Abstract
Studies of rare genetic bone disorders are often limited due to unavailability of tissue specimens and the lack of animal models fully replicating phenotypic features. Craniometaphyseal dysplasia (CMD) is a rare monogenic disorder characterized by hyperostosis of craniofacial bones concurrent with abnormal shape of long bones. Mutations for autosomal dominant CMD have been identified in the ANK gene (ANKH). Here we describe a simple and efficient method to reprogram adherent cells cultured from peripheral blood to human induced pluripotent stem cells (hiPSCs) from eight CMD patients and five healthy controls. Peripheral blood mononuclear cells (PBMCs) were separated from 5–7 mL of whole blood by Ficoll gradient, expanded in the presence of cytokines and transduced with Sendai virus (SeV) vectors encoding OCT3/4, SOX2, KLF4, and c-MYC. SeV vector, a cytoplasmic RNA vector, is lost from host cells after propagation for 10–13 passages. These hiPSCs express stem cell markers, have normal karyotypes, and are capable of forming embryoid bodies in vitro as well as teratomas in vivo. Further differentiation of these patient-specific iPSCs into osteoblasts and osteoclasts can provide a useful tool to study the effects CMD mutations on bone, and this approach can be applied for disease modeling of other rare genetic musculoskeletal disorders.
Introduction
Rapid advances in human induced pluripotent stem cell (iPSC) technology opened promising new avenues for medical research and disease modeling, drug screening, cell therapy, and genome editing (Cherry and Daley, 2013; Ding et al., 2013; Emborg et al., 2013). Although many concerns need to be resolved when iPSC-based strategies are used for clinical therapy, the current methods are sufficiently robust for the purpose of basic science research (Grabel, 2012). Patient-specific iPSCs have been established for several diseases, such as type I and type II diabetes, muscular dystrophy, amyotrophic lateral sclerosis, Parkinson's disease, and glioblastoma; however, no iPSCs derived from patients with rare genetic bone diseases have been published as of yet (Chen et al., 2013; Dimos et al., 2008; Kudva et al., 2012; Maehr et al., 2009; Park et al., 2008a; Teo et al., 2013). Rare genetic bone disorders have significant clinical relevance due to the sheer number of such disorders (more than 300) and due to the lifetime debilitating impact they have on many patients. Treatment for these diseases is often very limited because little is known about their pathogenesis. Obstacles for research in this field include unavailability of tissue specimens and lack of animal models. Genetic manipulation to generate a mouse model can be technically challenging and time-consuming, and in many cases mouse models fail to replicate the complete features of human disorders. These considerations highlight the importance for developing new systems to generate bone cells, osteoblasts, and osteoclasts for studying rare bone disorders. Research with iPSCs could in many cases enhance investigations for molecular mechanisms.
Craniometaphyseal dysplasia (CMD) is a rare monogenic bone disorder characterized by progressive thickening of craniofacial bones and widening of metaphyses in long bones (Jackson et al., 1954). CMD in many patients is associated with neurologic symptoms including facial palsy, visual impairment, and hearing loss, possibly due to narrowing of cranial foramina and neuronal compression caused by bony encroachment (Beighton et al., 1979; Franz et al., 1996; Richards et al., 1996). To date, treatment for CMD is limited to repetitive surgery for decompression of obstructed foramina, but severe complications have been reported (Puliafito et al., 1981; Satoh et al., 1994; Shea et al., 1981). Mutations in the ANKH gene were identified for the autosomal dominant form of CMD; however, the pathogenesis of CMD is largely unknown (Nurnberg et al., 2001; Reichenberger et al., 2001). ANKH transports intracellular pyrophosphate (PPi) to the extracellular matrix and prevents ectopic mineralization in soft tissues (Ho et al., 2000). Studies using a mouse model carrying a CMD mutation in ANK showed the involvement of osteoblasts and osteoclasts in the pathogenesis of CMD and suggested that CMD mutations in ANK may cause gain of function in addition to loss of PPi transporting activity (Chen et al., 2009; Chen et al., 2011). Here, we derived hiPSCs from CMD patients with three different mutations in ANKH. The availability of these CMD-hiPSCs is a step toward disease modeling of CMD in the human system, a valuable tool to confirm hypotheses generated from the CMD mouse model to better understand the molecular mechanism of human CMD.
Human somatic cells can be reprogrammed to hiPSCs through introduction of OCT3/4, SOX2, KLF4, and c-MYC or OCT3/4, SOX2, NANOG, and LIN28 (Park et al., 2008b; Takahashi et al., 2007; Yu et al., 2007). Many cell types have been reported to generate hiPSCs successfully, such as fibroblasts, keratinocytes, cord blood cells, T cells, dental pulp stem cells, and urinary cells (Aasen et al., 2008; Giorgetti et al., 2009; Seki et al., 2010; Takahashi et al., 2007; Yan et al., 2010; Zhou et al., 2012). Delivery of the pluripotency-inducing factors was achieved using retrovirus, lentivirus, adenovirus, piggyBac transposon, episomal vector, RNA, or protein (Sommer et al., 2009; Stadtfeld et al., 2008; Takahashi et al., 2007; Warren et al., 2010; Woltjen et al., 2009; Yu et al., 2009; Zhou et al., 2009).
Sendai virus (SeV) vector, a cytoplasmic RNA vector, was shown to deliver transgenes efficiently into several cell types (Tokusumi et al., 2002) and to generate hiPSCs free of genomic integration (Ban et al., 2011; Fusaki et al., 2009; Seki et al., 2010; Tokusumi et al., 2002). hiPSCs generated with retroviral or lentiviral methods carry viral transgene insertions in the host genome, and their safety is still a concern for clinical applications (Gonzalez et al., 2011). Here, we used SeV vectors encoding OCT3/4, SOX2, KLF4, and c-MYC to reprogram adherent cells from the peripheral blood mononuclear cell (PBMC) cultures, thus generating transgene free hiPSCs from CMD patients and healthy donors.
Materials and Methods
All studies involving human and animal protocols were approved by the Institutional Review Board and the Animal Care Committee of the University of Connecticut Health Center.
Cell culture
Peripheral blood was collected from eight CMD patients and five healthy controls whose written consent was obtained in accordance with guidelines of the University of Connecticut Health Center (UCHC) Institutional Review Board. Age, sex, and mutations in ANKH of donors are listed in Table 1. PBMCs were isolated as described (Susa et al., 2004). Briefly, 5–7 mL of peripheral blood was diluted with an equal amount of phosphate-buffered saline (PBS), layered over 10 mL of Ficoll with a density of 1.077 (Lymphoprep; Axis Shield, Oslo, Norway) and centrifuged at 800×g for 30 min (without break) to precipitate the red blood cells. The interface, containing mononuclear cells was collected, diluted with a three-fold volume of PBS, and centrifuged at 290×g for 10 min. PBMCs were grown on six-well non-tissue culture–treated plates (CytoOne, USA Scientific) and maintained in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Hyclone), 20 ng/mL recombinant human stem cell factor (rhSCF; Miltenyi Biotec), recombinant human thrombopoietin (rhTPO; Miltenyi Biotec), recombinant human interleukin-3 (rhIL-3; Miltenyi Biotec), rhIL-6 (R&D Systems), rhIL-6 receptor α (R&D Systems), rhFlt3 ligand (Miltenyi Biotec), recombinant human granulocyte macrophage colony-stimulating factor (rhGM-CSF; R&D Systems), and 50 ng/mL of rhM-CSF (Miltenyi Biotec) for 5 days.
