Abstract
Surface preparation is a key step for reliable and reproducible imaging of DNA and protein-DNA complexes with atomic force microscopy (AFM). This article describes the approaches for chemical functionalization of the mica surface. One approach utilizes 3-aminopropyl-trietoxy silane (APTES), enabling one to obtain a smooth surface termed AP-mica. This surface binds nucleic acids and nucleoprotein complexes in a wide range of ionic strengths, in the absence of divalent cations and in a broad range of pH. Another method utilizes aminopropyl silatrane (APS) to yield an APS-mica surface. The advantage of APS-mica compared with AP-mica is the ability to obtain reliable and reproducible time-lapse images in aqueous solutions. The chapter describes the methodologies for the preparation of AP-mica and APS-mica surfaces and the preparation of samples for AFM imaging. The protocol for synthesis and purification of APS is also provided. The applications are illustrated with a number of examples.
Keywords: Atomic force microscopy, AFM, Mica functionalization, Surface chemistry, Silanes, Silatranes, DNA structure and dynamics, Protein-DNA complexes
1. Introduction
The first results revealing reliable AFM imaging of long DNA molecules were reported in the early 1990s when a number of sample preparation methods were developed. The laboratory of Bustamante et al. (1) implemented the method of cationic treatment of mica (2). In this approach, the mica surface is treated with Mg2+ to increase the affinity of the negatively charged mica surface to DNA. Other metal cations such as Co2+, La3+, and Zr4+ can be used for mica pretreatment to obtain images of DNA (3). Later experiments showed that pretreatment of mica with cations was not necessary (4–7), as DNA adheres if Mg2+ cations are present in the buffer. In the laboratory of Z. Shao, an approach utilizing a modification of the well-known electron microscopic procedure for imaging DNA was developed (8, 9). This method involves spreading DNA onto a carbon-coated mica substrate following cytochrome c denaturation at the air–water interface. The same group showed that DNA can be absorbed onto a supported cationic bilayer surface and imaged with AFM in aqueous buffers (10). The authors used densely and uniformly packed DNA filaments, enabling them to resolve a periodic lateral modulation of 3.4 ± 0.4 nm that was in concordance with the known pitch of the double helix. Gold substrates can also be activated by self-assembled monolayers of thiols for reliable imaging of DNA (11) and nontreated cover glass appeared to be a good substrate for binding chromatin, although the rinsing step (to remove unbound material and salt components) needed to be done gently and the dried sample had to be imaged immediately (12). Among these methods, the cation-assisted technique has become the most widely used due to the simplicity of sample preparation. However, the requirement for multivalent cations is mandatory, and therefore limits the range of experimental conditions to buffers with a defined concentration of cations.
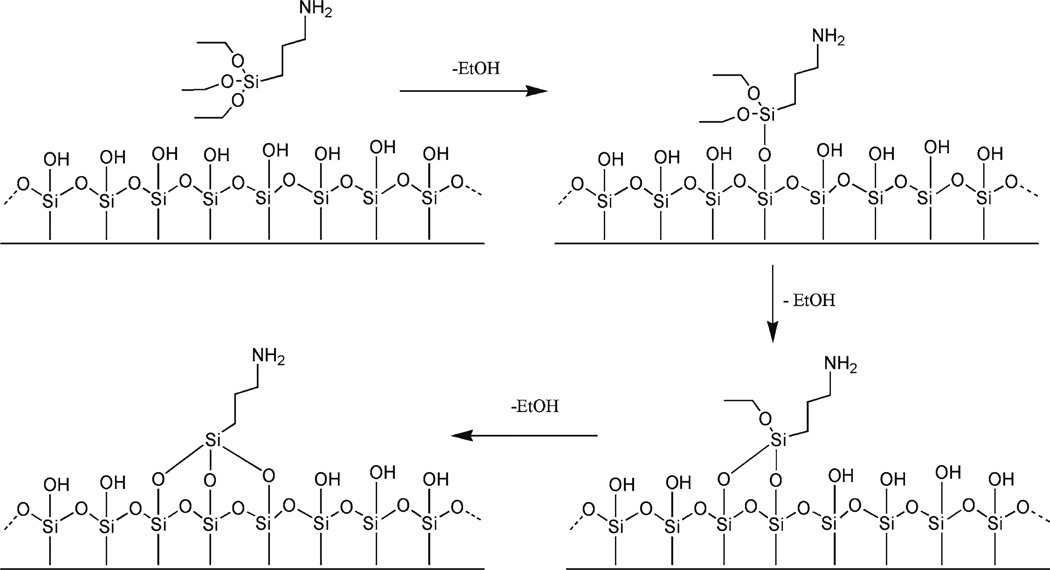
We (13–18) worked out a procedure for chemical functionalization of mica. A weak cationic surface is obtained if 3-aminopropyltriethoxy silane (APTES) is used to functionalize the mica surface with amino groups (AP-mica). The schematic for the chemical reaction of APTES with mica is shown in Fig. 1. As a result, functionalized surfaces remain positively charged at pH below pKa values (pKa = 10.4) and are capable of binding to negatively charged DNA in the pH range of stable DNA duplexes.
Fig. 1.
Scheme for the reaction of APTES with mica. APTES reacts with hydroxyl groups on the mica surface formed spontaneously after the mica is cleaved.
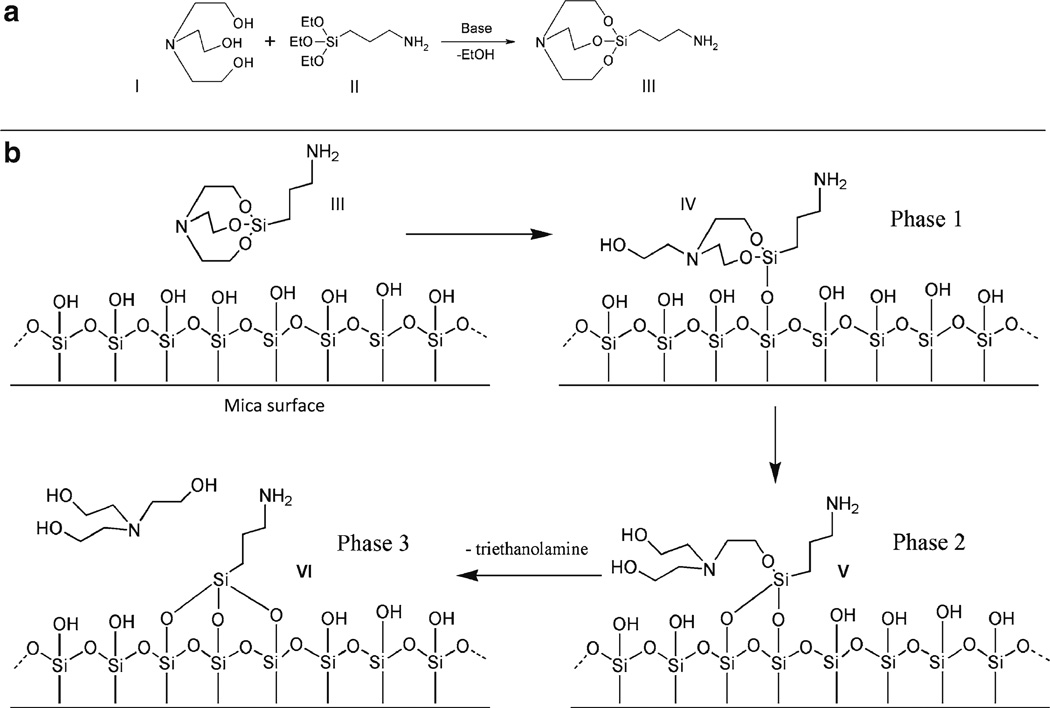
An analogous surface chemistry utilizes a more hydrolytically stable silatrane reagent 1-(3-aminopropyl) silatrane (APS) instead of silanes. Slow hydrolysis of the silatrane moiety allows one to avoid clumping and enable a smooth modification of the surface with amino groups achieving the results similar to vapor treatment. The schemes for the synthesis of APS and its reaction with the mica surface are shown in Fig. 2. The reaction with the surface (Fig. 2b) is apparently proceeding through several steps (phase 1–3) with eventual loss of triethanolamine molecule and covalent attachment of 3-aminopropyl siloxane group to mica. Both methods are robust and work reproducibly in various topographic studies involving DNA (19–32). The APS-mica methodology works reliably for force spectroscopy AFM applications (33–37). Sections below provide all specifics related to the preparation of both surfaces for AFM imaging.
