Abstract
Introduction
A 3D-nanofiber scaffold acts in a similar way to the extracellular matrix (ECM)/basement membrane that enhances the proliferation and self-renewal of stem cells. The goal of the present study was to investigate the effects of a poly L-lactic acid (PLLA) nanofiber scaffold on frozen-thawed neonate mouse spermatogonial stem cells (SSCs) and testis tissues.
Methods
The isolated spermatogonial cells were divided into six culture groups: (1) fresh spermatogonial cells, (2) fresh spermatogonial cells seeded onto PLLA, (3) frozen-thawed spermatogonial cells, (4) frozen-thawed spermatogonial cells seeded onto PLLA, (5) spermatogonial cells obtained from frozen-thawed testis tissue, and (6) spermatogonial cells obtained from frozen-thawed testis tissue seeded onto PLLA. Spermatogonial cells and testis fragments were cryopreserved and cultured for 3 weeks. Cluster assay was performed during the culture. The presence of spermatogonial cells in the culture was determined by a reverse transcriptase polymerase chain reaction for spermatogonial markers (Oct4, GFRα-1, PLZF, Mvh(VASA), Itgα6, and Itgβ1), as well as the ultrastructural study of cell clusters and SSCs transplantation to a recipient azoospermic mouse. The significance of the data was analyzed using the repeated measures and analysis of variance.
Results
The findings indicated that the spermatogonial cells seeded on PLLA significantly increased in vitro spermatogonial cell cluster formations in comparison with the control groups (culture of SSCs not seeded on PLLA) (P≤0.001). The viability rate for the frozen cells after thawing was 63.00% ± 3.56%. This number decreased significantly (40.00% ± 0.82%) in spermatogonial cells obtained from the frozen-thawed testis tissue. Both groups, however, showed in vitro cluster formation. Although the expression of spermatogonial markers was maintained after 3 weeks of culture, there was a significant downregulation for some spermatogonial genes in the experimental groups compared with those of the control groups. Furthermore, transplantation assay and transmission electron microscopy studies suggested the presence of SSCs among the cultured cells.
Conclusion
Although PLLA can increase the in vitro cluster formation of neonate fresh and frozen-thawed spermatogonial cells, it may also cause them to differentiate during cultivation. The study therefore has implications for SSCs proliferation and germ cell differentiation in vitro.
Keywords: PLLA nanofibers, tissue cryopreservation, testis
Introduction
Spermatogonial stem cells (SSCs) have the potential to self-renew and generate differentiated germ cells that will eventually lead to sperm.1 These cells can therefore play an important role in treating infertility, especially when it comes to cancer survivors who have been affected by the long-term adverse effects of cancer treatments: chemo- or radiotherapy.2 To store and preserve mature sperm prior to treatment is common practice for those other than prepubertal cancer patients. In such patients, however, the preservation of the male germ-line cells is a challenge.3,4 It should be noted here that testis tissue cryopreservation and then proliferation and autotransplantation of isolated SSCs can facilitate the medical application of these cells.5
The microenvironment of SSCs in the basal compartment of the seminiferous epithelium is important for the maintenance and self-renewal of these cells6,7 because Sertoli cells provide the growth factors necessary for self-renewal in this microenvironment.8–11 Glial cell line-derived neurotrophic factor (GDNF) is the most crucial factor in the balance between self-renewal and differentiation in the SSC pool12–14 as well as the promotion of SSCs’ self-renewal. Without this factor, either spermatogonial aggregates do not develop or SSCs perish.15 The generation process of SSCs and spermatogonia9,16,17 and also the localization of undifferentiated spermatogonia along specific portions of the basement membrane are done via stimuli from the vascular network and interstitial cells, ie, the peritubular myoid cells and the Leydig cells.18 On the other hand, the adhesion molecules of the basement membrane are anchored with SSCs.9,17,19,20
Nanofiber matrices mimic the architecture and size scale of the natural extracellular matrix (ECM).6 This scaffold provides more three-dimensional (3D) biomimicking topographical signals to seeded cells and results in a more physiologically relevant cellular phenotype21 compared with the two-dimensional substrates. It provides physical cues for cell orientation and spreading, and its pores make space for the remodeling of tissue structures.22
Recently, many electrospun nanofibrillar surfaces have been used for cell culture.23–26 Poly L-lactic acid (PLLA) is one of the most promising biodegradable, biocompatible, and US Food and Drug Administration-approved polymers27 and can easily be electrospun to form a 3D non-woven network.28,29 T o date, the proliferation of muscle-derived stem cells30 and the differentiation of hepatic cells from human mesenchymal stem cells31 have been established on PLLA. 3D soft agar culture system and electrospun polyamide nanofiber (Ultra-Web™; Corning Life Sciences, Tewksbury, MA, USA) also have been reported to yield complete in vitro spermatogenesis of mouse testicular germ cells32,33 as well as short-term culture of spermatogonial stem-like cell colonies.34
Culture and cryopreservation of SSCs are the most effective methods used for the long-term preservation of these cells. In addition, cryopreservation of cellular aggregates or tissues may allow for the isolation and culture of SSCs from frozen-thawed tissues in order to mimic conditions that will occur in oncology patients.35,36 To date, immature testicular pieces have been cryopreserved by slow-freezing and vitrification in both mice37 and humans38,39 and have been hetero-grafted beneath the tunica albuginea of a busulfan-treated recipient testis. These studies demonstrate that spermatogonia can survive in xenogeneic recipients after cryopreservation and therefore result in an offspring after sperm microinsemination.37 Our assumption here is that PLLA might provide an improved structural environment for the clonal expansion or differentiation of SSCs. While many efforts have been made to cryopreserve SSCs and testicular tissues in animals and humans,2,39–41 no report has yet been found with respect to the culture of SSCs obtained from frozen-thawed testis tissue on PLLA scaffolds.
For this reason, we have frozen and thawed immature testicular fragments following the removal of interstitial tissue with enzymatic digestion. We assumed that by combining the appropriate 3D scaffolds (provided by the biodegradable polymer scaffolds) with GDNF we could possibly create an appropriate environment for the proliferation of neonate mouse SSCs. The goal of the present study was to investigate the effects of a PLLA nanofiber scaffold on the frozen-thawed neonate mouse SSCs and testis tissue. We wanted to know whether in such a culture system, the SSCs would maintain the clonogenic and proliferation potential or instead differentiate.
Materials and methods
Animals
Eighty 3–6-day-old male mice from the National Medical Research Institute, initially from the original stocks of Razi Laboratory (Tehran, Iran), were used in the experiment. The animals were kept in plastic cages in a room maintained at a temperature range of 22°C–25°C, with a 12-hour light/dark cycle. The animals had free access to drinking water and standard laboratory pellets. The research was conducted in accordance with the National Research Council guidelines.
Isolation and cultivation of spermatogonial cells
Testes from the 3–6-day-old National Medical Research Institute mice were collected for the preparation of cell suspension following enzymatic digestions and purification steps. After decapsulation, the testes were minced and suspended in Dulbecco’s Modified Eagle Medium (DMEM; Life Technologies, Carlsbad, CA, USA) supplemented with 1.37 g/L NaHCO3 (Sigma-Aldrich, St Louis, MO, USA), single-strength nonessential amino acids, penicillin (100 IU/mL), streptomycin (100 μg/m), and gentamycin (40 μg/mL) (all from Life Technologies). Testicular cells were separated by the method of van Pelt et al42 with minor modifications. Briefly, minced testis pieces were suspended in DMEM containing 0.5 mg/mL collagenase/dispase, 0.5 mg/mL Trypsin, and 0.05 mg/mL DNAse, for 30 minutes (with shaking and a little pipetting) at 37°C. All enzymes were purchased from Sigma-Aldrich. For the next step, the interstitial cells were removed by washing in DMEM medium. A second digestion step was performed in DMEM media by adding a fresh enzyme solution into the seminiferous cord fragments as described above. After cell separation and filtration through 70-μm nylon filters, the collected cells were used for the culture cells. Immediately after cell isolation, the number of cells was determined using a hematocytometer. Viability was also assessed. Sertoli cells and myoid cells were also isolated through overnight differential plating in DMEM containing 10% fetal calf serum (FCS).
After the removal of the Sertoli and myoid cells, spermatogonia, which remained in suspension, were collected and cultured in DMEM containing 5% FCS and 10 ng/mL GDNF for 3 weeks. The cells were incubated at 32°C, 5% CO2 in a humidified atmosphere, and the medium was refreshed three times per week. The diameters and the number of clusters were determined every 7 days during the culture for 3 weeks. The cells were subcultured during cell culture, and cluster assay was carried out on the 7th, 14th, and 21st days of the culture. Using an inverted microscope (Carl Zeiss; Oberkochen, Germany), the number of clusters and their diameters were measured by Image J software (version 1.240; National Institutes of Health, Bethesda, MD, USA). The identity of the cultured cells was confirmed by the expression of spermatogonial genes, ultrastructural study of cell clusters, and SSC transplantation to a recipient mouse.
Cryopreservation and thawing procedure of SSCs and testis tissue
The isolated cells and testis tissue fragmentations were cryopreserved using a procedure described by Izadyar et al,43 with some modification. Briefly, cell viability was assessed immediately after cell isolation. Cell suspensions in 0.5 mL aliquots (2 × 106 cells per mL) were prepared. Then, an equal volume of 2× concentrated freezing medium was added dropwise to the Eppendorf vial containing the cell suspension, for a period of 10–15 minutes, and after gently mixing by inverting the vial, a sample was taken for viability assessment. The freezing media were based on DMEM supplemented with 10% (v/v) FCS, 1.4 M dimethyl sulfoxide (DMSO) and 0.07 M sucrose. Cryovials vials (1.8 mL; Nunc, Roskilde, Denmark) containing 1.0 mL of cell suspension in freezing medium were placed in −80°C for at least 1 day and then plunged into liquid nitrogen. The cells were thawed by swirling in a 38°C water bath for a period of 2 minutes. The contents of the vial was transferred to a tube and diluted slowly by adding two volumes, dropwise, of DMEM supplemented with 10% FCS. Then, the cells were pooled and centrifuged at 2000× g for 5 minutes, the supernatant was removed and the pellet was resuspended in DMEM/FCS. A sample was taken for viability assessment, and the remainder of the cells was used for culture experiments.
