Abstract
We describe here a protocol for digital transcriptome analysis in a single mouse blastomere using a deep sequencing approach. An individual blastomere was first isolated and put into lysate buffer by mouth pipette. Reverse transcription was then performed directly on the whole cell lysate. After this, the free primers were removed by Exonuclease I and a poly(A) tail was added to the 3′ end of the first-strand cDNA by Terminal Deoxynucleotidyl Transferase. Then the single cell cDNAs were amplified by 20 plus 9 cycles of PCR. Then 100-200 ng of these amplified cDNAs were used to construct a sequencing library. The sequencing library can be used for deep sequencing using the SOLiD system. Compared with the cDNA microarray technique, our assay can capture up to 75% more genes expressed in early embryos. The protocol can generate deep sequencing libraries within 6 days for 16 single cell samples.
INTRODUCTION
The identity and function of a cell is determined by its entire RNA component, which is called the transcriptome of a cell1,2. The transcriptome is the functional readout of the genome and epigenome. In an organism, essentially every cell has the same genome, while every cell type and potentially each individual cell has a unique transcriptome. Ideally, the transcriptome analysis should capture the exact quantity of all full length RNAs of all classes at single-base resolution in the smallest functional unit of an organism—an individual cell2. Eventually the transcriptome analysis may even become non-invasive, allowing us to read the sequences of every RNA molecule of a living cell without destroying the cell. During the past decade, the most successful and most widely used transcriptome analysis method has been the cDNA microarray3-8. Due to the development of the genome project, the cDNA microarray technique became available for most of the model organisms with a known genome. It is a powerful way to capture the expression pattern of tens of thousands of known genes by hybridization onto a tiny chip. However, it has significant drawbacks and limitations1,2, including (1) cross reactions between genes of similar sequences that occur due to the nature of hybridization on microarrays, (2) detection of expression levels only in the range of hundreds of folds, or three orders, despite the real dynamic range of gene expression in a cell being hundreds of thousands of folds, or six orders, (3) the exact length and sequence of the mRNAs analyzed is unknown, and (4) novel transcripts cannot be detected.
Using a tiling array can resolve some of these problems. The recently developed deep sequencing based transcriptome analysis, or RNA-Seq, can potentially overcome all of these problems1,9-12. RNA-Seq is sequencing based and can achieve single base resolution. The dynamic range of gene expression level that it can capture is theoretically unlimited, depending only on the depth of sequencing. More importantly, with the help of complete genome information, the exact length and sequence of all the RNAs analyzed can be captured accurately. During the past two years, people in the field have already witnessed the astonishingly fast development of the RNA-Seq technique and the deepening of our understanding of the complexity of the eukaryotic transcriptome, from yeast to human, from adult tissues to embryonic development13-21. However, due to the sensitivity of the method, it usually needs μg amounts of total RNAs, and most of the RNA-Seq studies used tissue, a mixture of different types of cells, or a cell line, which is at least a mixture of cells at different stages of the cell cycle. Also, recent progress on the stochastic nature of transcription and gene expression showed that even in the same cell type at the same cell cycle stage, the copy number of the mRNA of an expressed gene can be affected by both the microenvironment and the intrinsic noise of the transcription process22-27. Ideally the RNA-seq transcriptome analysis should be done using individual cells, or even the sub-compartment of a cell, such as the cytoplasm or nucleus. Also, for early embryonic development or stem cells in vivo, it is extremely difficult or even practically impossible to isolate millions of cells of a single type. RNA-Seq at single cell resolution will greatly promote the development of these fields by permitting comprehensive capture of the expression dynamics of all genes at all developmental stages.
During the past few years, people have already developed cDNA microarrays capable of using a small amount of starting material, or even a single cell through either in vitro transcription (IVT) based linear amplification, PCR-based exponential amplification, or a combination of the two methods28-38. However, these microarrays have inherited the limitations of the microarray technique. We improved a widely used single cell cDNA amplification strategy and combined it with SOLiD deep sequencing system to set up a digital transcriptome analysis method: single cell RNA-Seq38-40. By comparing the accuracy of single cell cDNA microarrays to our single cell RNA-Seq, we demonstrated that single cell RNA-Seq has greater accuracy. There are two reasons for this: (1) due to the higher sensitivity of the deep sequencing compared with the cDNA microarray, the IVT step is unnecessary to further amplify single cell cDNAs. This removes the amplification bias introduced by the IVT step. (2) With the better dynamic range of the deep sequencing method, the level of gene expression is more accurately captured by RNA-Seq. Using this method, we captured 5270 (75%) more genes than the cDNA microarray, expanding the transcriptome of a blastomere of a four-cell stage embryo from 7,050 expressed genes to 11,920 expressed genes. Around 10% of genes with multiple known transcript isoforms expressing more than two different transcript isoforms were detected in the same blastomere at the same time point, which has never been achieved by the single cell cDNA microarray. We also identified thousands of previously unknown splicing junctions from already known genes, indicating how limited our previous appreciation of the complexity of the eukaryotic transcriptome was based on the cDNA microarray.
This strategy has many possible future applications. It creates the possibility of isolating one or several cells from any of the organs or tumors of a patient and analyzing the transcriptome by RNA-seq, reducing the invasiveness of biopsies and clarifying the diagnosis of diseases. Potentially the sub-cellular distribution of mRNAs could be explored by comparing, for example, the axon and cell body of a neuron.
Current limitations of the single cell RNA-Seq include40:
The assay can only capture mRNAs with a poly(A) tail. Those mRNAs without poly(A) tails or other classes of RNAs such as small non-coding RNAs will not be detected.
The assay can not ascertain the strandedness of the transcript—that is, the sense or antisense transcripts can not be discriminated.
The assay can only capture about 3kb cDNA fragments from the 3′ end of a mRNA. For the genes with mRNAs longer than 3 kb, which constitute about 36% of all known genes, the 5′ end part of the mRNA can not be detected. We are currently improving the method to overcome these limitations.
MATERIALS
REAGENTS
Ac-BSA (Sigma, cat. no. B8894)
Acidic Tyrode’s solution (Sigma, cat. no. T1788)
1× PBS (pH 7.2) (Gibco, cat. no. 14249-95)
10× PCR Buffer II and 25 mM MgCl2 (Applied Biosystems, cat. no. 4379878)
Nonidet P-40 SP (Roche, cat. no. 11332473001)
SuperScript III Reverse Transcriptase with 0.1 M DTT (Invitrogen, cat. no. 18080-044 or 18080-085)
RNase Inhibitor (Cloned) (40 U μl−1) (Applied Biosystems, cat. no. AM2682)
SUPERase-In™ (20 U μl−1) (Applied Biosystems, cat. no. AM2694)
T4 gene 32 Protein (Roche, cat. no. 972983)
Exonuclease I (New England Biolabs, cat. no. M0293S)
Terminal Transferase (TdT) (Invitrogen, cat. no. 10533-065 or 10533-073)
100 mM dATP (Promega, cat. no. U1201)
Nuclease-free Water, 1 L (Applied Biosystems, cat. no. AM9932)
RNase H (Invitrogen, cat. no. 18021-014 or 18021-071)
TaKaRa Ex Taq™ HS (Includes: 10× ExTaq Buffer (mg2+ plus) and dNTP mixture) (Takara Bio Inc, cat. no. RR006A or RR006B)
QIAquick PCR Purification Kit (250) (Qiagen, cat. no. 28106)
QIAquick Gel Extraction Kit (250) (Qiagen, cat. no. 28706)
RNeasy Mini Kit (50) (Qiagen, cat. no. 74104)
PCR tubes, 0.5 ml (Eppendorf, cat. no. 951010057)
PCR tubes, 0.2 ml thin-wall (MLS)
Filtered pipettor tips (MLS)
SYBR Green PCR Mastermix (Applied Biosystems, cat. no. 4334973)
GeneAmp® dNTP Blend (100 mM) (Applied Biosystems, cat. no. N8080261)
3 M sodium acetate (pH 5.5) (Applied Biosystems/Ambion, cat. no. AM9740)
1 M Tris, pH 8.0 (100 ml) (Applied Biosystems/Ambion, AM9855G)
Nuclease-free Water (1 L) (Applied Biosystems/Ambion, cat. no. AM9932)
10× NEBuffer2 (New England BioLabs® Inc., B7002S)
Ethanol (Sigma-Aldrich®, cat. no. E7023)
Ethylene glycol (American Bioanalytical, cat. no. AB00455-01000)
Covaris microTUBE with AFA fiber and Snap-Cap with pre-slit Teflon/silicone/Teflon septa (Covaris™ Inc., cat. no. 520045)
End-It™ DNA End-Repair Kit (Epicentre®, cat. no. ER0720)
DNA Polymerase I (E. coli), (10U μl−1) (New England BioLabs® Inc., cat. no. M0209L)
Quick Ligation™ kit (New England Biolabs, cat. no. M2200L)
SYBR GreenER® qPCR SuperMix Universal (Invitrogen™ Corporation, cat. no. 11762-100) OR SYBR Green PCR Master Mix (Applied Biosystems, cat. no. 4309155)
Agilent DNA 1000 Kit (Agilent Technologies, cat. no. 5067-1504)
Agencourt® AMPure® 60 ml kit (Agencourt, cat. no. 000130)
AmpliTaq® DNA Polymerase, LD with Buffer I (Applied Biosystems, cat. no. N8080157)
Cloned Pfu polymerase (2.5 U μl−1) (Stratagene, cat. no. 600153)
Invitrogen Platinum® PCR SuperMix (Invitrogen™ Corporation, cat. no. 11306-016)
Quant-iT™ dsDNA HS Assay Kit, 100 assays (Invitrogen™ Corporation, cat. no. Q32854)
SOLiD™ Fragment Library Oligos Kit (Applied Biosystems, cat. no. 4401151)
EQUIPMENT
Brown-flaming micropipette puller (Sutter Instrument Co., Model P-80)
Covaris™ S2 System, (for system materials summary, see “Covaris™ S2 System Materials Summary,” SOLiD™ System 2.0 Site Preparation Guide.)
