Abstract
Background
National Surgical Adjuvant Breast and Bowel Project (NSABP) trial B-31 suggested the efficacy of adjuvant trastuzumab, even in HER2-negative breast cancer. This finding prompted us to develop a predictive model for degree of benefit from trastuzumab using archived tumor blocks from B-31.
Methods
Case subjects with tumor blocks were randomly divided into discovery (n = 588) and confirmation cohorts (n = 991). A predictive model was built from the discovery cohort through gene expression profiling of 462 genes with nCounter assay. A predefined cut point for the predictive model was tested in the confirmation cohort. Gene-by-treatment interaction was tested with Cox models, and correlations between variables were assessed with Spearman correlation. Principal component analysis was performed on the final set of selected genes. All statistical tests were two-sided.
Results
Eight predictive genes associated with HER2 (ERBB2, c17orf37, GRB7) or ER (ESR1, NAT1, GATA3, CA12, IGF1R) were selected for model building. Three-dimensional subset treatment effect pattern plot using two principal components of these genes was used to identify a subset with no benefit from trastuzumab, characterized by intermediate-level ERBB2 and high-level ESR1 mRNA expression. In the confirmation set, the predefined cut points for this model classified patients into three subsets with differential benefit from trastuzumab with hazard ratios of 1.58 (95% confidence interval [CI] = 0.67 to 3.69; P = .29; n = 100), 0.60 (95% CI = 0.41 to 0.89; P = .01; n = 449), and 0.28 (95% CI = 0.20 to 0.41; P < .001; n = 442; P interaction between the model and trastuzumab < .001).
Conclusions
We developed a gene expression–based predictive model for degree of benefit from trastuzumab and demonstrated that HER2-negative tumors belong to the moderate benefit group, thus providing justification for testing trastuzumab in HER2-negative patients (NSABP B-47).
Trastuzumab is a monoclonal antibody that is directed against HER2 protein overexpressed in approximately 20% of breast cancer patients with proven efficacy for both macro disease (metastatic and neoadjuvant setting) (1,2) and micrometastatic disease (adjuvant setting) (3,4).
The mechanisms responsible for trastuzumab response and resistance in adjuvant settings are not completely understood. National Surgical Adjuvant Breast and Bowel Project (NSABP) trial B-31 demonstrated the efficacy of adjuvant trastuzumab added to chemo-endocrine therapy not only for HER2-positive breast cancer but also, surprisingly, for HER2-negative breast cancer (4,5). Because HER2-positive tumors showed a high rate of pathologic complete response to neoadjuvant chemotherapy and complete responders tend to have favorable prognosis even without trastuzumab (6), in the adjuvant setting, where many patients may have already derived benefit from surgery and chemo-endocrine therapy, benefit from addition of trastuzumab could be determined through a complex interaction between HER2 and other confounding variables. In addition, more robust tumor cell response to trastuzumab could be expected in adjuvant vs advanced disease setting based on easier trastuzumab access to micrometastatic tumor cells (7), less compromised immune system favoring antibody-dependent cell-mediated cytotoxicity through trastuzumab (8), dependency of cancer stem cells on HER2 signaling pathway in the absence of HER2 overexpression (9,10), and overexpression of HER2 in bone metastasis in the absence of gene amplification (10).
Current clinical guidelines recommend that only HER2-positive patients be treated with trastuzumab. Because HER2 itself failed to show predictive interaction with trastuzmab in B-31, it is conceivable that not all HER2-positive patients receive benefit from trastuzumab while some HER2-negative patients may benefit. We attempted to use gene expression profiling of archived formalin-fixed paraffin-embedded tumor blocks from B-31 using the nCounter platform (NanoString Technologies, Seattle WA) (11) to develop a predictive model for the degree of benefit from adjuvant trastuzumab that would improve upon the current clinical guideline for trastuzumab treatment.