Fig. 2.
APS-mica preparation. (a) Scheme for synthesis of 1-(3-aminopropyl)silatrane (APS, III) using APTES (I) and triethanolamine (II). APS: Molecular weight 232.36; molecular formula C9H20N2O3Si. (b) Scheme for reaction of APS (III) with mica surface. Similarly to APTES, APS reacts with hydroxyl groups on mica surface formed spontaneously after the cleavage. Three different stages of the reaction with the formation of adducts IV and V are shown. Note the last stage, exposure of APS-functionalized mica to water; it leads to dissociation of triethanolamine yielding product VI.
2. Materials
2.1. General Equipment and Supplies
A vacuum cabinet or desiccator for storing samples. A Gravity Convention Utility Oven (VWR) is recommended.
Plastic tubes, 15 mL.
Eppendorf tubes, 1.5 mL.
Plastic cuvettes.
Scissors.
Razor blade.
2-L glass desiccators and vacuum line (50 mmHg is sufficient).
Pipettes with plastic tips for rinsing the samples.
Tweezers.
Gas tank with clean argon gas. Nitrogen gas can be used as well.
Vacuum distillation apparatus for distilling APTES.
Mica substrate. Any type of commercially available mica sheets (green or ruby mica) can be used. Asheville-Schoonmaker Mica Co (Newport News, VA) supplies thick and large (more than 5 × 7 cm) sheets (Grade 1) suitable for making the substrates of different thickness and sizes.
Deionized water filtered through 0.2-µm filter for mica functionalization and AFM sample preparation.
2.2. Reagents and Solutions
TE buffer solution: (20 mM Tris–HCl, pH 7.6, 1 mM EDTA) with 200 mM NaCl.
2.2.1. AP-Mica Preparation
3-Aminopropyltriethoxy silane for re-distillation to ensure a reliable and reproducible AP-mica preparation.
N,N-diisopropylethylamine (DIPEA) for AP-mica preparation.
2.2.2. APS-Mica Preparation
3-Aminopropyltriethoxy silane (APTES, TCI, USA) for APS-mica preparation.
Sodium hydroxide and triethanolamine for APS synthesis.
Methanol and toluene solvents for APS synthesis.
3. Methods
3.1. Synthesis of APS
The procedure described below is a modified version of our previously published method (38). We tested and verified that sodium hydroxide can be used as a catalyst instead of the previously recommended sodium metal. We also found that reproducibility and chance of crystallization after reaction completion substantially improve when precise stoichiometric amounts of triethanolamine and (3-aminopropyl) triethoxysilane are used. For this purpose, we recommend using a precision balance rather than graduated cylinders for measuring the volumes of these liquid reagents. The reaction is essentially quantitative and can be described by the scheme in Fig. 2a (see Notes 1 and 2).
Prepare the solution of sodium hydroxide in methanol (2 mg/mL) by adding sodium hydroxide granules to the calculated amount of methanol and stirring for a prolonged period of time until all sodium hydroxide is completely dissolved. Sonication or moderate heat can accelerate the process (Caution: sodium hydroxide solid and solutions can cause burns to the eyes and skin! Do not heat in a closed vial!).
Add triethanolamine (14.92 g, 0.1 M) to a 250 mL roundbottom flask followed by 2 mL of sodium hydroxide solution in methanol (2 mg/mL). A precise equivalent amount of (3-aminopropyl) triethoxysilane (22.14 g, 0.1 M) is measured in a separate flask and added to the reaction mixture. Complete transfer of the reagent is assured by washing the flask with two portions of methanol (2 × 10 mL), and adding the methanol washes to the reaction mixture.
Place the reaction flask on a rotary evaporator (see Note 1); methanol is evaporated at 40°C under a moderate vacuum (100 Torr). This part of the process takes approximately 10 min.
Lower the vacuum gradually to 1 Torr; raise the temperature of the water bath to 60°C. Make sure that the flask is rotated constantly and the vacuum is applied slowly to avoid bumping. Ethanol that forms in the reaction is evaporated off. At the end of the reaction, the product solidifies to a crystalline mass (see Note 2). The crystallization starts spontaneously after approximately 30 min. The product forms quantitatively; the yield is 23.22 g of colorless solid material. The melting point is 86–90°C. We have shown that this product is suitable for the preparation of APS-mica. Traces of sodium hydroxide in the product do not affect the performance or pH of solutions for AFM. Transfer of the crude solid product from the reaction flask into a storage container may be difficult due to its hardness.
Save a small portion of the solid product (10–20 mg) for seeding crystallization. The rest of the solid product dissolved in hot methanol (15 mL) and diluted with toluene (150 mL). The mixture is partially evaporated to approximately 1/2 of the volume. If the product does not crystallize during evaporation, the solution is seeded with a small portion of the saved crude solid product, stirred and crystallized. Crystallization can be accelerated by sonication or stirring and spreading the seed crystals with a spatula.
Cool the mixture by placing the flask on ice. Crystals of the product form a slurry.
The solid material is collected by filtration using a medium porosity sintered-glass filter and vacuum suction, washed with two 20-mL portions of ice-cold toluene, and dried under vacuum. Avoid prolonged suction of air through the solid product after the filtration and wash steps. The solid product can absorb moisture from the air and melt.
The final product (11.3 g) is a colorless powder. APS prepared and purified by this method has a melting point (m.p.) of 91–94°C (open capillary tube); literature m.p. 87.2−87.9°C (sealed capillary tube) (US Patent 3,118,921). 1H NMR (DMSO-d6), ppm: 0.08–0.14 (2H, m, SiCH2); 1.1 (2H, br. s, NH2); 1.28–1.37 (2H, m, CH2); 2.37 (2H, tJ = 7.2 Hz, NCH2); 2.77 (6H, tJ = 5.9 Hz, NCH2); 3.59 (6H, t J = 5.9 Hz, OCH2).
The APS reagent should be stored refrigerated over a desiccant. The reagent demonstrated good performance in AFM after 3 years of storage under such conditions.
3.2. Mica Functionalization with APS
Prepare a 50 mM APS stock solution in deionized water and store it in refrigerator. The stock solution can be kept for more than a year at 4°C (see Note 3).
Dissolve the APS from the stock in a 1:300 ratio in water (e.g., 45 µL of the stock to 15 mL deionized H2O) to make the working APS solution for mica modification; it can be stored at room temperature for several days.
