Abstract
Defects in the pyruvate dehydrogenase (PDH) complex result in severe neurological dysfunction, congenital lactic acidosis, growth retardation, and early death. Current treatments for PDH deficiency are administered postnatally and are generally unsuccessful. Because many patients with this disease are born with irreversible defects, a model system for the development of effective pre- and postnatal therapies would be of great value. In a behavioral genetic screen aimed to identify zebrafish with visual function defects, we previously isolated two alleles of the recessive lethal mutant no optokinetic response a (noa). Here we report that noa is deficient for dihydrolipoamide S-acetyltransferase (Dlat), the PDH E2 subunit, and exhibits phenotypes similar to human patients with PDH deficiency. To rescue the deficiency, we added ketogenic substrates to the water in which the embryos develop. This treatment successfully restored vision, promoted feeding behavior, reduced lactic acidosis, and increased survival. Our study demonstrates an approach for establishing effective therapies for PDH deficiency and other congenital diseases that affect early embryonic development.
Human diseases that affect tissues with high-energy requirements are often caused by defects in mitochondrial function (1). Pyruvate dehydrogenase (PDH) deficiency is a rare metabolic disorder that severely affects the central nervous system due to the high-energy demand of neurons and often results in developmental delay, feeding difficulties, lethargy, ataxia, blindness, and early death (2-4). PDH is a nuclear-encoded mitochondrial matrix multienzyme complex that provides the primary link between glycolysis and the tricarboxylic acid (TCA) cycle by catalyzing the irreversible conversion of pyruvate into acetyl-CoA (5). Defects in PDH result in decreased acetyl-CoA synthesis and the accumulation of pyruvate and lactate, causing reduced energy production and metabolic acidosis, respectively.
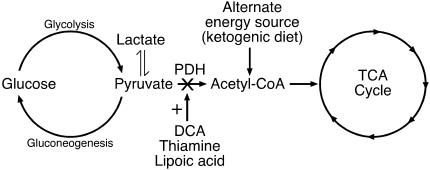
Current treatments for PDH deficiency, which include the activation of residual PDH with dichloroacetate (6), the administration of cofactors (lipoic acid and thiamine) (7, 8), and the provision of ketogenic diets (9, 10), have yielded only limited or variable success (see Fig. 1). The technical challenges of working with small numbers of human patients has made it difficult to include all of the necessary controls and to systematically evaluate therapeutic agents and diets with varying proportions of calories from fat, carbohydrate, and protein (9, 11). Furthermore, preexisting neurological disorders compound the difficulty of evaluating treatments. A therapy that bypasses the entire complex would potentially be of greatest benefit, because it could be used to treat patients with defects in any of the PDH subunits and varying degrees of deficiency.
Fig. 1.
The PDH complex. PDH is the primary link between glycolysis and the tricarboxylic acid (TCA) cycle. PDH deficiency results in reduced acetyl-CoA production and elevated levels of lactate and pyruvate. Dichloroacetate (DCA) inhibits PDH kinase, an inhibitor of PDH; thiamine is a cofactor of the E1α subunit; lipoic acid is a cofactor of the E2 subunit; and the ketogenic diet provides an alternate energy source for acetyl-CoA production.
An animal model for PDH deficiency would be of great value to improve current treatments and to allow the development of novel therapies. This model should exhibit phenotypic characteristics similar to those seen in human patients and provide an easy and reliable method for assessing treatment. Targeted disruption of the murine E1α and E3 subunits result in embryonic lethality (12, 13); thus, these mice are limited in their usefulness for improving and developing therapies. In contrast, a zebrafish model for this disease would have several important advantages. Zebrafish develop ex utero and produce large numbers of offspring, and therapeutic agents can easily be tested as supplements to the water in which the embryos develop.
As part of a behavioral screen for N-ethyl-N-nitrosourea-induced mutations that disrupt retinal function, two independent autosomal recessive alleles of the blind zebrafish mutant no optokinetic response a (noa) were isolated (14, 15). Histological characterization by light and electron microscopy revealed no gross abnormalities in the noa retina at 5-6 days postfertilization (dpf) (14, 16). Electroretinography revealed defects in synaptic transmission (14) and light adaptation (15) in cone photoreceptor cells (see Fig. 8 and Table 1, which are published as supporting information on the PNAS web site). Other characteristics of noa include expanded melanophores (14), absence of feeding behavior, lethargy, and premature death (17).
To gain further insight into these wide-ranging phenotypes, we used a positional cloning strategy to identify the defective gene in noa as dihydrolipoamide S-acetyltransferase (dlat), the PDH E2 subunit. Biochemical characterization of the mutants showed that Dlat protein levels are significantly reduced and that lactate and pyruvate levels are elevated. In addition, we developed a strategy for the metabolic rescue of the deficiency that bypasses PDH, resulting in restored vision, increased feeding behavior, reduced lactic acidosis, and prolonged survival. Our study demonstrates an approach for improving current treatments and evaluating novel therapies for patients with PDH deficiency and congenital lactic acidosis.
