Abstract
In bacteria, stalled ribosomes are rescued by transfer-mRNA (tmRNA) that catalyzes two steps. First, a non-encoded alanine is added to the incomplete polypeptide chain by the tRNAAla-like portion of tmRNA, and second, the ribosome switches to the mRNA-like domain of tmRNA, thus resuming protein synthesis. Mitochondrial DNA (mtDNA)-encoded mt-tmRNA is so far only known from jakobid protists, but we posit that the corresponding ssrA gene may also reside in other mtDNAs. Here we present a highly sensitive covariance model built from jakobid ssrA genes that identifies previously unrecognized ssrA homologs in mtDNAs of oomycetes. These genes, located in previously unassigned genomic regions, are circular permuted as in α-Protobacteria, implying that pre-tmRNA is processed and the two pieces are held together by non-covalent interactions. RNA-Seq data from Phytophthora sojae confirm predicted processing sites as well as post-transcriptional addition of 3′ CCA, a prerequisite for tmRNAs to be charged with alanine by alanyl-tRNA synthetase. Structure modeling of oomycete tmRNAs infers that the mRNA-like domain is lacking as in jakobids. Features of mitochondrial tmRNAs include the G-U pair at position three of the acceptor stem, a hallmark of bacterial tmRNAs, and a T-loop sequence that differs from that of standard tRNAs and most bacterial tmRNAs, forming alternative, virtually isosteric tertiary interactions with the D-loop. The anticodon stem has two additional G-A base pairs formed between the D-loop and the variable region, shortening the length of the variable region to a single nucleotide.
Keywords: Phytophthora, RNA-Seq analysis, covariance model, oomycetes, post-transcriptional modification, tmRNA processing, tmRNA tertiary structure
Introduction
During translation, amino acids are incorporated into the growing polypeptide chain until the last sense codon of the mRNA is reached. At this point, the P-site of the ribosome is occupied with a peptidyl-tRNA (attached to a polypeptide), while the empty A-site is opposite to the stop codon of the mRNA. In contrast to sense codons that are decoded by an elongation factor-tRNA complex, stop codons interact with a release factor to liberate the ribosome from polypeptide and mRNA. Therefore, mRNAs lacking a stop codon (e.g., due to RNA degradation) would cause stalling of translation, thus reducing the number of operational ribosomes in the cell. In bacteria, the deleterious effect of mRNAs without stop codon (“non-stop” mRNAs) is alleviated by transfer-mRNA (tmRNA, also termed 10Sa RNA or SsrA; for a review, see ref. 1), a peculiar molecule combining a tRNAAla-like domain with a short, protein-coding (mRNA-like) region.2-4
The tRNAAla-like domain of bacterial tmRNA allows the entry of the alanine-charged tmRNA into the A-site of a stalled ribosome (for addition of a non-encoded alanine), whereas the mRNA-like region allows translational template switching from the “non-stop” mRNA to the mRNA-like domain of tmRNA. This latter domain codes for a 10 residue-long amino acid sequence specifying a hydrophobic signal, which terminates with a regular stop codon and tags the incomplete polypeptide with a proteolysis signal (for a review, see references 5‒7). The tmRNA does not act alone; it is assisted by at least two proteins (SmpB and EF-Tu) to form a ribonucleoprotein (RNP) particle.8-10
The ssrA gene specifying tmRNA is found not only in the genome of bacteria, but also in plastid genomes of diverse photosynthetic eukaryotes.11 Mitochondrial ssrA genes are so far limited to jakobids,12 a group of free-living, bacterivorous protists13 whose mtDNAs have most bacteria-like features and the largest known gene repertoire.14,15
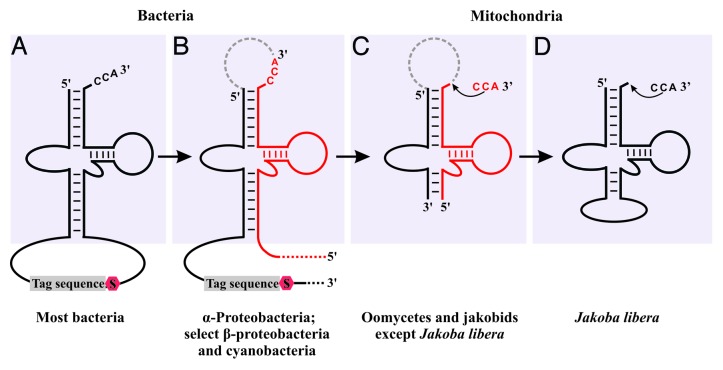
The majority of bacterial ssrA genes specify a single-piece tmRNA (Fig. 1A), as do their plastid homologs. However, in α-Proteobacteria (and subsets of other bacterial groups), the gene sequence is circularly permuted, with the 3′ portion of tmRNA encoded upstream of its 5′ portion. After transcription, the circularly permuted pre-tmRNA is processed, generating two tmRNA fragments. The two halves are held together and arranged in “correct” conformation by RNA base pairing and RNA-binding proteins,16,17 thus forming the canonical tmRNA secondary structure (Fig. 1B).