For tissue cryopreservation, tubule fragmentations obtained from the first enzymatic digestion were transferred into a cryovial and cryopreservation solution was added in the same manner as the cell cryopreservation procedure.
Fresh and cryopreserved spermatogonial cell culture on PLLA nanofibers
A layer of PLLA nanofiber was used to provide an environment that resembled as closely as possible that of in vivo. PLLA nanofibers composed of PLLA and collagen fabricated by the electrospinning technique were purchased from Stem Cells Technology (Tehran, Iran).31 The PLLA nanofiber was used in a culture system with both cryopreserved and fresh SSCs. After placing the nanofibers on the dishes, fresh and frozen-thawed spermatogonial cells were seeded (5 × 105 cells) on nanofiber and cultured in three groups: (1) fresh cells, (2) frozen-thawed cells, and (3) cells obtained from frozen-thawed testis tissue. In addition, fresh and frozen-thawed cells cultured on the plate without nanofibers were also considered as control groups. Cells were cultured for 3 weeks.44 The diameter and the number of colonies were determined every 7 days during the culture for 3 weeks. Cluster formation was assessed using the procedure described by Yeh et al.45
Identity confirmation of the spermatogonial cells
Ribonucleic acid (RNA) extraction and reverse transcription
The presence of spermatogonial cells during the culture was determined by the expression of spermatogonial genes based upon previous animal studies. Total RNA from the 6-day-old testis tissue (positive control) and cultured testicular cells from the entire culture dish were extracted using a standard RNA extraction kit (Qiagen, Hilden, Germany) per the manufacturer’s instructions. The purity and integrity of the RNA was checked by a 260/280 nm ratio measurement. In the reverse transcription reaction, 1 μg of total RNA was used with QuantiTect® Reverse Transcription Kit (Qiagen) per the manufacturer’s instructions.
Polymerase chain reaction (PCR)
The primers specific for promyelocytic leukemia zinc-finger (PLZF), octamer-binding transcription factor 4 (Oct4), GDNF family co-receptorα1 (GFRα-1), VASA homologue (Mvh), Itgß1 (ß1-integrin), Itgα6(α6-integrin), c-Kit, and GAPDH genes were designed using the previously described mouse sequences (Gene Bank) and Gene Runner software (version 3.02; Hastings Software Inc, New York, NY, USA) as shown in Table 1. GAPDH, a housekeeping gene, was included as an internal control to normalize the PCR reaction. Reverse-transcription PCR (RT-PCR) was performed using the prepared complementary deoxyribonucleic acid (cDNA), the primers, and with PCR Master Mix 2X kit (Fermentas, St. Leon-Rot, Germany) under the following conditions: 95°C for 3 minutes, followed by 35 cycles at 95°C for 30 seconds, under specific annealing temperature for each primer (PLZF, 55°C; Oct4, 60°C; GFRα−1.52°C; VASA, 62°C; Itgα6, 52°C; Itgß1, 55°C; c-Kit, 60°C; and GAPDH, 60°C) for 45 seconds, 72°C for 60 seconds, and a final extension of 72°C for 10 minutes. To separate PCR products, 1 μL of each sample was resolved on a 1.2% agarose gel, and electrophoresis was performed with Tris-Borate-EDTA (TBE) 1× loading buffer (Sigma-Aldrich) at a voltage of 95 for 45 minutes. The gels were stained with 0.1 μg/mL Gel Red™ (Biotium Inc, Hayward CA, USA) and the bands were visualized using Gel Logic (Carestream Health Inc., Rochester, NY, USA) and images were obtained. Semi-quantitative RT-PCR was carried out at the end of the third week for all culture groups.
Table 1.
The sequence of the designed primers used for reverse transcriptase polymerase chain reaction
| Gene | Primer sequence (5′–3′) | Annealing temperature (°C) | Size (bp) |
|---|---|---|---|
| Mvh(VASA) | F:5′ GAT AAT CAT TTA GCA CAG CCT C 3′ | 59–60 | 149 |
| R:5′ GTC AAC AGA TGC AAA CAC AG 3′ | |||
| Itgα6 | F:5′ CTC AGA ATA TCA AGC TCC CT 3′ | 60 | 148 |
| R:5′ AAA CAC TAA TAG AGC CAG CA 3′ | |||
| GFRα-1 | F:5′ AAT TGT CTG CGT ATC TAC TGG 3′ | 60 | 130 |
| R:5′ ACA TCT GAT ATG AAC GGG AC 3′ | |||
| Itgβ1 | F:5′ GAC ATT ACT CAG ATC CAA CCA 3′ | 60 | 115 |
| R:5′ AGG TAG TAG AGA TCA ATA GGG T 3′ | |||
| Oct4 | F:5′ GAA CTA GCA TTG AGA ACC GT 3′ | 60 | 129 |
| R:5′ CAT ACT CGA ACC ACA TCC TTC 3′ | |||
| PLZF | F:5′ CCCGTTGGGGGTCAGCTAGAA 3′ | 61 | 137 |
| R:5′ CTGCAAGGTGGGGCGGTGTAG 3′ | |||
| c-Kit/(CD117) | F:5′ CTA AAG ATG AAC CCT CAG CCT 3′ | 60 | 142 |
| R:5′ GCA TAA CAC ATG AAC ACT CCA 3′ | |||
| GAPDH | F:5′ CAA CTC CCA CTC TTC CAC TT 3′ | 60 | 125 |
| R:5′ GCA GCG AAC TTT ATT GAT GGT A 3′ |
Abbreviations: Mvh(VASA), mouse vasa-homologue; GFRα-1, GDNF family co-receptor α1; Itgα6, integrin-α6; Itgβ1, integrin-β1; Oct4, octamer-binding transcription factor 4; PLZF, promyelocytic leukemia zinc finger; c-Kit, proto-oncogene c-Kit or tyrosine-protein kinase Kit; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; bp, base pairs.
Quantifications of PCR products
Gene expression levels were examined by semi-quantitative RT-PCR. The PCR products were analyzed on a 1.2% agarose gel (Life Technologies BRL) and were visualized under ultraviolet transillumination after being stained with Gel Red. RT-PCR reaction was performed for SSCs and germ cell genes: PLZF, Oct4, GFRα-1, VASA, Itgα6, Itgß1, c-Kit, and GAPDH genes, and the intensity of each band was quantified using UVItec software (version 12.6 for Windows; UVItec Ltd Cambridge, UK). The ratios of the SSC-specific gene band intensities were compared with the corresponding GAPDH. All PCRs were independently replicated three times.
Immunohistological staining
For immunohistological staining, the clusters were fixed with 4% paraformaldehyde (pH 7.4) for 20 minutes and were then twice washed with 0.1% Tween-20 in phosphate buffered saline (PBS) solution prior to blocking. The clusters were blocked with normal goat serum (Vector, Burlingame, CA, USA) in PBS for 15 minutes and were incubated overnight at 4°C with primary antibody solutions, including: rat polyclonal anti-α6-integrin (1:100; Sigma-Aldrich), rat polyclonal anti-β1-integrin (1:100; Sigma-Aldrich), rat polyclonal anti-Oct (1:100; Sigma-Aldrich), and mouse polyclonal anti-Thy-1 antibody (1:100, Santa Cruz Biotechnology, Santa Cruz, CA, USA). The next day, the clusters were washed with 0.1% Tween-20 in PBS (three times for 5 minutes) and incubated with respect to the secondary antibody: goat fluorescein isothiocyanate (FITC)-conjugated anti-mouse or anti-rat immunoglobulin G, (Sigma-Aldrich) diluted 1:200 for 45 minutes at room temperature. Next, the clusters were washed with 0.1% Tween-20 in PBS (three times for 5 minutes). Finally, all clusters were counterstained with 4,6-diamidino-2-phenylindole (DAPI) in PBS (Santa Cruz Biotechnology) diluted 1:2 for 10 seconds. Labeled cells were examined with a fluorescent microscope (BX51; Olympus), and images were acquired using an Olympus D70 camera. Negative controls were treated without the primary antibody.
Ultrastructural study of cell clusters
SSC clusters from groups were washed with PBS, pre-fixed in 2.5% glutaraldehyde in PBS (pH 7.4) for 2 hours, and post-fixed with 1% osmium tetroxide in the same buffer for another 2 hours. After dehydration in an ascending series of ethanol, specimens were placed in propylene oxide and embedded in Epon 812 (TAAB, Berkshire, UK). Semi-thin sections (0.5 mm) were stained with toluidine blue for light microscopy and ultrathin sections (60–80 nm) were contrasted with uranyl acetate and lead citrate before being examined by transmission electron microscopy (LEO 906; Carl Zeiss). The cluster cells were identified as primitive or differentiated spermatogonia based on cellular morphologies previously reported.46–48
BrdU (5-bromo-2-deoxyuridine) cell labeling and transplantation into recipient mice
Seventy-two hours before transplantation, BrdU was added to the medium in order to label and trace cells in recipient mice. This was done after the SSC clusters and the underlying somatic cells were trypsinized at the end of the 3 weeks.
Subsequently, the spermatogonial cells were transplanted into the seminiferous tubules of recipient mice via the rete testis that was treated with 35 mg/kg busulfan prior to transplantation. The treated recipient mice were devoid of endogenous spermatogenesis at the time of transplantation (6 weeks after treatment).49 Adult recipient mice were anesthetized with 10% ketamine and 2% xylazine (Alfasan, Woerden, The Netherlands). Approximately, 105 of the cultured cells in 10 μL DMEM were injected into the seminiferous tubules in one testis of each recipient mouse, while the other testis served as an internal control. Transplantation was performed by retrograde injection through the efferent ducts.50
Statistical analysis
The repeated measures and analysis of variance followed by Tukey post-hoc tests were used to evaluate differences between the experimental and control groups as well as between different time points in a group. Data are given as means ± standard deviation. The results were assumed significant when P≤0.05.