Materials and Methods
Zebrafish Strains and Mutant Screening. The AB* and WIK strains were obtained from the University of Oregon, Eugene, and maintained as described (18). Two alleles of the noa mutation (a13 and m631) were isolated as described (14, 15). The a13 allele was originally called nrb (8). Embryos from heterozygous adults were maintained in embryo medium (18), and noa mutants were identified at 6 dpf by optokinetic response screening (14). The a13 allele and their wild-type siblings were used for all experiments unless otherwise stated. The experiments on live animals in this study were conducted in accordance with guidelines of the University of Washington Institutional Animal Care and Use Committee.
Genetic Mapping, Positional Cloning, and Gene Identification. Heterozygous AB* strain adults carrying the a13 allele were outcrossed with the WIK strain. Carriers of the a13 allele in the F1 generation were identified and crossed to produce a mapping panel of 2,000 mutants. Amplified fragment length polymorphism markers were generated as described (19) and mapped on two radiation hybrid (RH) panels (20, 21). Simple sequence-length polymorphism markers, Z20205 and Z7609, were used for recombination analysis by PCR and agarose gel electrophoresis. ESTs were examined for recombination by single-strand conformational polymorphisms. Bacterial artificial chromosomes (BACs) were identified by using a PCRable BAC library (Incyte). CHUNP69 was examined through a collaboration established at Children's Hospital, Boston (http://134.174.23.167/zonrhmapper). The coding regions of wild-type and mutant dlat were cloned by RT-PCR using primers designed against EST clones found in GenBank that contained the 5′ UTR (BG883925), ORF (AI965099), and the 3′ UTR (A1641410) of zebrafish dlat. Genotyping of noaa13 mutants and adult heterozygotes was performed by RFLP using the restriction enzyme NlaIII (New England Biolabs). DNA primers used for mapping, sequencing, and genotyping are available upon request.
Immunoblotting. Zebrafish embryos at 1 h postfertilization (hpf) and wild-type and mutant larvae at 6 dpf were homogenized in PBS with a protease inhibitor mixture (Roche Applied Science) and 1 mM EDTA on ice with a disposable homogenizer (Kontes). SDS sample buffer was added and the samples were boiled for 5 min. The equivalent of 1 larva per well was subjected to SDS/8% PAGE, transferred to a nitrocellulose membrane (Bio-Rad), and probed with anti-Dlat sera kindly provided by E. Gershwin (University of California, Davis). Porcine heart PDH (Sigma) was loaded as a control. The blot was stripped and re-probed with rabbit anti-bovine glial fibrillary acidic protein (Dako) as a loading control. A commercially available monoclonal antibody against the human PDH E2/E3-binding protein (Molecular Probes) was used to confirm results.
Quantification of Lactate, Pyruvate, ATP, and ADP. Lactate and pyruvate levels were quantified by standard spectrophotometric analysis (Sigma). Ten to 20 noaa13 mutants and wild-type siblings at 6 or 7 dpf were frozen on Dry Ice and stored at -70°C. Larvae were homogenized in ice-cold 1.2 M HClO4, proteins, and insoluble material were pelleted, exogenous lactate dehydrogenase (Sigma) and 5 mM NAD+ (for lactate measurements) or 83 μM NADH (for pyruvate measurements) were added, and the change in absorbance (340 nm) was measured after a 1 h incubation at 24°C. Each measurement was performed in triplicate or quadruplicate.
ATP and ADP levels were quantified by UV absorbance (260 nm) after eluting from a Whatman SAX ion-exchange HPLC column (22). The column was eluted with a (NH4)2HPO4·H2PO4 (pH 3.8) gradient from 0 to 0.6 M over 25 min, then 0.6 to 1.0 M over 10 min. Homogenates were prepared as above and adjusted to pH 3.5 with 1 M (NH4)2HPO4·H2PO4 (pH 3.8). Each measurement was performed in triplicate.
Ketogenic Diet Rescue. Equimolar amounts of palmitate, myristate, laurate, and caprate (Sigma) were emulsified by sonication in embryo medium containing phosphotidylcholine (1:2 wt/wt). The optimal fatty acid concentration for rescue and survival was determined to be 200 μM. Optokinetic responses were videotaped and digitized. The angle between the eye and body of individual larvae were measured in each frame by using nih image. Feeding and locomotion behavior were similarly videotaped and digitized and the position of each larvae was determined frame-by-frame by using NIH image. For survival curves, 40-60 wild-type and mutant larvae were raised in embryo medium (untreated), paramecia and larval food (normal), or fatty acids and paramecia (treated).