Methods
Study Design and Patient Cohort
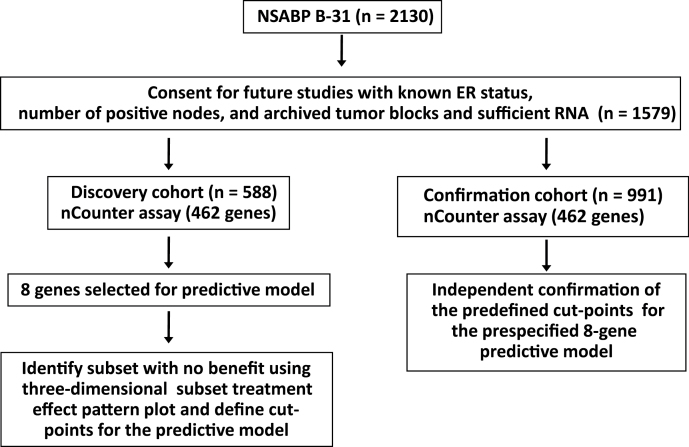
Among patients who participated in B-31 (n = 2130) (4), 1579 signed informed consent forms approved by a local Human Investigations Committee in accordance with assurances filed with and approved by the Department of Health and Human Services to permit use of banked tissue for future studies for cancer and clinical follow-up data (Figure 1). Information on estrogen receptor (ER) status and number of positive nodes was available for this cohort, as well as an adequate amount of extracted RNA for nCounter assays (see Supplementary Materials, File 1, available online).
Figure 1.
Study design and REMARK diagram: National Surgical Adjuvant Breast and Bowel Project (NSABP) B-31. See Supplementary Materials, File 1 (available online) for additional detail. ER, estrogen receptor.
The available samples were divided into candidate discovery (n = 588) and confirmation cohorts (n = 991). There were no major differences in clinical and pathological features between the two cohorts, as shown in Table 1.
Table 1.
The clinical and pathological characteristics of candidate discovery and confirmation cohorts*
| Characteristic | Discovery set (n = 588) No. (%) | Confirmation set (n = 991) No. (%) | P |
|---|---|---|---|
| Nodal status | |||
| 1–3 positive | 329 (56.0) | 567 (57.2) | .80† |
| ≥ 10 positive | 83 (14.1) | 129 (13.0) | |
| 4–9 positive | 176 (29.9) | 295 (29.8) | |
| ER status | |||
| Negative | 273 (46.4) | 464 (46.8) | .92† |
| Positive | 315 (53.6) | 527 (53.2) | |
| Treatment | |||
| Chemotherapy | 297 (50.5) | 500 (50.5) | 1.00† |
| Chemotherapy plus trastuzumab | 291 (49.5) | 491 (49.5) | |
| DFS status | |||
| Censored | 423 (71.9) | 724 (73.1) | .91‡ |
| Event | 165 (28.1) | 267 (26.9) | |
| Tumor size (cm) | |||
| Mean (SD) | 2.85 (1.68) | 2.93 (1.75) | .35§ |
| HER2 status | |||
| Negative | 72 (12.2) | 107 (10.8) | |
| Positive | 516 (87.8) | 884 (89.2) | .42† |
* DFS = disease-free survival; ER = estrogen receptor; SD = standard deviation.
† χ2 test.
‡ Log-rank test.
§ t test.
Based on analysis of nCounter assay data from 588 case subjects from the candidate discovery cohort, we committed to a single predictive model and cut points for each of the categories with varying degrees of expected benefit from trastuzumab. We then assessed these prespecified cut points in the remaining 991 case subjects (confirmation cohort), whose samples were not used for the selection of genes for the predictive model.
nCounter Assay
The nCounter platform was used for gene expression profiling because it uses short hybridization sequences and requires no enzymatic reactions, making it ideal for RNA extracted from formalin-fixed paraffin-embedded tumor block samples, which are prone to degradation and chemical modification (11). However, because the number of genes that could be included in an nCounter assay was limited to less than 500 and because we had no idea as to which genes might be predictive of trastuzumab benefit in the adjuvant setting, we had to rely on microarray-based screening of the discovery cohort for initial candidate gene discovery (Supplementary Materials, File 1, available online). We custom designed the nCounter assay with 462 probes to include candidate prognostic and predictive genes using microarray data from the same candidate discovery cohort, prognostic genes from unpublished microarray data from NSABP trial B-27 (12), PAM 50 genes (13), Oncotype DX genes (14), and internal reference genes. Because design of the nCounter assay was based on analysis of microarray data using 3-year clinical follow-up data at the time of unblinding, whereas predictive model building with nCounter data was performed using 7-year follow-up data with two times the number of events, the original selection criteria became irrelevant. Therefore, we do not describe microarray data and candidate gene selection steps in this report (see Supplementary Materials File 1, available online).