Table 1.
CMD Patients and Control Donors for hiPSCs Generation
| Sample no. | Ank mutation | Age | Gender |
|---|---|---|---|
| 1 | Ins Ala380 (exon 10) | 26 | M |
| 2 | Ser375del (exon 9) | 44 | F |
| 3 | Phe377del (exon 9) | 45 | F |
| 4 | Phe377del (exon 9) | 17 | F |
| 5 | Phe377del (exon 9) | 14 | F |
| 6 | Phe377del (exon 9) | 9 | F |
| 7 | Phe377del (exon 9) | 10 | F |
| 8 | Phe377del (exon 9) | 10 | M |
| 9 | None | 11 | M |
| 10 | None | 33 | F |
| 11 | None | 33 | F |
| 12 | None | 29 | M |
| 13 | None | 30 | F |
CMD, craniometaphyseal dysplasia; hiPSCs, human induced pluripotent cells; M, male; F, female.
Generation of iPSCs from human peripheral blood
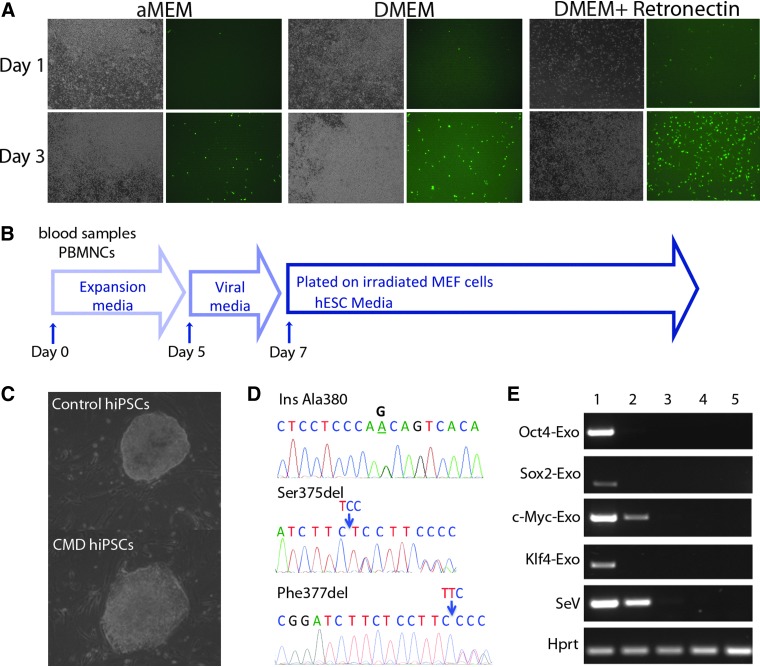
Reprogramming of PBMCs into iPSCs was performed 5 days after initial culture using SeV vectors encoding OCT3/4, SOX2, KLF4, and c-MYC (CytoTune-iPS kit, DNAVEC Corp., Japan). The floating cells were discarded and the adhesive cells were treated with 0.25% trypsin/EDTA and resuspended in the same medium described above. To transduce cells, SeV vectors containing four separate reprogramming factors were added to the cell suspension. Vector-transduced cells were immediately plated onto a 12-well plate that was precoated with 5 μg/cm2 RetroNectin (Takara Bio Inc.). The culture plate was centrifuged at 1000×g at 32°C for 45 min. The next day, medium was replaced with fresh medium. Two days later, cells were trypsinized and passed onto two 10-cm gelatin-coated culture dishes that had been seeded with irradiated mouse embryonic fibroblasts (irr-MEFs). The cultures were maintained in human embryonic stem cell (hESC) medium containing DMEM/F12 (Gibco), 20% KnockOut Serum Replacement (Gibco), 1% minimum essential medium (MEM) nonessential amino acid (Gibco), 1 mM L-glutamine (Gibco), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), and 8 ng/mL of basic fibroblast growth factor (bFGF; Millipore). hiPSC colonies were manually isolated based on morphology between day 14 to day 30 postinfection. hiPSC cultures were maintained on plates coated with Matrigel (BD Biosciences) in conditioned medium with 8 ng/mL of bFGF. The conditioned medium was collected from the supernatant of irr-MEF cultures fed with hESC medium without bFGF.
Gene expression analysis
Total RNA from iPSCs was isolated with TRIzol (Invitrogen), and RNA was treated with DNase I (Invitrogen). cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen). RT-PCR analysis for hESC marker genes OCT4, SOX2, NANOG and for the transgenes OCT4, SOX2, c-MYC, and KLF4 was performed. The presence of remaining transgenes was detected with SeV primers. Hypoxanthine guanine phosphoribosyl transferase (HPRT) was used as a loading control. PCR primer sequences are listed in Table 2. cDNA samples were also subjected to TaqMan Human Stem Cell Pluripotency Array (Life Technologies) to compare the global gene expression of iPSCs to hESCs H9.
Table 2.
Amplification Primers for RT-PCR
| Primer | Forward primer | Reverse primer | Size (bp) |
|---|---|---|---|
| Oct4-Endo | 5′-AGTTTGTGCCAGGGTTTTTG-3′ | 5′-ACTTCACCTTCCCTCCAACC-3′ | 113 |
| Sox2-Endo | 5′-AGTCTCCAAGCGACGAAAAA-3′ | 5′-TTTCACGTTTGCAACTGTCC-3′ | 189 |
| Nanog-Endo | 5′-TTTGGAAGCTGCTGGGGAAG-3′ | 5′-GATGGGAGGAGGGGAGAGGA-3′ | 194 |
| Oct3/4-Exo | 5′-CCCGAAAGAGAAAGCGAACCAG-3′ | 5′-AATGTATCGAAGGTGCTCAA-3′ | 483 |
| Sox2-Exo | 5′-ACAAGAGAAAAAACATGTATGG-3′ | 5′-ATGCGCTGGTTCACGCCCGCGCCCAGG-3′ | 591 |
| KLF4-Exo | 5′-ACAAGAGAAAAAACATGTATGG-3′ | 5′-CGCGCTGGCAGGGCCGCTGCTCGAC-3′ | 529 |
| c-Myc-Exo | 5′-TAACTGACTAGCAGGCTTGTCG-3′ | 5′-TCCACATACAGTCCTGGATGATGATG-3′ | 532 |
| SeV | 5′-GGATCACTAGGTGATATCGAGC-3′ | 5′-ACCAGACAAGAGTTTAAGAGATATGTATC-3′ | 181 |
ANKH mutation analysis
Genomic DNA was isolated from hiPSCs of CMD patients using the DNeasy kit (Qiagen) and was subjected to PCR of ANKH exons 9 and 10 to confirm the mutation sites identified in these patients before reprogramming. Primer sets for PCR amplification of ANKH exon 9 were: forward, 5′-CATG CCCACCACCCAGTCAG-3′, reverse, 5′-GCATCTTTCTAA GCCACAGTG-3′, and for exon 10 were: forward, 5′-CAG CGGGCTCAACAGCAAGGC-3′, reverse, 5′-GGATCCCAA GAGCCTCCACCTG-3′. The initial denaturation step of 5 min at 95°C was followed by 35 cycles of amplification at 95°C for 30 sec, annealing at 56°C (exon 9) and 60°C (exon 10) for 30 sec, and extension at 72°C for 90 sec, followed by final extension at 72°C for 10 min using GoTaq Flexi DNA polymerase (Promega). Unincorporated primers in PCR products were removed using ExoSAP-IT (USB Corp.) and samples were sent to an outside vendor (Agencourt Bioscience Corporation, Beverley, MA). Sequence results were analyzed with Chromas sequence editor (Technelysium, South Brisbane, QLD, Australia).