Cut both sides of the mica sheets to make strips of the needed size (typically 1.2 cm × 3 cm) and cleave the strips with a razor blade, or tape to make them as thin as 0.05–0.1 mm. Do not touch the cleaved mica surface (see Note 4).
Place the mica strips in appropriate plastic tubes.
Pour the working APS solution to cover the mica strip completely.
Leave the tubes/cuvettes on the bench for 30 min.
After 30 min discard the APS solution.
Rinse both sides of the mica with deionized water.
Completely dry both sides of the mica strips under argon flow. Put the dry mica strip into the clean dry cuvette for storage (see Note 5). The strips are now ready for the sample preparation. Additional storage in a vacuum for 1–2 h is recommended when the environment is humid.
3.3. Mica Functionalization with AP (Evaporation Method)
3.3.1 Vacuum Distillation of APTES
Assemble the distillation apparatus (see Fig. 3).
Fill the distillation flask by no more than 2/3 of the volume with APTES.
Carefully insert the capillary tube into the distillation flask to prevent bumping during the distillation process. Put a piece of flexible tubing onto the inlet portion of the capillary. Insert a piece of thin wire inside the tubing and squeeze the tubing and the wire part with a clamp (see Fig. 3, left). This technique enables a fine control of the gas flow into the capillary tube and at the same time prevents complete closing of the inlet.
Connect the inlet to the argon or nitrogen line.
Slowly apply the vacuum and increase the temperature in the oil bath. Monitor the distillation process. Discard the first 10% of the distillate. Distillation will be complete when approximately 10% of the initial amount of APTES remains in the distillation flask. Boiling temperature depends on the vacuum. Typically, the reagent distills at 103°C/20 mmHg. Refer to the boiling point calculators or Pressure–Temperature Nomograph when using other vacuums. The reference boiling point at atmospheric pressure is 217°C/760 mmHg.
Dispense the reagent into 1–2 mL screw cap vials, avoiding prolonged exposure to atmospheric moisture. The reagent showed good performance in AFM applications over 6 months when stored in frozen at −20°C.
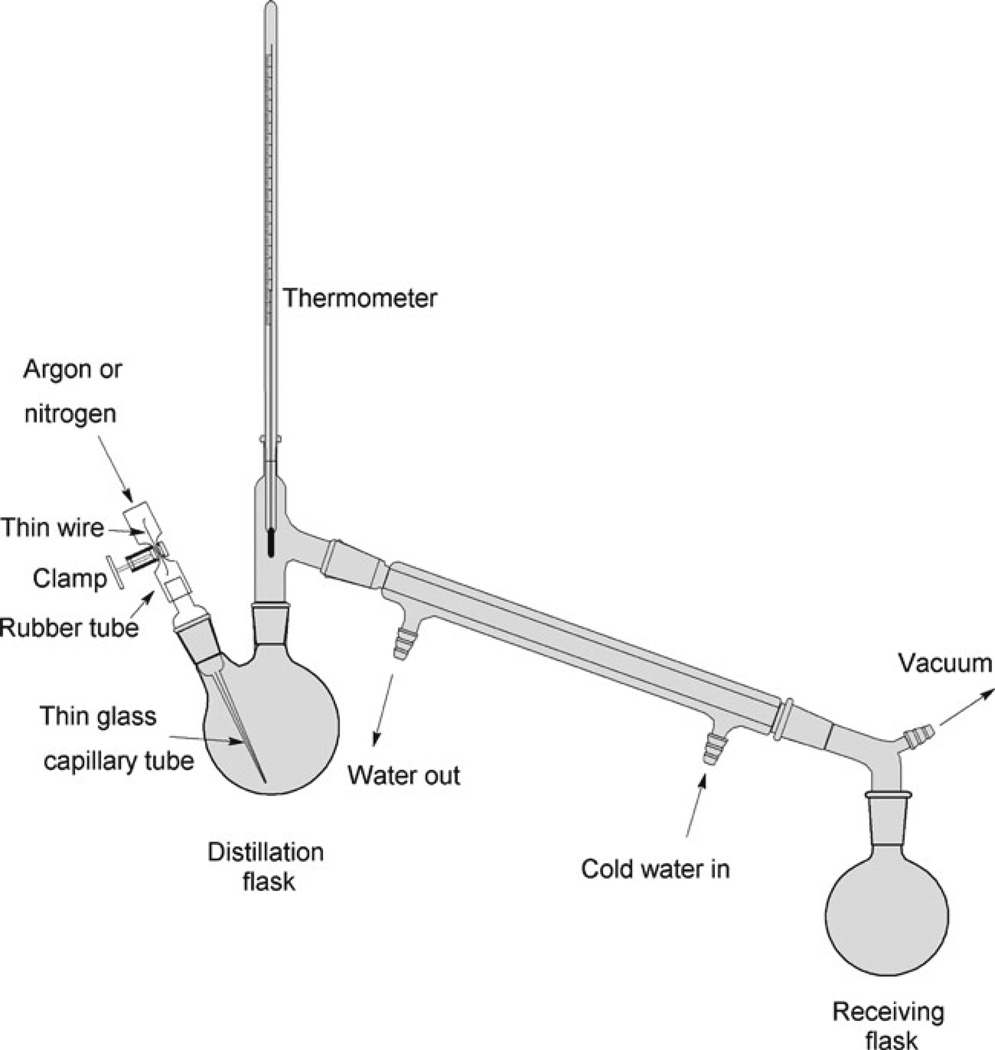
Fig. 3.
The scheme for the vacuum distillation apparatus of APTES. The distillation flask is immersed into the heated oil bath. A regular faucet aspirator (Nalgene) creates a necessary vacuum for the distillation process.
3.3.2. Preparation of AP-Mica
Place two plastic caps (cut them from regular 1.5 mL plastic Eppendorf tubes) on the bottom of a 2 L desiccator.
Place the desiccator under vacuum, and fill it with argon.
As described in step 3 from Subheading 3.2, cut the mica sheets to the required size with scissors; you can use the sheets as large as 5 cm × 5 cm, cleaving the large thick sheets with a razor blade. A thickness of ~0.1 mm is recommended.
Place 30 µL of APTES into one plastic cap in the desiccator and 10 µL of DIPEA into the other cap.
Mount the sheets at the top of the desiccator. Clip each mica sheet at the edge with a metal binder paper clip, use a glass or plastic rod to hold the clips and mount the rod at the top of the desiccator. The design in Fig. 4 accommodates five mica sheets that can be modified simultaneously.
Close the lid and allow the functionalization reaction to proceed for 2 h at room temperature.
Open the lid, remove the caps with reagents, and purge the desiccator with argon gas for 2 min.
Leave the mica sheets for 1–2 days in the desiccator to cure. The AP-mica is then ready for the sample deposition (see Notes 6 and 7).