Microcentrifuge 5417R, refrigerated, without rotor 120 V//60 Hz (Eppendorf, cat. no. 022621807)
96-Well GeneAmp® PCR System 9700 (thermal cycler) (Applied Biosystems, cat. no. N8050200)
Real-time PCR System with 96-well block (ABI PRISM 7000 real-time PCR system (Applied Biosystems)
NanoDrop ND1000 Spectrophotometer (computer required) (NanoDrop (Thermo Sci), cat. no. ND-1000)
Labquake Rotisserie Rotator (Barnstead/Thermolyne, cat. no. VWR 56264-312)
Qubit™ fluorometer (Invitrogen, cat. no. Q32857)
6 Tube Magnetic Stand (Applied Biosystems, cat. no. AM10055)
Agilent 2100 Bioanalyzer (Agilent Technologies, cat. no. G2938C)
REAGENT SETUP
PBS-BSA (1 mg ml−1) Dissolve Ac-BSA (20 mg ml−1) in PBS at 1mg ml−1 and aliquot into 1.5ml Eppendorf tubes. Store the solution at −20 °C.
PROCEDURE
Single cell lysis
Dilute the UP1 Primer to 0.5 μM by adding 1 μl of 100 μM UP1 Primer and 199 μl of nuclease-free water to a tube and mix well. (All of primer sequences are listed in Table 1).
- Prepare the Cell Lysis Buffer (4.45 μl per sample) in a 0.5 ml thin-wall PCR tube by combining and mixing the following components:
Component Stock
concentrationFinal
concentration
in RT (5 μl)×1
volume×12
volume10X PCR Buffer II (without MgCl2) 10× 0.9× 0.45 μl 5.4 μl 25 mM MgCl2 25 mM 1.35 mM 0.27 μl 3.24 μl 10% NP40 10% 0.45% 0.225 μl 2.7 μl 0.1 M DTT 100 mM 4.5 mM 0.225 μl 2.7 μl SUPERase-In (Ambion) 20 U μl−1 0.18 U μl−1 0.045 μl 0.54 μl RNase Inhibitor (Ambion) 40 U μl−1 0.36 U μl−1 0.045 μl 0.54 μl 0.5 μM UP1 Primer 500 nM 12.5 nM 0.125 μl 1.5 μl 2.5 mM each dNTP 2.5 mM 0.045 mM (each) 0.09 μl 1.08 μl Nuclease-free water — — 2.975 μl 35.7 μl Total volume — — 4.45 μl 53.4 μl
Table 1. All oligos needed for the single cell RNA-Seq protocol.
| Oligo Name | Oligo Sequence (all oligos sequence read 5′-->3′) |
|---|---|
| UP1 | ATATGGATCCGGCGCGCCGTCGACTTTTTTTTTTTTTTTTTTTTTTTT |
| UP2 | ATATCTCGAGGGCGCGCCGGATCCTTTTTTTTTTTTTTTTTTTTTTTT |
| AUP1 | (NH2)ATATGGATCCGGCGCGCCGTCGACTTTTTTTTTTTTTTTTTTTTTTTT |
| AUP2 | (NH2)ATATCTCGAGGGCGCGCCGGATCCTTTTTTTTTTTTTTTTTTTTTTTT |
| Fragment Library P1 PCR Primer | CCACTACGCCTCCGCTTTCCTCTCTATG |
| Fragment Library P2 PCR Primer | |
| Fragment Library P1 Adaptor -5′end | CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT |
| Fragment Library P1 Adaptor 3′ end | ATCACCGACTGCCCATAGAGAGGAAAGCGGAGGCGTAGTGGTT |
| Fragment Library P2 Adaptor 5′ end | AGAGAATGAGGAACCCGGGGCAGTT |
| Fragment Library P2 Adaptor 3′ end | CTGCCCCGGGTTCCTCATTCTCT |
| P1-Adaptor-5-end | ATCACCGACTGCCCATAGAGAGGTT |
| P1-Adaptor-3-end | CCTCTCTATGGGCAGTCGGTGAT |
| P2-Bar-1-5-end | CGCCTTGGCCGTACAGCAGGGGCTTAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-1-3-end | CTGCCCCGGGTTCCTCATTCTCTAAGCCCCTGCTGTACGGCCAAGGCG |
| P2-Bar-2-5-end | CGCCTTGGCCGTACAGCAGGGTGTGAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-2-3-end | CTGCCCCGGGTTCCTCATTCTCTCACACCCTGCTGTACGGCCAAGGCG |
| P2-Bar-3-5-end | CGCCTTGGCCGTACAGCAGAAGGGGAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-3-3-end | CTGCCCCGGGTTCCTCATTCTCTCCCCTTCTGCTGTACGGCCAAGGCG |
| P2-Bar-4-5-end | CGCCTTGGCCGTACAGCAGCCGATGAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-4-3-end | CTGCCCCGGGTTCCTCATTCTCTCATCGGCTGCTGTACGGCCAAGGCG |
| P2-Bar-5-5-end | CGCCTTGGCCGTACAGCAGCAACGAAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-5-3-end | CTGCCCCGGGTTCCTCATTCTCTTCGTTGCTGCTGTACGGCCAAGGCG |
| P2-Bar-6-5-end | CGCCTTGGCCGTACAGCAGGTGCCCAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-6-3-end | CTGCCCCGGGTTCCTCATTCTCTGGGCACCTGCTGTACGGCCAAGGCG |
| P2-Bar-7-5-end | CGCCTTGGCCGTACAGCAGGTCTGGAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-7-3-end | CTGCCCCGGGTTCCTCATTCTCTCCAGACCTGCTGTACGGCCAAGGCG |
| P2-Bar-8-5-end | CGCCTTGGCCGTACAGCAGACGGAGAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-8-3-end | CTGCCCCGGGTTCCTCATTCTCTCTCCGTCTGCTGTACGGCCAAGGCG |
| P2-Bar-9-5-end | CGCCTTGGCCGTACAGCAGGAAGGGAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-9-3-end | CTGCCCCGGGTTCCTCATTCTCTCCCTTCCTGCTGTACGGCCAAGGCG |
| P2-Bar-10-5-end | CGCCTTGGCCGTACAGCAGGACCGCAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-10-3-end | CTGCCCCGGGTTCCTCATTCTCTGCGGTCCTGCTGTACGGCCAAGGCG |
| P2-Bar-11-5-end | CGCCTTGGCCGTACAGCAGCTCAGGAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-11-3-end | CTGCCCCGGGTTCCTCATTCTCTCCTGAGCTGCTGTACGGCCAAGGCG |
| P2-Bar-12-5-end | CGCCTTGGCCGTACAGCAGAGCGTTAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-12-3-end | CTGCCCCGGGTTCCTCATTCTCTAACGCTCTGCTGTACGGCCAAGGCG |
| P2-Bar-13-5-end | CGCCTTGGCCGTACAGCAGCGGGTCAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-13-3-end | CTGCCCCGGGTTCCTCATTCTCTGACCCGCTGCTGTACGGCCAAGGCG |
| P2-Bar-14-5-end | CGCCTTGGCCGTACAGCAGCGTCTGAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-14-3-end | CTGCCCCGGGTTCCTCATTCTCTCAGACGCTGCTGTACGGCCAAGGCG |
| P2-Bar-15-5-end | CGCCTTGGCCGTACAGCAGTAGCGTAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-15-3-end | CTGCCCCGGGTTCCTCATTCTCTACGCTACTGCTGTACGGCCAAGGCG |
| P2-Bar-16-5-end | CGCCTTGGCCGTACAGCAGGCGTTTAGAGAATGAGGAACCCGGGGCAGTT |
| P2-Bar-16-3-end | CTGCCCCGGGTTCCTCATTCTCTAAACGCCTGCTGTACGGCCAAGGCG |
| 16Barcode Library PCR Primer-1 | CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT |
| 16Barcode Library PCR Primer-2 | CTGCCCCGGGTTCCTCATTCT |
Please refer to the web version for colour-coding.
Red – indicates barcode sequence
Green – 16Barcode Library PCR Primer-2 sequence for amplification during emulsion PCR
Blue – segment of 16Barcode Library PCR Primer-1 that matches P1 adapter
CRITICAL STEP Prepare master mix based on the number of the samples (here we show processing 10 samples as an example). To minimize pipetting error, prepare at least 12× worth of lysis buffer if the total number of samples is 10. The final volume of RT reaction is 5 μl per tube including 4.45 μl lysis buffer, about 0.1 μl PBS-BSA carryover (when picking single cells), and 0.45 μl RT mix. All final concentrations were calculated based on the volume of the 5 μl RT reaction.