Figure 1. Comparison of tmRNA structures from bacteria and mitochondria. (A and B) Bacterial tmRNAs. The 3′ CCA is genome-encoded in most but not all bacteria. (A) Standard one-piece bacterial tmRNA comprising two distinct functional portions, the tRNA-like domain (upper part, gray background) and the mRNA-like domain. Tag, nucleotide sequence encoding amino acids that specify the polypeptide degradation signal. S, stop codon. (B) Two-piece bacterial tmRNAs (e.g., in α-Proteobacteria) are encoded by a circularly permuted gene sequence. After the RNA processing steps that remove the intervening sequence (dashed ark), the two resulting pieces (marked black and red, respectively) remain together through base pairing. (C and D) Mitochondrial tmRNAs, lacking the mRNA domain. The 3′ CCA is added post-transcriptionally (as in mitochondrial tRNAs). (C) Common two-piece configuration of mt-tmRNA. (D) One-piece configuration of J. libera mt-tmRNA that most likely emerged secondarily through gene rearrangement.
In line with an α-proteobacterial origin of mitochondria, Williams and coworkers postulated a circularly permuted ssrA gene in the mitochondrial genome of the jakobid Reclinomonas americana.16 Subsequent studies in our laboratory have confirmed the presence of mitochondrial ssrA genes in R. americana and in eight out of nine other jakobids.12,15 The inferred jakobid mitochondrial tmRNAs (mt-tmRNAs) are two-piece molecules (Fig. 1C) except in Jakoba libera,12 where the gene has reverted to a one-piece, non-permuted conformation (Fig. 1D). Mitochondrial tmRNAs are characterized by non-canonical T-loop and D-loop regions, somewhat resembling the likewise unusual tRNAs in animal mtDNAs (for a recent review on this topic, see ref. 18). In vivo and in vitro studies in J. libera demonstrate that ssrA is transcribed, and that the transcript is processed, post-transcriptionally modified by CCA addition and charged with alanine by alanyl-tRNA synthetase.12 Remarkably, all jakobid ssrA genes lack the mRNA-like domain. This suggests that incomplete polypeptides are earmarked for degradation without specific tagging of the C terminus.
To identify ssrA genes in mtDNAs from organisms other than jakobids, we searched available mtDNAs with the tmRNA covariance models in Rfam10.1 that are based on bacterial sequences,19 but did neither detect all known jakobid ssrA genes nor new instances. Here we report the development of a covariance model for highly sensitive searches of mt-tmRNA genes, based on jakobid sequences. By scanning all available complete or nearly complete mtDNAs, we discovered with high confidence mt-tmRNA genes in oomycetes, which are phylogenetically very distant to jakobids, but not in similarly gene-rich mtDNAs of other eukaryotic groups. The search model will be made available via the Rfam database,20 and will also be included in the automated organelle genome annotation tool MFannot (megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl). This work is part of our research program aimed at the development of tools for annotating structural RNA genes in genome and transcriptome sequences.
Results
Discovery of mt-tmRNA genes in oomycete mitochondrial genomes
Since the first claim of a potential mtDNA-encoded tmRNA,16 only seven extra ssrA genes have been identified, including both one-piece and two-piece conformations.12,15 The four Rfam10.1 tmRNA covariance models, which are based on bacterial sequences,19 are not well suited for detecting mitochondrial counterparts. The α-proteobacterial model for two-piece tmRNAs finds only six out of the seven recognized jakobid genes15 (missing the Andalucia homolog), and reports none of the nine mt-tmRNAs in complete oomycete mtDNAs that are identified with a model based on jakobid mt-tmRNA sequences (see below). Further, the general, β-proteobacterial and cyanobacterial covariance models developed for one-piece tmRNAs report numerous false positives (overlapping identified tRNA sequences), but not the known J. libera sequence (for cut-off e-values as large as 1.0).