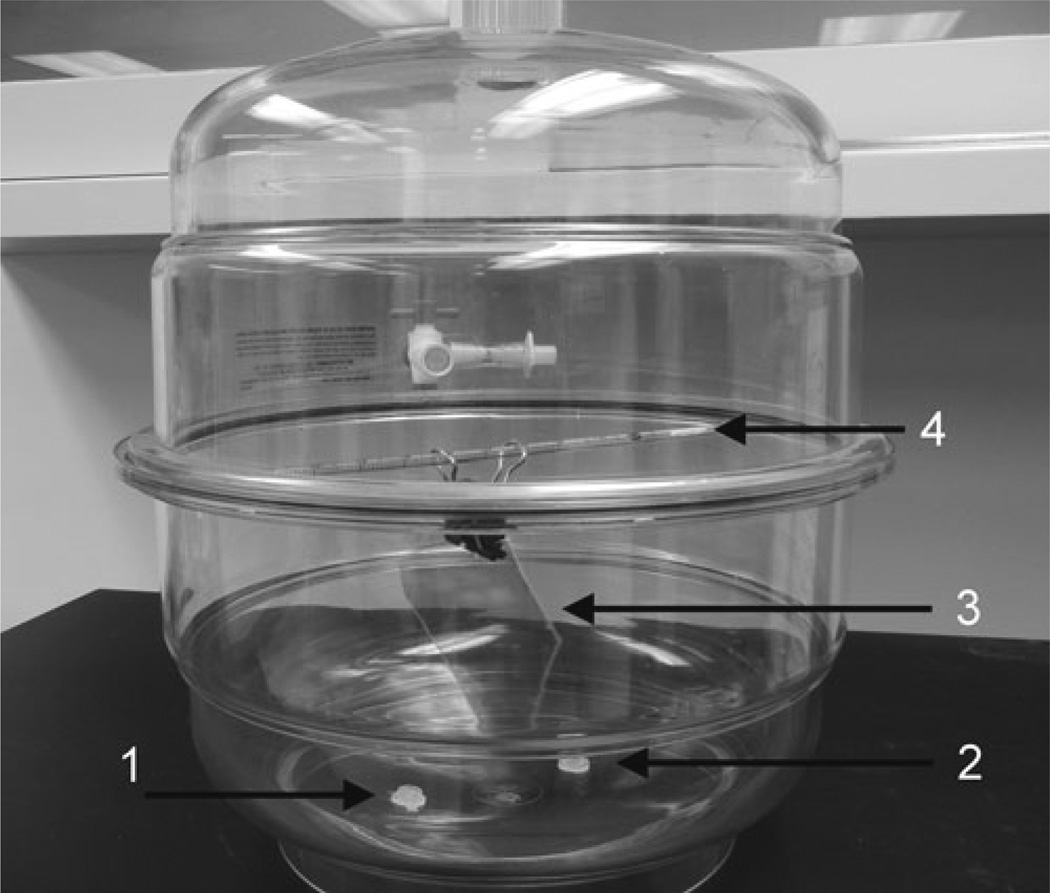
Fig. 4.
Photo of the setup used for the preparation of AP-mica. The mica sheet (3), clipped with the paper clip, is mounted at the top on a plastic rod (4); the two plastic caps with APTES (1) and DIPEA (2) reagents are placed at the bottom of the desiccator.
3.4. Sample Preparations for AFM Imaging
The sections below describe the procedures for the preparation of samples of DNA or protein-DNA complexes for AFM imaging. The procedures for AP-mica and APS-mica are similar; therefore, the type of functionalized mica is not specified unless it is required.
3.4.1 Sample Preparation for Imaging in Air
Droplet Procedure
Prepare the solution of the sample (DNA, RNA, protein-DNA complex) in an appropriate buffer. The DNA concentration should be between 0.8 and 0.01 µg/mL, depending on the size of the molecules. The concentration 0.2 µg/mL is recommended for PUC plasmid DNA and higher concentrations (0.8 µg/mL) are recommended for smaller, 1 Kb DNA fragments. Concentrations as low as 0.01 µg/mL were used for imaging of lambda DNA (~50 Kb) (14).
Cut the AP- or APS-mica substrates to a desired size (1 × 1 cm squares for the MultiMode (MM) AFM instrument) and place 5–10 µL of the solution in the middle of the substrate for 2 min.
Rinse the sample thoroughly with deionized water (2–3 mL per sample) to remove all buffer components. A 10 mL plastic syringe is useful for rinsing. Attach an appropriate plastic tip instead of a metal needle.
Dry the sample with clean argon gas. Additional drying of samples for an hour or two prior to imaging is recommended to ensure low tip adhesion. The samples can be stored in vacuum cabinets or desiccators filled with argon. The samples, as prepared, can be imaged many times provided that after imaging they are stored as described. Their shelf life is more than a month.
The Immersion Procedure
This procedure is recommended if the deposition should be performed at strictly controlled temperature conditions (0°C or elevated temperatures).
Prepare the solution (DNA, RNA, nucleoprotein complexes) in an appropriate tube and pre-incubate for 10–20 min to allow the temperature to equilibrate. The recommended concentration of DNA is between 0.8 and 0.01 µg/mL, depending on the DNA size (see Subheading 3.4.1.1 above) (see Notes 9 and 10).
Immerse a piece of functionalized mica into the tube for a defined time (2–10 min) to allow the sample to adhere to the surface.
Remove the mica strip, rinse with water thoroughly, and dry under argon flow as described above. The sample is then ready for imaging; however, it is recommended that the specimen be stored in a vacuum cabinet under argon for an hour to allow optimum sample drying.
3.5. AFM Imaging of the Samples
Two AFM microscopes, the MultiMode AFM system (Bruker-Nano/Veeco, Santa Barbara, CA) and the MFP3 (Asylum Research, Santa Barbara, CA) were our primary instruments employed. However, the procedures described below are general and can be adapted with minimal adjustments to any other AFM instrument (see Note 11).
3.5.1. Imaging in Air
AFM tips: For imaging in air, any type of tip with a spring constant of approximately 40 N/m and a resonant frequency between 300 and 340 kHz can be used. For example, Olympus silicon probes (Asylum Research, Santa Barbara, CA), with a spring constant of 40 N/m and a 300 kHz resonant frequency in air, work reliably in the Tapping/Oscillating Mode imaging in air. Probes with similar characteristics are currently manufactured by a large number of other vendors.
Mount the sample prepared as per Subheading 3.4 on the AFM stage.
Tune the AFM probe to find its resonance frequency.
Adjust the drive amplitude; for the MultiMode AFM, 6–8 mV is typical.
Set the image size to 100 × 100 nm and start approaching the surface.
Gradually reduce the set point until the surface of the sample is clearly seen. Increase the scan size and acquire the images.
