Abstract
MicroRNAs (miRNAs) are small non-coding RNA molecules, whose primary function is to regulate gene expression at the post-transcriptional/translational levels. MiRNAs play crucial roles in normal biological processes and are commonly dys-regulated in human diseases. Stem cells are regarded as the “mother” cells of all types of differentiated cells that comprise tissues and organs of the body. A novel hypothesis proposes that tumors are composed of heterogeneous cells derived from cancer stem cells, which have self-renewal and differentiation capabilities similar to those of normal stem cells. Cancer stem cells have been isolated and characterized from various tumors. Given recent studies supporting the critical regulatory roles of miRNAs in the self-renewal and differentiation of cancer stem cells, better understanding the functions of miRNAs will provide invaluable insights into the prevention of tumorigenesis and tumor progression. In this review, we will summarize the research progress in the study of miRNAs involved in the self-renewal and differentiation of cancer stem cells.
Keywords: cancer, stem cells, miRNAs, tumorigenesis, tumor progression, self-renewal, differentiation
Introduction
Stem cell research can be dated back to at least the 1960s, when Becker et al illustrated the presence of self-renewing cells in mouse bone marrow1. In 1998, Thomson et al successfully isolated and cultured human embryonic stem (ES) cells for the first time2; this work is considered to be a milestone study in human stem cell research. The concept that cancer might arise from a rare population of cells with stem cell-like properties was proposed more than 150 years ago3. However, cancer stem cells (CSCs) were not confirmed to exist until they were discovered in acute myeloid leukemia (AML) in 19974. CSCs have since been identified in most types of solid tumors5.
MicroRNAs (miRNAs) are non-coding regulatory RNA molecules that are approximately 22 nucleotides long6,7. After transcribed from the miRNA genes aided by RNA polymerase II or III8,9, pri-miRNAs with the hairpin structure are processed by Drosha and DiGeorge syndrome critical region gene 8 (DGCR8) to form pre-miRNAs, which are then exported out of the nucleus by Exportin-510,11,12. In the cytoplasm, pre-miRNAs are subsequently cleaved into mature miRNA sequences by Dicer13,14,15,16,17. By incorporating with the RNA-induced silencing complex (RISC), miRNAs exert the repressive function through the translational repression of target genes and/or mediation of the target mRNA transcripts cleavage18,19. Although the extent to which miRNAs regulate the human transcriptome has not yet been fully determined, increasing evidence now supports the crucial role of miRNAs in the regulation of gene expression.
In stem cell research, relatively fewer studies have examined non-coding miRNAs than protein-coding genes. Given the recent studies reporting that miRNAs play significant roles in the maintenance of stem cells in various cancers20, we will summarize the research on miRNAs in CSCs, focusing on the processes of self-renewal and differentiation.
Cancer stem cells
CSCs, also called tumor-initiating cells, have been identified in various types of cancers4,5. CSCs have the capacity to self-renew and produce the heterogeneous lineages of cancer cells that comprise the tumor21. Recent studies have found that CSCs account for resistance to chemotherapy in certain cancers, providing a novel insight into the mechanistic basis of chemoresistance22.
In 1997, Bonnet et al first isolated and identified CSCs from AML4, while subsequent studies found that solid tumors, including breast cancer, pancreatic cancer, colon cancer, brain cancer, liver cancer, head and neck cancer, ovarian cancer, and melanoma are also driven and sustained by CSCs23,24,25,26,27,28,29,30,31,32,33,34,35,36,37. Generally speaking, CSCs are only responsible for a very small portion of all tumor cells, although the percentage may vary depending on the tumor type. For instance, the CD133+ CSCs account for approximately 2.5% of the population of colorectal cancer cells34. However, recent studies support that CSCs play significant roles in tumor relapse and metastasis because they can differentiate into each of the diverse cell types that comprise the tumor through continuous self-renewal and differentiation5. As such, a better understanding of the CSC theory will shed light on the biology of tumorigenesis and aid in the development of novel therapeutic strategies to treat human cancer more efficaciously.
