apes and humans is not in doubt. Hylobates was not digitized for this study. We view it as a problematic outgroup for resolving the phylogeny of great apes and humans because of the great phenetic and size difference between it and the ingroup taxa.
The optimal trees were compared to the molecular tree of hominoid subspecies relationships based on nuclear and mitochondrial DNA sequences (1, 2). Support for each node was established by using the bootstrap procedure described in the Table 2 legend.
Table 2. Bootstrap results.
| NJ
|
LS
|
|||||
|---|---|---|---|---|---|---|
| Clade | Females | Males | Pooled-sex | Females | Males | Pooled-sex |
| Pongo | 98.6 | 98.9 | 99.9 | 98.9 | 99.4 | 99.9 |
| Gorilla | 84.7 | 88.3 | 91.8 | 83.4 | 81.8 | 88.6 |
| Pan + Homo | 82.2 | 98.8 | 98.7 | 83.9 | 99.1 | 98.7 |
| Pan | 96.2 | 66.9 | 85.3 | 98.3 | 80.1 | 92.3 |
| P. troglodytes | 86.8 | 82.2 | 92.8 | 42.8* | 44.3 | 32.9* |
| P. troglodytes schweinfurthii + P. troglodytes troglodytes | 93.6 | 99.9 | 99.4 | 96.2 | 99.9 | 99.8 |
Bootstrap support was determined by randomly sampling (1,000 times) 22 landmarks from the data set of taxon mean Procrustes coordinates and calculating Euclidean distances among taxa and corresponding trees each time. The results are the percentage of replications that support a given clade.
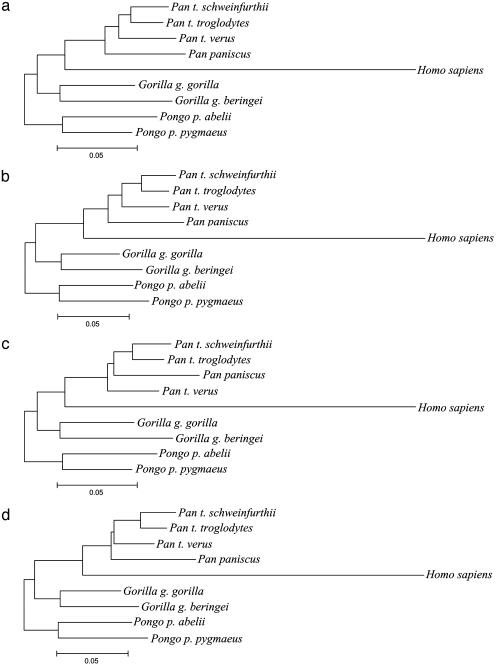
In a majority of LS analyses of females and the pooled-sex means, P. troglodytes is paraphyletic (see Fig. 1).
Results
Phylogenetic Results. In all trees based on shape distances, the length of the branch leading to humans illustrates a phenetic pattern in which the great apes are conservative in temporal bone shape and humans are very divergent (Fig. 1) (20). Differences between gorilla or orangutan subspecies are at least as pronounced as those between Pan paniscus and P. troglodytes. These differences are consistent with others that have led an increasing number of researchers to recognize two species within Gorilla (G. gorilla and G. beringei) and Pongo (P. pygmaeus and P. abelii) (31).
Fig. 1.
Phylograms representing preferred trees. (a) NJ tree using female means. (b) NJ tree using male means. (c) LS tree using female means. (d) LS tree using male means. The topology of a, b, and d is the same as the molecular phylogeny for these taxa, whereas that of c differs as described in the text. Analyses of pooled-sex means result in branching patterns identical to analyses of female means.
The NJ trees in this study correspond well to mitochondrial and nuclear DNA phylogenies of hominoid species and subspecies. Using either females or males, there is strong support for a Pan–Homo clade, with P. paniscus as the sister to P. troglodytes, and P. troglodytes verus as the sister to the eastern and central subspecies of P. troglodytes (Table 2). Gorilla is the sister group to chimpanzees and humans. Among all clades, NJ bootstrap values are lowest for Pan (67–96%, depending on the analysis). For 33% of bootstrapped male data sets, P. paniscus and Homo sapiens emerge as sister groups.