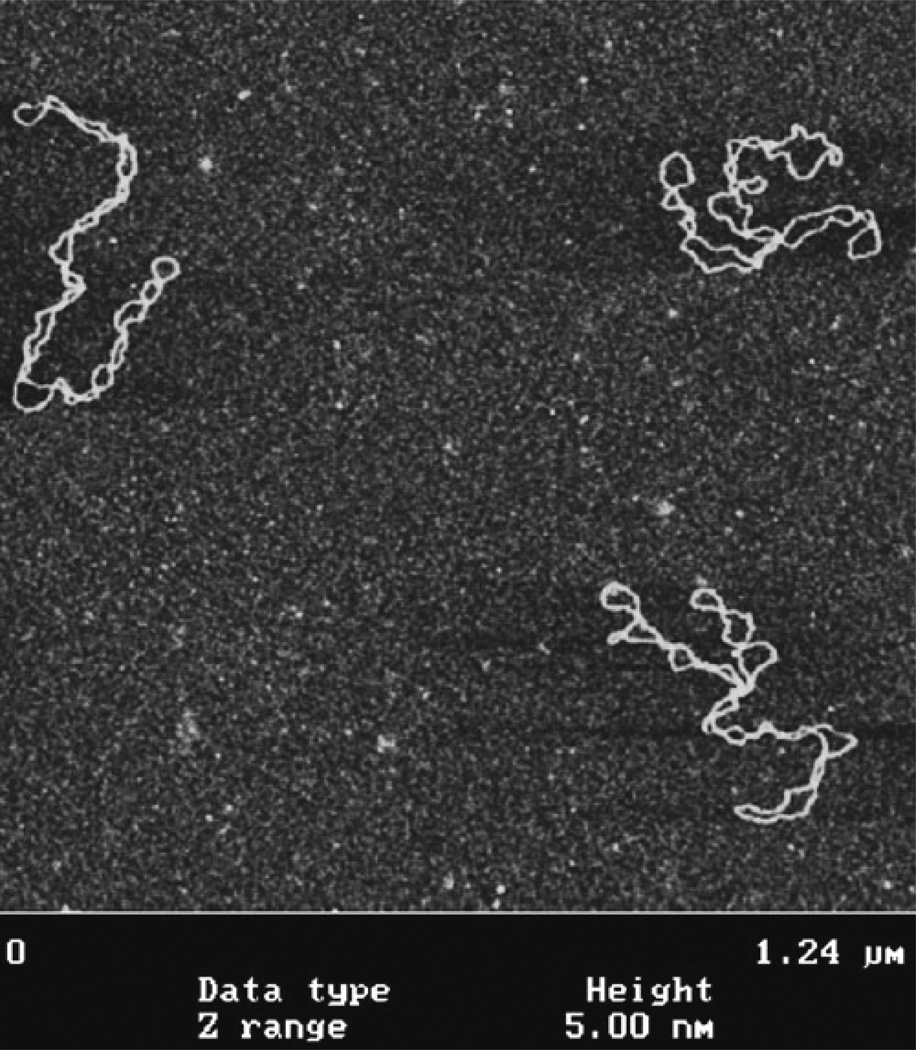
Typical AFM images of DNA obtained with the use of the AP-mica procedure are shown in Fig. 5. These are images of supercoiled plasmid DNA (5.6 kB) deposited on AP-mica from the TE buffer solution (20 mM Tris–HCl, pH 7.6, 1 mM EDTA) with 200 mM NaCl. These images highlight a number of important features of the AP-mica procedure. First, the background is smooth, enabling unambiguous visualization of DNA (15, 17). Second, the concentration of DNA was adjusted in such a way that the molecules are spread over the surface with no overlap. Third, supercoiled DNA molecules have plectonemic morphologies with clearly separated supercoiled loops. Two of the three molecules in Fig. 5a are branched, which is typical for molecules of this size. It was shown that the morphology of supercoiled DNA depends on ionic conditions (17, 24) and these observations are fully consistent with the data obtained in solution. Mg2+ cations further increase interwinding of plectonemic molecules (17, 24), which is in agreement with experimental studies in solution (reviewed in ref. (26)). Thus, the AP-mica procedure preserves the structure of supercoiled DNA. It is recommended to use this technique to image global DNA conformations and the structure and dynamics of negatively supercoiled DNA, which is the natural state of DNA at physiological conditions and within cells. It is important to note that other sample preparation methods for AFM failed to obtain supercoiled DNA with such morphologies; therefore precaution should be taken when using these procedures (26).
Fig. 5.
AFM images of supercoiled 5.6 Kb plasmid DNA deposited onto AP-mica. Images were acquired with the MultiMode AFM (Nanoscope III controller) operating in Tapping Mode.
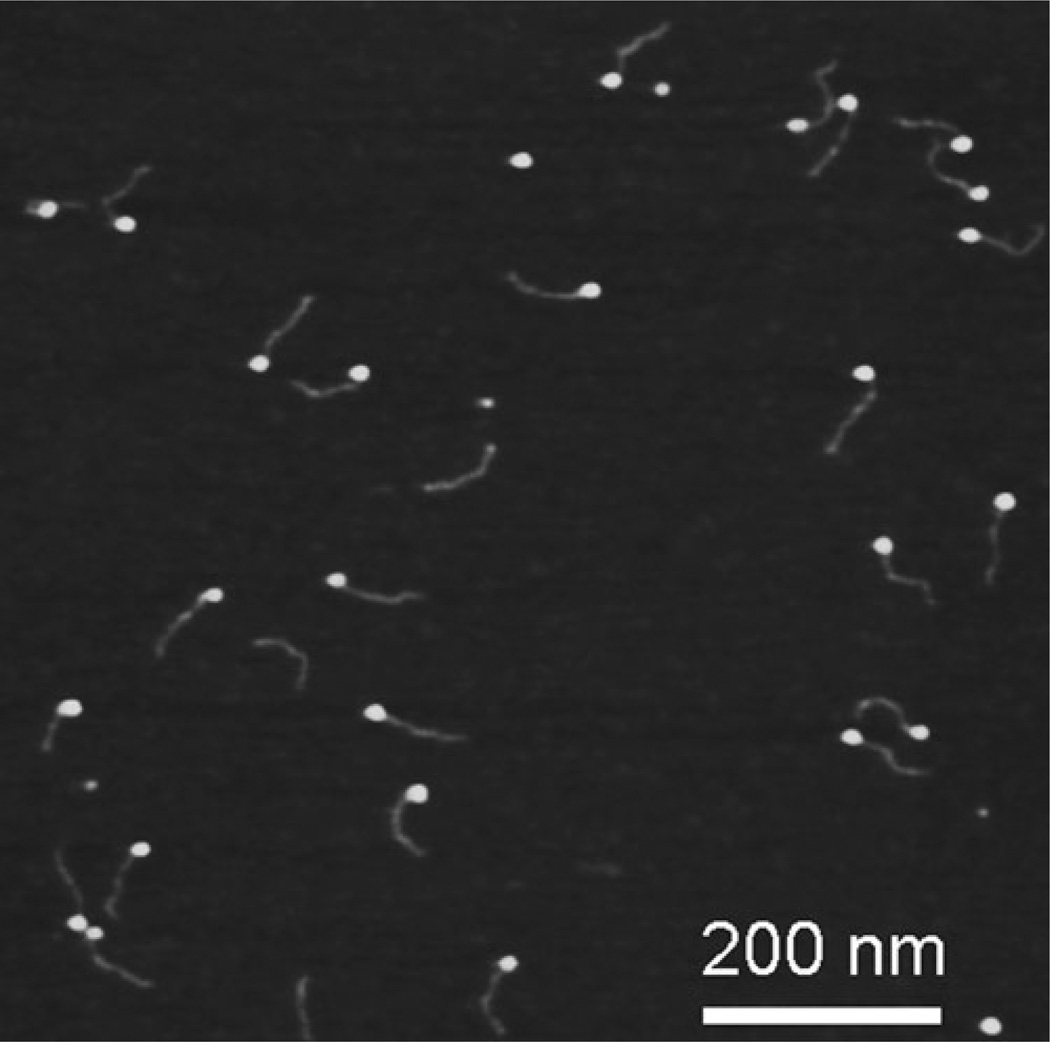
Figure 6 shows AFM images of DNA complexed with the single stranded DNA binding protein (SSB) obtained with the use of APS-mica (39). Similar to AP-mica, the APS-mica surface appears smooth, enabling unambiguous identification of the samples. The protein appears as a bright spherical particle at the end of the DNA molecules. Such an arrangement is due to a special design of the DNA substrate in which a single stranded region (69 nucleotides) is located at one of the ends of the DNA duplex. The end-specific location of the protein con firms its highly specific binding to single stranded DNA. The SSB-DNA complex needs to be purified to obtain such high quality images and the procedure is described in (39).
