Abstract
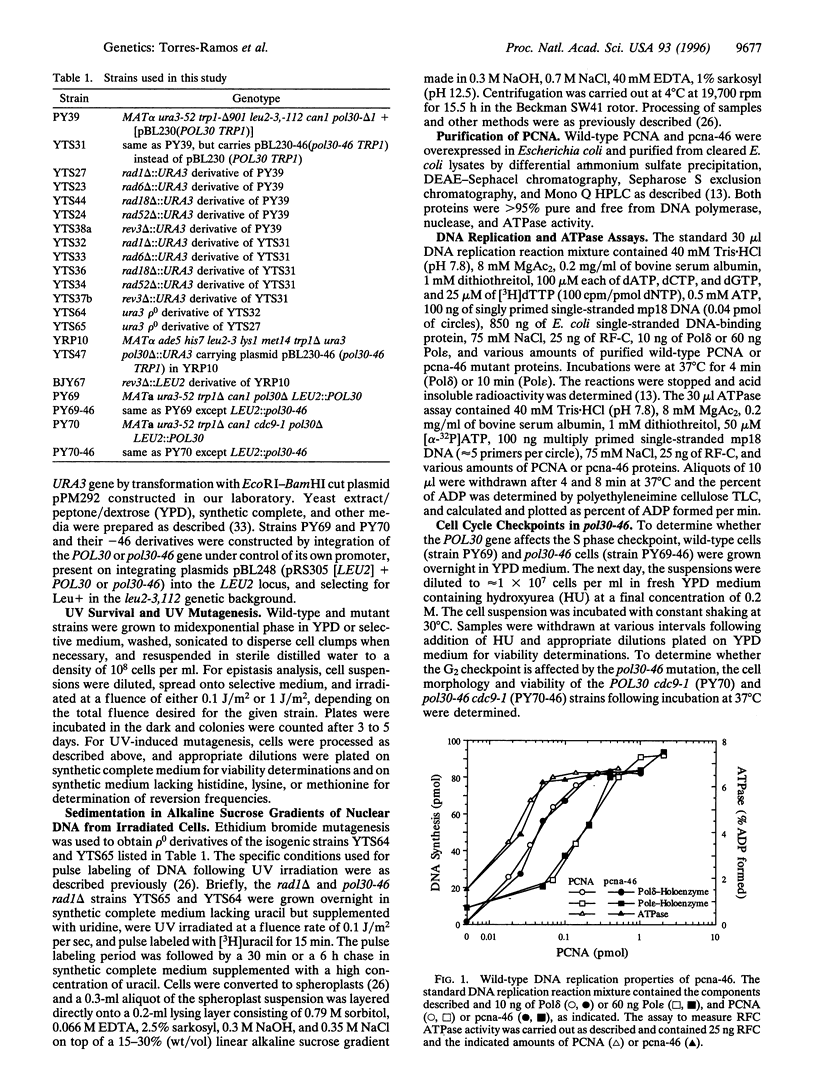
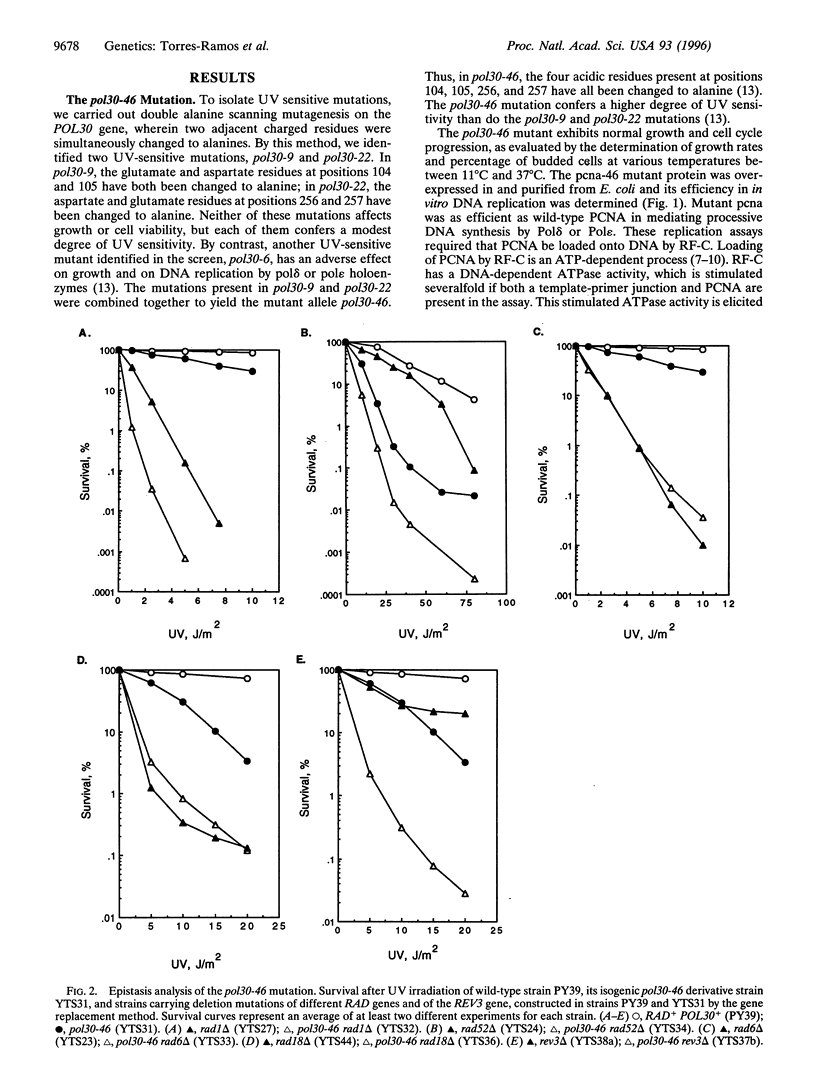
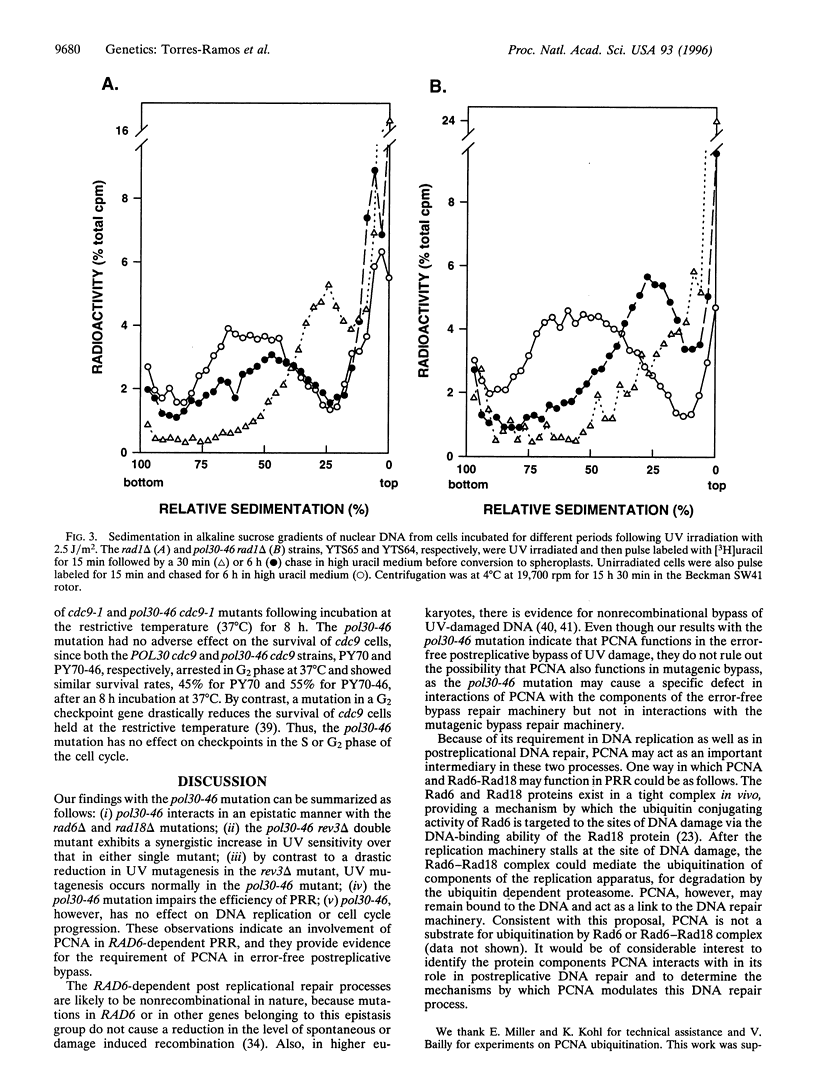
The proliferating cell nuclear antigen (PCNA) acts as a processivity factor for replicative DNA polymerases and is essential for DNA replication. In vitro studies have suggested a role for PCNA-in the repair synthesis step of nucleotide excision repair, and PCNA interacts with the cyclin-dependent kinase inhibitor p21. However, because of the lack of genetic evidence, it is not clear which of the DNA repair processes are in fact affected by PCNA in vivo. Here, we describe a PCNA mutation, pol30-46, that confers ultraviolet (UV) sensitivity but has no effect on growth or cell cycle progression, and the mutant pcna interacts normally with DNA polymerase delta and epsilon. Genetic studies indicate that the pol30-46 mutation is specifically defective in RAD6-dependent postreplicational repair of UV damaged DNA, and this mutation impairs the error-free mode of bypass repair. These results implicate a role for PCNA as an intermediary between DNA replication and postreplicational DNA repair.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong J. D., Chadee D. N., Kunz B. A. Roles for the yeast RAD18 and RAD52 DNA repair genes in UV mutagenesis. Mutat Res. 1994 Nov;315(3):281–293. doi: 10.1016/0921-8777(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Ayyagari R., Impellizzeri K. J., Yoder B. L., Gary S. L., Burgers P. M. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol. 1995 Aug;15(8):4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly V., Lamb J., Sung P., Prakash S., Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994 Apr 1;8(7):811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- Bauer G. A., Burgers P. M. Molecular cloning, structure and expression of the yeast proliferating cell nuclear antigen gene. Nucleic Acids Res. 1990 Jan 25;18(2):261–265. doi: 10.1093/nar/18.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P. M. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases delta and epsilon. J Biol Chem. 1991 Nov 25;266(33):22698–22706. [PubMed] [Google Scholar]