Results
The effect of cryopreservation on the percentage of viable cells in experimental groups
Cell viability was assessed after the isolation of testicular cells through enzymatic digestion. In this experiment, more than 89.25% ± 2.20% of the cells were determined viable in the fresh cell groups (control 1 and experimental 1). In the chemical toxicity test, however, adding freezing media did not seem to have a significant effect on viability, and more than 82.5% ± 4.2% remained viable. The viability rate of the frozen cells after thawing (control 2 and experimental 2) and spermatogonial cells obtained from frozen-thawed testis tissue (control 3 and experimental 3) were 63.00% ± 3.56% and 40.00% ± 0.82%, respectively. The viability rates decreased significantly in both groups when compared with the fresh cell groups (P≤0.001).
The effects of PLLA and cryopreservation on cluster formation of the SSCs in experimental groups
The clusters, ie, 3D aggregations of germ cells on a feeder layer, appeared 2–3 days after the primary culture. These clusters were clumpy and had individually recognizable cells, and once enzymatically dispersed and re-plated, their SSC content could start new clusters during 3 weeks of culture (Figure 1A).
Figure 1.
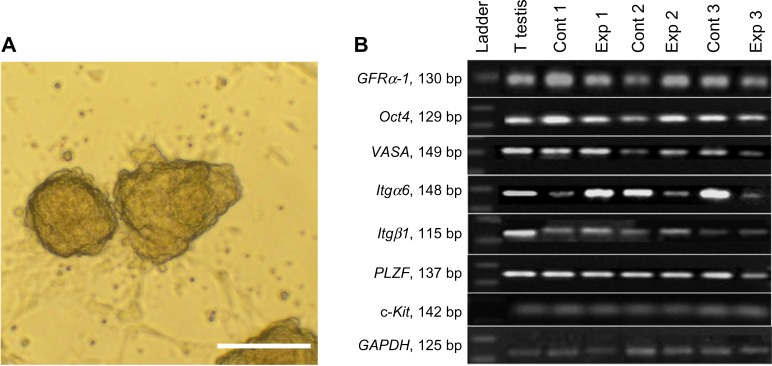
(A) The morphology of a typical cluster growing on top of a monolayer of somatic cells. (B) Molecular characterization of spermatogonial and germ cells at the RNA level after 3 weeks of culture.
Notes: RT-PCR was used to determine the expression of specific spermatogonia and germ cell markers. It showed that GFRα-1 (130 bp), Oct4 (129 bp), VASA (149 bp), Itgα6 (148 bp), Itgß1 (3115 bp), and PLZF (137 bp) genes, and c-Kit (142 bp) were expressed in: testis tissue (T testis), positive control (Cont 1), isolated testicular cells by two steps of enzymatic digestion before culture (Exp 1), fresh cells seeded on PLLA (Cont 2), frozen-thawed cells (Exp 2), frozen-thawed cells seeded on PLLA (Cont 3), frozen-thawed cells obtained from testis tissue (Exp 3), and frozen-thawed cells obtained from testis tissue seeded on PLLA groups after 3 weeks cultivation. GAPDH was used as a housekeeping gene (125 bp). As shown, all samples expressed specific spermatogonial and germ cell genes. PCR products were separated on 1.7% agarose gel. Scale bar = 100 μm.
Abbreviations: bp, base pairs; VASA mouse vasa-homologue; GFRα-1, GDNF family co-receptor α1; Itgα6, integrin-α6; Itgß1, integrin-ß1; Oct4, octamer-binding transcription factor 4; PLZF, promyelocytic leukemia zinc finger; c-Kit, proto-oncogene c-Kit or tyrosine-protein kinase Kit; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RNA, ribonucleic acid; RT-PCR, reverse transcriptase polymerase chain reaction; Cont, control; Exp, experimental.
To determine the effects of PLLA on cluster formation of SSCs in vitro, experimental groups (culture of SSCs with seeding onto PLLA) were compared with their respective control groups (culture of SSCs without seeding on PLLA). Overall, the results indicated that during the 3-week cultivation, the number of clusters for all of the experimental groups significantly increased compared with their respective control group (P≤0.001) (Figure 2A). Furthermore, a 3-week culture resulted in a significantly higher number of clusters compared with a 1- or 2-week cultivation (P<0.05).
Figure 2.
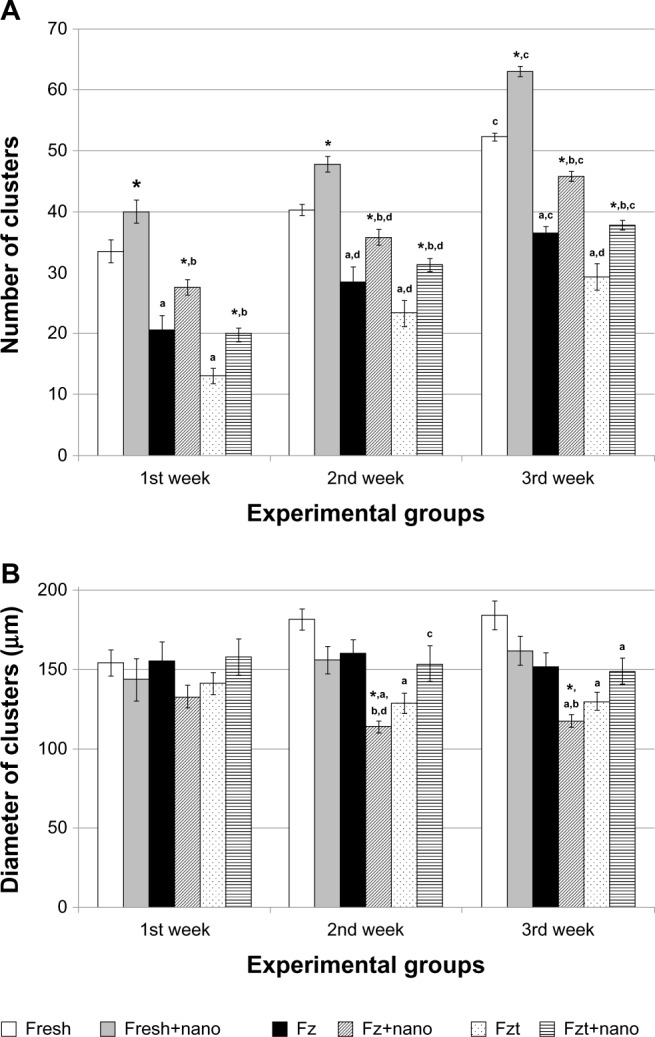
Comparison of (A) number and (B) diameter of clusters in experimental groups separately for each time point (per 5 × 105 primary cell number).
Notes: Results from five separate experiments were used for all groups. Values are the mean cluster numbers ± standard error at different times. *Significant difference versus culture of spermatogonial stem cells without seeding on PLLA in the same week (P≤0.05); asignificant difference versus fresh cells group in the same week (P≤0.05); bsignificant difference versus fresh cells seeded on PLLA group in the same week (P≤0.05); csignificant difference versus other weeks in the same group (P≤0.01); dsignificant difference versus first week in the same group (P≤0.05).
Abbreviations: Fresh, isolated testicular cells by two steps of enzymatic digestion before culture; Fresh+nano, fresh cells seeded on PLLA; Fz, frozen-thawed cells; Fz+nano, frozen-thawed cells seeded on PLLA; Fzt, frozen-thawed cells obtained from testis tissue; Fzt+nano, frozen-thawed cells obtained from testis tissue seeded on PLLA groups.
To determine the effects of cryopreservation on the cluster formation of SSCs in vitro, a frozen-thawed cell group was compared with a fresh cell culture group. During the third week of culture, the number of clusters observed in the frozen-thawed cell group (control 2) as well as those in the cell group obtained from the frozen-thawed testis tissue (control 3) were significantly lower than that of the fresh cell group (control 1) (Figure 2A).
As shown in Figure 2B, differences in the diameter of clusters (μm) did not vary significantly from one week to another. Although the differences in the diameter of clusters (μm) in the fresh cell groups (both in the presence or absence of PLLA) were not significant, in the culture of only frozen-thawed cells onto PLLA (experimental 2), this diameter was significantly lower compared with the control group (P≤0.01) (Figure 2B).
Identity confirmation of the spermatogonial cells
RT-PCR and immunocytochemistry
To analyze the expression of specific spermatogonial and germ cell markers in the isolated testicular cells and cultured cells, RT-PCR was performed after 3 weeks of culture. As shown in Figure 1B, all samples expressed specific genes of spermatogonial cells (PLZF, Oct4, GFRα-1, VASA, Itgα6, and Itgβ1) and c-Kit as a differentiated germ cell gene (Figure 1B).
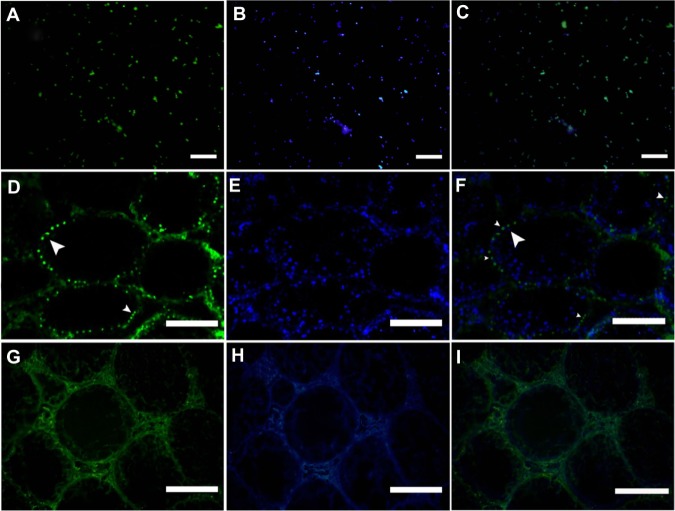
The presence of Itgα6 (Figure 3A–C), Itgβ1 (Figure 3G–I), Oct4 (Figure 3M–O) and thy-1 (Figure 3S–U) in cultured cells was confirmed by immunocytochemistry after 3 weeks of culture. Negative controls were treated without primary antibody (Figure 3D–F, J–L, P–R, and V–X).