Results and Discussion
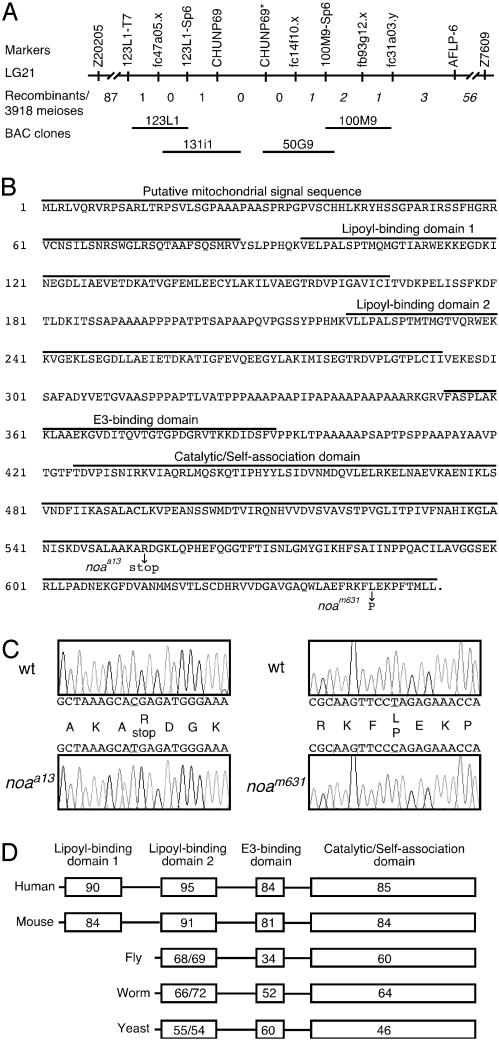
Positional Cloning of the noa Gene. We mapped the noa mutation near the centromere of linkage group 21 by generating amplified fragment length polymorphisms (19) and screening two RH panels (20, 21) with these markers (Fig. 2A). Flanking simple sequence-length polymorphisms (23) were identified and analyzed for recombination of 3,918 meioses. Fine mapping by single-strand conformational polymorphisms analysis showed that EST fc47a.05.x was 0.026 cM proximal and EST fb93g12.x was 0.077 cM distal to the noa gene. These ESTs were used to initiate a chromosomal walk. Concurrently, newly mapped ESTs and other markers were examined for recombination by single-strand conformational polymorphisms. EST fc14f10.x, corresponding to the 3′ untranslated region UTR of dlat, and the 3′ UTR of an unpublished marker, CHUNP69, showed no recombinants and were found to be located on bacterial artificial chromosomes 50G9 and 131i1, respectively. We designed primers to the 5′ UTR of CHUNP69 (CHUNP69*) and found that it bridged the gap of the critical interval and also showed no recombinants. Both dlat and CHUNP69 were examined as candidates for the noa gene. Sequence analysis of dlat cDNA uncovered mutations in both noa alleles. The a13 allele contains a C to T transition that results in a nonsense mutation at codon 554, and the m631 allele contains a T to C transition that results in a missense mutation at codon 644 (Fig. 2 B and C). No mutations were identified in CHUNP69.
Fig. 2.
Positional cloning of the noa gene. (A) Amplified fragment length polymorphism (AFLP) analysis and RH panel mapping placed marker AFLP-6 on linkage group 21. Simple sequence-length polymorphism markers (Z20205 and Z7609) were used for recombination analysis. The number of recombination events is shown with normal font on the proximal side and italics on the distal side. (B) Translated sequence of zebrafish Dlat. The protein domains are shown, and the mutations identified in the noaa13 and noam631 alleles are indicated. (C) Sequence chromatographs showing the C to T transition in the a13 allele and the T to C transition in the m631 allele. (D) Percent identities of the zebrafish Dlat domains compared with human (Homo sapiens), mouse (Mus musculus), fly (Drosophila melanogaster), worm (Caenorhabditis elegans), and yeast (Saccharomyces cerevisiae). For fly, worm, and yeast, the percent identity of the single lipoyl-binding domain is compared to both zebrafish lipoyl-binding domains.
Dlat, the PDH E2 subunit, forms the dodecahedral core of the PDH complex, which consists of 60 E2 subunits that serve as a scaffold for 22-30 E1 subunits (heterotrimers of 2α and 2β) and 6 E3 subunits (5, 24). The association between the E2 core and the E3 subunits is mediated by 12 molecules of the E3-binding protein (25). Whereas the majority of known human PDH deficiencies are caused by mutations in the E1α subunit, a significant number of patients show enzymatic and immunological evidence of E1α deficiency but no mutations in the E1 genes (26). This suggests that mutations in the other PDH subunits can also cause this disease. Indeed, defects have also been reported in the E2 subunit (27), E3 subunit (28), and the E3-binding protein (29).
The predicted zebrafish Dlat sequence reveals a putative mitochondrial signal sequence, two lipoyl-binding domains, an E3 subunit binding domain, a catalytic/self-association domain (Fig. 2B), and it shares strong sequence identities with higher vertebrates (Fig. 2D). The finding of a premature stop codon in the a13 allele strongly suggests that the mutant phenotype is caused by defective Dlat. In addition, the Leu-Pro substitution at position 644 in the m631 allele is likely to disrupt core formation and PDH activity, because the C terminus of Dlat is intimately involved in forming intertrimer contacts (30).