Immunofluorescence
Immunocytochemistry of hiPSCs was performed using the following antibodies based on manufacturers' protocols: Anti-human TRA-1-60 (StainAlive TRA-1-60, Stemgent), anti-human SSEA-4 (Santa Cruz Biotechnology), anti-human SOX2 (Santa Cruz Biotechnology), anti-human NANOG (Abcam), and anti-human OCT3/4 (Santa Cruz Biotechnology). In brief, cultured hiPSCs were fixed in 4% paraformaldehyde (PFA) for 10 min, followed by permeabilization (only required for detecting intracellular antigens) in 0.1% Triton X-100, blocking for 30 min, and incubation with primary antibodies at 4°C overnight and washed three times with PBS. Cells were incubated with corresponding fluorochrome-conjugated secondary antibodies at room temperature for 1 h and washed three times with PBS. Cells were examined and photographed under a Zeiss Imager Z1 fluorescence microscope with AxioVision software.
Embryoid body and teratoma formation assay of pluripotency
For in vitro embryoid body (EB) formation, hiPSCs were dissociated with 1 mg/mL collagenase IV (Invitrogen) and subsequently transferred to ultra-low-attachment T25 flasks in EB medium (hESC medium without bFGF). To test the pluripotency of hiPSC in vivo, hiPSCs were harvested from four wells of 80% confluent six-well plates using 1 mg/mL dispase and resuspended in 100 μL of hiPSC medium. Cells were injected intramuscularly into nonobese diabetic severe combined immunodeficient (NOD-SCID) mice (Jackson Laboratory, Bar Harbor, ME). Tumors were excised 10–16 weeks after injection, fixed in 10% formalin, and embedded in frozen embedding medium (Shandon Cryomatrix, Thermo Scientific, Pittsburgh, PA). Tissue sections were stained with Hematoxylin & Eosin (H&E).
Determination of karyotypes
Karyotype analysis of G-banded metaphase chromosomes was performed at the Induced Pluripotent Stem Cell and Chromosome Core at UCHC. A karyogram was produced for two cells per clonal line.
T cell receptor rearrangement assay
PCR reactions to examine T cell receptor (TCR) gene rearrangement were performed by a combination of TCR gamma-chain gene (TCRG) primers designed by McCarthy et al. and a commercial TCRG gene clonality assay (Invivoscribe Technologies) (Kuo et al., 2011; McCarthy et al., 1992). DNA from hiPSCs was extracted as described above. The McCarthy TCRG PCR assay consisted of two reactions (VJr12 and VJp12) with two V primers (Vr11 and Vr-101) and two J primers (Jr12 and JP12). The PCR mixture contained primers of 0.5 μmol of each of the V primers and 1 μmol of one of the J primers. PCR was performed with a denaturation step of 3 min at 95°C, 30 amplification cycles at 93°C for 1 min, annealing at 55°C for 1 min, and extension at 73°C for 1 min, followed by a final extension step at 73°C for 10 min using GoTaq Flexi DNA polymerase (Promega). PCR products were further denatured at 95°C for 10 min and transferred to 4°C for 60 min to promote duplex formation before loading on 6–8% non-denaturing polyacrylamide gels in Tris/borate/EDTA (TBE) buffer. The commercially available TCRG Gene Clonality Assay kit also contained the amplification control master mix that was used to confirm the quality of sample DNAs and the control DNA sample that served as a positive control. The commercial TCRG Clonality Assay was performed according to the manufacturer's instructions, and the PCR product was analyzed by capillary electrophoresis (Experion automated electrophoresis system, Bio-Rad).
Results
Generation of PBMC-derived hiPSCs using Sendai virus vectors
Previous methods to generate hiPSCs used retroviral vectors to introduce ectopic expression of OCT3/4, SOX2, KLF4, and c-MYC into fibroblasts (Lowry et al., 2008; Takahashi et al., 2007), but the efficiency of reprogramming of fibroblasts with retrovirus appeared to be very low. In addition, skin biopsies from individuals under 18 are difficult to obtain. Stem cells from human exfoliated teeth (SHED) can also be reprogrammed to hiPSCs; however, they are not readily available, especially from adult patients (Yan et al., 2010). In contrast, peripheral blood is a readily accessible cell source. Many of our study subjects are under 18 because CMD is usually diagnosed at early age. Therefore, we decided to generate hiPSCs from peripheral blood using a SeV vector, an RNA virus vector that can transduce cells with high efficiency and does not integrate into the host genome (Ban et al., 2011; Bitzer et al., 2003).
We hoped to develop a simple method to reprogram cells from whole blood without selecting specific population such as CD34+ or terminally differentiated T cells, as previously described (Seki et al., 2010; Ye et al., 2009). We first used a SeV vector carrying the green fluorescent protein (GFP) gene to optimize SeV vector transduction efficiency in cells from whole blood. After Ficoll separation, PBMCs were cultured on non-culture-treated plates in DMEM containing 10% FBS, 20 ng/mL hSCF, hTPO, hIL-3, hFlt-3L, IL-6, IL-6R, and GM-CSF and 50 ng/mL M-CSF for 5 days. The majority of cells were floating while a few cells attached on the plates. Both floating and adherent cells were collected and were subjected to SeV vector transduction. Transduced cells were grown on culture plates coated with or without Retronectin and were maintained in α-minimum essential medium (α-MEM) or DMEM containing 10% FBS, 20 ng/m hSCF, hTPO, hIL-3, hFlt-3L, IL-6, IL-6R, and GM-CSF and 50 ng/mL M-CSF for 2 days after viral infection.
Retronectin is a chimeric peptide containing a fragment of fibronectin, a cell adhesion glycoprotein that can significantly increase retroviral or lentiviral transduction efficiency by enhancing adhesion of target cells and co-localization of viral particles and target cells. We observed that the DMEM combined with Retronectin-coated groups had the most adherent cells, which were GFP positive and were morphologically heterogeneous, whereas many floating cells did not express GFP (Fig. 1A). On the basis of this preliminary data, we later collected only the adherent cells 5 days after the initial culture for generation of hiPSCs using SeV vectors from whole peripheral blood.