CSCs show greater tumorigenic potential than non-stem cancer cells and express specific markers. In 2003, Al-Hajj et al isolated and characterized CSCs from breast cancer cells based on the expression status of the specific cell surface markers CD44 and CD24, and this study was the first report showing the success of isolating CSCs from solid tumors23. Thereafter, CD133, CD166, epithelial cell adhesion molecule (EpCAM), and others were also used as the surface markers to identify and characterize CSCs in different tumors, such as brain cancer, prostate cancer, pancreatic cancer, colon cancer, and hepatocellular carcinoma25,34,38,39,40. Aldehyde dehydrogenase 1 (ALDH) was recently reported as a potential breast cancer stem/progenitor cell-specific marker30. In addition to identification of different CSCs in human tumors, the usage of these markers has been extended to evaluate the efficacy of chemotherapeutic drugs with the potential to target CSCs as well41,42.
The origin of CSCs remains elusive, but several hypotheses have been proposed. The cell fusion and horizontal gene transfer occurring in cell development and tissue repair process are considered to be the dominant origins of CSCs, although another opinion disputed that CSCs might arise from mutations in specific normal stem cells or early stem cell progenitors43. Interestingly, CSCs are also reported to be derived even from differentiated tumor cells in accordance to the report by Iliopoulos et al; they found that interleukin 6 (IL6) can convert non-stem cells to CSCs in breast and prostate cancer cell lines and in primary cells derived from human breast tumors44.
Based on the ability of stem cells to grow in serum-free and non-adherent suspensions as spherical clusters, the tumorsphere culture technique has been developed to isolate and characterize CSCs45,46. However, the ideal assay for CSC characterization would be serial transplantation in animal models in which cells are xenografted into an orthotopic site of an immunocompromised mouse for observing tumor formation. Given that there is very few drugs available that specifically target the unique machinery driving the renewal and differentiation of CSCs, the study of miRNAs in CSCs may provide a valuable insight into the development of novel strategies against human cancers.
MiRNAs are involved in the self-renewal and differentiation of cancer stem cells
Although the mechanism by which stem cells maintain self-renewal and differentiation remains unclear, it was shown that altered miRNA accumulation in murine ES cells with conditional knockout of Dicer1 and DGCR8 led to abnormalities in stem cell differentiation, suggesting that miRNAs may play important roles in stem cells47,48. It was also reported that miR-134, miR-296, and miR-470 can directly inhibit the self-renewing state by suppressing several factors with the documented effects on pluripotency maintenance, such as Nanog, octamer-binding transcription factor 4 (Oct4), and sex determining region Y-box 2 (Sox2)49. In human ES cells, miR-145 can promote cell differentiation by directly targeting the mRNA transcripts of Oct4, Sox2, and kruppel-like factor 4 (KLF4)50, and let-7 can translationally repress the expression of Lin28, which is a known factor to maintain cell pluripotency51,52. In addition, miR-290 and miR-302a are reported to promote G1-S transition that enables cellular rapid proliferation in human ES cells53,54. In proliferating ventral midbrain/hindbrain (vMH) neural progenitors, miR-200 is required to promote cell cycle exit and neuronal differentiation by targeting the expression of Sox2 and E2F transcription factor 3 (E2F3)55. These findings notably suggest that miRNAs can act as the upstream regulators of a panel of transcription factors that are involved in modulation of stem cell self-renewal and differentiation, such as Oct4, Sox2, KLF4, and E2F3.