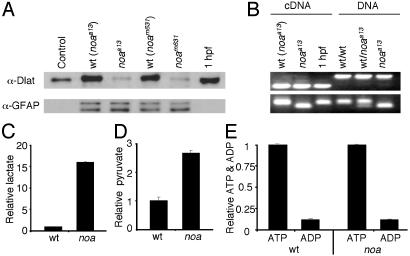
Dlat Protein Levels Are Reduced in noa. We hypothesized that both mutations would destabilize Dlat and cause it to be degraded. To examine the expression of Dlat in zebrafish larvae, we used a polyclonal antibody against the inner lipoyl-binding domain of human Dlat (a gift from E. Gershwin, University of California, Davis). Immunoblotting indicates that this antibody cross-reacts with zebrafish Dlat (Fig. 3A). We detected residual amounts of full-length Dlat from both noa alleles at 6 dpf. Dlat was present at similar levels in both noa alleles and was reduced >10-fold when compared to wild-type siblings. We did not detect a shortened form of Dlat in the a13 allele, suggesting that the truncated protein is completely degraded (data not shown). The same results were obtained by using a commercially available monoclonal antibody (Molecular Probes) against human PDH E2/E3-binding protein (data not shown), confirming that the reduced Dlat protein level in both alleles was most likely due to degradation of the subunit and not a result of reduced immunoreactivity.
Fig. 3.
Biochemical characterization of noa. (A) Immunoblots of porcine heart PDH (control), wild-type noaa13 siblings [wt (noaa13)], noaa13 mutants (noaa13), wild-type noam631 siblings [wt (noam631)], noam631mutants (noam631), and embryos at 1 hpf using anti-Dlat (Upper) or anti- glial fibrillary acidic protein as a loading control (Lower). (B Upper) RT-PCR (cDNA) and genomic DNA (DNA) PCR using primers that flank a small intron showing mRNA is maternal. (B Lower) PCR using primers that flank the C to T transition in the a13 allele followed by restriction digestion with NlaIII. Quantification of lactate (wt, 165 ± 12 pmol per larvae; noa, 2,655 ± 2 pmol per larvae) (C); pyruvate (wt, 101 ± 7 pmol per larvae; noa, 267 ± 13 pmol per larvae) (D); and ATP and ADP (ATP: wt, 65 ± 1 pmol per larvae; noa, 65 ± 1 pmol per larvae; and ADP: wt, 5.8 ± 0.6 pmol per larvae; noa, 5.9 ± 0.3 pmol per larvae) (E)at 6 dpf are shown. Graphs indicate relative values, and the error bars are the SEM.
Because the half-life of rat liver PDH is 8.1 days (31) and given that eggs inherit large numbers of maternal mitochondria, we speculated that the residual full-length Dlat protein in the mutants was maternally derived. Immunoblotting and RT-PCR confirmed that maternal Dlat protein and mRNA are present in embryos at 1hpf (Fig. 3 A and B). The Dlat levels at 1 hpf were also at least 10-fold higher than mutant levels at 6 dpf. We therefore suggest that the physiological effects of PDH deficiency in the mutants become evident when the maternal PDH levels fall below a critical level at 4-6 dpf. Interestingly, this is also near the developmental stage when the energy stores of the yolk sac become depleted. Thus, the reduced Dlat levels and the limited ketogenic energy sources from the yolk ultimately result in the symptomatology of PDH deficiency at this stage of development.
Lactate and Pyruvate Are Elevated and ATP and ADP Are Normal in noa. Elevated levels of lactate and pyruvate are diagnostic indicators of inborn errors of pyruvate metabolism. In fact, PDH deficiency is the most common biochemically proven cause of primary congenital lactic acidosis (32). To determine whether lactate and pyruvate levels are increased in the mutants, we measured these metabolites in both wild-type and mutant homogenates. In the mutants, we observed a 16.1-fold increase in lactate and a 2.6-fold increase in pyruvate (Fig. 3 C and D).
Previous studies have found normal levels of ATP in tissues from patients with various mitochondrial disorders (33, 34). To examine the level of ATP in PDH-deficient zebrafish, we quantified the total ATP and ADP content from wild-type and mutant homogenates, and observed normal levels and ratios of ATP and ADP (Fig. 3E). Interestingly, previous studies have shown that the rate of ATP production is significantly reduced in fibroblasts from patients with PDH deficiency (33, 35). These results indicate that the demand for ATP must be reduced to sustain normal ATP levels. This finding is corroborated by the observation that goldfish in anoxic environments maintain normal ATP levels by decreasing ATP consumption, resulting in reduced swimming activity and reduced visual function (36). Thus, taken together, these findings suggest that several of the clinical features of PDH deficiency, including hypotonia, ataxia, lethargy, and blindness, may be partially attributed to the maintenance of ATP/ADP homeostasis.
Given the genetic mutations in the dlat gene, the substantial reduction of Dlat protein, and the elevated levels of lactate and pyruvate, it is evident that the abnormal physiology and premature lethality of the noa mutant is caused by PDH deficiency. The fact that neurological function is so severely affected in this disease (2, 4) emphasizes the high-energy demand of neurons. Photoreceptor cells, in particular, are among the most energy-dependent cells in the body (37), and ophthalmologic symptoms are common to many mitochondrial diseases (38). Therefore, it is not surprising that noa was originally identified as a mutant with defective visual function (14).