Fig. 6.
AFM images of complexes of single stranded DNA binding protein (SSB) with DNA. The specially designed DNA substrate has a single stranded region (69 nucleotide) that binds the protein. The proteins appear as bright spherical features at one of the ends of the DNA substrate.
3.5.2. Imaging in Liquid
This mode of AFM imaging is attractive for two major reasons. First, imaging of the fully hydrated sample eliminates potential problems with the drying step. Second, imaging in liquid opens prospects for time-lapse visualization of sample dynamics and interactions. Importantly, due to AFM resolution at the nanometer scale, dynamics are observed at the single molecule level (see recent reviews in ref. (40–42) and references therein). Note in this regard, the advent and recent improvements of high-speed AFM, enabling the acquisition of the data at video rate (42–45). For imaging in liquid with a regular AFM, Si3N4, 100-µm-long probes (SNL, Bruker-Nano/Veeco, Santa Barbara, CA) with a spring constant approximately 0.06 N/m and a resonance frequency around 7–10 kHz are used. Tips with similar characteristics from other vendors are available. The protocols described below were developed with the use of MultiMode (MM) AFM (Bruker-Nano/Veeco), but they can be adapted to any type of AFM.
Mount the tip on the tip holder.
Place the stage with the attached AP-mica or APS-mica substrate on the instrument stage. Mica pieces 1 × 1 cm work well for MM AFM. Double sticky tape can be used for gluing the modified mica substrate to the metal discs. However, if glue is used, glue the mica and cleave it prior to the functionalization step. The glue vapors react with the mica surface resulting in complete deterioration.
Use the video camera to find the tip and approach the surface manually, leaving a 500–1,000 µm gap between the tip and the surface.
Place a droplet of the prepared sample solution and readjust the spot position. The spot changes due to the difference in the refractive indexes of air and water; for MM AFM, 50 µL of the solution is sufficient to fill the gap. Note that due to the elevated hydrophobicity of AP-mica compared to bare mica, the spot does not spread; therefore, an additional O-ring is not required to keep the solution in place.
Find a resonance peak. Typically, it is quite a broad peak, around 7–10 kHz, for the MM AFM instrument. Follow the recommendations provided in the manual for determining the peak in fluid.
Start the computer controlled approach. Operate with the setpoint voltage and drive amplitude parameters to improve the quality of images. Minimize the drive amplitude. The number varies from tip to tip, but amplitudes as low as 10 nm or less and a scanning rate of ~2 Hz provide better quality pictures.
Dried samples can be imaged in aqueous solutions as well. In this case, the procedure is similar, but instead of the sample solution, a buffer solution is placed on top of the substrate.
Figure 7 shows the dynamics of the cruciform structure within a supercoiled DNA, detected with the use of time-lapse AFM and the AP-mica procedure (46). The arms of the cruciform are indicated in the images with arrows and they are positioned at ~60° on the initial image (a). Over time, one arm changes its position, becoming almost invisible in image (b) and almost parallel to another arm in image (c). A large-scale segmental dynamics of supercoiled DNA was observed in (17) with the use of the same AP-mica approach. Note numerous small bright dots on the background of the images. These are not present on dried samples and appeared due to hydrolysis and aggregation of adsorbed APTES molecules (17). This disadvantage of the AP-mica procedure is eliminated if APS-mica is used (e.g., (19, 20, 24, 41, 42, 47, 48) and references therein). Figure 8 illustrates the smooth feature of the substrate, enabling us to follow the dynamics of nucleosomes. Three frames show the images acquired at different times of the unwrapping process (see Note 12 for alternative procedures).
Fig. 7.
Time-lapse AFM imaging of DNA cruciform dynamics captured in TE buffer with the use of AP-mica. The cruciform’s arms are indicated with arrows; the angle is ~60° in plate (a), one arm was in motion and is barely seen in plate (b), and both arms are almost parallel in plate (c). Images were acquired by the MultiMode AFM (Nanoscope IV) operating in Tapping Mode.
Fig. 8.
Time-lapse AFM imaging of the nucleosome unwrapping, with the use of APS-mica. Frames 1, 2, and 3 correspond to different times of the unwrapping process. Schematically, the structure of the nucleosome at each stage is indicated with insets. Images were acquired by the MultiMode AFM (Nanoscope IV) operating in Tapping mode.
Acknowledgements
The authors thank the members of the Lyubchenko lab for their contribution to different parts of the paper and T. Zaikova for useful comments on the APS synthesis. The work is supported by grants to YLL from the DOE (DE-FG02-08ER64579), NATO (SfP 983204), NIH (P01 GM091743, 1R01 GM096039), the Nebraska Research Initiative (NRI), National Institutes of Health Grants (1P01GM091743-01A1 and 1 R01 GM096039-01A1), US Department of Energy Grant DE-FG02-08ER64579, National Science Foundation (EPS—1004094), and the Nebraska Research Initiative grant to Y.L.L.
Footnotes
Although the APS synthesis can be performed in a round-bottom flask under vacuum with manual stirring, the best results are obtained using a rotary evaporator.
APS is typically crystallized as described above; however, both crystallized and non-crystallized reagents performed equally well in AFM experiments.
It is recommended to aliquot the APS stock solution (1 mL). The aliquots can be stored in a refrigerator (4°C) for more than a year.
Depending on the size of the mica strip, the plastic disposable 3-mL cuvettes or plastic 15 mL tubes are suitable for mica functionalization process and storage.
As prepared, the APS-mica sheets can be stored in dry (plastic tubes or cuvettes) in the argon atmosphere for at least a week.
A dry argon atmosphere is crucial for obtaining AP-mica substrates suitable for AFM studies of DNA and protein-DNA complexes. Allow the gas to flow while the desiccator is opened. With such precautions, the AP-mica substrates can be stored in the desiccator and retain their activity for 2 weeks. See Fig. 4 illustrating the setup used in the lab for the AP-mica preparation.
Both procedures described in Subheadings 3.2 and 3.3 yield weakly charged cationic surfaces with a uniform distribution of the charge.
AP-mica vs. APS-mica. There is no difference between the substrates when the samples are imaged in air. However, APS-mica is the preferred substrate for imaging in aqueous solutions.
DNA concentration: This parameter depends on the length of the molecules. If the molecules are as small as several hundred base pairs, the concentration of ~0.5–1 µg/mL is recommended to avoid intermolecular crossing. However, lower DNA concentrations are recommended for longer DNA molecules. For example, the concentration of lambda DNA (~48 kB), ~0.01 µg/mL, allowed us to image individual long DNA molecules without overlap (14).
DNA preparation: A very small amount of DNA is required for preparing the samples by the droplet procedure. Typically, 10 ng of DNA is sufficient for imaging plasmid DNA (~3 Kb long). Due to the fact that one band of DNA in agarose gels usually contains 100 ng of DNA, DNA extracted from the several bands is sufficient for the preparation of several samples. The gel purification procedure described in (47, 49) produces pure samples.
Imaging conditions: It is recommended to operate the instrument at the lowest possible drive amplitude. This recommendation is based on the following considerations. The oscillating tip transfers a large amount of energy to the sample. According to (17), the total energy applied to the sample by the oscillating tip can be as high as 10−16–10−17 J at 30 nm amplitude of oscillation. However, this value is almost three orders of magnitude lower if the microscope is operated at amplitude as low as ~3 nm. Such imaging conditions allow one to minimize the dragging effect of the tip, prevent damaging the tip, and enable one to acquire images with high contrast and facilitate the study of such dynamic processes as segmental DNA mobility or protein–DNA interactions (reviewed in ref. (42)).