-
3.
For picking up and transferring individual cells, we usually use a mouth tube to control a micropipette attached to it under a dissection microscope (also described in Ref. 39 & 41). First isolate four-cell stage embryos from the oviduct (see Ref. 42). Transfer them by micropipette to a drop of Acidic Tyrode’s solution to remove the zona pellucida. Then transfer the embryos into calcium free medium and gently pipette until the individual blastomeres separate. Transfer the blastomeres into three drops of PBS-BSA sequentially to wash them. Finally transfer the blastomeres into a final drop of PBS-BSA for picking.
CRITICAL STEP Be careful during all the transfer steps to make sure no bubbles are made in the drops by the micropipette.
-
4.
Seed a single cell (with PBS-BSA carryover) into each 0.5 ml thin-wall PCR tube containing 4.45 μl of Cell Lysis Buffer.
CRITICAL STEP Make sure the part of the micropipette outside the mouse tube is long enough (longer than 4 cm) to permit the tip of it to dip into the lysis buffer at the bottom of the 0.5 ml PCR tube with the mouse tube part not touching the lid and rim of the PCR tube to avoid any potential contamination. Also, during picking single cells using the micropipette gloves should be worn to avoid potential contaminations of the PCR tubes.
CRITICAL STEP First suck a small volume of PBS-BSA using the mouse pipette, then gently suck the single cell into the micropipette to keep the cell already inside the micropipette, but still near the tip of the micropipette. When pushing the cell out of the micropipette into the lysis, push out all the carryover PBS-BSA.
CRITICAL STEP It is crucial to make sure that the individual cell to be analyzed has actually been transferred into the lysis buffer from the drop of PBS-BSA by a micropipette. At this starting point the operator can practice transferring an individual cell into a drop of 4 - 5 μl PBS-BSA instead of lysis buffer on a 6 cm cell culture dish, and then recover it to ascertain if they can accurately transfer an individual cell into a small drop of the solution.
CRITICAL STEP Each micropipette can be used only once for an individual cell when picking the cell into the lysis buffer in the PCR tube. Never use the same micropipette repeatedly for transferring single cells into lysis buffer.
CRITICAL STEP It is essential to include a negative control sample without picking a single cell into it, but only pick a PBS-BSA solution equivalent to the carryover volume when picking a single cell into the lysis buffer tube. This negative control will ensure that all the steps from the start to the end of the whole procedure are not contaminated.
-
5.
Centrifuge for 30 sec at 7,500 g at 4 °C and put on ice immediately.
-
6.
Incubate at 70 °C for 90 sec and put on ice immediately.
CRITICAL STEP Use a thermal cycler with heated lid for all incubations throughout this protocol.
CRITICAL STEP The protocol also allows use of purified total RNA instead of single cells. If using total RNA, simply use total RNA to replace the single cell and keep the final volume of the reverse transcription reaction at 5 μl (although the volume of water in the lysis buffer should be adjusted). 20 pg to 1 ng of total RNA for each sample should work.
-
7.
Centrifuge the tubes for 30 sec at 7,500 g at 4 °C and put them on ice immediately for 1 min.
CRITICAL STEP After this step, all mRNAs from the single cell are released.
Reverse transcription
-
8.Prepare the Reverse Transcriptase (RT) mix (0.45 μl per sample) by combining and mixing the following components:
Component Stock
concentrationFinal
concentration×1
volume×12
volumeSuperScript III Reverse Transcriptase 200 U μl−1 13.2 U μl−1 0.33 μl 3.96 μl RNase Inhibitor (Ambion) 40 U μl−1 0.4 U μl−1 0.05 μl 0.6 μl T4 gene 32 protein 1-10 U μl−1
(5 U μl−1 in average)0.07 U μl−1 0.07 μl 0.84 μl Total volume — — 0.45 μl 5.4 μl
CRITICAL STEP Prepare master mix based on the number of the samples. To minimize pipetting error, prepare at least 12× worth of reaction mix if the total number of samples is 10.
-
9.
Add 0.45 μl RT mix to each tube. The total volume is now 5.0 μl.
-
10.
Incubate at 50 °C for 30 min.
-
11.
Inactivate the reverse transcriptase at 70 °C for 10 min.
-
12.
12. Centrifuge tubes for 30 sec at 7,500 g at 4 °C and immediately place on ice for 1 min.
CRITICAL STEP After this step, first strand cDNAs for all mRNAs are synthesized.
Free primer removal
-
13.Prepare the Exonuclease I mix by combining and mixing the following components:
Component Stock
concentrationFinal
concentration
in Cut (1 μL)×1
volume×12
volume10X Exonuclease I Buffer 10× 1× 0.1 μl 1.2 μl Nuclease-free water — — 0.8 μl 9.6 μl Exonuclease I 5 U μl−1 0.5 U μl−1 0.1 μl 1.2 μl Total volume — — 1 μl 12 μl
CRITICAL STEP Prepare master mix based on the number of the samples. To minimize pipetting error, prepare at least 12× worth of reaction mix if the total number of samples is 10.
-
14.
Add 1.0 μl of Exonuclease I mix to each reaction. Total volume in the tube is now 6 μl.
-
15.
Incubate at 37 °C for 30 min.
-
16.
Inactivate the Exonuclease I at 80 °C for 25 min.
-
17.
Centrifuge tubes for 30 sec at 7,500 g at 4 °C and immediately place on ice for 1 min.
CRITICAL STEP After this step, all free UP1 primers are destroyed and the 3′-end of cDNAs are shortened about 50 bp. The 5′-end of cDNAs (UP1 sequence) are intact.
3′-Poly-A tailing
-
18.Prepare terminal deoxynucleotidyl transferase (TdT) reaction mix by combining and mixing the following components:
Component Stock
concentrationFinal
concentration×1
volume×11
volume10X PCR Buffer II (without MgCl2) 10× 1× 0.6 μl 6.6 μl 25 mM MgCl2 25 mM 1.5 mM 0.36 μl 3.96 μl 100 mM dATP 100 mM 3 mM 0.18 μl 1.98 μl Nuclease-free water — — 4.26 μl 46.86 μl Terminal Transferase 15 U μl−1 0.75 U μl−1 0.3 μl 3.3 μl RNase H 2 U μl−1 0.1 U μl−1 0.3 μl 3.3 μl Total volume — — 6 μl 66 μl
CRITICAL STEP Prepare master mix based on the number of the samples. To minimize pipetting error, prepare at least 11× worth of reaction mix if the total number of samples is 10.
-
19.
Add 6.0 μl of TdT mixture to each reaction. Total volume is now 12.0 μl per tube.
-
20.
Incubate at 37 °C for 15 min.
-
21.
Inactivate the terminal deoxynucleotidyl transferase at 70 °C for 10 min.
-
22.
Centrifuge tubes for 30 sec at 7,500 g at 4 °C and immediately place on ice for 1 min.
CRITICAL STEP After this step, 3′-end of the first-stranded cDNAs has a poly(A) tail.
Second strand cDNA synthesis
-
23.Prepare 76 μl PCR Mix 1 for each reaction by combining and mixing the following components:
Component Stock
concentrationFinal
concentration×1
volume×11
volume10X Ex Taq Buffer (with MgCl2) 10× 1× 7.6 μl 83.6 μl 2.5 mM each dNTP 2.5 mM 0.25 mM 7.6 μl 83.6 μl UP2 primer (100 μM) 100 μM 1 μM 0.76 μl 8.36 μl Nuclease-free water — — 59.28 μl 652.08 μl TaKaRa Ex Taq™ HS 5 U μl−1 0.05 U μl−1 0.76 μl 8.36 μl Total volume — — 76 μl 836 μl -
24.
Divide the poly(A) tailed RT product (12 μl) into four empty 0.2ml thin wall PCR tubes (3 μl for each tube).
-
25.
Add 19 μl PCR Mix 1 (primer UP2) to each tube (final concentration of UP2 Primer is 0.86 μM). The total volume in the tube is 22 μl.
-
26.
Perform the following PCR program: one cycle of 95 °C for 3 min, 50 °C for 2 min, and 72 °C for 10 min.
-
27.
Put tubes on ice for 1 min.
-
28.
Centrifuge tubes for 30 sec at 7,500 g at 4 °C and immediately place on ice.
CRITICAL STEP After this step, the second-strand cDNAs are 5′-UP2-(T)n-cDNA-(A)n-UP1-3′
PCR amplification
-
29.Prepare 76 μl PCR Mix 2 for each reaction by combining and mixing the following components:
Component Stock
ConcentrationFinal
Concentration×1
Volume×11
Volume10X Ex Taq Buffer (with MgCl2) 10X 1 X 7.6 μl 83.6 μl 2.5 mM each dNTP 2.5 mM 0.25 mM 7.6 μl 83.6 μl 100 μM UP1 primer 100 μM 1 μM 0.76 μl 8.36 μl Nuclease-free water — — 59.28 μl 652.08 μl TaKaRa Ex Taq™ HS 5 U μl−1 0.05 U μl−1 0.76 μl 8.36 μl Total volume — — 76 μl 836 μl -
30.