FIG. 1.
Reprogramming of PBMCs by SeV vector transduction. (A) Optimization of SeV vector transduction efficiency in peripheral blood cultures using a SeV vector carrying the GFP gene. Representative images of bright field (left panel) and green fluorescence (right panel) were taken at days 1 and 3 after viral transduction. Note the relatively high GFP expression in adherent cells. (B) Schematic protocol of viral transduction. (C) Representative images of iPSC-like colonies from one healthy donor and one CMD patient. The PBMC-derived iPSCs have similar morphology to human embryonic stem cells. (D) Electropherograms of partial sequences of ANKH show mutations maintained in iPSCs: A heterozygous A→G transition in intron 9 leading to an Ala380 insertion, a heterozygous Ser375del, and Phe377del. (E) RT-PCR analysis for the transgenes OCT4, SOX2, c-MYC, KLF4, and SeV vector. HPRT was used as a loading control. Samples were: (1) PBMCs after SeV vector transduction; (2) hiPSCs passage 2; (3) hiPSCs passage 7; (4) hiPSC passage 13; (5) hESC H9. Color images available online at www.liebertpub.com/cell
We collected peripheral blood from a total of eight CMD patients and five healthy donors and generated PBMC-derived hiPSCs using SeV vectors encoding OCT3/4, SOX2, KLF4, and c-MYC. The schematic protocol of reprogramming of PBMCs by SeV vector was summarized (Fig. 1B). Briefly, PBMCs were cultured in media described above, and individual SeV vectors carrying OCT4, SOX2, c-MYC, and KLF4 transgenes were mixed and added to the cells. Adherent cells were transferred onto MEF cells 3 days after viral transduction. Starting from day 10, some small and loosely packed colonies appeared. These colonies did not show hESC-like morphology and were not fully reprogrammed iPSCs. Between 14 and 30 days, we picked up tightly packed colonies with similar morphology to hESCs (Fig. 1C). These PBMC-derived iPSC-like colonies can be passaged and grown on the Matrigel-coated plates without irradiated MEF cells.
To ensure that mutations of ANKH in CMD hiPSCs were preserved during reprogramming process, we sequenced the genomic DNA collected from PBMC-derived iPSCs from CMD patients. All three mutations were confirmed in hiPSCs (Fig. 1D). SeV vectors did not integrate into the host genome and gradually became diluted and disappeared during cell expansion (Ban et al., 2011). To monitor the residual amount of SeV vectors in PBMC-derived hiPSCs, we performed RT-PCR analysis for the transgenes OCT4, SOX2, c-MYC, and KLF4 as well as SeV vector backbone and showed that the majority of colonies were negative for SeV vector backbone or transduced transgenes after an average of 10–13 passages (Fig. 1E).
Characterization of PBMC-derived hiPSCs
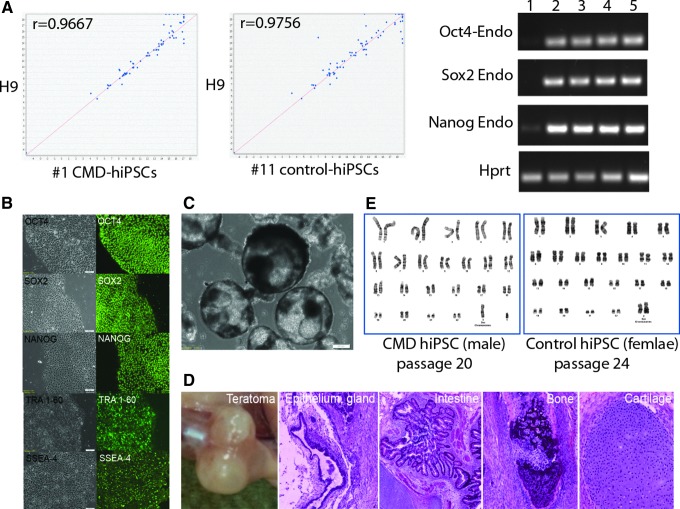
To examine the stemness of the PBMC-derived hiPSCs, we first performed RT-PCR for human stem cell markers OCT4, SOX2, and NANOG and immunostaining for TRA-1-60, SSEA-4, OCT4, SOX2, and NANOG. We also performed global expression analysis on one control and one CMD PBMC-derived hiPSC clone and compared to H9 hESCs. The PBMC-derived iPSCs expressed all markers examined at both RNA and protein levels (Fig. 2A, B). The human stem cell pluripotency array showed similar expression patterns between our hiPSCs and H9 cells (Fig. 2A).
FIG. 2.
Characterization of PBMC-derived iPSCs. (A) RT-PCR analysis for hESC marker genes OCT4, SOX2, NANOG, and HPRT as a loading control. Samples were: (1) PBMCs after SeV vector transduction; (2) hiPSCs passage2; (3) hiPSCs passage 7; (4) hiPSC passage 13; (5) hESC H9. Expression analysis (left panel) of 90 well-defined gene markers for pluripotency indicate similarity between PBMC-derived iPSCs and H9 hESCs. (B) Immunofluorescence staining for hESC markers OCT4, SOX2, NANOG, TRA-1-60, and SSEA-4 (right panel). Bright-field images of PBMC-derived iPSCs (left panel). (C) EBs from PBMC-derived hiPSCs growing in suspension cultures after 15 days. (D) Gross anatomy of different tissues derived from a teratoma of PBMC-derived hiPSCs (H&E). (E) Normal karyotype in both control PBMC-hiPSCs and CMD PBMC-iPSCs. Color images available online at www.liebertpub.com/cell
To further examine their in vitro differentiation potential, the PBMC-derived hiPSCs were dissociated and grown in ultra-low-attachment flasks with EB medium. They were capable of forming typical well-rounded EBs in suspension cultures (Fig. 2C). To study the differentiation potential in vivo, these hiPSCs were injected intramuscularly into NOD-SCID mice, and teratomas were histologically evaluated. We observed various tissues formed from three germ layers, such as epithelium (ectoderm), intestine (endoderm), bone, and cartilage (mesoderm) in teratomas (Fig. 2D). After more than 20 passages, these hiPSCs still maintained normal karyotypes of either 46 XX or 46 XY (Fig. 2E). Results from these assays suggested that the PBMC-derived hiPSCs generated with SeV vectors were free of vector-derived transgenes and present the required characteristics of pluripotent stem cells.