- Cassier-Chauvat C., Fabre F. A similar defect in UV-induced mutagenesis conferred by the rad6 and rad18 mutations of Saccharomyces cerevisiae. Mutat Res. 1991 May;254(3):247–253. doi: 10.1016/0921-8777(91)90063-u. [DOI] [PubMed] [Google Scholar]
- Fien K., Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol Cell Biol. 1992 Jan;12(1):155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Rozas H., Kelman Z., Dean F. B., Pan Z. Q., Harper J. W., Elledge S. J., O'Donnell M., Hurwitz J. Cdk-interacting protein 1 directly binds with proliferating cell nuclear antigen and inhibits DNA replication catalyzed by the DNA polymerase delta holoenzyme. Proc Natl Acad Sci U S A. 1994 Aug 30;91(18):8655–8659. doi: 10.1073/pnas.91.18.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987 Sep 10;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Jones J. S., Weber S., Prakash L. The Saccharomyces cerevisiae RAD18 gene encodes a protein that contains potential zinc finger domains for nucleic acid binding and a putative nucleotide binding sequence. Nucleic Acids Res. 1988 Jul 25;16(14B):7119–7131. doi: 10.1093/nar/16.14.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna T. S., Kong X. P., Gary S., Burgers P. M., Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994 Dec 30;79(7):1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W., Christensen R. B. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J Bacteriol. 1979 Sep;139(3):866–876. doi: 10.1128/jb.139.3.866-876.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. W. Mutagenesis in Saccharomyces cerevisiae. Adv Genet. 1982;21:173–254. doi: 10.1016/s0065-2660(08)60299-0. [DOI] [PubMed] [Google Scholar]
- Lee S. H., Hurwitz J. Mechanism of elongation of primed DNA by DNA polymerase delta, proliferating cell nuclear antigen, and activator 1. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5672–5676. doi: 10.1073/pnas.87.15.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Pan Z. Q., Kwong A. D., Burgers P. M., Hurwitz J. Synthesis of DNA by DNA polymerase epsilon in vitro. J Biol Chem. 1991 Nov 25;266(33):22707–22717. [PubMed] [Google Scholar]