Add 19 μl of the PCR Mix 2 to each tube (the final concentration of UP1 and UP2 is 0.46 μM each). The total volume in the tube is 41 μl.
-
31.
Perform the following PCR program: 95 °C for 3 min, then 20 cycles of 95 °C for 30 sec, 67 °C for 1 min, and 72 °C for 6 min (+ 6 sec each cycle).
PAUSE POINT The single cell cDNA PCR product can be saved at −80 °C for 6 months.
CRITICAL STEP The PCR amplification cycles can be adjusted according to the size (or mRNA content) of the single cell. For a blastomere from a four-cell stage embryo, we use 20 cycles of PCR. For a mature oocyte, we use 18 cycles of PCR.
CRITICAL STEP 1st cycle = 6 minutes and 6 seconds, 2nd cycle = 6 minutes and 12 seconds, 3rd cycle = 6 minutes and 18 seconds, etc.
CRITICAL STEP After this step, all cDNAs have been amplified.
Optional DNA purification step
-
32.
Mix the divided PCR product together (41 μl × 4 = 164 μl for each sample).
-
33.
Take 10 μl of single cell PCR products, add 90 μl Nuclease-free water to dilute ten folds. Take 1 μl (or 2 μl) as template to run a 20 μl SYBR Green real-time PCR reaction to check the expression of house-keeping genes (such as GAPDH, HPRT, β-actin) or specific marker genes (such as H1foo, oog1 for mature oocytes).
-
34.
Purify the remaining 154 μl PCR product with QIAquick PCR Purification Kit, and elute with 50 μl EB buffer.
-
35.
Store at −80 °C.
PAUSE POINT The purified PCR product can be saved at −80 °C for 6 months.
2nd round of PCR and QC steps
-
36.2nd PCR amplification – Set up 4 of the following 90 μl PCR reactions per sample. (Adjust the number of reactions according to the final yield of the cDNA after gel purification.)
Component Stock
ConcentrationFinal
Concentration×1
VolumePurified 1st PCR products — — 1.2 μl 10X Ex Taq Buffer 10X 1X 9 μl 2.5 mM each dNTP 2.5 mM (each) 0.25 mM (each) 9 μl Amine-blocked Universal Primer 1 (AUP1) 100 μM 1 μM 0.9 μl Amine-blocked Universal Primer 2 (AUP2) 100 μM 1 μM 0.9 μl Nuclease-free water — — 68.1 μl Ex Taq HS 5 U μl−1 0.05 U μl−1 0.9 μl Total volume — — 90 μl
CRITICAL STEP In our experience, with one blastomere of four-cell stage embryo (or 20 pg of input total RNA), four reactions at this step will produce at least 100 ng of gel purified cDNA in the next step.
-
37.
Run the following PCR program: 95 °C for 3 min, then 9 cycles of 95 °C for 30 sec, 67 °C for 1 min, and 72 °C for 6 min (+ 6 sec each cycle).
PAUSE POINT The 2nd round PCR product can be saved at −80 °C for 6 months.
CRITICAL STEP The template amount for the 2nd round of PCR can be flexible. 1 μl to 4 μl of the purified 1st round PCR product can be used for each 90 μl PCR reaction. The PCR amplification cycles can also be adjusted according to the final amount of cDNA needed. The PCR cycles can be between 6 to 12. And cycles of PCR should be minimized to reduce bias. Also, the combined cycles of PCR from the 1st and 2nd rounds of PCR should be less than 30 cycles.
-
38.
Optional quality check: Combine the four 90 μl PCR reactions. Take 20 μl of the 2nd round PCR product to run electrophoresis on a 2% E-Gel.
-
39.
For the remaining 340 μl PCR product, purify the cDNA using Qiaquick PCR purification kit. Elute in 30 μl EB buffer.
-
40.
Store at −80 °C.
PAUSE POINT The purified PCR product can be saved at −80 °C for 6 months.
Gel purification
-
41.
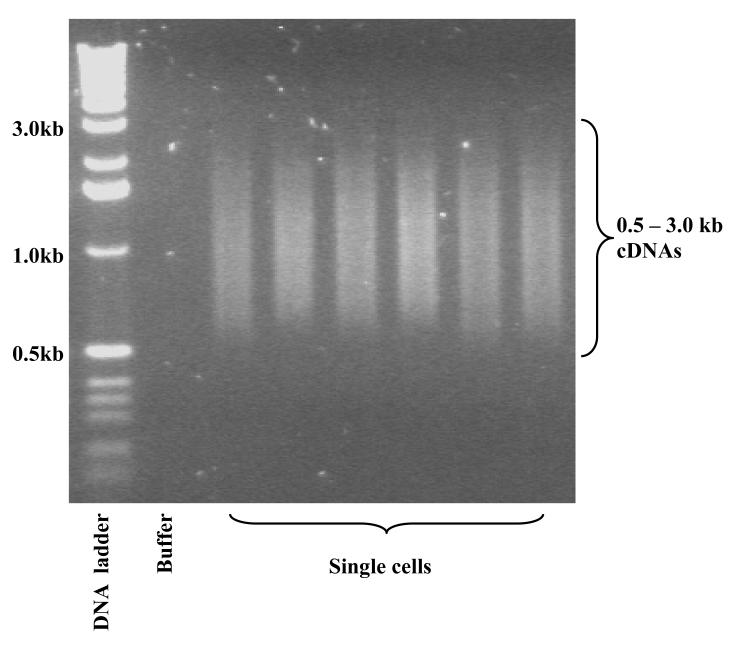
Load the cDNA on 1% agarose gel. Run the gel until the BPB dye is 2 cm from the well. Excise the cDNA from 0.5 kb to 3 kb. Purify the cDNA using Qiaquick gel extraction kit. Elute the cDNA in 30 ul EB buffer. Store at −80 °C. Quantify the cDNA using Qubit and Quant-it HS dsDNA kit.
PAUSE POINT The gel purified PCR product can be saved at −80 °C for 6 months.
CRITICAL STEP The cDNA is weakly stained because the primer dimer band saturates the ethidium bromide. If this happens, simply cut the gel between 0.5 kb to 3 kb and proceed to gel extraction. Or the operator can stain the gel with ethidium bromide to see the cDNA smear.
CRITICAL STEP The AUP1 and AUP2 primers have a NH2 modification at their 5′-ends that prevents the ligation of the 5′-end fragments of the double-stranded cDNA (after the shearing) to the SOLiD™ library P1/P2 adaptors. Because of this, any undesired small amplification products that may have been formed will not be carried into the library being generated.
-
42.
Optional quality check: If you have enough cDNA to spare and want to check the quality of the gel purified cDNA, run 50 – 100 ng of the purified cDNA on a 2% agarose gel (see Fig. 1).
Figure 1. Typical agarose gel electrophoresis for single cell cDNAs after 20 + 9 cycles of PCR and gel purification.
SOLiD System Express Library Preparation for Fragment Libraries and Multiplexed Fragment Libraries
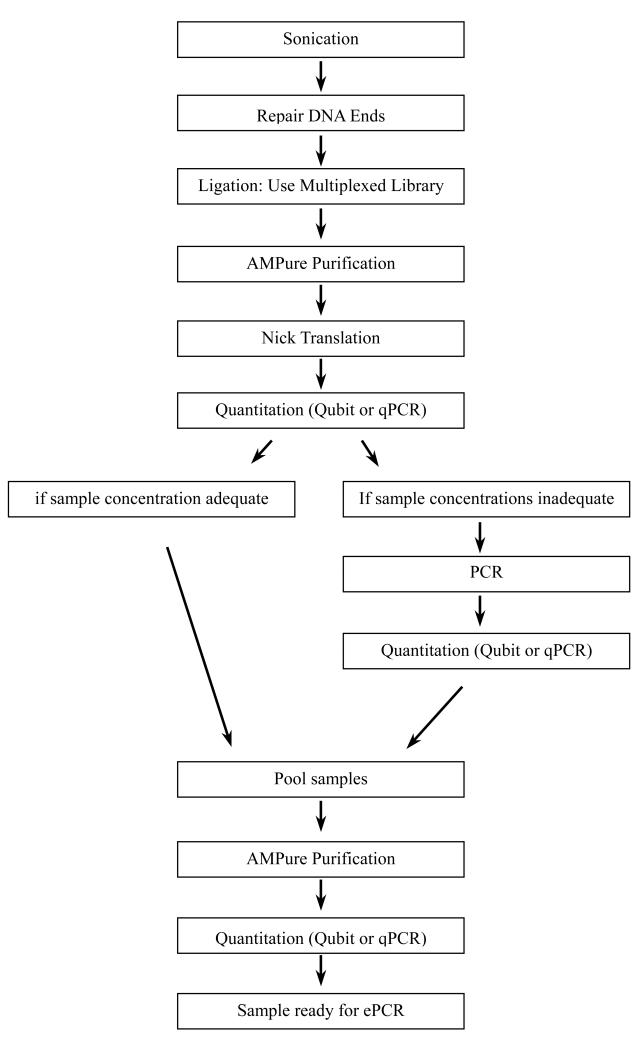
Workflow (see Fig. 2).
Figure 2. Workflow of the SOLiD System Express Library Preparation for Fragment Libraries and Multiplexed Fragment Libraries.