Analysis of T cell receptor gene rearrangement
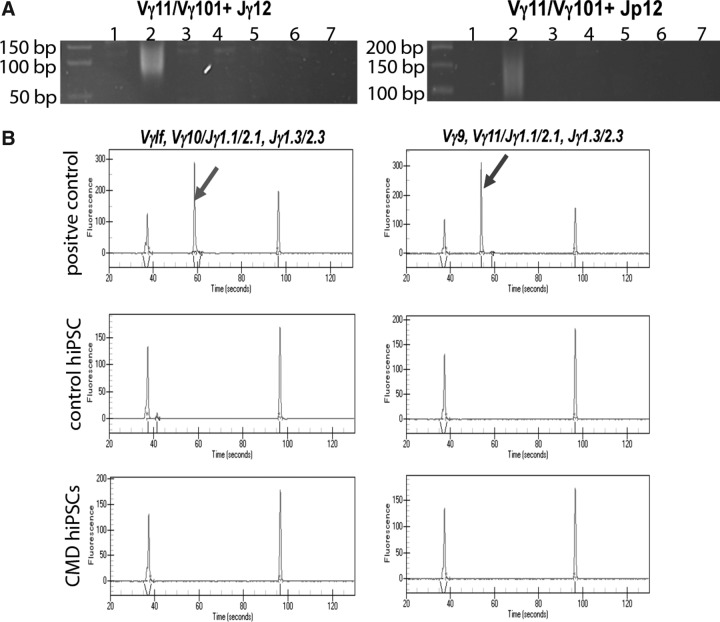
In some previous studies, terminally differentiated T cells were preferentially selected to be reprogrammed into hiPSCs after culturing the peripheral blood with plate-bound anti-CD3 antibody and rIL-2 or expanded in IL-7 (Seki et al., 2010; Staerk et al., 2010). Methods using T cells from peripheral blood to reprogram into hiPSCs are advantageous for research of blood or immunologic disorders. We aim to establish hiPSC disease modeling for rare genetic bone disorders, thus selection of T cell populations in peripheral blood would not be useful for our studies. Our simplified method is likely to exclude T cells as a cell source for reprogramming, but we wished to evaluate whether iPSC lines that we produced were derived from T cells. Genetic DNA rearrangement of TCR genes occurs during T cell development and leads to a variety of T cell repertoires to adapt to diverse antigen response requirements. Examining the existence of TCR gene rearrangement allowed us to assess whether our hiPSCs originated from T cells. We studied the rearrangement of TCR γ loci with a combination of McCarthy's in-house primers and TCRG BIOMED-2 primers purchased from Invivoscribe (McCarthy et al., 1992). Genomic DNA isolated from fibroblasts (negative control), peripheral blood (polyclonal control), and iPSCs was subjected to PCR with four primer combinations (Vγ11/Vγ101+Jγ12; Vγ11/Vγ101+Jp12) in two reactions (McCarthy et al., 1992). The rearranged bands are expected to be in the size range of 75–95 and 80–110 bp for VJγ12 and VJp12, respectively. The peripheral T cells were polyclonal and thus showed a smeared band, whereas fibroblasts and our hiPSCs did not have bands as shown on polyacrylamide gels (Fig. 3A). The PCR products with BIOMED-2 primers were subjected to capillary electrophoresis. The positive control provided by Invivoscribe showed a specific peak, whereas all of our examined hiPSCs did not have this specific peak (Fig. 3B). In conclusion, we did not detect a TCRG rearrangement in our PBMC-derived hiPSCs using either the McCarthy or BIOMED-2 primer sets.
FIG. 3.
T cell receptor gene rearrangement assay. (A) PBMC-derived iPSC lines show no TCR rearrangement in TCRG loci with McCarthy primers. Genomic DNA from: (1) Fibroblasts; (2) peripheral blood; (3–6) iPSCs; (7) no DNA. (B) TCRG examined by BIOMED-2 primer sets and capillary electrophoresis. Specific peaks shown by arrows are seen in positive control (clonal DNA) but not control iPSCs and CMD iPSCs. The two peripheral peaks in each plot are DNA ladders.
Discussion
Research focusing on rare genetic skeletal disorders is not only beneficial for future treatment of those patients, but contributes to the understanding of important biological mechanisms in bone development and remodeling. Many of these diseases are still understudied due to lack of human specimens and proper study models. We previously studied CMD using a knockin mouse model replicating many characteristics of human CMD and could show that CMD is caused by a complex mechanism involving impaired osteoblastogenesis and osteoclastogenesis (Chen et al., 2009; Chen et al., 2011). Ultimately, we plan to investigate the cellular and molecular basis of CMD in the human system, which is only possible with a sufficient supply of human cells. A simple method to generate patient-specific, viral-free hiPSCs from an easily accessible cell source, such as peripheral blood, is a significant improvement to study this disease pathology. hiPSCs without transgene integration may also serve as a model for preclinical trials and as a relatively safe cell source for possible therapeutical use in future.
We demonstrated a simple and efficient method to generate virus/transgene-free hiPSCs from peripheral blood using SeV vectors. The resulting hiPSCs had the required properties of pluripotent stem cells, as indicated by the uniform expression of the pluripotency marker genes and proteins, the ability to form EBs and teratomas, and a normal karyotype after more than 20 passages. We initially had generated hiPSCs from dermal fibroblasts and SHEDs by retroviral and lentiviral transduction. Retroviruses can transduce dividing cells and are usually silenced in embryonic cells and during the reprogramming process by de novo methylation (Jahner et al., 1982; Wernig et al., 2007). Incomplete transgene silencing is considered an indication that an iPSC is not successfully reprogrammed and may have a negative impact on iPSC differentiation (Maherali and Hochedlinger, 2008; Papapetrou et al., 2009). The use of a lentiviral stem cell cassette (STEMCCA) allows reprogramming of both dividing and nondividing cells with a single viral integration rather than by four separate retroviral transgenes (Sommer et al., 2009). Although this can improve efficiency of hiPSC generation, an additional step to excise the reprogramming vector should be done to eliminate the possibility of incomplete silencing of the transgene (Sommer et al., 2010).
CMD is an early-onset disease, and skin biopsies are difficult to obtain from children. Peripheral blood is a more convenient source of somatic cells. In addition, cells from peripheral blood can be reprogrammed several days after initial culture, unlike dermal fibroblasts and other cell types, which may take weeks for establishing primary cell cultures. We have used lentiviral vectors to transfect blood cells but could not produce hiPSCs consistently, and the efficiency of reprogramming was five to ten times lower than by using a SeV vector (data not shown). A SeV vector was chosen because of its high transduction efficiency and because it can produce iPSCs free of vector integration into chromosomes.
Several papers have reported the generation of hiPSCs from CD34+ cells, mature T cells, myeloid cells, or nonlymphoid cells in peripheral blood with retroviral, lentiviral, or SeV vector methods (Kunisato et al., 2011; Merling et al., 2013; Seki et al., 2010; Staerk et al., 2010; Ye et al., 2009). We directly reprogrammed the adherent cells from the initial peripheral blood cultures to generate hiPSCs without preselection or sorting of specific cell populations. Our method largely excluded T cells that were floating cells in cultures and excluded B cells, which would need IL4/CD40 ligand to promote their survival and expansion (von Bergwelt-Baildon et al., 2002).