On the other hand, miRNAs can also be regulated by some transcription factors and serve as downstream effectors in the signaling pathways associated with stem cell self-renewal and differentiation. For example, Lin et al reported that in ES cells, the expression of the miR-200 family was regulated by c-Myc. The transcriptional induction of these miRNAs by c-Myc significantly attenuated the down-regulation of pluripotency markers, which indicates that in ES cells, c-Myc acts, at least in part, through the miR-200 family to attenuate differentiation56. In addition, Wang et al found that during the reprogramming of somatic cells, Oct4 and Sox2 can induce the transcriptional activation of the miR-200 family, which can in turn promote mesenchymal-epithelial transition (MET) and generation of the induced pluripotent stem cells (iPSCs) by targeting zinc finger E-box binding homeobox 2 (ZEB2)57. It is also notable that Oct4 and Sox2 can transcriptionally regulate the expression of miR-302a that is involved in the cell cycle progression in human ES cells by targeting cyclin D154.
Based on their roles, those functional miRNAs can be sorted to two subgroups: pluripotent miRNAs and pro-differentiation miRNAs. Pluripotent miRNAs are able to promote the self-renewal and proliferation of stem cells but inhibit cell differentiation. This class of miRNAs includes miR-137, miR-184, miR-200, miR-290, miR-302, and miR-954,56,58,59,60,61,62. The pro-differentiation miRNAs that can initiate or stabilize differentiation include let-7, miR-122, miR-134, miR-145, miR-181, miR-296, and miR-47049,50,52,63,64,65,66. These two types of miRNAs and their targets that have been validated to be involved in the self-renewal and differentiation of CSCs are summarized in Table 1. Our recent study reported that miR-181 direct downregulation of Lin28 can promote the megakaryocytic differentiation by disrupting the let-7/Lin28 negative feedback loop in which let-7 translationally suppresses the expression of Lin28, whereas Lin28 controls the maturation of let-752. However, a recent report found that the overexpression of miR-122 could promote hepatic differentiation and maturation in murine ES cells through a miR-122/forkhead box protein A1(FoxA1)/ hepatocyte nuclear factor 4 alpha (HNF4α) positive feedback loop66. Interestingly, FoxA1 and HNF4α are not directly putative target genes of miR-122 but both of them play crucial roles in promoting the differentiation of hepatocytes. FoxA1 can induce HNF4α that further enables upregulation of miR-122 through the transcriptional modulation; miR-122 can indirectly elevate the expression of FoxA1 but the mechanism of such an action is still uncovered. The positive regulatory effects on the interactions among miR-122, FoxA1, and HNF4α lead to hepatic differentiation and maturation unremittingly66. Likely, these results support that the regulatory circuits consisting of miRNAs and pluripotency factors can provide more useful insights into understanding the molecular mechanisms by which the cells maintain the balance between stemness and differentiation.
Table 1. MiRNAs involved in stem cell pluripotency maintenance and differentiation promotion.
| Subgroup |
miRNAs |
Validated target genes that are involved in the self-renewal and differentiation of CSCs |
| Pluripotent miRNAs | miR-137 | Mib1 |
| miR-184 | Numbl | |
| miR-200 | ZEB1; ZEB2 | |
| miR-290 | CDKN1a | |
| miR-302 | Cyclin D1; AOF1; AOF2; MECP1-p66; MECP2 | |
| miR-9 | Stathmin | |
| Pro-differentiation miRNAs | let-7 | Lin28; Lin28B; IMP-1; HRAS; HMGA2 |
| miR-122 | Not determined | |
| miR-134 | Nanog; LRH1; Sox2 | |
| miR-145 | Oct4; Sox2; KLF4 | |
| miR-181 | Lin28 | |
| miR-296 | Nanog | |
| miR-470 | Nanog; Oct4 |
Abbreviations: Mib1 (Mind bomb 1); Numbl (Numblike); ZEB (Zinc finger E-box binding homeobox); CDKN1a (Cyclin-dependent kinase inhibitor 1a); AOF1 (Lysine-specific demethylase 1B); AOF2 (Lysine-specific demethylase 1A); MECP1-p66 (Methyl CpG binding protein 1-p66 beta component); MECP2 (Methyl CpG binding protein 2); IMP-1 (Insulin-like growth factor 2 mRNA binding protein 1); HRAS (v-Ha-ras Harvey rat sarcoma viral oncogene homolog); HMGA2 (High mobility group AT-hook 2); LRH1 (Nuclear receptor subfamily 5, group A, member 2), Sox2 (Sex determining region Y-box 2); Oct 4 (Octamer-binding transcription factor 4); KLF4 (Kruppel-like factor 4).