Figure 3.
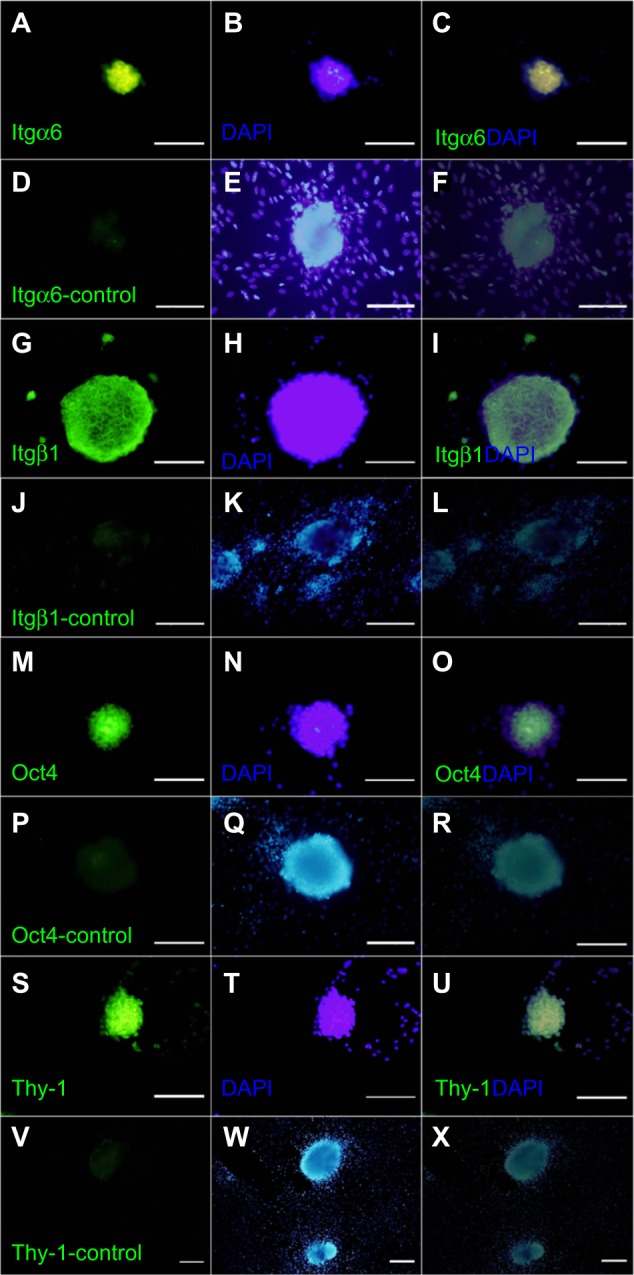
Immunofluorescent staining of spermatogonial cells in clusters. Integrin-α6 (A–C), integrin-β1 (G–I), Oct4 (M–O), and Thy-1 (S–U) were detected within the cluster cells (first column). DAPI staining in the second column and a merge of FITC and DAPI in the third column. Negative controls were treated without primary antibody (D–F, J–L, P–R and V–X).
Note: Scale bars = 200 μm.
Abbreviations: DAPI, 4,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate; Itgα6, integrin-α6; Itgβ 1, integrin-β 1; Oct4, octamer-binding transcription factor 4; Thy-1, thymus cell antigen 1.
Functional assay of the transplanted cluster cells
Germ cells were labeled with BrdU before transplantation. Immunofluorescence indicated that before transplantation, approximately 70% of cells had been labeled with BrdU (Figure 4A–C). After a month of cultivation, 105 cells from the third experimental group were injected into the seminiferous tubules through the rete testis of the recipient testes. This was done in order to confirm the presence of SSCs in clusters as well as to assess SSC colonization in the testis. One month after transplantation, those cells whose nuclei were positively stained with FITC for BrdU were considered the transplanted cells (Figure 4D–F).44,51 Transplantation studies revealed that compared with the non-transplant control group (Figure 4G–I), the transplanted cells localized as single cells only in the basal membrane of the seminiferous tubules of the recipient testes.
Figure 4.
Functional assay of spermatogonial stem cells into recipient mouse testis. (A) BrdU was added, and staining was examined in cultured spermatogonial cells before transplantation. (D) Transplanted cells on the base membrane of mouse seminiferous tubules 1 month after transplantation (arrowhead). These cells were traced in the recipient testes by BrdU staining. (G) The non-transplanted right testis was considered as the control group. (B, E and H) DAPI staining and (C, F and I) merge of FITC and DAPI.
Note: Scale bars = 200 μm.
Abbreviations: BrdU, 5-bromo-2-deoxyuridine; DAPI, 4,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate.
Ultrastructural characterization of cluster cells
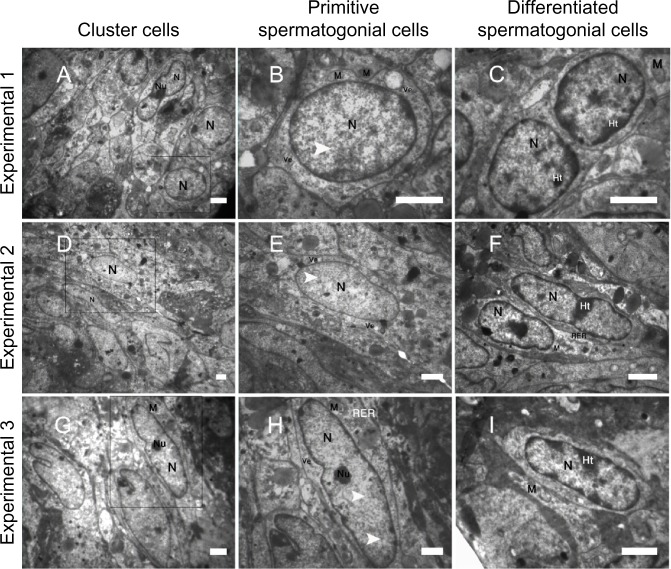
The ultrastructural characteristics of spermatogonial cell clusters were examined via transmission electron microscopy. After 3 weeks of cultivation, the electron micrograph showed that cell clusters from the experimental culture groups (Figure 5A, D, and G) had the typical morphology of spermatogonial cells (Figure 5B, E, and H); however, cell differentiation was also observed among cell clusters obtained from the PLLA groups (Figure 5C, F, and I).
Figure 5.
Representative transmission electron micrographs from spermatogonial stem cell clusters (A–I). The electron micrograph showed cells from spermatogonial stem cell clusters in culture groups (A, D and G) on nanofiber scaffolds (Experimental 1–3) had morphology typical of spermatogonial cells. The nucleus shown contains a mottled appearance with dark speckles of heterochromatin (arrowhead; B, E and H). The cytoplasm was characterized by organelles, eg, mitochondria (M), rough endoplasmic reticulum (RER) and vesicles (Ve) that were mostly located in the perinuclear region. A portion of a small compact nucleolus (Nu) is visible and highly reticulated. In addition, the nucleus (N) of some cluster cells had marginal heterochromatin (Ht) and their morphology was similar to differentiated cells (C, F and I). The heterochromatin ratio increases in differentiated spermatogonial cells.
Note: Scale bar = 5 μm.
As shown in Figure 5B, E, and H, cell clusters had large spherical nuclei that contained one or two prominent nucleoli located either along the nuclear membrane or in the center of the nucleus. The shape of the cells was flat with either a long and regular or a round nucleus. In primitive spermatogonial cells, both the nucleus and cytoplasm are spherical. The spherical nucleus contains scattered flakes of heterochromatin. The nucleoli had a prominent reticulated nucleolonema, and an irregular shape, and occupied an eccentric position in the nucleus. The cytoplasm was characterized by organelles, eg, mitochondria, rough endoplasmic reticulum, and vesicles that were mostly located in the perinuclear region. The mitochondria were clumped, interconnected by bars of cementing substance, and possessed parallel cristae. Spherical mitochondria were found single in relatively high numbers. The Golgi complex was poorly developed. In the differentiated spermatogonia, the nucleus was more heterochromatin; however, the shape of the heterochromatin was less dentate compared with the precursor cells.
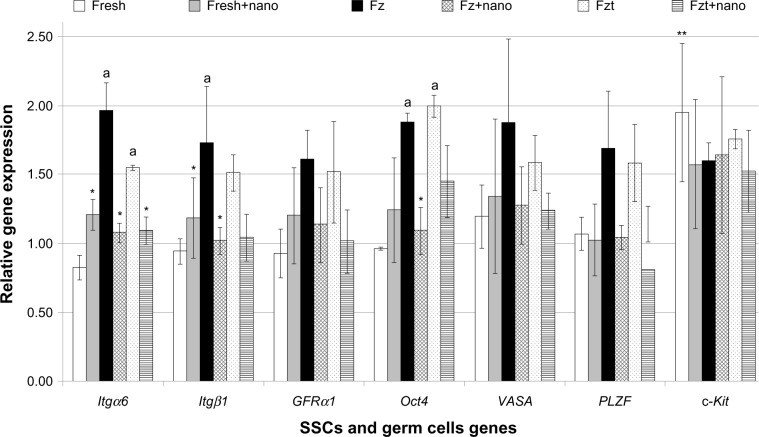
The effects of PLLA nanofiber scaffold on SSCs genes expression in culture
Gene expression of various genes was assessed and normalized based on GAPDH as a reference gene. Results showed lower gene expression of Itgα6, Itgβ1, and Oct4 in the second experimental group compared with the second control group (P≤0.05); however, no significant differences were observed in other gene expressions (PLZF, GFRα-1, VASA, and c-Kit) among the groups (Figure 6). High expression of Itgα6 and Itgβ1 in the first experimental group was observed in the culture as opposed to the first control group (P≤0.05). Also, gene expression of c-Kit in the fresh cell group significantly increased in comparison with the other genes.