One hundred nanograms of total RNA were used for the assay. The data for each tumor were normalized for technical variability with the sum of the positive controls inherent to nCounter assay and within-sample reference normalized with the geometric mean of four internal reference genes (ACTB, RPLP0, SNRP70, H2AFY). In detail, nCounter raw counts consisting of a separate file for each sample were compiled in R (Development Core Team, Statistical Computing, Vienna, Austria). The raw counts were first corrected for technical variability by normalizing to the sum of six synthetic positive controls, supplied by the manufacturer, which were spiked into the mix before hybridization. The median of these sums was approximately 16500 in the discovery set, so a technical normalization factor was generated for each sample: 16500/sum[POS_A(128) POS_B(32) POS_C(8) POS_D(2) POS_E(0.5) POS_F(0.125)]. Raw counts for each sample were multiplied by these factors. The data were then adjusted for biological variation by reference gene normalization. The median geometric mean of the four control genes (ACTB, H2AFY, RPLP0, and SNRP70) in the discovery cohort was approximately 6000, so a normalization factor for each sample was generated according to: 6000/geometric mean (ACTB, H2AFY, RPLP0, SNRP70). The technical normalized counts for each sample were multiplied by these factors. Finally, the data were log transformed. The constants from the discovery set (16 500 and 6000) were applied to the confirmation set.
An application for depositing the anonymized data file to the dbGap database together with clinical data has been submitted to the National Cancer Institute (NCI). Data will be submitted to dbGAP as soon as NCI issues a dbGAP accession number.
Statistical Analysis
We included follow-up information up to June 2010. Patients from the control arm who crossed over to receive trastuzumab were censored at the time of cross-over. The definition of the primary endpoint for this analysis (disease-free survival) was previously described (4). Disease-free survival events included local, regional, and distant recurrence; contralateral breast cancer, including ductal carcinoma in situ; other second primary cancers; and death before recurrence or a second primary cancer.
We categorized gene expression values into quartiles for screening possible predictive genes because many genes showed nonlinearity of their association with treatment effect upon initial review of the data. The gene-by-treatment interaction was tested in the Cox proportional hazard models using the cross-product term of indicator variables for trastuzumab treatment and each marker status with adjustment for nodal status. For single markers other than ER, analyses were adjusted for ER and nodal status. Correlations between variables were assessed with Spearman’s correlation coefficient (r).
The principal component analysis was performed on the final set of selected genes to determine the first two components that would capture most of the variation in the data. Once the two principal components were chosen, interactions between treatments and the first two principal components (PC1 and PC2) of the candidate predictive genes from nCounter assay were evaluated by the Cox model as well as by means of the nonparametric subpopulation treatment effect pattern plot (15), which was extended for three dimensions (see Supplementary Materials, File 2, available online, for detailed methods and code). The three-dimensional surface plot was drawn with spline interpolation to smooth the plot using S-PLUS version 8.1 (TIBCO Software, Palo Alto, CA). All statistical analyses were done with SAS version 9.2 (SAS Institute, Cary, NC). The proportional hazards assumption for the Cox model was assessed by using the method developed by Lin et al., which was implemented in PROC PHREG (SAS version 9.2) (16). The proportionality assumptions between two treatment groups adjusted for nodal status were satisfied both in the discovery and validation cohorts. All statistical tests were two-sided.
Results
Results of the nCounter Assay in the Candidate Discovery Cohort (n=588) and Development of a Prediction Model
Perhaps because of the confounding by adjuvant chemo-endocrine therapy, which is quite efficacious for HER2-positive breast cancer (6), we found that gene-by-trastuzumab interaction was nonlinear, making it difficult to build a predictive model based purely on statistical methods.
Initially we attempted to build a model by selecting genes strictly based on statistical criteria using gene-by-treatment interaction terms in Cox models and identifying genes by 10-fold jack-knifing and other criteria described in the Supplementary Materials, File 3 (available online). However, clustering of these or any other combination of genes selected purely based on statistical significance did not allow us to robustly identify clinically meaningful subsets with differential benefit from trastuzumab. In light of this failure, we decided to attempt a biological approach to identify subsets with differential benefit from trastuzumab.