Fragment Library Construction Oligonucleotides
CRITICAL STEP All library construction oligonucleotides needed for fragment library construction are included in the SOLiD™ Fragment Library Oligo Kit. All multiplexed oligonucleotides must be ordered and hybridized to create double-stranded adapters (see Table 1).
CRITICAL STEP Adjust microcentrifuge speeds and times according to the g-forces specified in the protocols. Applied Biosystems recommends the Eppendorf 5417R tabletop microcentrifuge.
CRITICAL STEP All steps requiring the use of 1.5-ml tubes should be done with the 1.5-ml Lo-Bind tubes (Eppendorf, catalog number 022431021).
Hybridization of Oligonucleotides
-
43.
Prepare 1 mM stock of individual oligonucleotides.
-
44.
Mix equal volumes of 1 mM oligonucleotide A and B. Add enough 5X Ligase buffer for a final concentration of 1X Ligase buffer.
-
45.Hybridize the oligonucleotides by running the following program on a PCR machine:
Temperature Time 95 °C 5 min 72 °C 5 min 60 °C 5 min 50 °C 3 min 40 °C 3 min 30 °C 3 min 20 °C 3 min 10 °C 3 min 4 °C Forever -
46.
After hybridization, store hybridized oligonucleotides at −20 °C until ready for use.
DNA Shearing using the Covaris™ S2 System
CRITICAL STEP Fine tuning the shearing protocol may be necessary for certain DNA samples. However, the following procedures can be used as a guideline when using Covaris™ S2 System. These conditions have been tested for shearing 50 ng- 10 μg of DNA in a total volume of 100 μl in a Covaris™ S2 System. Be sure to use the appropriate tube holder, and program the instrument correspondingly.
CRITICAL STEP Set the chiller temperature to between 2-5 °C to ensure the temperature reading in the water bath displays 5 °C. The circulated water chiller should be supplemented with 20% ethylene glycol.
Shear the DNA
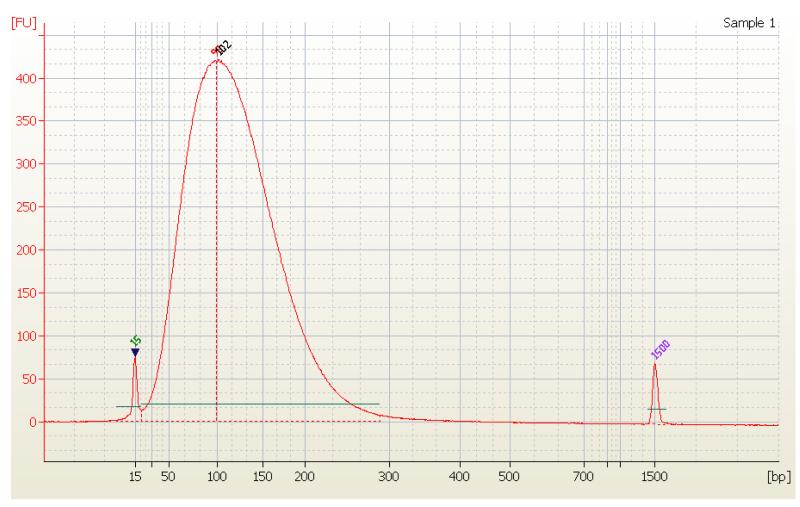
The sonication protocol generates DNA fragments with mean size 100-110 bp (see Fig. 3)
Figure 3. Electrophoregram of DNA sheared in C4011-10 tubes generated using Bioanalyzer (Agilent Technologies, Inc.).
-
47.In borosilicate glass microTUBE dilute desired number of micrograms of DNA in 100 μl of 10 mM Tris, pH 7.5.
Component Volume 1-2 μg DNA x μl 1M Tris, pH 7.5 1 μl Nuclease-free water Variable Total volume 100 μl -
48.Shear the DNA using the Covaris™ S2 System shearing program described below:
- No. of cycles: 15
- Bath Temperature: 5 °C
- Bath temp limit: 30 °C
- Mode: Frequency sweeping
- Water Quality Testing Function: Off
- Treatment 1:
- Duty Cycle: 20%
- Intensity: 5
- Cycles/burst: 200
- Time: 60 sec
-
49.
Transfer the sheared DNA into a clean 1.5 ml LoBind tube.
-
50.
For quality control, 2 μl of the reaction should be saved and used to confirm that shearing has created the desired range of fragment sizes using the Agilent 2100 Bioanalyzer.
Repair the DNA Ends
-
51.Add to the tube containing sheared DNA the following components:
Component Volume Sheared DNA 98 μl 10× End-it Buffer 14 μl End-it ATP (10 mM) 7 μl End-it dNTPs (2.5 mM) 14 μl End-it Enzyme Mix 2 μl Nuclease-free water 5 μl Total volume 140 μl -
52.
Incubate at room temperature for 30 min.
-
53.
Inactivate End-it Enzymes by transferring tube to the heating block and incubating for 20 min at 65 °C. Centrifuge the tube for 5 sec at 7,500 g at 4 °C.
Ligate P1 (ds) and P2 (ds) Adapters to End-Repaired DNA
-
54.
Thaw the adapters on ice or at room temperature.
-
55.Use the information below to calculate the pmoles of adapter needed for the reaction.
- 1 μg DNA × 106 pg μg−1 × 1 pmol 660pg−1 × 1/(average insert fragment size) = X pmoles
- (# μg DNA) × (X pmoles DNA) = (# pmoles DNA for adapter ligation)
- (# pmoles DNA for adapter ligation) × (30) = (# pmoles adapters needed)
- (# pmoles adapters needed) / (# pmoles/μl stock) = (# μl adapter needed)
-
56.In a tube containing the end-repaired DNA from the previous step combine and mix the components below. For DNA inputs higher than 2 μg, scale-up the total volume of the reaction and amount of components (buffer, adaptors, ATP, ligase) correspondingly. Add 1 μl of Quick Ligase per 20 μl of reaction volume. Use one adapter set per sample.
Component Volume i.e. Volume
for 2 μg input
DNAP1 (ds) Adapter (500 pmol μl−1) As determined above 1.8 μl P2 (ds) Adapter (500 pmol μl−1) As determined above 1.8 μl 10× End-it Buffer 2 μl 2 μl End-it ATP (10 mM) 2 μl 2 μl Quick Ligase Enzyme (NEB) 8 μl 8 μl End-repaired DNA 140 μl 140 μl Nuclease-Free Water Variable 4.4 μl Total volume 160 μl 160 μl -
57.
Incubate at room temperature for 10-15min.
Purification of ligated DNA using Agencourt’s Solid Phase Reversible Immobilization (SPRI) purification system
CRITICAL STEP Sample purification at the post-ligation step is recommended with the Agencourt® AMPure® Kit. The Agencourt® AMPure® PCR purification system utilizes solid-phase paramagnetic bead technology to purify DNA. The reaction conditions are optimized to selectively bind DNA 100 bp and larger. Excess oligonucleotides, nucleotides, salts and enzymes can be removed by a simple washing procedure. Agencourt® AMPure® purification will remove the 68-bp primer-dimer contamination and reduce carryover of this product into the fragment libraries.
-
58.
Purify ligated DNA using the Agencourt® AMPure® Kit.
-
a)
Add 1.8× volumes of Agencourt® AMPure® beads to the sample (~160 μl) and incubate for 5 min at room temperature on a rotator
-
b)
Prepare 70% ethanol (~650 μl per sample).
CRITICAL STEP Using freshly prepared and precisely 70% ethanol is critical, as a higher percentage will result in inefficient washing of smaller-sized molecules and using lower than 70% ethanol could cause loss of sample.
-
c)
Place the tube of beads in the magnetic rack to separate beads from solution. Wait for the solution to clear before proceeding to the next step.
-
d)
Remove the supernatant and discard.
-
e)
Dispense 200 μl of freshly prepared 70% ethanol, vortex the tube thoroughly, and incubate for 30 sec at room temperature.
-
f)
Place the tube of beads in the magnetic rack to separate beads from solution. Wait for the solution to clear before proceeding to the next step.
-
g)
Aspirate out the ethanol and discard.
-
h)
Repeat steps d–g two more times.
-
i)
Place the tube of beads in the magnetic rack and remove the supernatant and pulse-spin to remove the residual ethanol.
-
j)
Repeat step i 2-3 more times to remove the residual ethanol.
-
k)
Dry the beads at room temperature for approximately 5 min.
-
l)
Elute the DNA by adding 36 μl 10 mM Tris pH 8, vortexing for 10 sec and ensuring homogeneity by pipetting the solution up and down several times.
-
m)
Place the tube of beads in the magnetic rack to separate beads from solution. Wait for the solution to clear before proceeding to the next step.
-
n)
Save the eluant in a 1.5 ml LoBind tube.
-
o)
Place the eluted sample in the magnetic rack again to separate any remaining beads from solution. Wait for the solution to clear before proceeding to the next step.
-
p)
Save the eluted sample in a new 1.5 ml LoBind tube.
-
q)
Repeat steps p-q once.
-
r)
Store DNA in a 1.5 ml LoBind tube.
-
s)
Save 2 μl of the AMPure®-purified ligated library DNA. The aliquot can be used for running a library QC for troubleshooting purposes.
PAUSE POINT The sample(s) may be stored at −20°C.