Figure 6.
Comparison between the relative gene expression of SSCs and germ cell genes in the experimental groups.
Notes: Values are mean ± standard deviation. The experiments were replicated at least three times. *Significant difference versus culture of SSCs without seeding on PLLA in the same gene (P≤0.05); **significant difference versus all genes in the same group (P≤0.05); asignificant difference versus fresh cells group in the same gene (P≤0.05).
Abbreviations: Fresh, isolated testicular cells by two steps of enzymatic digestion before culture; Fresh+nano, fresh cells seeded on PLLA; Fz, frozen-thawed cells; Fz+nano, frozen-thawed cells seeded on PLLA; Fzt, frozen-thawed cells obtained from testis tissue; Fzt+nano, frozen-thawed cells obtained from testis tissue seeded on PLLA groups; PLLA, poly L-lactic acid; SSC, spermatogonial stem cell; Itgα6, integrin-α6; Itgβ1, integrin-β1; VASA, mouse vasa-homologue; GFRα-1, GDNF family co-receptorα1; Oct4, octamer-binding transcription factor 4; PLZF, promyelocytic leukemia zinc finger; c-Kit, proto-oncogene c-Kit or tyrosine-protein kinase Kit; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Discussion
In this study, we were able to show that the culture of fresh or frozen-thawed SSCs as well as SSCs obtained from frozen-thawed testis tissue seeded on PLLA can increase the colony formation of SSCs in the culture system when compared with the control groups (without seeding the cells on PLLA). SSCs obtained from frozen-thawed testis tissue can also form many clusters after the freezing procedure in vitro.
In this study, we used 3–6-day-old mice testes, because in these animals, spermatogenesis begins immediately after birth.52 Only germ cells in the newborn mouse testis can be the gonocyte or prospermatogonia, located in the center of the seminiferous tubule.53,54 By 6 days postpartum, these cells migrate to the basal membrane and produce undifferentiated type A spermatogonia, which begin to differentiate in a stepwise manner.55,56
Since SSCs are rare in rodent testes (only 0.03% of all germ cells),57 and there are no clear surface specific markers for SSCs isolation, SSC culture and proliferation can provide new tools to investigate molecular mechanisms and signaling pathways that regulate SSC functions.19 In vitro culture may increase the rate of successful transplantation, even with a small number of dissected seminiferous tubules.56 One way to reach these goals may be to co-culture these cells within the presence of ECM components on 3D scaffolds. In this study, PLLA nanofiber scaffolds were used to mimic the structure of ECM. In recent years, PLLA has been widely explored as a biomaterial scaffold because of its impressive biocompatibility, biodegradability, minimal inflammatory reaction, and excellent mechanical properties.58 The electrospinning method has allowed the successful preparation of PLLA scaffolds with various topographical structures.59,60 The morphology and architecture of the electrospun structure is similar to that of the natural ECM.30 Indeed, randomly oriented PLLA nanofibers can enhance cellular infiltration within the in vivo scaffold.61
To date, one established clinical option to preserve fertility is cryopreservation of sperm. There is no clinical technique currently available to preserve fertility in the prepubertal male who is likely to be sterilized by chemotherapy or radiotherapy, because he has no mature sperm. To overcome this problem, testicular tissues could be harvested before treatment and cryopreserved in these patients. Immature germ cells could somehow be matured either by autotransplantation, in vitro maturation, or xenografting.62
In this study, the cryopreservation method of Izadyar et al43 was used to isolate neonate testicular cells as well as to fragment seminiferous tubules. Goossens et al63 cryopreserved mouse tissue pieces in ethylene glycol and DMSO as cryoprotectant while their protocols had already been used for freezing testicular stem cell suspensions by Frederickx et al.64 They obtained the best morphology of the basal compartment when the cryoprotective medium contained DMSO.63 Honaramooz et al65 investigated the effects of cooling or cryopreservation with DMSO on the testis fragments of pigs before grafting and observed complete spermatogenesis. Although some studies37,39,66–70 cryopreserved tissue pieces and successfully carried out grafting, they did not show isolated SSCs in order to increase cell number.
The survival rate of the frozen-thawed testicular cells (control 2) and the isolated cells after the freezing-thawing of the testicular tissue (control 3) were 63.00% ± 3.56% and 40.00% ± 0.82%, respectively, which decreased after cryopreservation in both groups. The SSCs that survived after cryopreservation were also able to form clusters in vitro. The survival rates of the frozen-thawed testicular cells obtained from our experiments were rather similar to the rates reported by Izadyar et al43 for prepubertal bovines (68%). Our study showed, however, that these rates decreased for those cells isolated after the freezing-thawing of the testicular tissue. Very likely, the decline may be the result of the diminished cell recovery following the freeze-thaw procedure. Previous studies have demonstrated that cryopreservation will preserve purified spermatogonial cells, because these cells are relatively resistant to freezing solutions compared with other spermatogenic cells.71 Other investigations have shown similar results for the cryopreservation of non-pure spermatogonia in other species including rodents23,24 and domestic animals20,25 as well as humans.72
Since germ cell clusters have a distinct 3D structure, it may be possible to quantitatively analyze SSCs in vitro by counting clusters.44 In other words, by simply counting the number of colonies, the number of functional SSCs can be determined. In this study, we showed that the PLLA nanofiber significantly increased the number of clusters during the 3 weeks of cultivation. Additionally, the number of clusters in all groups significantly increased after the 3-week culture compared with a 1- or 2-week culture. Our finding is in line with Shakeri et al,34 who observed an increase of spermatogonial stem-like cell colonies during a short-term culture.
We also examined the effects of a PLLA nanofiber scaffold in combination with GDNF on the SSCs. Although several reports have described culturing various stem cells within the 3D scaffolds,23,73–77 the result of such cultures has most commonly been stem cell differentiation. Nur et al23 cultured embryonic stem cells on a 3D nanofibrillar surface and observed proliferation with self-renewal. Because FGF-2 can promote the proliferation of embryonic stem cells, they also used FGF-2 with the nanofiber to proliferate these cells.78 Recent studies have shown that some soluble growth factors, especially GDNF, can have a long-term positive effect on SSC maintenance and may also stimulate SSC division in animals.10,11,44,79–82 We conclude that the presence of GDNF in combination with a nanofiber scaffold and the presence of somatic cells in the culture likely creates a testis-like microenvironment in which proliferation is promoted.
During cell culture, the number of clusters for both the frozen-thawed cell group and the cell group obtained from the frozen-thawed testis tissue were significantly lower compared with the fresh cell group. However, SSCs were probably more resistant to the freezing solution and the freezing-thawing procedure compared with Sertoli cells83, which could not survive and died earlier in the procedure. Previous studies have demonstrated that somatic cells are able to either differentiate84 or support SSCs in both mice44,85–87 and human cultures.2,88,89 Because Sertoli cells can secrete various growth factors or cytokines, they can therefore create a microenvironment that will promote the maintenance, survival, and proliferation of spermatogonia.72,90 We presume that the decline of the somatic cells may have reduced contact between Sertoli cells and SSCs in vitro and therefore decreased the chance of survival.
In spite of the decrease in cluster formation, SSCs may still remain alive in the culture without the ability to form clusters. This would be in accordance with our previous results that showed an increase in the gene expression of spermatogonial cells (Itgα6, Itgβ1, and Oct4) in frozen-thawed groups compared with the fresh cells. Previous studies have shown that β1-integrin is dominantly expressed in rodent SSCs and germ cells as a surface molecule.91,92 In this study, we conclude that spermatogonial cells are probably not as sensitive to the freezing–thawing procedure as are Sertoli cells,83 and so the number of these cells that can combine with different cell types that have no stem cell potential (Sertoli cells) are high in the culture system. In other words, the high expression of β1-integrin is possibly related to the presence of Sertoli cells in testis.
In this study it was also demonstrated that gene expressions of spermatogonial cells decreased in cells cultured on PLLA. In contrast, the expression of the differentiation marker gene (c-Kit) increased in culture systems after the use of PLLA. Electron microscopy studies also confirmed the presence of differentiated cells in experimental groups. It is possible that the use of PLLA provides a structure for germ cells that allows for a disruption in the differentiation arrest of the spermatogonial cells in the culture system, and therefore cell differentiation is initiated in the experimental groups. Thus the expression of spermatogonial genes is decreased compared with the control groups.
Other studies have demonstrated that there are no specific biochemical or morphological markers for SSCs,93,94 but a combination of the expression of multiple markers can provide important information about spermatogonial cell types in rodents and other species.94,95 Therefore, in order to confirm the presence of spermatogonial cells during cultivation, an RT-PCR using spermatogonial and germ cells markers (Oct4, GFRα-1, PLZF, VASA, Itgα6, Itgβ1, and c-Kit) in all culture groups was performed. PLZF and GFRa-1 are markers for spermatogonial stem/progenitor cells and are well known spermatogonial-specific markers in many species2,84,96 and are considered to be markers of SSCs in rodents.7Oct4 is a general marker for stem cells and is also expressed in mouse spermatogonial stem/progenitor cells.8,10,44,97,98Itgα6 and Itgβ1 are cell surface receptors for spermatogonial stem/progenitor cells and mediate cell–cell and cell–ECM attachments and are expressed in germ cells of rodents.20,99Mvh(VASA) as a marker of germ cell is expressed in all spermatogenic cells. The expression of c-Kit, the receptor for stem cell factor, is low or absent in As, Apr, and early Aal spermatogonia and enhanced in late Aal and further differentiated spermatogonia.100
In our study, the expression of Oct4, GFRα-1, PLZF, VASA, Itgα6 and Itgβ1 markers of spermatogonial and germ cell identification were observed in isolated testicular cells and all culture groups. Our findings are in line with results of previous research and support the aforementioned studies. Also c-Kit as a differentiated spermatogonia marker was observed in all culture groups, suggesting that the clusters contained differentiating germ cells. Kanatsu-Shinohara et al101 showed weak expressions for c-Kit in some colonies.