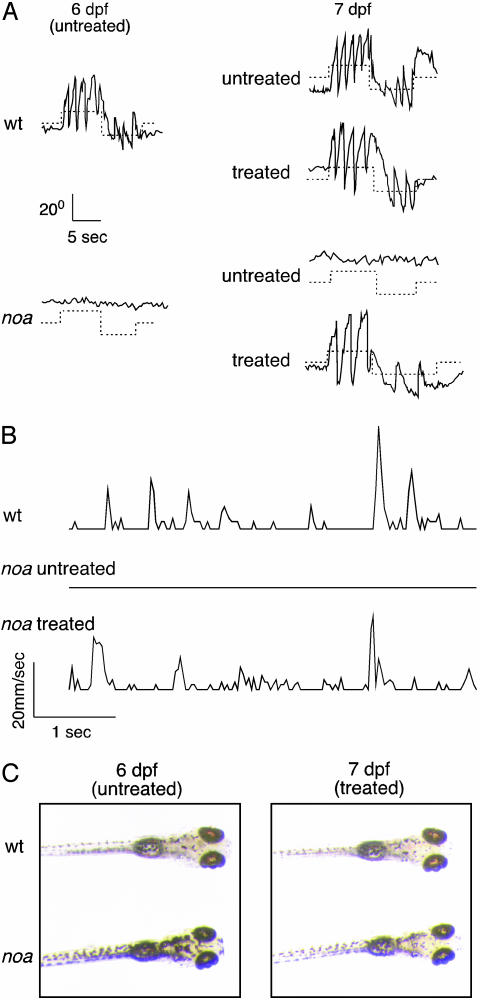
Rescue of PDH Deficiency Using a Ketogenic Diet. Although there is no proven treatment for PDH deficiency, ketogenic diets have benefited some patients and have been included in the standard care for many of these patients (9-11). To further establish noa as a model for PDH deficiency, we attempted to rescue the mutants by using a ketogenic diet consisting of medium- to long-chain fatty acids in an emulsion of phosphatidylcholine (see Fig. 1). We first isolated mutants from their wild-type siblings by optokinetic response (OKR) screening (14). Wild-type larvae have a robust visual response at 6 dpf, whereas the mutants show no response (Fig. 4A). We then placed the wild-type and mutant larvae in an emulsion of fatty acids overnight and reevaluated their OKR performance at 7 dpf. Remarkably, fatty acid-treated (FAT) mutants displayed a visual response that is similar to wild-type larvae (Fig. 4A). Approximately 70% of FAT-mutants showed a strong to moderate visual response, 25% showed a weak response, and 5% showed no response (n = 128) (data not shown). We also found that FAT mutants have a strong optomotor response that is similar to wild-type larvae (data not shown). To further examine the rescue of visual function in FAT mutants, we performed electroretinography and detected responses similar to wild-type larvae (see Fig. 9, which is published as supporting information on the PNAS web site, and Table 1). These data indicate that severe neurological dysfunction, such as blindness, is reversible when treated early with the ketogenic diet.
Fig. 4.
Rescue of noa using a ketogenic diet. (A) Representative optokinetic responses of untreated wild-type (wt) and mutant (noa) larvae at 6 dpf (Left), and untreated or FAT wild-type (wt) and mutant (noa) larvae at 7 dpf (Right). (B) Feeding and locomotion behavior of wild-type (wt), untreated mutant, and FAT-mutant larvae at 7 dpf. Movements of larvae are plotted as linear velocity vs. time. The traces illustrate the lunging activity of larvae feeding on paramecia. (C) Pigmentation phenotype of untreated wild-type (wt) and mutant (noa) larvae at 6 dpf (Left), and the same larvae after 24 h in fatty acids (Right).
We also observed that FAT mutants display feeding and swimming behaviors that are similar to wild-type larvae. Untreated mutants lie lethargically, often floating on their side or back, whereas FAT mutants swim energetically and actively pursue and eat paramecia (Fig. 4B). Movies of the rescued optokinetic response and feeding behavior are shown in Movies 1-6, which are published on the PNAS web site.
Another striking phenotype of noa is dark pigmentation due to expanded melanophores (14), a trait common to zebrafish vision mutants (39). Zebrafish have the ability, as do many other cold-blooded vertebrates, to adjust melanin pigment levels in response to light or dark background. The mutants appear abnormally dark in pigmentation under room light, presumably because they do not detect light and respond as though the background is dark. In contrast, when mutants are treated with fatty acids, the melanin aggregates as in wild-type larvae, demonstrating that the FAT mutants respond and adapt to background light (Fig. 4C).
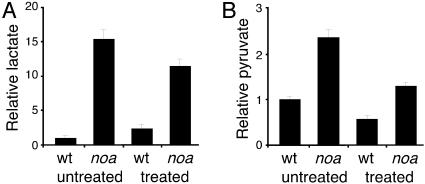
Because studies of PDH-deficient patients have reported reduced lactic acidosis after high fat/low carbohydrate diets (9, 10), we analyzed the effect of the ketogenic diet on lactate and pyruvate levels in mutants after a 24-h treatment. We observed that the treatment reduced lactate by 26% in the mutants, whereas the same treatment increases lactate by 59% in wild-type larvae (Fig. 5A). Pyruvate decreased by 45% in the mutants and 43% in wild-type larvae (Fig. 5B). In addition, the ratio of lactate to pyruvate significantly improved in the mutants compared to wild-type larvae. These data demonstrate that the ketogenic diet has a positive effect in reducing lactic acidosis in our model as reported for human patients.