From among all of the results of gene assessment we had performed, we noticed that the top predictive genes included several ER-associated genes—CA12 (mean P interaction = .006), GATA3 (P = .007), PIK3A (P = .04)—as well as genes from the HER2 amplicon—ERBB2 (P = .049) and C17orf37 (P = .04). Using this information and the facts that ER status has been associated with lower rates of complete pathological response in several published studies (2,17) and that HER2 (ERBB2) is the target for trastuzumab, we decided to select genes whose expression levels were correlated with ESR1 mRNA or with ERBB2 mRNA as the basis to develop a predictive model. The top genes correlated with ESR1 and ERBB2 are shown in Table 2. From this pool, eight genes met the criteria of a Spearman correlation coefficient greater than 0.7 and a minimum interaction P value less than .10. These genes included ESR1, NAT1, GATA3, CA12, IGFR1, ERBB2, c17orf37, and GRB7.
Table 2.
Top genes correlated with ERBB2 and ESR1 and their minimum, two-sided P interaction values for trastuzumab when examined as a categorical variable (quartiles)
| Gene symbol | Correlation with ERBB2 | Minimum P intercation |
|---|---|---|
| ERBB2* | 1 | .03 |
| GRB7* | 0.912 | .06 |
| C17orf37* | 0.833 | .0003 |
| KRT7 | 0.498 | .047 |
| TMEM45B | 0.453 | .29 |
| ORMDL3 | 0.448 | .08 |
| C1orf93 | 0.427 | .10 |
| SPDEF | 0.4 | .01 |
| VEGFA | 0.395 | .24 |
| FGFR4 | 0.347 | .35 |
| Correlation with ESR1 | ||
| ESR1* | 1 | .06 |
| TBC1D9 | 0.757 | .49 |
| CA12* | 0.733 | .002 |
| IGF1R* | 0.731 | .04 |
| GATA3* | 0.727 | .004 |
| THSD4 | 0.727 | .12 |
| NAT1* | 0.701 | .08 |
| SLC39A6 | 0.685 | .21 |
| SCUBE2 | 0.637 | .47 |
| SIAH2 | 0.632 | .19 |
* Genes selected for prediction model.
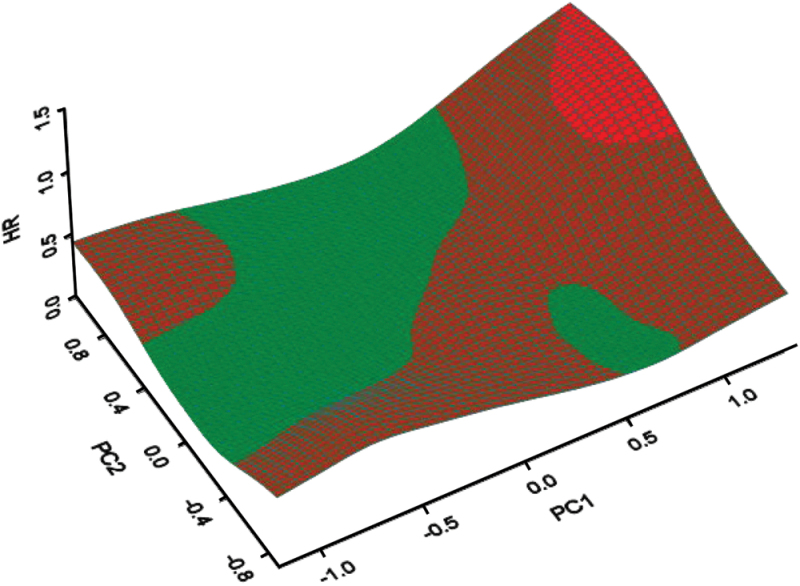
In a principal component analysis, the first two principal components of these genes accounted for 78.6% of the total variance (Supplementary Materials, File 3, available online). To identify subsets with different degree of benefit from trastuzumab while accommodating the nonlinearity of interaction between genes and trastuzumab, we used the first two principal components (PC1 and PC2) obtained from the eight selected predictive genes to create a three-dimensional subset treatment effect pattern plot with spline interpolation to smooth the plot with hazard ratio (HR) for trastuzumab on the Z-axis (Figure 2). Hazard ratios were color coded as green if equal to or less than 0.5 (large benefit from trastuzumab), brown if 0.5 to 1.0 (moderate benefit), or red if equal to or more than1.0 (no benefit). This plot readily identified subsets with differential benefit from trastuzumab. We derived cut points for two principal components (PC1 and PC2), which defined three subsets based on three-dimensional subset treatment effect pattern plot and the event rate in each subgroup. To derive best cut points, we moved cut points back and forth and checked the goodness of fit and discrimination when we applied internal cross-validation to the discovery set. We ignored the small green region in Figure 2 because it seemed that this region showed green because of error in smoothing.