The Pan–Homo clade also is strongly supported in the LS trees (Table 2). Differences between NJ and LS trees occur in the relationships among the taxa of Pan. In the LS tree of females, P. troglodytes verus is the sister of a weakly supported clade containing P. paniscus and two subspecies of P. troglodytes (Fig. 1). The LS and NJ trees for males coincide, but bootstrapping suggests weak support for the position of P. paniscus in the LS tree (Table 2).
The positions of P. paniscus and P. troglodytes verus reveal differences in the algorithms of phylogenetic reconstruction. P. troglodytes verus is more similar to gorillas and orangutans than is P. paniscus, P. troglodytes schweinfurthii, or P. troglodytes troglodytes (see Table 4, which is published as supporting information on the PNAS web site). This similarity is clearly homoplastic and affects the LS results more than those from the NJ analyses.
In other results, minimum evolution algorithms (28) provided optimal trees that are equivalent to the ordinary LS trees. Weighted LS algorithms (29) differed only in the male tree, in which P. troglodytes verus was positioned as the sister group to all other Pan. To evaluate the position of P. troglodytes verus further, we also conducted analyses of only Pan taxa by using Homo as an outgroup. In these analyses, the monophyly of P. troglodytes was well supported by every analysis.
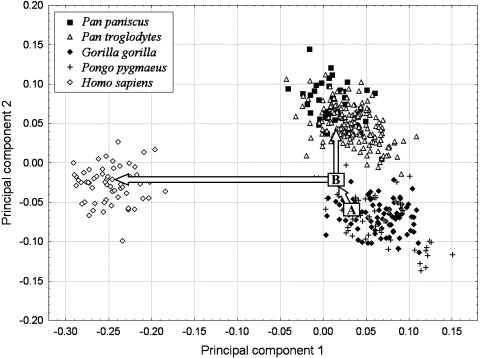
Shape and Size. Shape differences among taxa suggest that the Pan–Homo ancestor was phenetically in the range of modern great apes and more similar to gorillas than to chimpanzees (Fig. 2). In other words, the common ancestor that is reconstructed by linear parsimony falls nearest to gorillas, suggesting that Pan is autapomorphic in many respects. This conclusion is consistent with qualitative observations on fossil hominin temporal bones (32).
Fig. 2.
Principal components analysis of Procrustes coordinates for all hominoid males and females (see also ref. 20). The analysis was done by using full tangent space projection in morphologika (21). The first principal component (39.4% of the total variance) primarily distinguishes humans from apes, whereas the second (12.8%) distinguishes chimpanzees and bonobos from gorillas and orangutans. Both of these axes are important in supporting the Pan–Homo clade, because similarities between gorillas and orangutans are reconstructed as primitive (the third principal component separates gorillas and orangutans). This plot also illustrates the positions estimated for the common ancestor of Homo and Pan (B) and the common ancestor of African hominoids (A). Values for these ancestral nodes were estimated only for illustrative purposes by using linear parsimony analysis of female operational taxonomic unit means. The values show that a shift from the primitive condition to the derived Pan–Homo condition involves change in characters related to both principal components 1 and 2. Some of the characters that influence these axes are reduced projection of the entoglenoid process, medial position of the temporomandibular joint, less projecting postglenoid process, a shorter tympanic element, and reduced relative size of the temporomandibular joint (20).
The pattern of these results raises the question of whether the (apparently plesiomorphic) similarity between gorilla and orangutan temporal bones is related to size. However, the impression that orangutans are large-bodied is based only on male size. Orangutan and chimpanzee females are similar in both centroid size (Table 1) and body mass (33). Bonobos and humans are smaller in centroid size, whereas human females are larger, on average, in body mass than all female apes except gorillas (33). Thus, gorillas and orangutans are similar in some aspects of shape despite differences in size.
We further evaluated the effect of this size distribution by Mantel comparisons (mxcomp in nt-sys 2.02j, with 1,000 permutations) of female centroid size difference matrices with the shape distance matrices. Using the entire sample, centroid size differences are not significantly correlated with shape distances (Mantel r = 0.20; P = 0.22). However, if humans are excluded, size differences among the apes are significantly correlated with shape distances (Mantel r = 0.49; P = 0.02). This result is consistent with the view that size contributes significantly to shape variation among great apes. However, it demonstrates that the Pan–Homo affinity is not due to size.