SSCs are the only cells capable of being recolonized in the seminiferous tubules of infertile animals.102,103 Therefore, cultured testicular cells were transplanted into a mouse busulfan azoospermic model through the efferent ducts, in order to examine the functionality of SSCs among the culture cells. A month later, colonization of spermatogonial cells in recipient mouse testes and SSCs was detected via BrdU staining. On the other hand, tubules with homing spermatogenesis were considered as colonized seminiferous tubules. In these tubules, spermatogonial cells were seen on the basal membrane of the cross-sectioned tubule of the recipient testes, which is in accordance with previous reports.44,104,105
Moreover, an ultrastructure study of the cluster cells was performed. In rodents and humans alike, the relative amount of heterochromatin lining the nucleus is characteristic of specific types of spermatogonial cells.46 According to previous studies, the primitive spermatogonial group (As, A pr, and Aal) showed the large nucleus to cytoplasm ratio, intensive nucleolus, and low dense heterochromatin regions, whereas the heterochromatin face increased in differentiated spermatogonial group.46,50,106–108 Our findings are in line with previous studies.
Results of our study suggest that PLLA nanofiber scaffolds are useful in tissue engineering and can support the cell proliferation process. Frozen-thawed SSCs obtained from testis tissue can also form clusters after the freezing procedure in vitro. The self-renewal of SSCs in our cultural system enables this system to be utilized for the proliferation or differentiation of these cells in clinical applications, tissue engineering applications, cell replacement therapy, and tissue regeneration
Acknowledgments
We appreciate the contributions of Arghavan Janan for technical support and Marzieh Rostami for editing the manuscript. This study was funded by a grant from the Deputy of Research in Tehran University of Medical Sciences (TUMS), Tehran, Iran (number: 90-1-30-13036) and all experiments have been performed in the Cellular and Molecular Research Center, IUMS, Tehran, Iran.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Aponte PM, Soda T, Teerds KJ, Mizrak SC, van de Kant HJ, de Rooij DG. Propagation of bovine spermatogonial stem cells in vitro. Reproduction. 2008;136(5):543–557. doi: 10.1530/REP-07-0419. [DOI] [PubMed] [Google Scholar]
- 2.Sadri-Ardekani H, Mizrak SC, van Daalen SK, et al. Propagation of human spermatogonial stem cells in vitro. JAMA. 2009;302(19):2127–2134. doi: 10.1001/jama.2009.1689. [DOI] [PubMed] [Google Scholar]
- 3.Aslam I, Fishel S, Moore H, Dowell K, Thornton S. Fertility preservation of boys undergoing anti-cancer therapy: a review of the existing situation and prospects for the future. Hum Reprod. 2000;15(10):2154–2159. doi: 10.1093/humrep/15.10.2154. [DOI] [PubMed] [Google Scholar]
- 4.Hovatta O, Foudila T, Siegberg R, Johansson K, von Smitten K, Reima I. Pregnancy resulting from intracytoplasmic injection of spermatozoa from a frozen-thawed testicular biopsy specimen. Hum Reprod. 1996;11(11):2472–2473. doi: 10.1093/oxfordjournals.humrep.a019140. [DOI] [PubMed] [Google Scholar]
- 5.Brinster RL. Male germline stem cells: from mice to men. Science. 2007;316(5823):404–405. doi: 10.1126/science.1137741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X, Cao HQ, Shi LY, Ng SY, Stanton LW, Chew SY. Nanofiber topography and sustained biochemical signaling enhance human mesenchymal stem cell neural commitment. Acta Biomater. 2012;8(3):1290–1302. doi: 10.1016/j.actbio.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Kolasa A, Misiakiewicz K, Marchlewicz M, Wiszniewska B. The generation of spermatogonial stem cells and spermatogonia in mammals. Reprod Biol. 2012;12(1):5–23. doi: 10.1016/s1642-431x(12)60074-6. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann MC, Braydich-Stolle L, Dym M. Isolation of male germ-line stem cells; influence of GDNF. Dev Biol. 2005;279(1):114–124. doi: 10.1016/j.ydbio.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kostereva N, Hofmann MC. Regulation of the spermatogonial stem cell niche. Reprod Domest Anim. 2008;43(Suppl 2):386–392. doi: 10.1111/j.1439-0531.2008.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2004;101(47):16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng X, Lindahl M, Hyvonen ME, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287(5457):1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 12.Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol. 2008;24:263–286. doi: 10.1146/annurev.cellbio.24.110707.175355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmann MC. Gdnf signaling pathways within the mammalian spermatogonial stem cell niche. Mol Cell Endocrinol. 2008;288(1–2):95–103. doi: 10.1016/j.mce.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato T, Aiyama Y, Ishii-Inagaki M, et al. Cyclical and patch-like GDNF distribution along the basal surface of Sertoli cells in mouse and hamster testes. PLoS One. 2011;6(12):e28367. doi: 10.1371/journal.pone.0028367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huleihel M, Abuelhija M, Lunenfeld E. In vitro culture of testicular germ cells: regulatory factors and limitations. Growth Factors. 2007;25(4):236–252. doi: 10.1080/08977190701783400. [DOI] [PubMed] [Google Scholar]
- 16.de Rooij DG. The spermatogonial stem cell niche. Microsc Res Tech. 2009;72(8):580–585. doi: 10.1002/jemt.20699. [DOI] [PubMed] [Google Scholar]
- 17.Hermo L, Pelletier RM, Cyr DG, Smith CE. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes. Microsc Res Tech. 2010;73(4):241–278. doi: 10.1002/jemt.20783. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida S, Sukeno M, Nabeshima Y. A vasculature-associated niche for undifferentiated spermatogonia in the mouse testis. Science. 2007;317(5845):1722–1726. doi: 10.1126/science.1144885. [DOI] [PubMed] [Google Scholar]
- 19.Phillips BT, Gassei K, Orwig KE. Spermatogonial stem cell regulation and spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365(1546):1663–1678. doi: 10.1098/rstb.2010.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shinohara T, Avarbock MR, Brinster RL. beta1- and alpha6-integrin are surface markers on mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 1999;96(10):5504–5509. doi: 10.1073/pnas.96.10.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delgado-Rivera R, Harris SL, Ahmed I, et al. Increased FGF-2 secretion and ability to support neurite outgrowth by astrocytes cultured on polyamide nanofibrillar matrices. Matrix Biol. 2009;28(3):137–147. doi: 10.1016/j.matbio.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Vacanti JP, Langer R. Tissue engineering: the design and fabrication of living replacement devices for surgical reconstruction and transplantation. Lancet. 1999;354(Suppl 1):SI32–SI34. doi: 10.1016/s0140-6736(99)90247-7. [DOI] [PubMed] [Google Scholar]
- 23.Nur EKA, Ahmed I, Kamal J, Schindler M, Meiners S. Three- dimensional nanofibrillar surfaces promote self-renewal in mouse embryonic stem cells. Stem Cells. 2006;24(2):426–433. doi: 10.1634/stemcells.2005-0170. [DOI] [PubMed] [Google Scholar]
- 24.Nur EKA, Ahmed I, Kamal J, Schindler M, Meiners S. Three dimensional nanofibrillar surfaces induce activation of Rac. Biochem Biophys Res Commun. 2005;331(2):428–434. doi: 10.1016/j.bbrc.2005.03.195. [DOI] [PubMed] [Google Scholar]
- 25.Schindler M, Ahmed I, Kamal J, et al. A synthetic nanofibrillar matrix promotes in vivo-like organization and morphogenesis for cells in culture. Biomaterials. 2005;26(28):5624–5631. doi: 10.1016/j.biomaterials.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Srouji S, Kizhner T, Suss-Tobi E, Livne E, Zussman E. 3-D Nanofibrous electrospun multilayered construct is an alternative ECM mimicking scaffold. J Mater Sci Mater Med. 2008;19(3):1249–1255. doi: 10.1007/s10856-007-3218-z. [DOI] [PubMed] [Google Scholar]
- 27.Schofer MD, Roessler PP, Schaefer J, et al. Electrospun PLLA nanofiber scaffolds and their use in combination with BMP-2 for reconstruction of bone defects. PLoS One. 2011;6(9):e25462. doi: 10.1371/journal.pone.0025462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dersch R, Liu T, Schaper A, Greiner A, Wendorff J. Electrospun nanofibers: internal structure and intrinsic orientation. J Polym Sci A Polym Chem. 2003;41(4):545–553. [Google Scholar]
- 29.Jun Z, Hou H, Schaper A, Wendorff J, Greiner A. Poly-L-lactide nanofibers by electrospinning-Influence of solution viscosity and electrical conductivity on fiber diameter and fiber morphology. e-Polymers. 2003;9:1–9. [Google Scholar]
- 30.Lee BN, Kim da Y, Kang HJ, et al. In vivo biofunctionality comparison of different topographic PLLA scaffolds. J Biomed Mater Res A. 2012;100(7):1751–1760. doi: 10.1002/jbm.a.34135. [DOI] [PubMed] [Google Scholar]
- 31.Ghaedi M, Soleimani M, Shabani I, Duan Y, Lotfi AS. Hepatic differentiation from human mesenchymal stem cells on a novel nanofiber scaffold. Cell Mol Biol Lett. 2011;17(1):89–106. doi: 10.2478/s11658-011-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahmoud H. Concise review: spermatogenesis in an artificial three-dimensional system. Stem Cells. 2012;30(11):2355–2360. doi: 10.1002/stem.1238. [DOI] [PubMed] [Google Scholar]