Fig. 5.
Quantification of lactate and pyruvate after ketogenic diet treatment. (A) Lactate levels of untreated larvae (wt, 139 ± 50 pmol per larvae; noa, 2,137 ± 182 pmol per larvae) and treated larvae (wt, 337 ± 79 pmol per larvae; noa, 1,588 ± 142 pmol per larvae). (B) Pyruvate levels of untreated larvae (wt, 129 ± 7 pmol per larvae; noa, 303 ± 22 pmol per larvae) and treated larvae (wt, 74 ± 10 pmol per larvae; noa, 167 ± 9 pmol per larvae). Graphs indicate relative values, and the error bars are the SEM.
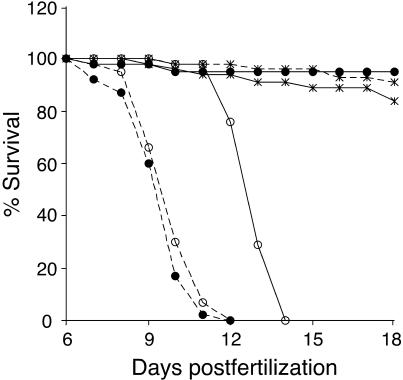
To determine whether the ketogenic diet prolongs survival, as has been reported for human patients, we raised larvae in embryo medium (untreated), paramecia and larval food (normal), or fatty acids and paramecia (treated) (Fig. 6). We observed that untreated mutants die 3 days before untreated wild-type larvae. On a diet of paramecia and larval food, wild-type larvae survived to adulthood, whereas the mutants expired at the same rate as untreated mutants, probably because they did not eat (see Fig. 4B). In contrast, FAT mutants survived as long as FAT wild-type larvae, showing that the ketogenic diet rescues embryonic lethality and prolongs survival of noa. To date, we have had mutants surviving to 40 dpf before killing the fish (see Fig. 7).
Fig. 6.
Survival curves. Wild-type (solid line) and mutant (dashed line) larvae raised in embryo medium (open circle), paramecia and larval food (filled circle), or fatty acids and paramecia (star) are shown.
Fig. 7.
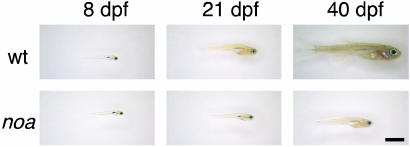
Growth retardation in noa. Wild-type (wt) and mutant (noa) larvae were raised on the ketogenic diet, and their growth was assessed at 8, 21, and 40 dpf. Representative pictures of the fish are shown (n = 8 for each group). Note that mutants at 40 dpf are approximately the same size as wild-type fish at 21 dpf. (Bar = 2 mm.)
Histological Analysis of noa. Human patients with PDH deficiency develop symptoms soon after birth, and exhibit either a metabolic or neurological presentation of the disease (2). Patients with the metabolic form present with severe lactic acidosis, which is usually refractory to treatment, and often die early in the neonatal period. Surprisingly, patients with the metabolic form of the disease may show remarkably little neuropathology in comparison to patients with the neurological form of the disease (2, 40). To determine whether noa exhibits neuropathology, we histologically examined the mutants for brain abnormalities at 8 dpf, but did not uncover any obvious defects. We then raised wild-type and mutant larvae to 21 and 40 dpf by using the ketogenic diet and examined brain cross-sections at 3-μm intervals. Other than the obvious growth retardation (see Fig. 7 and below), we did not observe any neurological defects in the mutants (see Fig. 10, which is published as supporting information on the PNAS web site). This finding suggests that, if treated early enough, the ketogenic diet may help prevent the development of brain defects. We also examined skeletal muscle and other tissues for defects but did not uncover any obvious abnormalities.
In an attempt to induce developmental abnormalities, we removed the ketogenic diet from the mutants. Unfortunately, the mutants died 2-4 days after removing the diet, emphasizing the complete reliance on the diet for survival. We hypothesize that this is most likely due to the severity of the deficiency in the mutants. The lack of residual PDH activity is incompatible with life in the absence of the ketogenic diet, and most likely causes death before the formation of developmental defects; a result that has been observed in mice with targeted disruptions of the E1α and E3 subunits (12, 13).
Growth Retardation of noa. Human patients with PDH deficiency most often exhibit developmental delay and growth retardation (2, 41). To address this issue in noa, we raised larvae on the ketogenic diet and assessed their growth rates. The diet promoted normal growth and survival of wild-type larvae; however, the growth rate of the mutant was significantly reduced (Fig. 7). At 8 dpf, the sizes of wild-type and mutant larvae were indistinguishable. As time progressed, the growth retardation of the mutants became increasingly noticeable. By 40 dpf, the mutants were significantly smaller than the wild-type fish and were approximately the same size as wild-type fish at 21 dpf. Therefore, even under dietary conditions that promoted a dramatic rescue of several phenotypes, the developmental growth of mutants was delayed. This finding suggests that the inability of ketogenic diets to completely rescue human patients (4-6) may not solely be due to preexisting conditions at the onset of treatment, and that novel therapies are needed to totally cure the disease.