The effect of size is seen instead in some of the poorly supported trees that occur at low frequencies in the bootstrap evaluation. As might be expected, this effect is most evident in the analysis of males, in which a third of the bootstrapped NJ analyses support a clade of bonobos and humans, the two taxa with the smallest male centroid size. The bonobo–human clade is only supported in 3.8% of the female analyses, whereas a clade consisting of humans and gorillas is a more common error. The pattern of errors demonstrates the effect of size and further shows that the shape data support the correct tree despite the potentially confounding effects of allometry.
Discussion and Conclusions
The use of morphological data in phylogenetic reconstruction has been criticized because of the failure to account for genetic and developmental correlations among characters. In addition, progress in using quantitative morphological data in primate phylogenetics has been limited, perhaps because of the difficulty of fitting quantitative data into a framework designed for discrete characters. In light of these concerns, the reasons why our analysis accurately recovers hominoid relationships are potentially several: (i) geometric morphometric analyses of densely sampled 3D landmarks capture shape differences among taxa more effectively than do other morphometric methods; (ii) distance-based methods of phylogenetic reconstruction may be more appropriate for continuous data than the transformation of these data into discrete character states; and (iii) the temporal bone's role in a variety of functional systems maximizes the number of independent influences on morphology and minimizes the effect of homoplasy in any one of those functional systems. These explanations are not mutually exclusive, and their relative impact can be evaluated by geometric morphometric analyses of other sets of skeletal data in a phylogenetic context.
Another conclusion that emerges from this study is that Pan is autapomorphic in temporal bone shape. Although Pan and Homo do share some aspects of temporal bone shape, the common ancestor of these taxa was probably more similar to gorillas (Fig. 2), which retain more primitive features in the temporal bone (20, 32). This conclusion is testable through incorporation of fossil taxa and of course does not imply that other parts of the skull follow a similar pattern.
More generally, our findings emphasize that paleoanthropologists need not shy away from addressing phylogenetic questions by using morphological data acquired from the hominoid fossil record (4). The congruence of our results with the consensus molecular tree, and the strong bootstrap support for the Pan–Homo clade, reaffirms the potential of skeletal evidence to recover hominoid relationships. We have shown that the temporal bone is particularly informative, and because temporal bone anatomy figures prominently in discussions of major transitions in human evolution (ape to hominin, Australopithecus to Homo, and the Pleistocene evolution of Homo), we encourage further detailed work on this anatomical element (34). Similarly targeted analyses of other cranial regions will expand the applicability of morphometric evidence to paleoanthropological questions.
Supplementary Material
Acknowledgments
We thank the following curators and museums for access to collections: B. Latimer and L. Jellema, Cleveland Museum of Natural History (Cleveland, OH); W. van Neer, Royal Museum for Central Africa (Tervuren, Belgium); R. Thorington and L. Gordon, National Museum of Natural History (Washington, DC); D. Pilbeam, Peabody Museum (Harvard University, Cambridge, MA); and J. Harrison and M. Harman, Powell–Cotton Museum (Birchington, Kent, United Kingdom). We also thank D. Begun, M. Collard, D. Pilbeam, Y. Rak, M. Rosenberg, J. Scott, D. Slice, F. Spoor, D. Strait, W. Sweitzer, M. Tocheri, B. Villmoare, and B. Wood for comments on prior versions of the manuscript. Financial support was provided by U.S. National Science Foundation Grant BCS-9982022 and a faculty grant-in-aid from Arizona State University.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LS, least squares; NJ, neighbor joining.
References
- 1.Ruvolo, M. (1997) Mol. Biol. Evol. 14, 248–265. [DOI] [PubMed] [Google Scholar]
- 2.Satta, Y., Klein, J. & Takahata, N. (2000) Mol. Phylogenet. Evol. 14, 259–275. [DOI] [PubMed] [Google Scholar]
- 3.Groves, C. P. (1986) in Comparative Primate Biology I: Systematics, Evolution, and Anatomy, eds. Erwin, J. & Swindler, D. R. (Liss, New York), pp. 187–217.