- 33.Stukenborg JB, Wistuba J, Luetjens CM, et al. Coculture of spermatogonia with somatic cells in a novel three-dimensional soft-agar-culture-system. J Androl. 2008;29(3):312–329. doi: 10.2164/jandrol.107.002857. [DOI] [PubMed] [Google Scholar]
- 34.Shakeri M, Kohram H, Shahverdi A, et al. Behavior of mouse spermatogonial stem-like cells on an electrospun nanofibrillar matrix. J Assist Reprod Genet. 2013;30(3):325–332. doi: 10.1007/s10815-012-9916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyns C, Curaba M, Vanabelle B, Van Langendonckt A, Donnez J. Options for fertility preservation in prepubertal boys. Hum Reprod Update. 2010;16(3):312–328. doi: 10.1093/humupd/dmp054. [DOI] [PubMed] [Google Scholar]
- 36.Tournaye H, Goossens E, Verheyen G, et al. Preserving the reproductive potential of men and boys with cancer: current concepts and future prospects. Hum Reprod Update. 2004;10(6):525–532. doi: 10.1093/humupd/dmh038. [DOI] [PubMed] [Google Scholar]
- 37.Shinohara T, Inoue K, Ogonuki N, et al. Birth of offspring following transplantation of cryopreserved immature testicular pieces and in-vitro microinsemination. Hum Reprod. 2002;17(12):3039–3045. doi: 10.1093/humrep/17.12.3039. [DOI] [PubMed] [Google Scholar]
- 38.Poels J, Van Langendonckt A, Many MC, Wese FX, Wyns C. Vitrification preserves proliferation capacity in human spermatogonia. Hum Reprod. 2013;28(3):578–589. doi: 10.1093/humrep/des455. [DOI] [PubMed] [Google Scholar]
- 39.Wyns C, Van Langendonckt A, Wese FX, Donnez J, Curaba M. Long-term spermatogonial survival in cryopreserved and xenografted immature human testicular tissue. Hum Reprod. 2008;23(11):2402–2414. doi: 10.1093/humrep/den272. [DOI] [PubMed] [Google Scholar]
- 40.Sadri-Ardekani H, Akhondi MA, van der Veen F, Repping S, van Pelt AM. In vitro propagation of human prepubertal spermatogonial stem cells. JAMA. 2011;305(23):2416–2418. doi: 10.1001/jama.2011.791. [DOI] [PubMed] [Google Scholar]
- 41.Gouk SS, Loh YF, Kumar SD, Watson PF, Kuleshova LL. Cryopreservation of mouse testicular tissue: prospect for harvesting spermatogonial stem cells for fertility preservation. Fertil Steril. 2011;95(7):2399–2403. doi: 10.1016/j.fertnstert.2011.03.035. [DOI] [PubMed] [Google Scholar]
- 42.van Pelt AM, Morena AR, van Dissel-Emiliani FM, et al. Isolation of the synchronized A spermatogonia from adult vitamin A-deficient rat testes. Biol Reprod. 1996;55(2):439–444. doi: 10.1095/biolreprod55.2.439. [DOI] [PubMed] [Google Scholar]
- 43.Izadyar F, Matthijs-Rijsenbilt JJ, den Ouden K, Creemers LB, Woelders H, de Rooij DG. Development of a cryopreservation protocol for type A spermatogonia. J Androl. 2002;23(4):537–545. [PubMed] [Google Scholar]
- 44.Koruji M, Movahedin M, Mowla SJ, Gourabi H, Arfaee AJ. Efficiency of adult mouse spermatogonial stem cell colony formation under several culture conditions. In Vitro Cell Dev Biol Anim. 2009;45(5–6):281–289. doi: 10.1007/s11626-008-9169-y. [DOI] [PubMed] [Google Scholar]
- 45.Ye h JR, Zhang X, Nagano MC. Establishment of a short-term in vitro assay for mouse spermatogonial stem cells. Biol Reprod. 2007;77(5):897–904. doi: 10.1095/biolreprod.107.063057. [DOI] [PubMed] [Google Scholar]
- 46.Russell LD, Ettlin RA, Sinha-Hikim AP, Clegg ED. Mammalian spermatogenesis. In: Russell LD, Ettlin RA, Sinha-Hikim AP, Clegg ED, editors. Histological and Histopathological Evaluation of the Testis. Clearwater, FL: Cache River Press; 1990. pp. 1–40. [Google Scholar]
- 47.Chiarini-Garcia H, Russell LD. Characterization of mouse spermatogonia by transmission electron microscopy. Reproduction. 2002;123(4):567–577. doi: 10.1530/rep.0.1230567. [DOI] [PubMed] [Google Scholar]
- 48.Dettin L, Ravindranath N, Hofmann MC, Dym M. Morphological characterization of the spermatogonial subtypes in the neonatal mouse testis. Biol Reprod. 2003;69(5):1565–1571. doi: 10.1095/biolreprod.103.016394. [DOI] [PubMed] [Google Scholar]
- 49.Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proc Natl Acad Sci U S A. 1994;91(24):11303–11307. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogawa T, Arechaga JM, Avarbock MR, Brinster RL. Transplantation of testis germinal cells into mouse seminiferous tubules. Int J Dev Biol. 1997;41(1):111–122. [PubMed] [Google Scholar]
- 51.Koruji M, Shahverdi A, Janan A, Piryaei A, Lakpour MR, Gilani Sedighi MA. Proliferation of small number of human spermatogonial stem cells obtained from azoospermic patients. J Assist Reprod Genet. 2012;29(9):957–967. doi: 10.1007/s10815-012-9817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shinohara T, Orwig KE, Avarbock MR, Brinster RL. Remodeling of the postnatal mouse testis is accompanied by dramatic changes in stem cell number and niche accessibility. Proc Natl Acad Sci U S A. 2001;98(11):6186–6191. doi: 10.1073/pnas.111158198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clermont Y, Perey B. Quantitative study of the cell population of the seminiferous tubules in immature rats. Am J Anat. 1957;100(2):241–267. doi: 10.1002/aja.1001000205. [DOI] [PubMed] [Google Scholar]
- 54.Nagano M, Brinster RL. Spermatogonial transplantation and reconstitution of donor cell spermatogenesis in recipient mice. APMIS. 1998;106(1):47–55. doi: 10.1111/j.1699-0463.1998.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 55.McCarrey JR. Development of the germ cell. In: Desjardins C, Ewing LL, editors. Cell and Molecular Biology of the Testis. New York: Oxford University Press; 1993. pp. 58–89. [Google Scholar]
- 56.McLean DJ, Friel PJ, Johnston DS, Griswold MD. Characterization of spermatogonial stem cell maturation and differentiation in neonatal mice. Biol Reprod. 2003;69(6):2085–2091. doi: 10.1095/biolreprod.103.017020. [DOI] [PubMed] [Google Scholar]
- 57.Tegelenbosch RA, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res. 1993;290(2):193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- 58.Niu X, Luo Y, Li Y, Fu C, Chen J, Wang Y. Design of bioinspired polymeric materials based on poly(D,L-lactic acid) modifications towards improving its cytocompatibility. J Biomed Mater Res A. 2008;84(4):908–916. doi: 10.1002/jbm.a.31360. [DOI] [PubMed] [Google Scholar]
- 59.Dong Y, Liao S, Ngiam M, Chan CK, Ramakrishna S. Degradation behaviors of electrospun resorbable polyester nanofibers. Tissue Eng Part B Rev. 2009;15(3):333–351. doi: 10.1089/ten.TEB.2008.0619. [DOI] [PubMed] [Google Scholar]
- 60.Smith LA, Liu X, Ma PX. Tissue engineering with nano-fibrous scaffolds. Soft Matter. 2008;4(11):2144–2149. doi: 10.1039/b807088c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Usas A, Huard J. Muscle-derived stem cells for tissue engineering and regenerative therapy. Biomaterials. 2007;28(36):5401–5406. doi: 10.1016/j.biomaterials.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fujita K, Ohta H, Tsujimura A, et al. Transplantation of spermatogonial stem cells isolated from leukemic mice restores fertility without inducing leukemia. J Clin Invest. 2005;115(7):1855–1861. doi: 10.1172/JCI24189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goossens E, Frederickx V, Geens M, De Block G, Tournaye H. Cryosurvival and spermatogenesis after allografting prepubertal mouse tissue: comparison of two cryopreservation protocols. Fertil Steril. 2008;89(3):725–727. doi: 10.1016/j.fertnstert.2007.03.044. [DOI] [PubMed] [Google Scholar]
- 64.Frederickx V, Michiels A, Goossens E, De Block G, Van Steirteghem AC, Tournaye H. Recovery, survival and functional evaluation by transplantation of frozen-thawed mouse germ cells. Hum Reprod. 2004;19(4):948–953. doi: 10.1093/humrep/deh154. [DOI] [PubMed] [Google Scholar]
- 65.Honaramooz A, Snedaker A, Boiani M, Scholer H, Dobrinski I, Schlatt S. Sperm from neonatal mammalian testes grafted in mice. Nature. 2002;418(6899):778–781. doi: 10.1038/nature00918. [DOI] [PubMed] [Google Scholar]
- 66.Curaba M, Verleysen M, Amorim CA, et al. Cryopreservation of prepubertal mouse testicular tissue by vitrification. Fertil Steril. 2009;95(4):1229–1234. e1. doi: 10.1016/j.fertnstert.2010.04.062. [DOI] [PubMed] [Google Scholar]
- 67.Travers A, Milazzo JP, Perdrix A, et al. Assessment of freezing procedures for rat immature testicular tissue. Theriogenology. 2011;76(6):981–990. doi: 10.1016/j.theriogenology.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 68.Unni S, Kasiviswanathan S, D’Souza S, et al. Efficient cryopreservation of testicular tissue: effect of age, sample state, and concentration of cryoprotectant. Fertil Steril. 2012;97(1):200–208. e1. doi: 10.1016/j.fertnstert.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 69.Keros V, Hultenby K, Borgstrom B, Fridstrom M, Jahnukainen K, Hovatta O. Methods of cryopreservation of testicular tissue with viable spermatogonia in pre-pubertal boys undergoing gonadotoxic cancer treatment. Hum Reprod. 2007;22(5):1384–1395. doi: 10.1093/humrep/del508. [DOI] [PubMed] [Google Scholar]