To further assess whether our hiPSCs were derived from T cells, the main cell type reported in previous publications, we examined the hallmark of T cells, the TCR gene rearrangement that occurs as sequential rearrangement of variable (V), diversity (D), and joining (J) gene segments during T cell development. We adapted the cost-effective and efficient strategy to combine primers published by McCarthy and colleagues and commercial TCRG BIOMED-2 primers (Kuo et al., 2011). The detection rate of TCR gene rearrangement by this method reaches 91%, whereas 85% can be achieved by all six BIOMED-2 reactions and 94% by combining the primers used by McCarthy et al. and all BIOMED-2 reactions (Kuo et al., 2011). As expected, our results showed negative TCRG in a total of nine hiPSCs that we tested. Additional tests with primer sets for TCRβ, TCRδ loci, and immunoglobulin heavy chain were not used in our study but can be considered if identifying the source of somatic cells becomes critical to a specific study. SeV-transduced adherent cells in PBMC cultures are heterogeneous with various morphologies (Fig. S1) (Supplementary Data are available at www.liebertpub.com/cell/.). The majority of cells are spherical or “fried-egg”-shaped, which presumably corresponds to monocytes and macrophages, respectively (Young and Deane, 2005). Additionally, we found spindle-shaped cells with morphology similar to circulating fibrocytes, as reported previously (Fang et al., 2012; Yang et al., 2002). Last, there are a few cells possessing thin projections resembling dendritic cells (Knight et al., 1992; Zhou and Tedder, 1995).
It has been reported that adherent cells in PBMC cultures can express markers for mesenchymal stem cells (MSCs) with differentiation potentials to adipocytes, osteocytes, and chondrocytes under the stimulation of differentiation factors (Ab Kadir et al., 2012; Kim et al., 2012). More characteristic and functional studies will be needed to further categorize these attached and transduced cells derived from peripheral blood. On the basis of our assays and observations, we suggest that the PBMC-derived iPSCs likely originate from a mixed population of nonlymphoid cells.
Reprogramming somatic cells from patients into iPSCs and differentiating them into cells of interest provides an excellent tool for studying human diseases. To use these patient-specific hiPSCs to study CMD, our next steps are to differentiate these hiPSCs to skeletal cell types, including bone-forming (osteoblasts) and bone-resorbing cells (osteoclasts). Differentiation of hiPSCs into MSC–like populations that have osteogenic potential has been reported in vitro and in vivo (Villa-Diaz et al., 2012). hiPSCs were reported to be differentiated into osteoclasts through an EB protocol or through OP9-hiPSC co-cultures (Choi et al., 2011; Grigoriadis et al., 2010). Both protocols resulted in tartrate-resistant acid phosphatase (TRAP)-positive multinucleated cells with resorption capability. We had initial success of generation of MSCs and osteoclast-like cells from H9 hESCs using both published protocols (data not shown). We will apply the same methodology to differentiate PBMC-derived hiPSCs into MSCs and osteoclasts. We expect this tool will allow us to study molecular mechanisms by which ANKH mutations cause impaired osteoblastogenesis and osteoclastogenesis in CMD patients.
In summary, we have successfully generated hiPSCs from the peripheral blood of CMD patients and healthy donors. We expect that combining hypotheses generated from the CMD mouse model and findings from differentiated hiPSCs together with genome editing techniques will significantly increase our understanding of the CMD pathology and possibly lead to discovery of therapeutic targets. By using CMD as a paradigm, we hope to establish novel tools for studying other rare genetic skeletal disorders.
Supplementary Material
Acknowledgments
The project was supported by National Institutes of Health (NIH) grant K99DE021442 (NIDCR) to I.P.C. and Connecticut State Grant 09SCBUCHC20 and R21DEO19892 to A.L. and E.R.
Author Disclosure Statement
I.P.C., N.F., K.F., A.L., and E.R. declare that no conflicting financial interests exist. A.I. is an employee of DNAVEC Corporation. M.H. is a founder of DNAVEC Corporation.
References
- Aasen T. Raya A. Barrero M.J. Garreta E. Consiglio A. Gonzalez F. Vassena R. Bilić J. Pekarik V. Tiscornia G. Edel M. Boué S. Izpisúa Belmonte J.C. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Ab Kadir R. Zainal Ariffin S.H. Megat Abdul Wahab R. Kermani S. Senafi S. Characterization of mononucleated human peripheral blood cells. Scientific World Journal. 2012;2012:843843. doi: 10.1100/2012/843843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban H. Nishishita N. Fusaki N. Tabata T. Saeki K. Shikamura M. Takada N. Inoue M. Hasegawa M. Kawamata S. Nishikawa S. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc. Natl. Acad. Sci. USA. 2011;108:14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beighton P. Hamersma H. Horan F. Craniometaphyseal dysplasia—variability of expression within a large family. Clin. Genet. 1979;15:252–258. doi: 10.1111/j.1399-0004.1979.tb00976.x. [DOI] [PubMed] [Google Scholar]
- Bitzer M. Armeanu S. Lauer U.M. Neubert W.J. Sendai virus vectors as an emerging negative-strand RNA viral vector system. J. Gene Med. 2003;5:543–553. doi: 10.1002/jgm.426. [DOI] [PubMed] [Google Scholar]
- Chen C. Wang Y. Goh S.S. Yang J. Lam D.H. Choudhury Y. Tay F.C. Du S. Tan W.K. Purwanti Y.I. Fan W. Wang S. Inhibition of neuronal nitric oxide synthase activity promotes migration of human-induced pluripotent stem cell-derived neural stem cells toward cancer cells. J. Neurochem. 2013;126:318–330. doi: 10.1111/jnc.12199. [DOI] [PubMed] [Google Scholar]