Alternative procedures for AFM sample preparation: This chapter describes protocols for substrate preparation, utilizing chemical functionalization of mica, and outlines the major features of these methods. Other methods were developed, and among the techniques applied to AFM studies of DNA, the method based on using multivalent metal cation was proposed (1, 3, 50). This approach is attractive by the simplicity of the procedure, but the mandatory requirement for the presence of multivalent metal ions limits the use of the cation-assisted method. Other problems with this method were identified and discussed in (26, 41). On the contrary, the AP-mica and APS-mica procedures are much more flexible. They do not require additional ions for DNA immobilization and they work in a broad range of ionic strengths, pH, and temperatures. Alternative AFM sample preparation techniques do not have these features.
References
- 1.Bustamante C, Vesenka J, Tang CL, Rees W, Guthold M, Keller R. Circular DNA molecules imaged in air by scanning force microscopy. Biochemistry. 1992;31:22–26. doi: 10.1021/bi00116a005. [DOI] [PubMed] [Google Scholar]
- 2.Brack C. DNA electron microscopy. CRC Crit Rev Biochem. 1981;10:113–169. doi: 10.3109/10409238109114551. [DOI] [PubMed] [Google Scholar]
- 3.Thundat T, Allison DP, Warmack RJ, Brown GM, Jacobson KB, Schrick JJ, Ferrell TL. Atomic force microscopy of DNA on mica and chemically modified mica. Scanning Microsc. 1992;6:911–918. [PubMed] [Google Scholar]
- 4.Hansma HG, Vesenka J, Siegerist C, Kelderman G, Morrett H, Sinsheimer RL, Elings V, Bustamante C, Hansma PK. Reproducible imaging and dissection of plasmid DNA under liquid with the atomic force microscope. Science. 1992;256:1180–1184. doi: 10.1126/science.256.5060.1180. [DOI] [PubMed] [Google Scholar]
- 5.Bezanilla M, Manne S, Laney DE, Lyubchenko YL, Hansma HG. Adsorption of DNA to mica, silylated mica, and minerals: characterization by atomic force microscopy. Langmuir. 1995;11:655–659. [Google Scholar]
- 6.Bustamante C, Rivetti C. Visualizing protein-nucleic acid interactions on a large scale with the scanning force microscope. Annu Rev Biophys Biomol Struct. 1996;25:395–429. doi: 10.1146/annurev.bb.25.060196.002143. [DOI] [PubMed] [Google Scholar]
- 7.Bustamante C, Rivetti C, Keller DJ. Scanning force microscopy under aqueous solutions. Curr Opin Struct Biol. 1997;7:709–716. doi: 10.1016/s0959-440x(97)80082-6. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Takeyasu K, Shao Z. Atomic force microscopy of DNA molecules. FEBS Lett. 1992;301:173–176. doi: 10.1016/0014-5793(92)81241-d. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Tamm LK, Tillack TW, Shao Z. New approach for atomic force microscopy of membrane proteins. The imaging of cholera toxin. J Mol Biol. 1993;229:286–290. doi: 10.1006/jmbi.1993.1033. [DOI] [PubMed] [Google Scholar]
- 10.Mou J, Czajkowsky DM, Zhang Y, Shao Z. High-resolution atomic-force microscopy of DNA: the pitch of the double helix. FEBS Lett. 1995;371:279–282. doi: 10.1016/0014-5793(95)00906-p. [DOI] [PubMed] [Google Scholar]
- 11.Hegner M, Wagner P, Semenza G. Immobilizing DNA on gold via thiol modification for atomic force microscopy imaging in buffer solutions. FEBS Lett. 1993;336:452–456. doi: 10.1016/0014-5793(93)80854-n. [DOI] [PubMed] [Google Scholar]
- 12.Allen MJ, Dong XF, O’Neill TE, Yau P, Kowalczykowski SC, Gatewood J, Balhorn R, Bradbury EM. Atomic force microscope measurements of nucleosome cores assembled along defined DNA sequences. Biochemistry. 1993;32:8390–8396. doi: 10.1021/bi00084a002. [DOI] [PubMed] [Google Scholar]
- 13.Lyubchenko YL, Gall AA, Shlyakhtenko LS, Harrington RE, Jacobs BL, Oden PI, Lindsay SM. Atomic force microscopy imaging of double stranded DNA and RNA. J Biomol Struct Dyn. 1992;10:589–606. doi: 10.1080/07391102.1992.10508670. [DOI] [PubMed] [Google Scholar]
- 14.Lyubchenko Y, Shlyakhtenko L, Harrington R, Oden P, Lindsay S. Atomic force microscopy of long DNA: imaging in air and under water. Proc Natl Acad Sci U S A. 1993;90:2137–2140. doi: 10.1073/pnas.90.6.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyubchenko YL, Jacobs BL, Lindsay SM, Stasiak A. Atomic force microscopy of nucleoprotein complexes. Scanning Microsc. 1995;9:705–724. discussion 724–707. [PubMed] [Google Scholar]
- 16.Lyubchenko YL, Blankenship RE, Gall AA, Lindsay SM, Thiemann O, Simpson L, Shlyakhtenko LS. Atomic force microscopy of DNA, nucleoproteins and cellular complexes: the use of functionalized substrates. Scanning Microsc Suppl. 1996;10:97–107. discussion 107–109. [PubMed] [Google Scholar]
- 17.Lyubchenko YL, Shlyakhtenko LS. Visualization of supercoiled DNA with atomic force microscopy in situ. Proc Natl Acad Sci U S A. 1997;94:496–501. doi: 10.1073/pnas.94.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyubchenko YL, Gall AA, Shlyakhtenko LS. Atomic force microscopy of DNA and protein-DNA complexes using functionalized mica substrates. Methods Mol Biol. 2001;148:569–578. doi: 10.1385/1-59259-208-2:569. [DOI] [PubMed] [Google Scholar]
- 19.Shlyakhtenko LS, Potaman VN, Sinden RR, Gall AA, Lyubchenko YL. Structure and dynamics of three-way DNA junctions: atomic force microscopy studies. Nucleic Acids Res. 2000;28:3472–3477. doi: 10.1093/nar/28.18.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyubchenko YL, Shlyakhtenko LS, Potaman VP, Sinden RR. Global and Local DNA Structure and Dynamics. Single molecule studies with AFM. Microsc Microanal. 2002;8:170–171. [Google Scholar]
- 21.Yodh JG, Woodbury N, Shlyakhtenko LS, Lyubchenko YL, Lohr D. Mapping nucleosome locations on the 208–12 by AFM provides clear evidence for cooperativity in array occupation. Biochemistry. 2002;41:3565–3574. doi: 10.1021/bi011612e. [DOI] [PubMed] [Google Scholar]
- 22.Kato M, Hokabe S, Itakura S, Minoshima S, Lyubchenko YL, Gurkov TD, Okawara H, Nagayama K, Shimizu N. Interarm interaction of DNA cruciform forming at a short inverted repeat sequence. Biophys J. 2003;85:402–408. doi: 10.1016/S0006-3495(03)74484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potaman VN, Bissler JJ, Hashem VI, Oussatcheva EA, Lu L, Shlyakhtenko LS, Lyubchenko YL, Matsuura T, Ashizawa T, Leffak M, Benham CJ, Sinden RR. Unpaired structures in SCA10 (ATTCT)n. (AGAAT)n repeats. J Mol Biol. 2003;326:1095–1111. doi: 10.1016/s0022-2836(03)00037-8. [DOI] [PubMed] [Google Scholar]