The roles of miRNAs in various cancers have been examined in dozens of studies, but their functions in CSCs have not yet been observed intensively. The first study of miRNA expression in CSCs was carried out by Yu et al67, who found that several miRNAs appeared at lower levels in breast CSCs, including let-7, miR-200a/b/c, miR-16, miR-107, miR-128, and miR-20b67. Among these small RNA molecules, let-7 emerged as the most consistently and significantly reduced miRNA, suggesting that let-7 acts to suppress CSC self-renewal. Shimono et al later identified 37 miRNAs that were differentially expressed in breast CSCs; the miR-200 family and the miR-183-96-182 cluster were among those significantly downregulated68. Notably, all five members of the miR-200 family (miR-200a, -200b, -200c, -141, and -429) were downregulated in human breast cancer stem cells and in normal human and murine mammary stem/progenitor cells68.
The miRNA profiles in CSCs have been examined in various tumor types in addition to breast cancer. In glioblastoma multiforme, the levels of miRNAs including miR-451, miR-486, miR-425, miR-16, miR-103, miR-107, and miR-185 were decreased in the stem cell (CD133+) population compared to the non-stem (CD133-) cell populations69. The overexpression of miR-451 inhibited neurosphere formation and cell growth69. In hepatocellular carcinoma, CSCs show a unique miRNA signature characterized by the upregulation of miR-181a-1, miR-181a-2, miR-181b-1, miR-181b-2, miR-181c, miR-17, miR-20a, miR-25, miR-92, miR-93, and miR-106b70. The inhibition of miR-181 led to a reduction in the CSC (EpCAM+) cell number and tumor initiating ability, whereas the expression of exogenous miR-181 resulted in an enrichment of CSCs (EpCAM+)70. In prostate cancer stem or progenitor cell populations that are enriched for CD44, CD133, or α2β1, the expression of miR-34a was low71. However, the forced expression of miR-34a can obviously inhibit prostate cell proliferation, tumor regeneration and metastasis by directly repressing CD4471. In colon CSCs (CD133+), 11 miRNAs (miR-16-2*, miR-744, miR-185, miR-455-3p, miR-155, miR-455-5p, miR-105, miR-494, miR-1826, miR-423-5p, and miR-181b) were upregulated, whereas 8 miRNAs (miR-221, miR-548d-5p, miR-636, miR-31, miR-320d, miR-151-3p, miR-429, and miR-151-5p) were downregulated72. Nam et al evaluated the miRNA expression profiles of ovarian CSCs (CD133+) and found that 34 miRNAs were significantly upregulated and 3 miRNAs were downregulated73. These differentially expressed miRNAs in CSCs indicated the crucial regulatory roles of miRNA in CSCs biological processes (Table 2), while let-7 and miR-200 are noted to be the mostly studied miRNAs in CSCs.