Figure 2.
Three-dimensional subset treatment effect pattern plot analysis of candidate discovery set (n = 588). Cut points were determined by moving the cut points back and forth and checking the goodness of fit and discrimination by applying internal cross-validation to the discovery set. The cut points for two principal components of the eight predictive genes (PC1 and PC2) that defined the three subsets were determined as follows: no benefit group if PC1 > 0.6 and PC2 > 0.1; large benefit group if −0.12 < PC1 ≤ 0.6 and 0.1 < PC2 ≤ 0.6 and PC2 > PC1 + 0.22, if −0.6 < PC1 ≤ 0.6 and PC2 ≥ 0.6, or if PC1 ≤ −0.12 and −0.55 <PC2 < 0.6. Remaining patients were classified as the moderate benefit group. Red indicates case subjects no benefit from trastuzumab (hazard ratio [HR] ≥1). Brown indicates case subjects with moderate benefit (0.5 < HR < 1). Green indicates case subjects with large benefit (HR ≤ 0.5).
The cut points for two principal components (PC1 and PC2) that defined these three subsets were determined as follows: no benefit group with HR of 1.56 (if PC1 > 0.6 and PC2 > 0.1); large benefit group with hazard ratio of 0.27 (if −0.12 < PC1 ≤ 0.6 and 0.1 < PC2 ≤ 0.6 and PC2 > PC1 + 0.22, if − 0.6 <PC1 ≤ 0.6 and PC2 ≥ 0.6, or if PC1 ≤ −0.12 and −0.55 < PC2 < 0.6). Remaining patients were classified as the moderate benefit group with hazard ratio of 0.56 (see Supplementary Materials, File 3, available online, for Kaplan–Meier plots of the discovery cohort based on these cut points).
Assessment of the Predefined Cut Points for the Prediction Model in the Confirmation Cohort
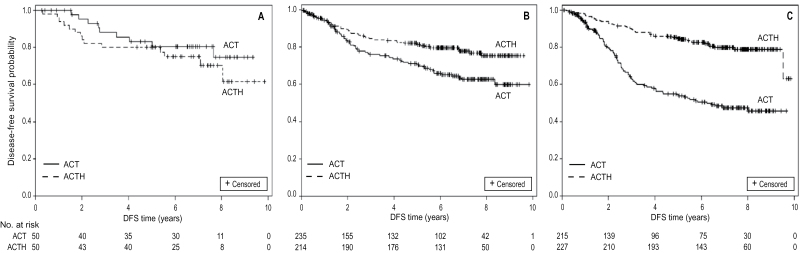
We assessed the predefined cut points from the eight-gene prediction model described above in the confirmation cohort (n = 991) B-31 patients not included in the discovery phase for whom specimens were available. Because the predictive model has not yet been developed into a formal clinical test, we did not develop a formal NCI registered date-stamped protocol before proceeding to the cut points assessment. We created Kaplan–Meier plots based on the predefined cut point values for the two principal components created by applying the eigenvector coefficients from the candidate discovery set to the confirmation dataset. As shown in Figure 3, A–C, applying the predefined cut points for the 8-gene prediction model readily identified the following: a subset with no benefit from trastuzumab (Group 1) with hazard ratio of 1.58 (95% confidence interval [CI] = 0.67 to 3.69; P = .29; n = 100) (Figure 3A); a subset with moderate benefit (Group 2) with hazard ratio of 0.60 (95% CI = 0.41 to 0.89; P = .01; n = 449) (Figure 3B); and a subset with large benefit (Group 3) with hazard ratio of 0.28 (95% CI = 0.20 to 0.41; P < .001; n = 442) (Figure 3C). The P value for the interaction between predictive algorithm and trastuzumab was <.001.
Figure 3.