For Multiplexed Samples
CRITICAL STEP Using Qubit (following manufacturer’s protocol) or qPCR (see Steps 66-71) determine the concentration of each of the multiplexed samples. Pool equal concentrations of the samples and purify with AMPure beads as above, eluting with 34 μl of 10 mM Tris, pH 8 solution and continue with nick translation. If necessary, after nick translation PCR amplification can be performed on individual samples, or on pooled samples, as long as the pooled samples are equal in concentration to each other.
CRITICAL STEP For fragment libraries or multiplexed libraries that are not yet pooled, the operator may skip quantitation and go directly on to nick translation—pooling before nick translation is primarily to conserve reagents.
Nick-Translation of the DNA
-
59.Combine and mix the following components in a LoBind tube:
Component Volume P1 (ds) and P2 (ds) adapter ligated fragment DNA 34 μl 10× NEBuffer 2 4 μl GeneAmp® dNTP Blend (100mM) 0.8 μl DNA Polymerase I (10 U μl−1) 1.0 μl Nuclease-free water Variable Total volume 40 μl -
60.
Incubate at 16 °C for 30 min.
-
61.
Stop the reaction by adding 0.5μl 0.5 M EDTA pH8.0
-
62.
62. Purify the nick-translated DNA with the the Agencourt® AMPure® Kit as before, eluting with 40 μl of a 10 mM Tris, pH 8.0 solution.
CRITICAL STEP Save 2 μl of the AMPure®-purified ligated library DNA. The aliquot can be used for running a library QC for troubleshooting purposes.
PAUSE POINT The sample(s) may be stored at −20°C.
CRITICAL STEP Proceed to qPCR to quantify library molecules bearing P1 and P2 adaptors. If the library concentration is too low for emulsion PCR (it should be above 1 ng μl −1) continue with PCR, otherwise begin emulsion PCR.
Quantification of library by qPCR
CRITICAL STEP Quantitative PCR is performed using real-time SYBR Green assay. For multiplexed samples that need to be pooled, normalize the Ct values and pool accordingly. For determining relative concentrations of the samples, two titration points are adequate per multiplexed sample.
CRITICAL STEP For the final pooled multiplexed library, Qubit may be used to determine the final concentration of the pooled samples, following the manufacturers’ protocol. For a fragment library sample, either Qubit or qPCR may be used to determine the final concentration. For qPCR of fragment libraries, a Standard Library is used to generate a standard curve.
CRITICAL STEP For quantitating samples for pooling, make sure all samples are done on the same plate to avoid variation that can occur between plates and instruments.
-
63.Prepare serial 10-fold dilutions of the library eluted in EB:
- 10−1: 3 μl of eluted library + 27 μl H2O,
- 10−2: 5 μl of 10−1 + 45 μl H2O,
- etc. down to 10−6
-
64.
Prepare Standard samples containing 390 fg, 39 fg, 3.9 fg, 0.39 fg, and 0.039 fg of Standard Library (E.coli DH10B fragment library, 150 bp mean fragment size) in 5 μl H2O. These values should be used for absolute quantification when programming the 7900HT Fast Real-Time PCR System to generate a Standard Curve.
CRITICAL STEP Both AB’s and Invitrogen’s SYBR Green Mixes work well. Assay is performed in a standard 96-well plate from Applied Biosystems.
qPCR Assay Pre-Mix using SYBR Green PCR Master Mix (AB, Cat. No 4309155)*
For one reaction:
| Final concentration | ×1 volume | |
|---|---|---|
| 2× SYBR Green Master Mix (ABI) | 1× | 15 μl |
| qPCR primer Fw (50 μM) | 0.9 μM | 0.54 μl |
| qPCR primer Rev (50 μM) | 0.9 μM | 0.54 μl |
| H2O | 8.92 μl | |
| Total volume | 25 μl |
ROX already pre-mixed in Master Mix
-
65.
Distribute 25 μl Pre-Mix in corresponding wells. Then add 5 μl of template DNA (or H2O for NTC). Seal the plate with adhesive optical cover. The total volume of PCR reaction is 30 μl.
-
66.When using Absolute Quantification with 7900HT Fast Real-Time PCR System, set up the following thermal profile:
95°C 2min 95°C 15sec 40 cycles 62°C 15 sec 70°C 1min - Also set:
- Thermal Cycler Protocol Mode: Standard
- Data Collection: at 70 °C (extension step)
-
67.
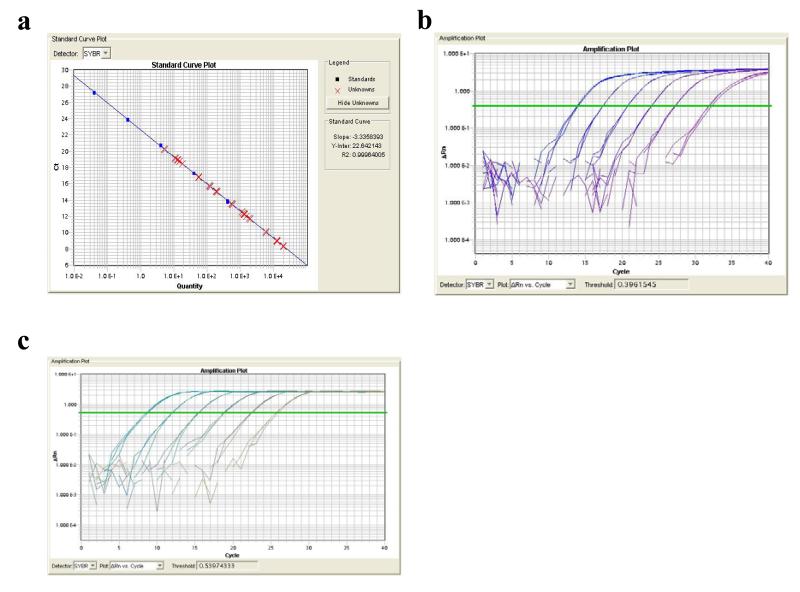
Run qPCR and quantify the amount of library DNA/number of P1-insert-P2 molecules. Typical Standard curve and Amplification plots for Standards and Library samples are shown in Fig. 4.
-
68.
Proceed to emulsion PCR step if sample concentration(s) adequate. Otherwise continue with PCR amplification, steps 69-71.
Figure 4. Standard curve of the cDNAs.
(a) Standard curve. (b) Standard plot. (c) Library plot.
(Optional) PCR Amplification
-
69.Perform minimal cycles of PCR as follows below. 25 μl of a sample, either a multiplexed pool of samples or a single multiplexed sample, is amplified using the following protocol:
Component Volume Multiplexed sample 25 μl Invitrogen™ SuperMix 75 μl 16Barcode Library PCR Primer 1 1.5 μl 16Barcode Library PCR Primer 2 1.5 μl Pfu polymerase 0.25 μl AmpliTaq 0.2 μl Total volume 103.45 μl Temperature Time Condition/note 95 °C 5 min Hold 95 °C 15 sec 8-16 cycles 62 °C 15 sec 70 °C 1 min 70 °C 5 min Hold 4 °C Forever Hold
CRITICAL STEP The number of cycles should be decided based on the amount of starting material used for shearing. Minimal cycling is desirable for avoiding over-amplification. Use the table below as a guideline to determine the number of PCR cycles based on the amount of input DNA:
| Starting Amount of DNA | Number of PCR Cycles |
|---|---|
| 1 μg to 2 μg | 8-10 |
| 100 ng to 1 μg | 10-12 |
| 20 ng to 100 ng | 13-16 |
-
70.
To determine if amplification was adequate, load and run 4 μl of sample on a 2.2% Lonza FlashGel® cassette for 6 min at 275 V to ensure amplification after a minimal number of cycles as described above.
-
71.If fairly robust amplification products are visible, proceed with Step 72. If little or no amplification products are observed at this point, return the tubes to the thermal cycler and run the PCR cycling program below:
Temperature Time Condition 95 °C 5 min Hold 95 °C 15 sec X 2-3 cycles 62 °C 15 sec 70 °C 1 min 70 °C 5 min Hold 4 °C Forever Hold -
72.
Repeat AMPure purification as in step 58.
-
73.
Quantify sample(s) with either Qubit quantitation or qPCR.