Table 2. Aberrant expression of miRNAs in various human CSCs.
| CSC | Expression | miRNAs | References |
|---|---|---|---|
| Breast cancer | Down | let-7a, let-7b, let-7c, let-7d, let-7e, let-7g, let-7i, miR-103, miR-107, miR-10a, miR-128a, miR-128b, miR-130a, miR-138, miR-141, miR-15a, miR-15b, miR-16, miR-17, miR-181b, miR-182, miR-183, miR-193b, miR-196a, miR-200a, miR-200a*, miR-200b, miR-200c, miR-20b, miR-210, miR-215, miR-22, miR-96 | Yu et al67; Shimono et al68 |
| Up | miR-125b, miR-127, miR-132, miR-142-3p, miR-146b, miR-150, miR-155, miR-199a, miR-199a*, miR-199b, miR-212, miR-214, miR-221, miR-222, miR-223, miR-299-5p, miR-31, miR-409-3p, miR-432, miR-495 | Gal et al69 | |
| Glioblastoma | Down | miR-103, miR-107, miR-16, miR-185, miR-425-5p, miR-451, miR-486 | Ji et al70 |
| Up | N/A | ||
| Hepatic cancer | Down | N/A | |
| Up | miR-106b, miR-17, miR-181a, miR-181b, miR-181c, miR-20a, miR-25, miR-92, miR-93 | Liu et al71 | |
| Prostate cancer | Down | miR-34a | |
| Up | N/A | ||
| Ovarian cancer | Down | miR-1181, miR-1202, miR-1207-5p | Nam et al73 |
| Up | let-7f, miR-100, miR-107, miR-135b, miR-146a, miR-181a, miR-183, miR-193a-3p, miR-200a, miR-200b, miR-205, miR-21, miR-210, miR-26b, miR-29b, miR-33a, miR-34a, miR-340, miR-340*, miR-365, miR-424, miR-425, miR-449a, miR-455-3p, miR-494, miR-516a-5p, miR-517a, miR-517c, miR-522, miR-7, miR-886-3p, miR-96 | ||
| Colon cancer | Down | miR-151-3p, miR-151-5p, miR-221, miR-31, miR-320d, miR-429, miR-548d-5p, miR-636 | Zhang et al72 |
| Up | miR-105, miR-155, miR-16-2*, miR-181b, miR-1826, miR-185, miR-423-5p, miR-455-3p, miR-455-5p, miR-494, miR-744 |
Underlined miRNAs represent those miRNAs that show similar dysregulation (up or down) in more than one type of cancer stem cells.
As described above, the expression of let-7 was significantly reduced in breast CSCs compared to non-stem cancer cells67. The upregulation of let-7 in breast CSCs reduced proliferation, mammosphere formation, the proportion of undifferentiated cells in vitro, and tumor formation and metastasis in vivo, while the downregulation of let-7 enhanced the in vitro self-renewal of non-stem cancer cells67. Further research indicated that let-7 targets v-Ha-ras Harvey rat sarcoma viral oncogene homolog (HRAS) and high mobility group AT-hook 2 (HMGA2); the silencing of HRAS in breast CSCs reduced self-renewal with little effect on differentiation, whereas the silencing of HMGA2 enhanced differentiation but not self-renewal67.
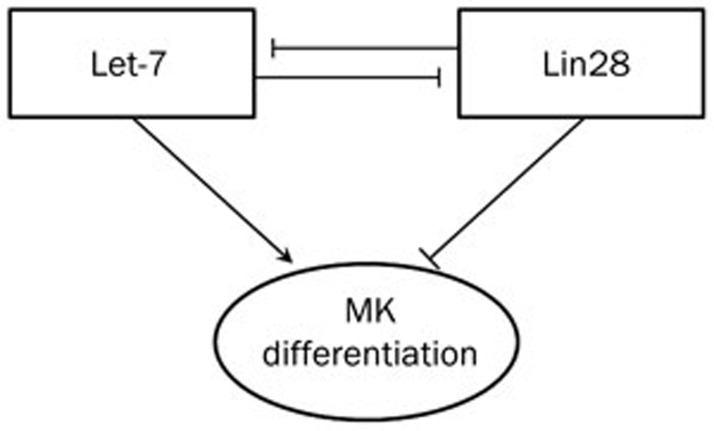
In one of our recent studies, we investigated the mechanism by which let-7 regulates cell differentiation using bipotent K562 human leukemia cells and human CD34+ hematopoietic progenitor cells as research models52. We found that let-7 and Lin28 appear to play contrary roles in megakaryocytic (MK) differentiation and maintain a dynamic balance through a reciprocal regulatory loop (Figure 1). As discussed earlier, Lin28 is one of the direct targets of let-7 and can also influence the biogenesis of let-7 by recruiting terminal uridylyl transferase-4 (TUT4) to add a uracil residue to the 3′ end of pre-let-7; this modification results in the degradation of pre-let-7 and a blockade of let-7 maturation74,75. Interestingly, when miR-181 is introduced to translationally downregulated Lin28, the let-7/Lin28 loop is disrupted and let-7 expression is thereby induced that lead to the promotion of MK differentiation. Our results are consistent with the observation that miRNAs play important roles in the control of cell differentiation.