In conclusion, we have demonstrated that the zebrafish mutant noa exhibits many disease characteristics similar to those of human patients with PDH deficiency. The availability of this mutant can provide simple and reliable methods for assessing the efficacy of therapeutic treatments and for resolving issues regarding the pathophysiology and the course of treatment for this disease. In addition, it provides the opportunity for high-throughput testing of multiple ketogenic compounds, metabolic substrates, and potentially therapeutic drugs either individually or in combination.
Supplementary Material
Acknowledgments
We thank E. Gershwin at University of California, Davis, for providing the Dlat antibody; R. Palmiter, J. Saari, and V. Ramamurthy for critical review of the manuscript; M. Kushmerick for helpful discussions; G. Niemi for help with bacterial artificial chromosome screening; A. Scotti for help with amplified fragment length polymorphism analysis; Glenda Froelick and Dan Possin for help with histology; L. Swaim for help with fish maintenance; and M. Hunsley for help with manuscript preparation. This work was supported by National Institutes of Health Grant EY12373 (to S.E.B.) and U.S. Public Health Service National Research Grant T32 GM07270 (to M.R.T.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PDH, pyruvate dehydrogenase; dpf, days postfertilization; hpf, hours post-fertilization; FAT, fatty acid-treated.
Data deposition: The cDNA sequence for zebrafish dihydrolipoamide S-acetyltransferase has been deposited in the GenBank database (accession no. AY188775).
References
- 1.Wallace, D. C. (1999) Science 283, 1482-1488. [DOI] [PubMed] [Google Scholar]
- 2.Brown, G. K., Otero, L. J., LeGris, M. & Brown, R. M. (1994) J. Med. Genet. 31, 875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Vivo, D. C. (1998) Neurology 51, 1247-1249. [DOI] [PubMed] [Google Scholar]
- 4.DiMauro, S. & De Vivo, D. C. (1996) Ann. Neurol. 40, 5-7. [DOI] [PubMed] [Google Scholar]
- 5.Patel, M. S. & Roche, T. E. (1990) FASEB J. 4, 3224-3233. [DOI] [PubMed] [Google Scholar]
- 6.Morten, K. J., Beattie, P., Brown, G. K. & Matthews, P. M. (1999) Neurology 53, 612-616. [DOI] [PubMed] [Google Scholar]
- 7.Byrd, D. J., Krohn, H. P., Winkler, L., Steinborn, C., Hadam, M., Brodehl, J. & Hunneman, D. H. (1989) Eur. J. Pediatr. 148, 543-547. [DOI] [PubMed] [Google Scholar]
- 8.Naito, E., Ito, M., Yokota, I., Saijo, T., Ogawa, Y. & Kuroda, Y. (2002) J. Neurol. Sci. 201, 33-37. [DOI] [PubMed] [Google Scholar]
- 9.Wexler, I. D., Hemalatha, S. G., McConnell, J., Buist, N. R., Dahl, H. H., Berry, S. A., Cederbaum, S. D., Patel, M. S. & Kerr, D. S. (1997) Neurology 49, 1655-1661. [DOI] [PubMed] [Google Scholar]
- 10.Wijburg, F. A., Barth, P. G., Bindoff, L. A., Birch-Machin, M. A., van der Blij, J. F., Ruitenbeek, W., Turnbull, D. M. & Schutgens, R. B. (1992) Neuropediatrics 23, 147-152. [DOI] [PubMed] [Google Scholar]
- 11.Weber, T. A., Antognetti, M. R. & Stacpoole, P. W. (2001) J. Pediatr. 138, 390-395. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, M. T., Yang, H. S., Magnuson, T. & Patel, M. S. (1997) Proc. Natl. Acad. Sci. USA 94, 14512-14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, M. T., Mahmood, S., Hyatt, S. L., Yang, H. S., Soloway, P. D., Hanson, R. W. & Patel, M. S. (2001) Mol. Genet. Metab. 74, 293-302. [DOI] [PubMed] [Google Scholar]
- 14.Brockerhoff, S. E., Hurley, J. B., Janssen-Bienhold, U., Neuhauss, S. C., Driever, W. & Dowling, J. E. (1995) Proc. Natl. Acad. Sci. USA 92, 10545-10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brockerhoff, S. E., Dowling, J. E. & Hurley, J. B. (1998) Vision Res. 38, 1335-1339. [DOI] [PubMed] [Google Scholar]
- 16.Allwardt, B. A. (1999) Doctoral dissertation (Harvard University, Cambridge, MA).