- Chen I.P. Wang C.J. Strecker S. Koczon-Jaremko B. Boskey A. Reichenberger E.J. Introduction of a Phe377del mutation in ANK creates a mouse model for craniometaphyseal dysplasia. J. Bone Min. Res. 2009;24:1206–1215. doi: 10.1359/JBMR.090218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.P. Wang L. Jiang X. Aguila H.L. Reichenberger E.J. A Phe377del mutation in ANK leads to impaired osteoblastogenesis and osteoclastogenesis in a mouse model for craniometaphyseal dysplasia (CMD) Hum. Mol. Genet. 2011;20:948–961. doi: 10.1093/hmg/ddq541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry A.B. Daley G.Q. Reprogrammed cells for disease modeling and regenerative medicine. Annu. Rev. Med. 2013;64:277–290. doi: 10.1146/annurev-med-050311-163324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.D. Vodyanik M. Slukvin Hematopoietic differentiation and production of mature myeloid cells from human pluripotent stem cells. Nat. Protoc. 2011;6:296–313. doi: 10.1038/nprot.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos J.T. Rodolfa K.T. Niakan K.K. Weisenthal L.M. Mitsumoto H. Chung W. Croft G.F. Saphier G. Leibel R. Goland R. Wichterle H. Henderson C.E. Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Ding Q. Lee Y.K. Schaefer E.A. Peters D.T. Veres A. Kim K. Kuperwasser N. Motola D.L. Meissner T.B. Hendriks W.T. Trevisan M. Gupta R.M. Moisan A. Banks E. Friesen M. Schinzel R.T. Xia F. Tang A. Xia Y. Figueroa E. Wann A. Ahfeldt T. Daheron L. Zhang F. Rubin L.L. Peng L.F. Chung R.T. Musunuru K. Cowan C.A. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emborg M.E. Liu Y. Xi J. Zhang X. Yin Y. Lu J. Joers V. Swanson C. Holden J.E. Zhang S.C. Induced pluripotent stem cell-derived neural cells survive and mature in the nonhuman primate brain. Cell Rep. 2013;3:646–650. doi: 10.1016/j.celrep.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L. Moore X.L. Chan W. White D.A. Chin-Dusting J. Dart A.M. Decreased fibrocyte number is associated with atherosclerotic plaque instability in man. Cardiovasc. Res. 2012;95:124–133. doi: 10.1093/cvr/cvs156. [DOI] [PubMed] [Google Scholar]
- Franz D.C. Horn K.L. Aase J. Craniometaphyseal dysplasia: Operative findings and treatment. Am. J. Otol. 1996;17:283–287. [PubMed] [Google Scholar]
- Fusaki N. Ban H. Nishiyama A. Saeki K. Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Japan Acad. Ser. B, Phys. Biol. Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti A. Montserrat N. Aasen T. Gonzalez F. Rodríguez-Pizà I. Vassena R. Raya A. Boué S. Barrero M.J. Corbella B.A. Torrabadella M. Veiga A. Izpisua Belmonte J.C. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell. 2009;5:353–357. doi: 10.1016/j.stem.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F. Boue S. Izpisua Belmonte J.C. Methods for making induced pluripotent stem cells: Reprogramming a la carte. Nat. Rev. Genet. 2011;12:231–242. doi: 10.1038/nrg2937. [DOI] [PubMed] [Google Scholar]
- Grabel L. Prospects for pluripotent stem cell therapies: Into the clinic and back to the bench. J. Cell. Biochem. 2012;113:381–387. doi: 10.1002/jcb.23364. [DOI] [PubMed] [Google Scholar]
- Grigoriadis A.E. Kennedy M. Bozec A. Brunton F. Stenbeck G. Park I.H. Wagner E.F. Keller G.M. Directed differentiation of hematopoietic precursors and functional osteoclasts from human ES and iPS cells. Blood. 2010;115:2769–2776. doi: 10.1182/blood-2009-07-234690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A.M. Johnson M.D. Kingsley D.M. Role of the mouse ank gene in control of tissue calcification and arthritis. Science. 2000;289:265–270. doi: 10.1126/science.289.5477.265. [DOI] [PubMed] [Google Scholar]
- Jackson W.P. Albright F. Drewry G. Hanelin J. Rubin M.I. Metaphyseal dysplasia, epiphyseal dysplasia, diaphyseal dysplasia, and related conditions. I. Familial metaphyseal dysplasia and craniometaphyseal dysplasia; their relation to leontiasis ossea and osteopetrosis; disorders of bone remodeling. AMA Arch. Int. Med. 1954;94:871–885. doi: 10.1001/archinte.1954.00250060005001. [DOI] [PubMed] [Google Scholar]
- Jahner D. Stuhlmann H. Stewart C.L. Harbers K. Lohler J. Simon I. Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- Kim J. Shin J.M. Jeon Y.J. Chung H.M. Chae J.I. Proteomic validation of multifunctional molecules in mesenchymal stem cells derived from human bone marrow, umbilical cord blood and peripheral blood. PloS One. 2012;7:e32350. doi: 10.1371/journal.pone.0032350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.C. Stagg A. Hill S. Fryer P. Griffiths S. Development and function of dendritic cells in health and disease. J. Invest. Dermatol. 1992;99:33S–38S. doi: 10.1111/1523-1747.ep12668601. [DOI] [PubMed] [Google Scholar]
- Kudva Y.C. Ohmine S. Greder L.V. Dutton J.R. Armstrong A. De Lamo J.G. Khan Y.K. Thatava T. Hasegawa M. Fusaki N. Slack J.M. Ikeda Y. Transgene-free disease-specific induced pluripotent stem cells from patients with type 1 and type 2 diabetes. Stem Cells Transl. Med. 2012;1:451–461. doi: 10.5966/sctm.2011-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisato A. Wakatsuki M. Shinba H. Ota T. Ishida I. Nagao K. Direct generation of induced pluripotent stem cells from human nonmobilized blood. Stem Cells Dev. 2011;20:159–168. doi: 10.1089/scd.2010.0063. [DOI] [PubMed] [Google Scholar]
- Kuo S.Y. Liu H. Liao Y.L. Chang S.T. Hsieh Y.C. Bandoh B.A. Du M.Q. Chuang S.S. A parallel comparison of T-cell clonality assessment between an in-house PCR assay and the BIOMED-2 assay leading to an efficient and cost-effective strategy. J. Clin. Pathol. 2011;64:536–542. doi: 10.1136/jcp.2010.086637. [DOI] [PubMed] [Google Scholar]
- Lowry W.E. Richter L. Yachechko R. Pyle A.D. Tchieu J. Sridharan R. Clark A.T. Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl. Acad. Sci. USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehr R. Chen S. Snitow M. Ludwig T. Yagasaki L. Goland R. Leibel R.L. Melton D.A. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc. Natl. Acad. Sci. USA. 2009;106:15768–15773. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N. Hochedlinger K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;3:595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- McCarthy K.P. Sloane J.P. Kabarowski J.H. Matutes E. Wiedemann L.M. A simplified method of detection of clonal rearrangements of the T-cell receptor-gamma chain gene. Diagn. Mol. Pathol. 1992;1:173–179. [PubMed] [Google Scholar]
- Merling R.K. Sweeney C.L. Choi U. De Ravin S.S. Myers T.G. Otaizo-Carrasquero F. Pan J. Linton G. Chen L. Koontz S. Theobald N.L. Malech H.L. Transgene-free iPSCs generated from small volume peripheral blood nonmobilized CD34+ cells. Blood. 2013;121:e98–e107. doi: 10.1182/blood-2012-03-420273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberg P. Thiele H. Chandler D. Hohne W. Cunningham M.L. Ritter H. Leschik G. Uhlmann K. Mischung C. Harrop K. Goldblatt J. Borochowitz Z.U. Kotzot D. Westermann F. Mundlos S. Braun H.S. Laing N. Tinschert S. Heterozygous mutations in ANKH, the human ortholog of the mouse progressive ankylosis gene, result in craniometaphyseal dysplasia. Nat. Genet. 2001;28:37–41. doi: 10.1038/ng0501-37. [DOI] [PubMed] [Google Scholar]