TIMING
Day 1: Step 1 - 2: 1 h
Step 3 - 11: 1 h
Step 12 - 16: 1.5 h
Step 17 - 21: 1 h
Step 22 - 27: 0.5h
Step 28 - 30: 5h
Day 2: Step 31 - 34: 3 - 7 h
Day 3: Step 35 - 36: 3 h
Step 37 - 39: 3h
Day 4: Step 40: 5h
Step 41: 1.5h
Day 5:
Step 43-46: 0.5 h
Step 47-50: 0.5-1 h if Bioanalyzer used
Step 51-53: 1 h
Step 54-57: 0.3 h
Step 58: Time depends on sample numbers. For 16 samples, about 1 h
Step 59-61: 0.5 h
Step 62: Time depends on sample numbers. For 16 samples, about 1 h
Step 63-68: 2 h
Day 6:
Step 69-71 (optional): 1 h
Step 72: Time depends on sample numbers. For 16 samples, about 1 h
Step 73: 2 h if using qPCR
TROUBLESHOOTING
| Problem | Possible reason | Solution |
|---|---|---|
| Cell not transferred into lysis tube | Cell stuck to the inside wall of the glass capillary (micropipette) | First suck a small volume of PBS-BSA using the mouse pipette, then suck the single cell into the glass capillary gently to keep the cell already inside the capillary but still near the tip of the capillary. When pushing the cell out of the capillary into the lysis, push all the carryover PBS-BSA until a bubble is visible from the capillary into the lysis buffer. |
| Picking buffer only control shows positive signal when using real-time PCR to check expression of house-keeping genes | PBS-BSA drop is contaminated by lysed cells | Make sure the PBS-BSA drop for holding single cells is not contaminated by lysed cells. Wash the single cells through several PBS-BSA drops before picking them. Aliquot all reagents for lysis, RT, cutting, tailing, and PCR steps in small batches. Each aliquot is only used once and the remainder is discarded. Also only load the exact number of single cells that will be picked into the final PBS-BSA drop. After picking each of them, check the number of remaining single cells to make sure every cell is correctly picked and no cell is by chance lysed or incorrectly picked. |
| The expression level of house-keeping genes is much lower than expected in some single cell cDNA samples. | Some dead or partially damaged single cells were picked | Before picking, try to check the quality of the single cells by either morphology or trypan blue staining. Only pick healthy single cells. |
| RNase contamination during the lysate and reverse transcription step | Keep the bench and surrounding area very clean. Wear a dusk mask to avoid breathing contaminates into reaction reagents. Change gloves regularly during these steps. Try to do all manipulations in a clean hood. | |
| Loss of activity of some enzymes and reagents | Some of the reagents are not very stable, such as dATP, dNTP, or primers at low concentrations. Aliquot them into small batches. Avoid repeated freeze/thaws. Make sure reagents are not expired. | |
| Low recovery of gel purification | Gel fragment recovered more than the kit requested | Try to cut the gel fragment containing the cDNA smear more accurately. If too much gel is already cut, split them into two gel purification columns and finally combine the purified cDNAs. Each QIAGEN column can hold up to 400 mg of gel. |
| Primer dimers present on Lonza Gel in step 70 | Too many cycles of PCR were done, or the amount of adaptors added was too large relative to sample size | Gel purification must be done to remove primer dimers. SOLiD Fragment Library protocol recommends the use of 3% or 4% agarose gel run in 1X TAE, using a 25 bp Track-It ladder as reference. Cut the gel between 125 bp and 200 bp and purify using QIAGEN MinElute Gel Extraction Kit following manufacturer’s instructions. |
| Size distribution of fragment after shearing is not in the desired range | Sonication protocol was not calibrated correctly for the sample | Readjust shearing protocol and redo shearing. Make sure the Covaris S2 System is properly set up. |
| Low yield after AMPure purification | Ethanol was not freshly prepared, and/or was not at 70% | If there is adequate sample remaining, PCR can be done to increase the quantity, see steps 69-72. If not, protocol must be redone, using freshly prepared 70% ethanol. |
| Overall yield is low | Sample quality was poor | Confirm quality of sample before beginning Express protocol using a housekeeping gene assay, or use Qubit to confirm that the sample’s concentration is adequate for input. |
| Sample loss occurred during AMPure purification | See above. Check QC aliquots using Qubit or NanoDrop to determine where loss occurred. | |
| Loss of sample can occur during steps when sample is transferred or purified | Take care to transfer the entire volume during purification steps and minimize transferring of samples. Sample input may need to be increased if initial input was low and loss occurs. |
ANTICIPATED RESULTS
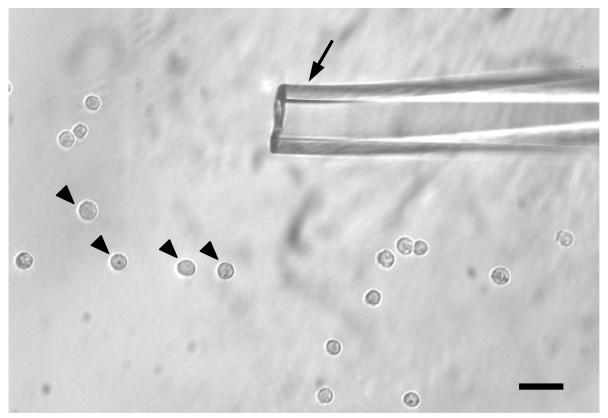
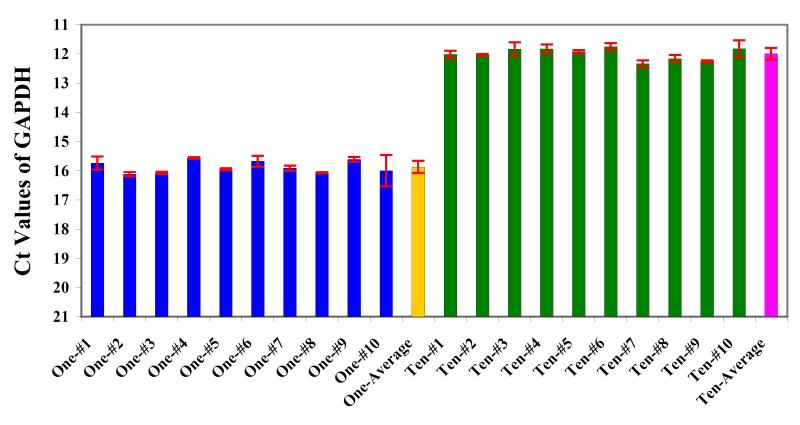
We first used relatively small cells—mouse embryonic stem (ES) cells—to show that the method is sensitive and reproducible, and we picked into each tube either one individual ES cell or ten ES cells together (Fig. 5). Then we ran the single cell cDNA amplification procedure and ran real-time PCR to analyze the expression of house-keeping genes, GAPDH. (Fig. 6).
Figure 5. The single ES cells in PBS-BSA drop.
Black arrowheads indicate individual ES cells. Black arrow indicates the tip of micropipette for picking single cells. The inner diameter of the micropipette is about 2 – 3 fold of the diameter of an individual ES cell. Scale bar: 30um.
Figure 6. GAPDH expression in single and ten mouse ES cells measured by real-time PCR.
One or ten ES cells were picked into each tube to amplify cDNAs by 20 cycles of PCR. Then the PCR product was diluted 10 fold (20ul into 200ul) and take 2ul as template for a 20ul real-time PCR reaction.
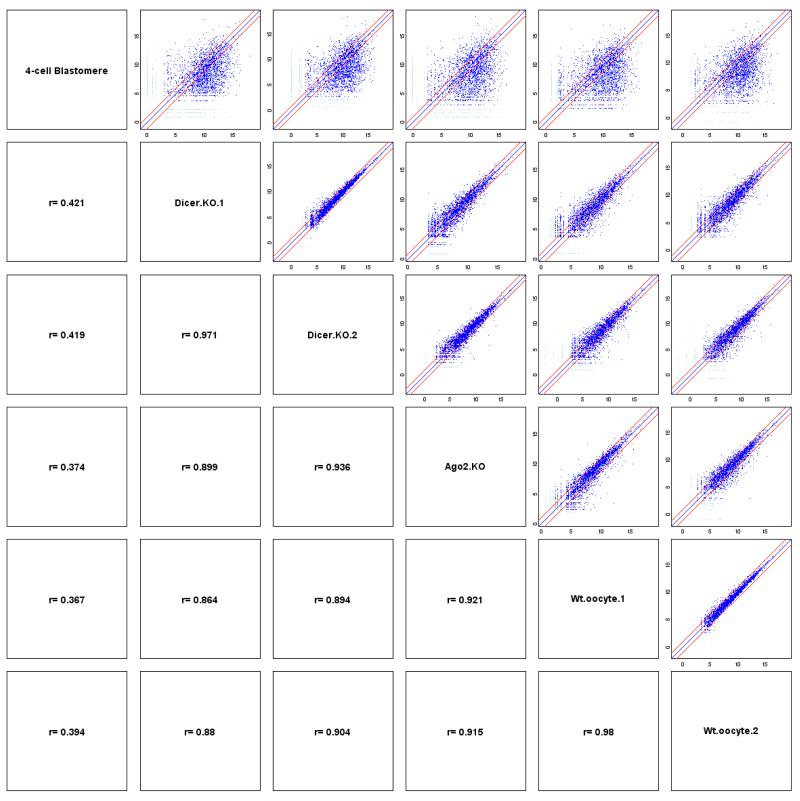
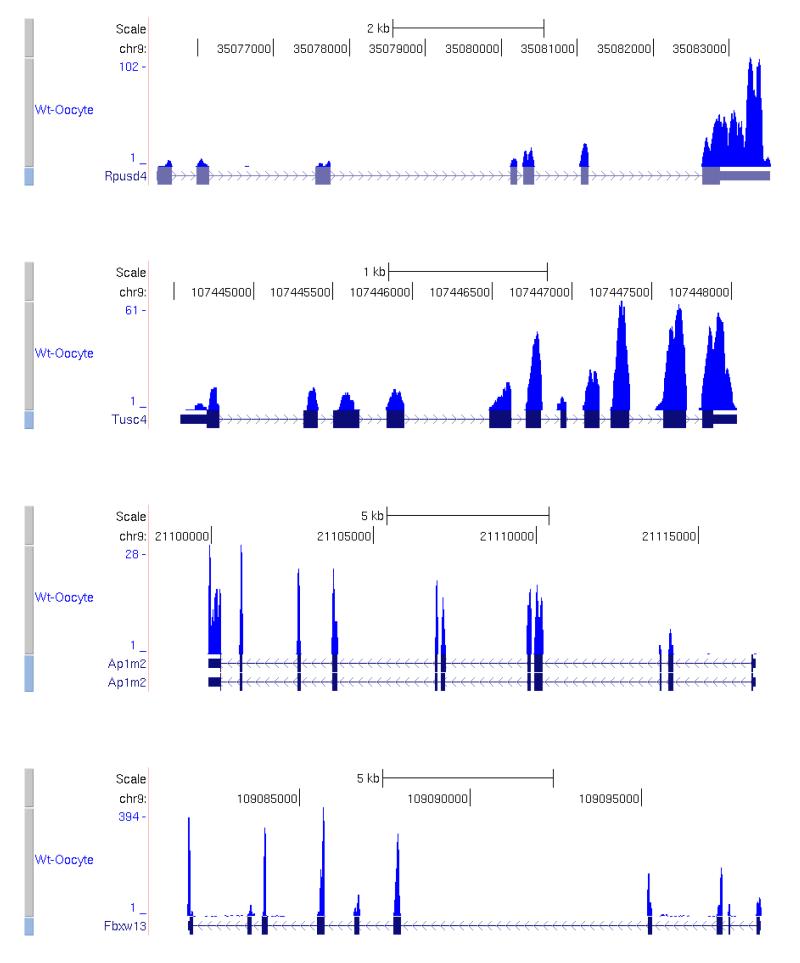
Then we confirmed the accuracy of the method by analyzing the correlation efficiency between two separately processed biological replicates. Based on our previous single cell cDNA microarray results (unpublished) and results from other labs, undifferentiated ES cells have strong subpopulations with dramatic differences in gene expression43-45. So we analyzed biological replicates of mature oocytes, as we know that mature oocytes are a much more homogeneous population40. We can achieve correlation coefficients as high as 0.986 (Fig. 7). The coverage plots40 of several individual genes are shown in Fig. 8. The top 20 most abundant genes40 in the mature oocyte and the blastomere of a 4-cell stage embryo respectively are shown in Table 2.