- 17.Taylor, M. R., Van Epps, H. A., Kennedy, M. J., Saari, J. C., Hurley, J. B. & Brockerhoff, S. E. (2000) Methods Enzymol. 316, 536-557. [DOI] [PubMed] [Google Scholar]
- 18.Westerfield, M. (1995) The Zebrafish Book (Univ. of Oregon Press, Eugene).
- 19.Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M., et al. (1995) Nucleic Acids Res. 23, 4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisler, R., Rauch, G. J., Baier, H., van Bebber, F., Brobeta, L., Dekens, M. P., Finger, K., Fricke, C., Gates, M. A., Geiger, H., et al. (1999) Nat. Genet. 23, 86-89. [DOI] [PubMed] [Google Scholar]
- 21.Hukriede, N., Fisher, D., Epstein, J., Joly, L., Tellis, P., Zhou, Y., Barbazuk, B., Cox, K., Fenton-Noriega, L., Hersey, C., et al. (2001) Genome Res. 11, 2127-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auger, K. R., Serunian, L. A. & Cantley, L. C. (1990) in Methods in Inositide Research, ed. Irvine, R. F. (Raven, New York), pp. 159-166.
- 23.Shimoda, N., Knapik, E. W., Ziniti, J., Sim, C., Yamada, E., Kaplan, S., Jackson, D., de Sauvage, F., Jacob, H. & Fishman, M. C. (1999) Genomics 58, 219-232. [DOI] [PubMed] [Google Scholar]
- 24.Zhou, Z. H., McCarthy, D. B., O'Connor, C. M., Reed, L. J. & Stoops, J. K. (2001) Proc. Natl. Acad. Sci. USA 98, 14802-14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanderson, S. J., Khan, S. S., McCartney, R. G., Miller, C. & Lindsay, J. G. (1996) Biochem. J. 319, 109-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown, R. M., Otero, L. J. & Brown, G. K. (1997) Hum. Mol. Genet. 6, 1361-1367. [DOI] [PubMed] [Google Scholar]
- 27.Robinson, B. H., MacKay, N., Petrova-Benedict, R., Ozalp, I., Coskun, T. & Stacpoole, P. W. (1990) J. Clin. Invest. 85, 1821-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerna, L., Wenchich, L., Hansikova, H., Kmoch, S., Peskova, K., Chrastina, P., Brynda, J. & Zeman, J. (2001) Med. Sci. Monit. 7, 1319-1325. [PubMed] [Google Scholar]
- 29.Brown, R. M., Head, R. A. & Brown, G. K. (2002) Hum. Genet. 110, 187-191. [DOI] [PubMed] [Google Scholar]
- 30.Zhou, Z. H., Liao, W., Cheng, R. H., Lawson, J. E., McCarthy, D. B., Reed, L. J. & Stoops, J. K. (2001) J. Biol. Chem. 276, 21704-21713. [DOI] [PubMed] [Google Scholar]
- 31.Weinberg, M. B. & Utter, M. F. (1979) J. Biol. Chem. 254, 9492-9499. [PubMed] [Google Scholar]
- 32.Robinson, B. H. & Sherwood, W. G. (1984) J. Inherit. Metab. Dis. 7 Suppl. 1, 69-73. [DOI] [PubMed] [Google Scholar]
- 33.Vazquez-Memije, M. E., Shanske, S., Santorelli, F. M., Kranz-Eble, P., Davidson, E., DeVivo, D. C. & DiMauro, S. (1996) J. Inherit. Metab. Dis. 19, 43-50. [DOI] [PubMed] [Google Scholar]
- 34.Lofberg, M., Lindholm, H., Naveri, H., Majander, A., Suomalainen, A., Paetau, A., Sovijarvi, A., Harkonen, M. & Somer, H. (2001) Neuromuscul. Disord. 11, 370-375. [DOI] [PubMed] [Google Scholar]
- 35.Saada, A., Aptowitzer, I., Link, G. & Elpeleg, O. N. (2000) Biochem. Biophys. Res. Commun. 269, 382-386. [DOI] [PubMed] [Google Scholar]
- 36.Nilsson, G. E. (2001) News Physiol. Sci. 16, 217-221. [DOI] [PubMed] [Google Scholar]
- 37.Cooper, L. L., Hansen, R. M., Darras, B. T., Korson, M., Dougherty, F. E., Shoffner, J. M. & Fulton, A. B. (2002) Arch. Ophthalmol. 120, 1055-1062. [DOI] [PubMed] [Google Scholar]
- 38.Johns, D. R. (1995) Semin. Ophthalmol. 10, 295-302. [DOI] [PubMed] [Google Scholar]
- 39.Neuhauss, S. C., Biehlmaier, O., Seeliger, M. W., Das, T., Kohler, K., Harris, W. A. & Baier, H. (1999) J. Neurosci. 19, 8603-8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen, L. L., Brown, G. K., Kirby, D. M. & Dahl, H. H. (1991) J. Inherit. Metab. Dis. 14, 140-151. [DOI] [PubMed] [Google Scholar]
- 41.Rahman, S., Blok, R. B., Dahl, H. H., Danks, D. M., Kirby, D. M., Chow, C. W., Christodoulou, J. & Thorburn, D. R. (1996) Ann. Neurol. 39, 343-351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.