- Papapetrou E.P. Tomishima M.J. Chambers S.M. Mica Y. Reed E. Menon J. Tabar V. Mo Q. Studer L. Sadelain M. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc. Natl. Acad. Sci. USA. 2009;106:12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.H. Arora N. Huo H. Maherali N. Ahfeldt T. Shimamura A. Lensch M.W. Cowan C. Hochedlinger K. Daley G.Q. Disease-specific induced pluripotent stem cells. Cell. 2008a;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.H. Zhao R. West J.A. Yabuuchi A. Huo H. Ince T.A. Lerou P.H. Lensch M.W. Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008b;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Puliafito C.A. Wray S.H. Murray J.E. Boger W.P., 3rd Optic atrophy and visual loss in craniometaphyseal dysplasia. Am. J. Ophthalmol. 1981;92:696–701. doi: 10.1016/s0002-9394(14)74664-1. [DOI] [PubMed] [Google Scholar]
- Reichenberger E. Tiziani V. Watanabe S. Park L. Ueki Y. Santanna C. Baur S.T. Shiang R. Grange D.K. Beighton P. Gardner J. Hamersma H. Sellars S. Ramesar R. Lidral A.C. Sommer A. Raposo do Amaral C.M. Gorlin R.J. Mulliken J.B. Olsen B.R. Autosomal dominant craniometaphyseal dysplasia is caused by mutations in the transmembrane protein ANK. Am. J. Hum. Genet. 2001;68:1321–1326. doi: 10.1086/320612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A. Brain C. Dillon M.J. Bailey C.M. Craniometaphyseal and craniodiaphyseal dysplasia, head and neck manifestations and management. J. Laryngol. Otol. 1996;110:328–338. doi: 10.1017/s0022215100133560. [DOI] [PubMed] [Google Scholar]
- Satoh K. Iwata T. Ikeda H. Unsuccessful consequence of optic canal decompression for a case of craniometaphyseal dysplasia. Plastic Reconstr. Surg. 1994;94:705–708. doi: 10.1097/00006534-199410000-00022. [DOI] [PubMed] [Google Scholar]
- Seki T. Yuasa S. Oda M. Egashira T. Yae K. Kusumoto D. Nakata H. Tohyama S. Hashimoto H. Kodaira M. Okada Y. Seimiya H. Fusaki N. Hasegawa M. Fukuda K. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7:11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Shea J. Gerbe R. Ayani N. Craniometaphyseal dysplasia: the first successful surgical treatment for associated hearing loss. The Laryngoscope. 1981;91:1369–1374. doi: 10.1288/00005537-198108000-00019. [DOI] [PubMed] [Google Scholar]
- Sommer C.A. Stadtfeld M. Murphy G.J. Hochedlinger K. Kotton D.N. Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C.A. Sommer A.G. Longmire T.A. Christodoulou C. Thomas D.D. Gostissa M. Alt F.W. Murphy G.J. Kotton D.N. Mostoslavsky G. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 2010;28:64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M. Nagaya M. Utikal J. Weir G. Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staerk J. Dawlaty M.M. Gao Q. Maetzel D. Hanna J. Sommer C.A. Mostoslavsky G. Jaenisch R. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell. 2010;7:20–24. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susa M. Luong-Nguyen N.H. Cappellen D. Zamurovic N. Gamse R. Human primary osteoclasts:I vitro generation and applications as pharmacological and clinical assay. J. Transl. Med. 2004;2:6. doi: 10.1186/1479-5876-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Teo A.K. Windmueller R. Johansson B.B. Dirice E. Njolstad P.R. Tjora E. Raeder H. Kulkarni R.N. Derivation of human induced pluripotent stem cells from patients with maturity onset diabetes of the young. J. Biol. Chem. 2013;288:5353–5356. doi: 10.1074/jbc.C112.428979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokusumi T. Iida A. Hirata T. Kato A. Nagai Y. Hasegawa M. Recombinant Sendai viruses expressing different levels of a foreign reporter gene. Virus Res. 2002;86:33–38. doi: 10.1016/s0168-1702(02)00047-3. [DOI] [PubMed] [Google Scholar]
- Villa-Diaz L.G. Brown S.E. Liu Y. Ross A.M. Lahann J. Parent J.M. Krebsbach P.H. Derivation of mesenchymal stem cells from human induced pluripotent stem cells cultured on synthetic substrates. Stem Cells. 2012;30:1174–1181. doi: 10.1002/stem.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bergwelt-Baildon M.S. Vonderheide R.H. Maecker B. Hirano N. Anderson K.S. Butler M.O. Xia Z. Zeng W.Y. Wucherpfennig K.W. Nadler L.M. Schultze J.L. Human primary and memory cytotoxic T lymphocyte responses are efficiently induced by means of CD40-activated B cells as antigen-presenting cells: Potential for clinical application. Blood. 2002;99:3319–3325. doi: 10.1182/blood.v99.9.3319. [DOI] [PubMed] [Google Scholar]
- Warren L. Manos P.D. Ahfeldt T. Loh Y.H. Li H. Lau F. Ebina W. Mandal P.K. Smith Z.D. Meissner A. Daley G.Q. Brack A.S. Collins J.J. Cowan C. Schlaeger T.M. Rossi D.J. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M. Meissner A. Foreman R. Brambrink T. Ku M. Hochedlinger K. Bernstein B.E. Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Woltjen K. Michael I.P. Mohseni P. Desai R. Mileikovsky M. Hämäläinen R. Cowling R. Wang W. Liu P. Gertsenstein M. Kaji K. Sung H.K. Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X. Qin H. Qu C. Tuan R.S. Shi S. Huang G.T. iPS cells reprogrammed from human mesenchymal-like stem/progenitor cells of dental tissue origin. Stem Cells Dev. 2010;19:469–480. doi: 10.1089/scd.2009.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. Scott P.G. Giuffre J. Shankowsky H.A. Ghahary A. Tredget E.E. Peripheral blood fibrocytes from burn patients: identification and quantification of fibrocytes in adherent cells cultured from peripheral blood mononuclear cells. Lab. Invest. 2002;82:1183–1192. doi: 10.1097/01.lab.0000027841.50269.61. [DOI] [PubMed] [Google Scholar]
- Ye Z. Zhan H. Mali P. Dowey S. Williams D.M. Jang Y.Y. Dang C.V. Spivak J.L. Moliterno A.R. Cheng L. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009;114:5473–5480. doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L.J. Deane E.M. Culture and characterisation of peripheral blood monocytes and monocyte-derived adherent cells of the tammar wallaby, Macropus eugenii. Immunol. Lett. 2005;96:253–259. doi: 10.1016/j.imlet.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Yu J. Vodyanik M.A. Smuga-Otto K. Antosiewicz-Bourget J. Frane J.L. Tian S. Nie J. Jonsdottir G.A. Ruotti V. Stewart R. Slukvin I.I. Thomson J.A. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu J. Hu K. Smuga-Otto K. Tian S. Stewart R. Slukvin Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. Wu S. Joo J.Y. Zhu S. Han D.W. Lin T. Trauger S. Bien G. Yao S. Zhu Y. Siuzdak G. Schöler H.R. Duan L. Ding S. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L.J. Tedder T.F. Human blood dendritic cells selectively express CD83, a member of the immunoglobulin superfamily. J. Immunol. 1995;154:3821–3835. [PubMed] [Google Scholar]
- Zhou T. Benda C. Dunzinger S. Huang Y. Ho J.C. Yang J. Wang Y. Zhang Y. Zhuang Q. Li Y. Bao X. Tse H.F. Grillari J. Grillari-Voglauer R. Pei D. Esteban M.A. Generation of human induced pluripotent stem cells from urine samples. Nat. Protoc. 2012;7:2080–2089. doi: 10.1038/nprot.2012.115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.