Figure 7. Pearson coefficient plots for single cell RNA-Seq of one blastomere of a 4-cell stage embryo, two wildtype mature oocytes, two Dicer knockout mature oocytes, and one Ago2 knockout mature oocyte.
Figure 8. Coverage plots of RNA-Seq reads in a single wildtype mature oocyte.
Table 2. Top 20 most abundant genes in the mature oocyte and the blastomere of 4-cell stage embryos, respectively.
| Gene name | RefSeq ID | Relative counts in mature oocyte |
Relative counts in 4-cell Blastomere |
|---|---|---|---|
| H1foo | NM_138311 | 209,872 | 2,117 |
| Bcl2l10 | NM_013479 | 205,060 | 1,027 |
| Spin1 | NM_011462 | 200,333 | 23,757 |
| Gdf9 | NM_008110 | 165,482 | 14,100 |
| Obox1 | NM_027802 | 149,944 | 2,247 |
| Oog1 | NM_178657 | 136,086 | 100,086 |
| Tcl1b2 | NM_013775 | 131,326 | 76,552 |
| Tcl1 | NM_009337 | 129,237 | 22,122 |
| Omt2b | NM_205822 | 122,678 | 258,088 |
| Tcl1b1 | NM_013773 | 118,580 | 74,377 |
| Omt2a | NM_001111286 | 113,360 | 243,955 |
| Obox5 | NM_145709 | 110,924 | 2,669 |
| E330034G19Rik | NM_001033214 | 106,956 | 1,162 |
| Zbed3 | NM_028106 | 106,540 | 3,005 |
| Slc45a3 | NM_145977 | 98,658 | 14,813 |
| Tcl1b5 | NM_013776 | 95,977 | 68,387 |
| Khdc1b | NM_001113187 | 95,938 | 23,715 |
| Oosp1 | NM_133353 | 93,718 | 14,502 |
| Bpgm | NM_007563 | 86,249 | 4,611 |
| EG194588 | NM_001038676 | 84,998 | 2,979 |
| Map1lc3b | NM_026160 | 984 | 279,836 |
| Omt2b | NM_205822 | 12,2678 | 258,088 |
| Omt2a | NM_001111286 | 113,360 | 243,955 |
| Oaz1 | NM_008753 | 30,967 | 165,822 |
| Rplp0 | NM_007475 | 1,844 | 151,774 |
| Pdxk | NM_172134 | 0 | 126,122 |
| Akp5 | NM_007433 | 17 | 121,700 |
| Ubb | NM_011664 | 21,871 | 118,549 |
| Sp110 | NM_175397 | 1,238 | 115,350 |
| Klf17 | NM_029416 | 24,531 | 112,781 |
| H3f3b | NM_008211 | 47,128 | 112,188 |
| Dppa3 | NM_139218 | 29,844 | 106,509 |
| Rps5 | NM_009095 | 129 | 105,279 |
| Tubb2c | NM_146116 | 3,013 | 101,600 |
| Oog1 | NM_178657 | 136,086 | 100,086 |
| EG547109 | NM_001034906 | 0 | 98,682 |
| Serf2 | NM_011354 | 17,828 | 95,696 |
| Impdh2 | NM_011830 | 27 | 90,365 |
| Prps1 | NM_021463 | 20 | 88,663 |
| Ccnb1 | NM_172301 | 35,328 | 87,846 |
References
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cloonan N, Grimmond SM. Transcriptome content and dynamics at single-nucleotide resolution. Genome Biol. 2008;9:234. doi: 10.1186/gb-2008-9-9-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 4.DeRisi J, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat. Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 5.Lockhart DJ, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 6.Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat. Genet. 1999;21:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- 7.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat. Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 8.Blackshaw S, Livesey R. Applying genomics technologies to neural development. Curr. Opin. Neurobiol. 2002;12:110–114. doi: 10.1016/s0959-4388(02)00298-2. [DOI] [PubMed] [Google Scholar]
- 9.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Wold B, Myers RM. Sequence census methods for functional genomics. Nat. Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 11.Schuster SC. Next-generation sequencing transforms today’s biology. Nat. Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 12.Shendure J. The beginning of the end for microarrays? Nat. Methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 13.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 14.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 16.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 18.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 19.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 21.Li H, et al. Determination of tag density required for digital transcriptome analysis: application to an androgen-sensitive prostate cancer model. Proc. Natl. Acad. Sci. USA. 2008;105:20179–20184. doi: 10.1073/pnas.0807121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blake WJ, KAErn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 23.Raser JM, O’Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arias AM, Hayward P. Filtering transcriptional noise during development: concepts and mechanisms. Nat. Rev. Genet. 2006;7:34–44. doi: 10.1038/nrg1750. [DOI] [PubMed] [Google Scholar]
- 25.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shahrezaei V, Swain PS. The stochastic nature of biochemical networks. Curr. Opin. Biotechnol. 2008;19:369–374. doi: 10.1016/j.copbio.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 28.Kawasaki ES. Microarrays and the gene expression profile of a single cell. Ann. N Y Acad. Sci. 2004;1020:92–100. doi: 10.1196/annals.1310.010. [DOI] [PubMed] [Google Scholar]
- 29.Livesey FJ. Strategies for microarray analysis of limiting amounts of RNA. Brief. Funct. Genomic Proteomic. 2003;2:31–36. doi: 10.1093/bfgp/2.1.31. [DOI] [PubMed] [Google Scholar]
- 30.Iscove NN, et al. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat. Biotechnol. 2002;20:940–943. doi: 10.1038/nbt729. [DOI] [PubMed] [Google Scholar]
- 31.Klein CA, et al. Combined transcriptome and genome analysis of single micrometastatic cells. Nat. Biotechnol. 2002;20:387–392. doi: 10.1038/nbt0402-387. [DOI] [PubMed] [Google Scholar]
- 32.Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 33.Wang QT, et al. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell. 2004;6:133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- 34.Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev. Biol. 2004;272:483–96. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 35.Hartmann CH, Klein CA. Gene expression profiling of single cells on large-scale oligonucleotide arrays. Nucleic Acids Res. 2006;34:e143. doi: 10.1093/nar/gkl740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen KB, Watt FM. Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc. Natl. Acad. Sci. U S A. 2006;103:11958–11963. doi: 10.1073/pnas.0601886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bontoux N, et al. Integrating whole transcriptome assays on a lab-on-a-chip for single cell gene profiling. Lab Chip. 2008;8:443–450. doi: 10.1039/b716543a. [DOI] [PubMed] [Google Scholar]
- 38.Kurimoto K, et al. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 2006;34:e42. doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurimoto K, Yabuta Y, Ohinata Y, Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc. 2007;2:739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- 40.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 41.Tang F, et al. 220-plex microRNA expression profile of a single cell. Nat. Protoc. 2006;1:1154–1159. doi: 10.1038/nprot.2006.161. [DOI] [PubMed] [Google Scholar]
- 42.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the mouse embryo. 3rd ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. pp. 194–200. [Google Scholar]
- 43.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 44.Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 45.Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]