- Lemontt J. F. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971 May;68(1):21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. J., Deshaies R. J. Exercising self-restraint: discouraging illicit acts of S and M in eukaryotes. Cell. 1993 Jul 30;74(2):223–226. doi: 10.1016/0092-8674(93)90413-k. [DOI] [PubMed] [Google Scholar]
- Li R., Waga S., Hannon G. J., Beach D., Stillman B. Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature. 1994 Oct 6;371(6497):534–537. doi: 10.1038/371534a0. [DOI] [PubMed] [Google Scholar]
- Morrison A., Christensen R. B., Alley J., Beck A. K., Bernstine E. G., Lemontt J. F., Lawrence C. W. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989 Oct;171(10):5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Miller E. J., Prakash L. Domain structure and functional analysis of the carboxyl-terminal polyacidic sequence of the RAD6 protein of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Mar;8(3):1179–1185. doi: 10.1128/mcb.8.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas T. A., Zhou Z., Elledge S. J. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995 Jan 13;80(1):29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- Ng L., McConnell M., Tan C. K., Downey K. M., Fisher P. A. Interaction of DNA polymerase delta, proliferating cell nuclear antigen, and synthetic oligonucleotide template-primers. Analysis by polyacrylamide gel electrophoresis-band mobility shift assay. J Biol Chem. 1993 Jun 25;268(18):13571–13576. [PubMed] [Google Scholar]
- Nichols A. F., Sancar A. Purification of PCNA as a nucleotide excision repair protein. Nucleic Acids Res. 1992 Jul 11;20(13):2441–2446. doi: 10.1093/nar/20.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z. Q., Reardon J. T., Li L., Flores-Rozas H., Legerski R., Sancar A., Hurwitz J. Inhibition of nucleotide excision repair by the cyclin-dependent kinase inhibitor p21. J Biol Chem. 1995 Sep 15;270(37):22008–22016. doi: 10.1074/jbc.270.37.22008. [DOI] [PubMed] [Google Scholar]
- Pines J. Cell cycle. p21 inhibits cyclin shock. Nature. 1994 Jun 16;369(6481):520–521. doi: 10.1038/369520a0. [DOI] [PubMed] [Google Scholar]
- Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol Gen Genet. 1981;184(3):471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- Prakash S., Sung P., Prakash L. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu Rev Genet. 1993;27:33–70. doi: 10.1146/annurev.ge.27.120193.000341. [DOI] [PubMed] [Google Scholar]