Figure 1.
The reciprocal regulatory loop of let-7 and Lin28 in the control of megakaryocytic (MK) differentiation.
The first report of miR-200 in stem cells was published in Cell in 200968. In this study, the authors found that all the miR-200 family members (miR-200a, -200b, -200c, -141, and -429) were downregulated in breast CSCs compared to non-stem cancer cells. By targeting BMI1 polycomb ring finger oncogene (BMI1), miR-200c can inhibit the clonal expansion of breast cancer cells and suppress the growth of embryonal carcinoma cells in vitro. Moreover, miR-200c can strongly suppress tumor formation driven by breast CSCs in vivo68. In support of these results, Iliopoulos et al reported that miR-200b can suppress the formation and maintenance of mammospheres in vivo, which may, at least in part, attribute to the repression of the target gene named suppressor of zeste 12 homolog (Suz12)76. Moreover, Lim et al reported that the conversion of immortalized human mammary epithelial cells from a non-stem phenotype to a stem-like phenotype was accompanied by the loss of miR-200 expression. The restoration of miR-200 expression in these cells decreased their stem-like properties while promoting their transition to an epithelial phenotype, suggesting a negative role of miR-200 in CSC tumorigenesis77.
Conclusion and perspective
MiRNAs, as the master post-transcriptional and translational regulators on gene expression, have been reported to play important roles in stem cells and tumorigenesis. CSCs are now believed to be responsible for tumor relapse and chemotherapy failure in many cancers. Recent studies show that miRNAs are significantly involved in the CSC self-renewal and differentiation. Given that the dysregulation of miRNAs has been intimately implicated in tumor development, the modulation of CSC properties may contribute to the underlying mechanisms by which miRNAs regulate tumorigenesis. For examples, let-7 controls the cell cycle progression and differentiation of CSCs, miR-200c modulates the self-renewal of CSCs by targeting BMI1, and miR-34a restricts the migratory and invasive properties of prostate CSCs by directly repressing CD44, which have been discussed earlier in details. These findings support the crucial roles of miRNAs in the regulation of CSCs. As such, further studies on this topic are expected to provide more insights into our understanding of tumorigenesis and aid in the development of new strategies against chemoresistance by targeting CSCs.
Acknowledgments
This study is supported by an American Cancer Society Research Scholar Grant (RSG-13-265-01-RMC, to Yaguang XI) and an NIH/NCI R21 Grant (5R21CA160280, to Yaguang XI). We sincerely apologize to those colleagues whose work have not be cited due to time and space constraints.