- 4.Begun, D. R. (1992) Science 257, 1929–1933. [DOI] [PubMed] [Google Scholar]
- 5.Begun, D. R. (1994) Yearb. Phys. Anthropol. 37, 11–63. [Google Scholar]
- 6.Shoshani, J., Groves, C. P., Simons, E. L. & Guralnick, R. P. (1996) Mol. Phylogenet. Evol. 5, 102–154. [DOI] [PubMed] [Google Scholar]
- 7.Gibbs, S., Collard, M. & Wood, B. (2000) Proc. Natl. Acad. Sci. USA 97, 11130–11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collard, M. & Wood, B. (2000) Proc. Natl. Acad. Sci. USA 97, 5003–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collard, M. & Wood, B. (2001) J. Hum. Evol. 41, 167–194. [DOI] [PubMed] [Google Scholar]
- 10.Pilbeam, D. (1997) in Function, Phylogeny, and Fossils: Miocene Hominoid Evolution and Adaptations, eds. Begun, D. R., Ward, C. V. & Rose, M. D. (Plenum, New York), pp. 13–28.
- 11.Morin, P. A., Moore, J. J., Chakraborty, R., Jin, L., Goodall, J. & Woodruff, D. S. (1994) Science 265, 1193–1201. [DOI] [PubMed] [Google Scholar]
- 12.Xu, X. & Arnason, U. (1996) J. Mol. Evol. 43, 431–437. [DOI] [PubMed] [Google Scholar]
- 13.Gonder, M. K., Oates, J. F., Disotell, T. R., Forstner, M. R. J., Morales, J. C. & Melnick, D. J. (1997) Nature 388, 337 (lett.). [DOI] [PubMed] [Google Scholar]
- 14.Gagneux, P., Wills, C., Gerloff, U., Tautz, D., Morin, P. A., Boesch, C., Fruth, B., Hohmann, G., Ryder, O. A. & Woodruff, D. S. (1999) Proc. Natl. Acad. Sci. USA 96, 5077–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groves, C. P. (1970) J. Zool. (Lond.) 161, 287–300. [Google Scholar]
- 16.Braga, J. (1995) Ph.D. thesis (Univ. of Bordeaux, Talence, France).
- 17.Uchida, A. (1996) Craniodental Variation Among the Great Apes. Peabody Museum Bulletin 4 (Harvard Univ. Press, Cambridge, MA).
- 18.Shea, B. T., Leigh, S. R. & Groves, C. P. (1993) in Species, Species Concepts, and Primate Evolution, eds. Kimbel, W. H. & Martin, L. B. (Plenum, New York), pp. 265–296.
- 19.Guy, F., Brunet, M., Schmittbuhl, M. & Viriot, L. (2003) Am. J. Phys. Anthropol. 121, 198–218. [DOI] [PubMed] [Google Scholar]
- 20.Lockwood, C. A., Lynch, J. M. & Kimbel, W. H. (2002) J. Anat. 201, 447–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Higgins, P. & Jones, N. (1998) J. Anat. 193, 251–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohlf, F. J. & Slice, D. E. (1990) Syst. Zool. 39, 40–59. [Google Scholar]
- 23.Goodall, C. (1991) J. R. Stat. Soc. 53, 285–339. [Google Scholar]
- 24.Rohlf, F. J. (1998) tpssmall (State Univ. of New York, Stony Brook), Version 1.18.
- 25.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 26.Cavalli-Sforza, L. L. & Edwards, A. W. F. (1967) Am. J. Hum. Genet. 19, 233–257. [PMC free article] [PubMed] [Google Scholar]
- 27.Felsenstein, J. (1993) phylip (Univ. of Washington, Seattle), Version 3.5c.
- 28.Nei, M. & Kumar, S. (2000) Molecular Evolution and Phylogenetics (Oxford Univ. Press, Oxford).
- 29.Fitch, W. M. & Margoliash, E. (1967) Science 155, 279–284. [DOI] [PubMed] [Google Scholar]
- 30.Felsenstein, J. (1984) Evolution (Lawrence, Kans.) 38, 16–24. [DOI] [PubMed] [Google Scholar]
- 31.Groves, C. P. (2001) Primate Taxonomy (Smithsonian Institution, Washington, DC).
- 32.Kimbel, W. H., Rak, Y. & Johanson, D. C. (2004) The Skull of Australopithecus afarensis (Oxford Univ. Press, Oxford).
- 33.Smith, R. J. & Jungers, W. L. (1997) J. Hum. Evol. 32, 523–559. [DOI] [PubMed] [Google Scholar]
- 34.Harvati, K. (2003) Am. J. Phys. Anthropol. 120, 323–338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.