- Prelich G., Tan C. K., Kostura M., Mathews M. B., So A. G., Downey K. M., Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987 Apr 2;326(6112):517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Saparbaev M., Prakash L., Prakash S. Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics. 1996 Mar;142(3):727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivji K. K., Kenny M. K., Wood R. D. Proliferating cell nuclear antigen is required for DNA excision repair. Cell. 1992 Apr 17;69(2):367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- Spivak G., Hanawalt P. C. Translesion DNA synthesis in the dihydrofolate reductase domain of UV-irradiated CHO cells. Biochemistry. 1992 Jul 28;31(29):6794–6800. doi: 10.1021/bi00144a021. [DOI] [PubMed] [Google Scholar]
- Sung P., Prakash S., Prakash L. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 1988 Nov;2(11):1476–1485. doi: 10.1101/gad.2.11.1476. [DOI] [PubMed] [Google Scholar]
- Tan C. K., Castillo C., So A. G., Downey K. M. An auxiliary protein for DNA polymerase-delta from fetal calf thymus. J Biol Chem. 1986 Sep 15;261(26):12310–12316. [PubMed] [Google Scholar]
- Thomas D. C., Kunkel T. A. Replication of UV-irradiated DNA in human cell extracts: evidence for mutagenic bypass of pyrimidine dimers. Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7744–7748. doi: 10.1073/pnas.90.16.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Functions of replication factor C and proliferating-cell nuclear antigen: functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991 Jan 25;266(3):1950–1960. [PubMed] [Google Scholar]
- Waga S., Hannon G. J., Beach D., Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994 Jun 16;369(6481):574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- Waga S., Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994 May 19;369(6477):207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Kiser G. L., Hartwell L. H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994 Mar 15;8(6):652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]