Confirmation of predictive model and its cut points (n = 991). A) Disease-free survival (DFS) of patients treated with chemo-endocrine therapy (adriamycin cyclophosphamide followed by taxol [ACT]; solid line) vs those treated with trastuzumab added to chemo-endocrine therapy (ACT + herceptin [ACTH]; dashed line) among the no-benefit subgroup (n = 100) identified using the cut point from the candidate discovery set. Hazard ratio for trastuzumab was 1.58 (95% confidence interval [CI] = 0.67 to 3.69; P = .29 by Kaplan–Meier analysis). All statistical tests were two-sided. B) DFS of patients treated with chemo-endocrine therapy (ACT; solid line) vs those treated with trastuzumab added to chemo-endocrine therapy (ACTH; dashed line) among the moderate-benefit subgroup (n = 449) identified using the cut point from the candidate discovery set. Hazard ratio for trastuzumab was 0.60 (95% CI = 0.41 to 0.89; P = .01 by Kaplan–Meier analysis). All statistical tests were two-sided. C) DFS of patients treated with chemo-endocrine therapy (ACT; solid line) vs those treated with trastuzumab added to chemo-endocrine therapy (ACTH; dashed line) among the large-benefit subgroup (n = 442) identified using the cut point from the candidate discovery set. Hazard ratio for trastuzumab was 0.28 (95% CI = 0.20 to 0.41; P < .001 by Kaplan–Meier analysis). All statistical tests were two-sided.
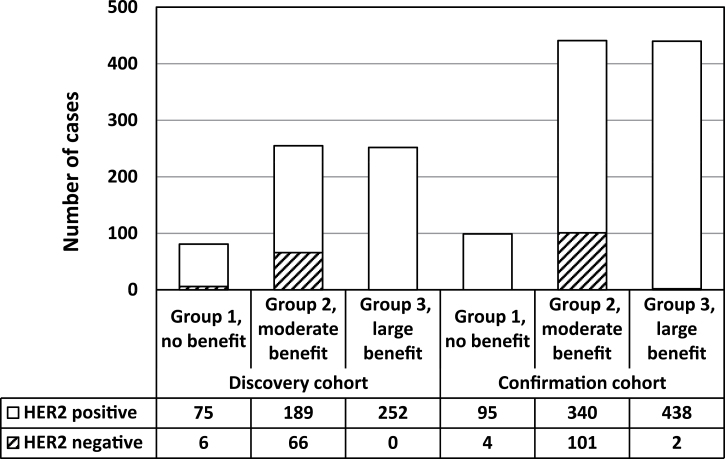
Distribution of Central HER2 Assay Negative Cases among Categories Defined by the Prediction Model
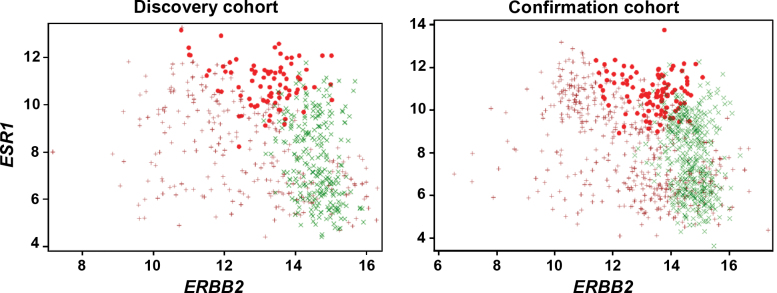
Because HER2 is the target for trastuzumab, it was expected that Group 1 with no benefit should express the lowest levels of ERBB2 mRNA. Figure 4 shows the result of a correlation analysis between ERBB2 and ESR1 mRNA levels in which each subgroup defined by the eight-gene prediction model is color coded. Surprisingly, the subset with no benefit expressed high levels of ESR1 mRNA and intermediate (but overexpressed) levels of ERBB2 mRNA rather than the lowest levels in both candidate discovery and confirmation cohorts (Figure 4).
Figure 4.
Nonlinear interaction between expression levels of ESR1 and ERBB2 and trastuzumab benefit. Tumors from patients with no benefit expressed moderate levels of ERBB2 mRNA and high levels of ESR1 mRNA. Red circles indicate Group 1, no benefit; brown crosses indicate Group 2, moderate benefit; Green Xs indicate Group 3, large benefit.
Previously we have reported an unexpected finding from the B-31 trial that central HER2 assay–negative patients also derived benefit from trastuzumab (5). Because the eight-gene prediction model was developed independently of the knowledge of centrally performed HER2 testing results, we tested whether central HER2 assay–negative cases belong to Group 1 defined by the predictive model as having no expected benefit. When central HER2-negative results were overlaid on these subsets, only a few HER2-negative patients belonged to the subgroup with no benefit, whereas a majority belonged to the moderate-benefit subgroup (Figure 5).
Figure 5.