- 70.Baert Y, Goossens E, van Saen D, Ning L, in’t Veld P, Tournaye H. Orthotopic grafting of cryopreserved prepubertal testicular tissue: in search of a simple yet effective cryopreservation protocol. Fertil Steril. 2012;97(5):1152–1157. e1–e2. doi: 10.1016/j.fertnstert.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 71.Redden E, Davey R, Borjigin U, et al. Large quantity cryopreservation of bovine testicular cells and its effect on enrichment of type A spermatogonia. Cryobiology. 2009;58:190–195. doi: 10.1016/j.cryobiol.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 72.Mirzapour T, Movahedin M, Tengku Ibrahim TA, Haron AW, Nowroozi MR. Evaluation of the effects of cryopreservation on viability, proliferation and colony formation of human spermatogonial stem cells in vitro culture. Andrologia. 2013;45(1):26–34. doi: 10.1111/j.1439-0272.2012.01302.x. [DOI] [PubMed] [Google Scholar]
- 73.Garreta E, Genove E, Borros S, Semino CE. Osteogenic differentiation of mouse embryonic stem cells and mouse embryonic fibroblasts in a three-dimensional self-assembling peptide scaffold. Tissue Eng. 2006;12(8):2215–2227. doi: 10.1089/ten.2006.12.2215. [DOI] [PubMed] [Google Scholar]
- 74.Al-Salihi KA, Samsudin AR. Bone marrow mesenchymal stem cells differentiation and proliferation on the surface of coral implant. Med J Malaysia. 2004;59(Suppl B):45–46. [PubMed] [Google Scholar]
- 75.Borden M, Attawia M, Laurencin CT. The sintered microsphere matrix for bone tissue engineering: in vitro osteoconductivity studies. J Biomed Mater Res. 2002;61(3):421–429. doi: 10.1002/jbm.10201. [DOI] [PubMed] [Google Scholar]
- 76.Smith LA, Liu X, Hu J, Wang P, Ma PX. Enhancing osteogenic differentiation of mouse embryonic stem cells by nanofibers. Tissue Eng Part A. 2009;15(7):1855–1864. doi: 10.1089/ten.tea.2008.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saito E, Liao EE, Hu WW, Krebsbach PH, Hollister SJ. Effects of designed PLLA and 50:50 PLGA scaffold architectures on bone formation in vivo. J Tissue Eng Regen Med. 2013;7(2):99–111. doi: 10.1002/term.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nur EKA, Ahmed I, Kamal J, Babu AN, Schindler M, Meiners S. Covalently attached FGF-2 to three-dimensional polyamide nanofibrillar surfaces demonstrates enhanced biological stability and activity. Mol Cell Biochem. 2008;309(1–2):157–166. doi: 10.1007/s11010-007-9654-8. [DOI] [PubMed] [Google Scholar]
- 79.Kanatsu-Shinohara M, Miki H, Inoue K, et al. Long-term culture of mouse male germline stem cells under serum-or feeder-free conditions. Biol Reprod. 2005;72(4):985–991. doi: 10.1095/biolreprod.104.036400. [DOI] [PubMed] [Google Scholar]
- 80.Ebata KT, Ye h JR, Zhang X, Nagano MC. Soluble growth factors stimulate spermatogonial stem cell divisions that maintain a stem cell pool and produce progenitors in vitro. Exp Cell Res. 2011;317(10):1319–1329. doi: 10.1016/j.yexcr.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 81.Nagano M, Ryu BY, Brinster CJ, Avarbock MR, Brinster RL. Maintenance of mouse male germ line stem cells in vitro. Biol Reprod. 2003;68(6):2207–2214. doi: 10.1095/biolreprod.102.014050. [DOI] [PubMed] [Google Scholar]
- 82.Koruji M, Movahedin M, Mowla SJ, Gourabi H. Colony formation ability of frozen thawed spermatogonial stem cell from adult mouse. Iranian Journal of Reproductive Medicine. 2007;5(3):109–115. [Google Scholar]
- 83.Kanatsu-Shinohara M, Ogonuki N, Inoue K, Ogura A, Toyokuni S, Shinohara T. Restoration of fertility in infertile mice by transplantation of cryopreserved male germline stem cells. Hum Reprod. 2003;18(12):2660–2667. doi: 10.1093/humrep/deg483. [DOI] [PubMed] [Google Scholar]
- 84.Aponte PM, van Bragt MP, de Rooij DG, van Pelt AM. Spermatogonial stem cells: characteristics and experimental possibilities. APMIS. 2005;113(11–12):727–742. doi: 10.1111/j.1600-0463.2005.apm_302.x. [DOI] [PubMed] [Google Scholar]
- 85.Kim J, Seandel M, Falciatori I, Wen D, Rafii S. CD34+ testicular stromal cells support long-term expansion of embryonic and adult stem and progenitor cells. Stem Cells. 2008;26(10):2516–2522. doi: 10.1634/stemcells.2008-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang YH, Chin CC, Ho HN, et al. Pluripotency of mouse spermatogonial stem cells maintained by IGF-1-dependent pathway. FASEB J. 2009;23(7):2076–2087. doi: 10.1096/fj.08-121939. [DOI] [PubMed] [Google Scholar]
- 87.Mohamadi SM, Movahedin M, Koruji SM, Jafarabadi MA, Makoolati Z. Comparison of colony formation in adult mouse spermatogonial stem cells developed in Sertoli and STO coculture systems. Andrologia. 2012;44(Suppl 1):431–437. doi: 10.1111/j.1439-0272.2011.01201.x. [DOI] [PubMed] [Google Scholar]
- 88.Liu S, Tang Z, Xiong T, Tang W. Isolation and characterization of human spermatogonial stem cells. Reprod Biol Endocrinol. 2011;9:141. doi: 10.1186/1477-7827-9-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mirzapour T, Movahedin M, Tengku Ibrahim TA, et al. Effects of basic fibroblast growth factor and leukaemia inhibitory factor on proliferation and short-term culture of human spermatogonial stem cells. Andrologia. 2012;44:41–55. doi: 10.1111/j.1439-0272.2010.01135.x. [DOI] [PubMed] [Google Scholar]
- 90.Izadyar F, Den Ouden K, Stout TA, et al. Autologous and homologous transplantation of bovine spermatogonial stem cells. Reproduction. 2003;126(6):765–774. [PubMed] [Google Scholar]
- 91.Clark AT. The stem cell identity of testicular cancer. Stem Cell Rev. 2007;3(1):49–59. doi: 10.1007/s12015-007-0002-x. [DOI] [PubMed] [Google Scholar]
- 92.Kanatsu-Shinohara M, Kato M, Takehashi M, et al. Production of transgenic rats via lentiviral transduction and xenogeneic transplantation of spermatogonial stem cells. Biol Reprod. 2008;79(6):1121–1128. doi: 10.1095/biolreprod.108.071159. [DOI] [PubMed] [Google Scholar]
- 93.Brinster RL. Germline stem cell transplantation and transgenesis. Science. 2002;296(5576):2174–2176. doi: 10.1126/science.1071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McLean DJ. Spermatogonial stem cell transplantation, testicular function, and restoration of male fertility in mice. Methods Mol Biol. 2008;450:149–162. doi: 10.1007/978-1-60327-214-8_11. [DOI] [PubMed] [Google Scholar]
- 95.Hermann BP, Sukhwani M, Hansel MC, Orwig KE. Spermatogonial stem cells in higher primates: are there differences from those in rodents? Reproduction. 2010;139(3):479–493. doi: 10.1530/REP-09-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hermann B, Sukhwani M, Hansel M, Orwig K. Spermatogonial stem cells in higher primates: are there differences to those in rodents? Reproduction. 2010;139(3):479–493. doi: 10.1530/REP-09-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ohbo K, Yoshida S, Ohmura M, et al. Identification and characterization of stem cells in prepubertal spermatogenesis in mice small star, filled. Dev Biol. 2003;258(1):209–225. doi: 10.1016/s0012-1606(03)00111-8. [DOI] [PubMed] [Google Scholar]
- 98.Ohmura M, Yoshida S, Ide Y, Nagamatsu G, Suda T, Ohbo K. Spatial analysis of germ stem cell development in Oct-4/EGFP transgenic mice. Arch Histol Cytol. 2004;67(4):285–296. doi: 10.1679/aohc.67.285. [DOI] [PubMed] [Google Scholar]
- 99.Kubota H, Avarbock MR, Brinster RL. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proc Natl Acad Sci U S A. 2003;100(11):6487–6492. doi: 10.1073/pnas.0631767100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schrans-Stassen BH, van de Kant HJ, de Rooij DG, van Pelt AM. Differential expression of c-kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology. 1999;140(12):5894–5900. doi: 10.1210/endo.140.12.7172. [DOI] [PubMed] [Google Scholar]
- 101.Kanatsu-Shinohara M, Ogonuki N, Inoue K, et al. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69(2):612–616. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- 102.Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci U S A. 1994;91(24):11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shinohara T, Orwig KE, Avarbock MR, Brinster RL. Spermatogonial stem cell enrichment by multiparameter selection of mouse testis cells. Proc Natl Acad Sci U S A. 2000;97(15):8346–8351. doi: 10.1073/pnas.97.15.8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Anjamrooz SH, Movahedin M, Tiraihi T, Mowla SJ. In vitro effects of epidermal growth factor, follicle stimulating hormone and testosterone on mouse spermatogonial cell colony formation. Reprod Fertil Dev. 2006;18(6):709–720. doi: 10.1071/rd05126. [DOI] [PubMed] [Google Scholar]
- 105.Nagano M, Avarbock MR, Brinster RL. Pattern and kinetics of mouse donor spermatogonial stem cell colonization in recipient testes. Biol Reprod. 1999;60(6):1429–1436. doi: 10.1095/biolreprod60.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bellve AR, Cavicchia JC, Millette CF, O’Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74(1):68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Morena AR, Boitani C, Pesce M, De Felici M, Stefanini M. Isolation of highly purified type A spermatogonia from prepubertal rat testis. J Androl. 1996;17(6):708–717. [PubMed] [Google Scholar]
- 108.Chiarini-Garcia H, Russell LD. High-resolution light microscopic characterization of mouse spermatogonia. Biol Reprod. 2001;65(4):1170–1178. doi: 10.1095/biolreprod65.4.1170. [DOI] [PubMed] [Google Scholar]