- 24.Shlyakhtenko LS, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003;97:279–287. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 25.Lushnikov AY, Brown BA, 2nd, Oussatcheva EA, Potaman VN, Sinden RR, Lyubchenko YL. Interaction of the Zalpha domain of human ADAR1 with a negatively supercoiled plasmid visualized by atomic force microscopy. Nucleic Acids Res. 2004;32:4704–4712. doi: 10.1093/nar/gkh810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyubchenko YL. DNA structure and dynamics: an atomic force microscopy study. Cell Biochem Biophys. 2004;41:75–98. doi: 10.1385/CBB:41:1:075. [DOI] [PubMed] [Google Scholar]
- 27.Tiner WJ, Sr, Potaman VN, Sinden RR, Lyubchenko YL. The structure of intramolecular triplex DNA: atomic force microscopy study. J Mol Biol. 2001;314:353–357. doi: 10.1006/jmbi.2001.5174. [DOI] [PubMed] [Google Scholar]
- 28.Kato M, McAllister CJ, Hokabe S, Shimizu N, Lyubchenko YL. Structural heterogeneity of pyrimidine/purine-biased DNA sequence analyzed by atomic force microscopy. Eur J Biochem. 2002;269:3632–3636. doi: 10.1046/j.1432-1033.2002.03063.x. [DOI] [PubMed] [Google Scholar]
- 29.Dahlgren PR, Karymov MA, Bankston J, Holden T, Thumfort P, Ingram VM, Lyubchenko YL. Atomic force microscopy analysis of the Huntington protein nanofibril formation. Dis Mon. 2005;51:374–385. doi: 10.1016/j.disamonth.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Lonskaya I, Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem. 2005;280:17076–17083. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- 31.Lushnikov AY, Potaman VN, Lyubchenko YL. Site-specific labeling of supercoiled DNA. Nucleic Acids Res. 2006;34:e111. doi: 10.1093/nar/gkl642. (111–117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lushnikov AY, Potaman VN, Oussatcheva EA, Sinden RR, Lyubchenko YL. DNA Strand Arrangement within the SfiI-DNA complex: atomic force microscopy analysis. Biochemistry. 2006;45:152–158. doi: 10.1021/bi051767c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McAllister C, Karymov MA, Kawano Y, Lushnikov AY, Mikheikin A, Uversky VN, Lyubchenko YL. Protein interactions and misfolding analyzed by AFM force spectroscopy. J Mol Biol. 2005;354:1028–1042. doi: 10.1016/j.jmb.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 34.Kransnoslobodtsev AV, Shlyakhtenko LS, Ukraintsev E, Zaikova TO, Keana JF, Lyubchenko YL. Nanomedicine and protein misfolding diseases. Nanomedicine. 2005;1:300–305. doi: 10.1016/j.nano.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyubchenko YL, Sherman S, Shlyakhtenko LS, Uversky VN. Nanoimaging for protein misfolding and related diseases. J Cell Biochem. 2006;99:53–70. doi: 10.1002/jcb.20989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krasnoslobodtsev AV, Shlyakhtenko LS, Lyubchenko YL. Probing interactions within the synaptic DNA-SFII complex by AFM force spectroscopy. J Mol Biol. 2007;365:1407–1418. doi: 10.1016/j.jmb.2006.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shlyakhtenko LS, Yuan B, Emadi S, Lyubchenko YL, Sierks MR. Single-molecule selection and recovery of structure-specific antibodies using atomic force microscopy. Nanomedicine. 2007;3:192–197. doi: 10.1016/j.nano.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Lyubchenko YL, Shlyakhtenko LS, Gall AA. Atomic force microscopy imaging and probing of DNA, proteins, and protein DNA complexes: silatrane surface chemistry. Methods Mol Biol. 2009;543:337–351. doi: 10.1007/978-1-60327-015-1_21. [DOI] [PubMed] [Google Scholar]
- 39.Shlyakhtenko LS, Lushnikov AY, Li M, Lackey L, Harris RS, Lyubchenko YL. Atomic force microscopy studies provide direct evidence for dimerization of the HIV restriction factor APOBEC3G. J Biol Chem. 2011;286:3387–3395. doi: 10.1074/jbc.M110.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyubchenko YL, Shlyakhtenko LS. AFM for analysis of structure and dynamics of DNA and protein-DNA complexes. Methods. 2009;47:206–213. doi: 10.1016/j.ymeth.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyubchenko YL. Preparation of DNA and nucleoprotein samples for AFM imaging. Micron. 2011;42:196–206. doi: 10.1016/j.micron.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lyubchenko YL, Shlyakhtenko LS, Ando T. Imaging of nucleic acids with atomic force microscopy. Methods. 2011;54:274–283. doi: 10.1016/j.ymeth.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto D, Uchihashi T, Kodera N, Yamashita H, Nishikori S, Ogura T, Shibata M, Ando T. High-speed atomic force microscopy techniques for observing dynamic biomolecular processes. Methods Enzymol. 2010;475:541–564. doi: 10.1016/S0076-6879(10)75020-5. [DOI] [PubMed] [Google Scholar]
- 44.Ando T, Uchihashi T, Kodera N, Yamamoto D, Miyagi A, Taniguchi M, Yamashita H. High-speed AFM and nano-visualization of biomolecular processes. Pflugers Arch. 2008;456:211–225. doi: 10.1007/s00424-007-0406-0. [DOI] [PubMed] [Google Scholar]
- 45.Ando T, Uchihashi T, Kodera N, Yamamoto D, Taniguchi M, Miyagi A, Yamashita H. High-speed atomic force microscopy for observing dynamic biomolecular processes. J Mol Recognit. 2007;20:448–458. doi: 10.1002/jmr.843. [DOI] [PubMed] [Google Scholar]
- 46.Shlyakhtenko LS, Potaman VN, Sinden RR, Lyubchenko YL. Structure and dynamics of supercoil-stabilized DNA cruciforms. J Mol Biol. 1998;280:61–72. doi: 10.1006/jmbi.1998.1855. [DOI] [PubMed] [Google Scholar]
- 47.Lushnikov AY, Bogdanov A, Lyubchenko YL. DNA recombination: holliday junctions dynamics and branch migration. J Biol Chem. 2003;278:43130–43134. doi: 10.1074/jbc.M308228200. [DOI] [PubMed] [Google Scholar]
- 48.Shlyakhtenko LS, Lushnikov AY, Lyubchenko YL. Dynamics of nucleosomes revealed by time-lapse atomic force microscopy. Biochemistry. 2009;48:7842–7848. doi: 10.1021/bi900977t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shlyakhtenko LS, Gilmore J, Kriatchko AN, Kumar S, Swanson PC, Lyubchenko YL. Molecular mechanism underlying RAG1/RAG2 synaptic complex formation. J Biol Chem. 2009;284:20956–20965. doi: 10.1074/jbc.M109.028977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vesenka J, Guthold M, Tang CL, Keller D, Delaine E, Bustamante C. Substrate preparation for reliable imaging of DNA molecules with the scanning force microscope. Ultramicroscopy. 1992;42–44(Pt B):1243–1249. doi: 10.1016/0304-3991(92)90430-r. [DOI] [PubMed] [Google Scholar]