HER2-negative tumors belonging to the moderate-benefit group rather than no-benefit group. Distribution of HER2 FISH–positive (blank) and –negative (diagonal lines) cases according to trastuzumab benefit group.
These results support the hypothesis that HER2-negative patients may derive benefit from trastuzumab.
Discussion
Using multiplexed gene expression profiling with RNA extracted from archived formalin-fixed paraffin-embedded tumor blocks from NSABP trial B-31, we were able to develop a predictive algorithm for the degree of benefit from trastuzumab added to adjuvant chemo-endocrine therapy of HER2-positive breast cancer. In the internal confirmation set of 991 patients, this algorithm and predefined cutpoints were confirmed with interaction P value <.001. The model identified approximately 10% of the clinically HER2-positive patients who may not benefit from adding trastuzumab to adjuvant chemo-endocrine therapy.
Our data demonstrate a complex relationship between HER2 and ER as determinants of clinical benefit from trastuzumab added to adjuvant chemo-endocrine therapy. ERBB2 mRNA–by-trastuzumab interaction was not linear and was also modulated by other genes, especially those from the ER pathway. Most surprisingly, the identified subgroup with no clinical benefit from adjuvant trastuzumab actually expressed intermediately overexpressed—not the lowest—levels of ERBB2 mRNA, together with the highest levels of ESR1-associated genes. This subgroup also had an excellent baseline prognosis, which was similar to the prognosis of others treated with trastuzumab.
There could be two explanations for the lack of benefit in this subgroup. In NSABP trial B-14, we observed that ESR1 mRNA level is a linear predictor of the degree of benefit from tamoxifen (18). Therefore, one explanation may be that patients with tumors that express the highest levels of ESR1 and its associated mRNAs may have already derived maximum clinical benefit from antiestrogen therapy. An alternative explanation is that such tumors are biologically resistant to trastuzumab. A lower rate of complete pathological response to neoadjuvant trastuzumab in ER-positive tumors compared with ER-negative tumors supports the second interpretation (2,17). It is possible that ER is directly responsible by inducing antiapoptotic proteins such as Bcl-2 or IGF1R. Overexpressed IGF1R can heterotrimerize with HER2 and EGFR and cause resistance to trastuzumab in vitro and in vivo (19,20). In reality, because of a close association of expression levels among these genes, it is impossible to separate them.
Regardless of the mechanisms responsible for no clinical benefit, therapeutic strategies to improve the outcome of this subgroup need to be developed because, although their prognosis is favorable, patients still suffer from greater than 10% recurrence rates in 5 years, which is not improved by the addition of trastuzumab. A combination of HER2, ER, and IGF1R targeting, HER2 targeting combined with complete blockage of ER pathway using fulvestrant (because IGF1R is induced by ER) (21), or a SRC inhibitor (22) may be potential strategies.
There are some limitations in our study. Because the assays were performed using research-grade nCounter reagents, the commercial-grade assay needs to be developed and analytically validated for our results to be clinically applied. It will be also prudent to validate our findings in an independent, randomized clinical trial for trastuzumab using the analytically validated assay.
Our data support the hypothesis based on central HER2 testing results from B-31 that HER2-negative patients may benefit from adjuvant trastuzumab (5). Because HER2-negative patients belong to Group 2, approximately 40% reduction in recurrences is expected from the addition of trastuzumab to adjuvant chemotherapy with minor side effects. This hypothesis is currently being tested through a randomized clinical trial (NSABP protocol B-47: NCT01275677).
Funding
This work was supported by the National Cancer Institute, Department of Health and Human Services, Public Health Service (grants U10-CA-12027, U10-CA-69651, U10-CA-37377, and U10-CA-69974) and by a grant from the Pennsylvania Department of Health. The latter Department specifically disclaims responsibility for any analysis, interpretations, or conclusions.
Supplementary Material
KL Pogue-Geile and C. Kim contributed equally in design, conduct, and analyses of the data.
The study sponsor played no role in the design, collection of data, analysis or interpretation of the study, nor in the writing of the manuscript nor in the decision to submit the manuscript for publication.
We thank Melanie Finnigan and Melanie Prior for data and tissue block management; William J. Hiller and Theresa A. Oeler for histology; Teresa L. Bradley, PhD, Ethan Barry, and Joyce Mull for regulatory affairs; Barbara Harkins and Frances Fonzi for protocol development; Barbara C. Good, PhD, and Wendy L. Rea for manuscript editing and submission; and Christine I. Rudock for graphics. We thank Drs Jo Anne Jujewski, Lisa McShane, and Jeffrey Abrams from CTEP/NCI for independent review of the study results. We also thank NSABP members who contributed tissue blocks and patients enrolled in the study.