References
- Becker AJ, Mc CE, Till JE. Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature. 1963;197:452–4. doi: 10.1038/197452a0. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz–Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea — a paradigm shift. Cancer Res. 2006;66:1883–90. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–6. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–9. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–71. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–8. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Liu C, Tang DG. MicroRNA regulation of cancer stem cells. Cancer Res. 2011;71:5950–4. doi: 10.1158/0008-5472.CAN-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia MHP. Great potential of microRNA in cancer stem cell. J Cancer Mol. 2008;4:79–89. [Google Scholar]
- Sakariassen PO, Immervoll H, Chekenya M. Cancer stem cells as mediators of treatment resistance in brain tumors: status and controversies. Neoplasia. 2007;9:882–92. doi: 10.1593/neo.07658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–8. [PubMed] [Google Scholar]
- O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–9. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–66. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Bednar F, Simeone DM. Pancreatic cancer stem cells and relevance to cancer treatments. J Cell Biochem. 2009;107:40–5. doi: 10.1002/jcb.22093. [DOI] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–52. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapat SA, Mali AM, Koppikar CB, Kurrey NK. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65:3025–9. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–37. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–51. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–56. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 2009;69:7507–11. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang T, Korkaya H, Liu S, Lee HF, Newman B, et al. Sulforaphane, a dietary component of broccoli/broccoli sprouts, inhibits breast cancer stem cells. Clin Cancer Res. 2010;16:2580–90. doi: 10.1158/1078-0432.CCR-09-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Y, Cao D. The origin of cancer stem cells. Front Biosci (Schol Ed) 2012;4:819–30. doi: 10.2741/s302. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci U S A. 2011;108:1397–402. doi: 10.1073/pnas.1018898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–83. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–5. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–52. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang J, Gao L, McClellan S, Finan MA, Butler TW, et al. MiR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell Death Differ. 2012;19:378–86. doi: 10.1038/cdd.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–54. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–38. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Li N, Ng YK, Zhang J, Meier F, Theis FJ, et al. A unilateral negative feedback loop between miR-200 microRNAs and Sox2/E2F3 controls neural progenitor cell-cycle exit and differentiation. J Neurosci. 2012;32:13292–308. doi: 10.1523/JNEUROSCI.2124-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28:3157–70. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Guo X, Hong W, Liu Q, Wei T, Lu C, et al. Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc Natl Acad Sci U S A. 2013;110:2858–63. doi: 10.1073/pnas.1212769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Blelloch R. MicroRNA regulation of embryonic stem cell self-renewal and differentiation. Adv Exp Med Biol. 2010;695:105–17. doi: 10.1007/978-1-4419-7037-4_8. [DOI] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–96. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, et al. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–35. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–70. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1–S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–83. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–6. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–57. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- Deng XG, Qiu RL, Wu YH, Li ZX, Xie P, Zhang J, et al. Overexpression of miR-122 promotes the hepatic differentiation and maturation of mouse ESCs through a miR-122/FoxA1/HNF4a-positive feedback loop Liver Int 2013 Jul 5. doi: 10.1111/liv.12239 [DOI] [PubMed]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal H, Pandi G, Kanner AA, Ram Z, Lithwick-Yanai G, Amariglio N, et al. MIR-451 and imatinib mesylate inhibit tumor growth of glioblastoma stem cells. Biochem Biophys Res Commun. 2008;376:86–90. doi: 10.1016/j.bbrc.2008.08.107. [DOI] [PubMed] [Google Scholar]
- Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–80. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–5. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li W, Nan F, Ren F, Wang H, Xu Y, et al. MicroRNA expression profile of colon cancer stem-like cells in HT29 adenocarcinoma cell line. Biochem Biophys Res Commun. 2011;404:273–8. doi: 10.1016/j.bbrc.2010.11.106. [DOI] [PubMed] [Google Scholar]
- Nam EJ, Lee M, Yim GW, Kim JH, Kim S, Kim SW, et al. MicroRNA profiling of a CD133(+) spheroid-forming subpopulation of the OVCAR3 human ovarian cancer cell line. BMC Med Genomics. 2012;5:18. doi: 10.1186/1755-8794-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–5. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–72. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YY, Wright JA, Attema JL, Gregory PA, Bert AG, Smith E, et al. Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J Cell Sci. 2013;126:2256–66. doi: 10.1242/jcs.122275. [DOI] [PubMed] [Google Scholar]