Clinical Trials Registration: NSABP B-31: NCT00004067.
References
- 1. Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. [DOI] [PubMed] [Google Scholar]
- 2. Untch M, Rezai M, Loibl S, et al. Neoadjuvant treatment with trastuzumab in HER2-positive breast cancer: results from the GeparQuattro study. J Clin Oncol. 2010;28(12):2024–2031. [DOI] [PubMed] [Google Scholar]
- 3. Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353(16):1659–1672. [DOI] [PubMed] [Google Scholar]
- 4. Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353(16):1673–1684. [DOI] [PubMed] [Google Scholar]
- 5. Paik S, Kim C, Wolmark N. HER2 status and benefit from adjuvant trastuzumab in breast cancer. N Engl J Med. 2008;358(3):1409–1411. [DOI] [PubMed] [Google Scholar]
- 6. Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13(8):2329–2334. [DOI] [PubMed] [Google Scholar]
- 7. Barok M, Isola J, Palyi-Krekk Z, et al. Trastuzumab causes antibody-dependent cellular cytotoxicity-mediated growth inhibition of submacroscopic JIMT-1 breast cancer xenografts despite intrinsic drug resistance. Mol Cancer Ther. 2007;6(7):2065–2072. [DOI] [PubMed] [Google Scholar]
- 8. Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytoxicity against tumor targets. Nat Med. 2000;6(4):443–446. [DOI] [PubMed] [Google Scholar]
- 9. Nakanishi T, Chumsri S, Khakpour N, et al. Side-population cells in luminal-type breast cancer have tumour-initiating cell properties, and are regulated by HER2 expression and signalling. Br J Cancer. 2010;102(5): 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ithimakin S, Day KC, Malik F, et al. HER2 drives luminal breast cancer stem cells in the absence of HER2 amplification: implications for efficacy of adjuvant trastuzumab. Cancer Res. 2012;73(5):1635–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. [DOI] [PubMed] [Google Scholar]
- 12. Bear HD, Anderson S, Smith RE, et al. Sequential preoperative or postoperative docetaxel added to preoperative doxorubicin plus cyclophosphamide for operable breast cancer: National Surgical Adjuvant Breast and Bowel Project Protocol B-27. J Clin Oncol. 2006;24(13):2019–2027. [DOI] [PubMed] [Google Scholar]
- 13. Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. [DOI] [PubMed] [Google Scholar]
- 15. Bonetti M, Gelber RD. Patterns of treatment effects in subsets of patients in clinical trials. Biostatistics (Oxford, England). 2004;5(3):465–481. [DOI] [PubMed] [Google Scholar]
- 16. Lin DY, Wei LJ, Ying Z. Checking the Cox model with cumulative sums of Martingale-based residuals. Biometrika. 1993;80(3):557–572. [Google Scholar]
- 17. Bhargava R, Dabbs DJ, Beriwal S, et al. Semiquantitative hormone receptor level influences response to trastuzumab-containing neoadjuvant chemotherapy in HER2-positive breast cancer. Mod Pathol. 2011;24(3):367–374. [DOI] [PubMed] [Google Scholar]
- 18. Kim C, Tang G, Pogue-Geile KL, et al. Estrogen receptor (ESR1) mRNA expression and benefit from tamoxifen in the treatment and prevention of estrogen receptor-positive breast cancer. J Clin Oncol. 2011;29(31):4160–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang X, Gao L, Wang S, et al. Heterotrimerization of the growth factor receptors erbB2, erbB3, and insulin-like growth factor-I receptor in breast cancer cells resistant to herceptin. Cancer Res. 2010;70(3):1204–1214. [DOI] [PubMed] [Google Scholar]
- 20. Lu Y, Zi X, Zhao Y, Mascarenhas D, Pollak M. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (herceptin). J Natl Cancer Inst. 2001;93(24):1852–1857. [DOI] [PubMed] [Google Scholar]
- 21. Osborne CK, Wakeling A, Nicholson RI. Fulvestrant: an oestrogen receptor antagonist with a novel mechanism of action. Br J Cancer. 2004;90(Suppl 1):S2–S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang S, Huang WC, Li P, et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med. 2011;17(4):461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.