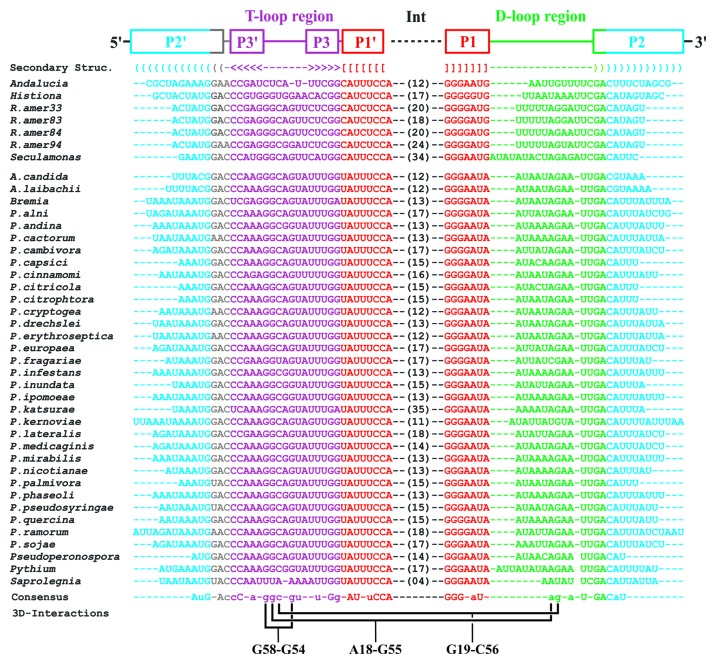
Now, with the availability of nine complete jakobid mtDNA sequences,15 and more sensitive tools for searching secondary structure motifs,21-23 we set out to build a covariance model based on jakobid mitochondrial tmRNAs using the cmbuild component of the Infernal package.23 Employing version 1.1 of cmsearch,19 we screened all known complete mitochondrial genomes (listed at the NCBI website; 3,328 in total, see Materials and Methods) from organisms spanning all major eukaryotic groups. As expected, we detected ssrA genes in jakobid mtDNAs, but also in all nine complete mtDNAs from oomycetes representing three different genera: Phytophthora, Pythium and Saprolegnia (Fig. 2; Table S1). However, we did not find ssrA in mtDNAs from other eukaryotes with a similarly large gene complement as oomycetes, not even after including the newly found oomycete sequences in an extended search model. As oomycete tmRNAs are fairly similar at the primary sequence level, a Blast search of GenBank (non-redundant nucleotide sequence section) identified another 25 mitochondrial ssrA genes from additional oomycetes, in sequence entries of partial mtDNA. This increases the total number of currently known mt-tmRNAs to 42 (Table S2).
Figure 2. Conservation of mitochondrial two-piece tmRNAs. Sequence alignment of two-piece mt-tmRNAs (upper block from jakobid, lower block from oomycete mtDNAs). The conserved paired regions (P1, P2 and P3), loop regions (T-loop and D-loop) and the intervening sequence (Int) are indicated. The sequence consensus shows identical positions in upper case and positions with at least 90% identity in lower case. Tertiary interactions that conform perfectly with those known from tRNAs (for a review, see ref. 28) are indicated in the bottom line (for a graphical representation of these interactions, see Fig. 5).
Throughout all examined oomycetes, the ssrA gene is flanked by two tRNA genes [trnG(gcc) and trnG(ucc)], except in S. ferax, where the adjacent genes are rnl and rps7 (encoding the large subunit rRNA and the mito-ribosomal protein S7, respectively). Pythium and Saprolegnia have two identical ssrA copies that are located in repeat regions. Notably, the mRNA-like domain is absent from oomycete mt-tmRNAs, as previously noted for jakobids.12,15
Transcription and RNA processing of oomycete tmRNAs
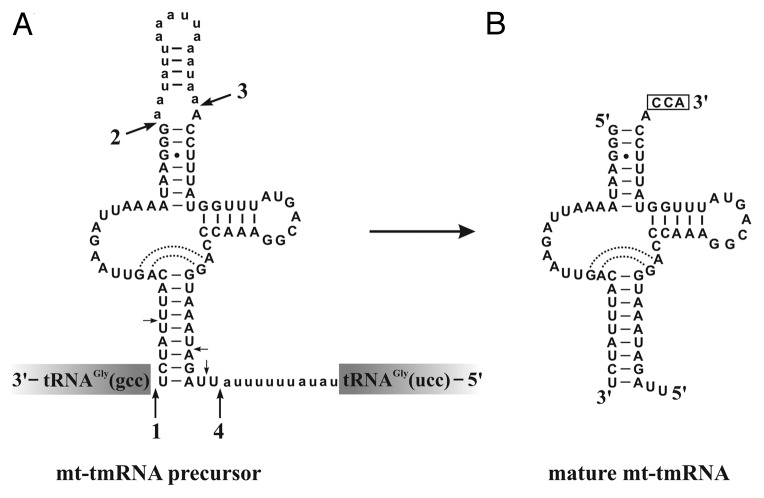
To obtain insight into transcription and processing of oomycete mt-tmRNAs, we analyzed previously published RNA-Seq data from a library of total small RNAs from the type species Phytophthora sojae, strain P6497.24 The data show that the predicted ssrA gene region is transcribed as a precursor molecule, with an overall transcript abundance similar to that of the flanking tRNAGly. The most frequent tmRNA termini coincide with the four predicted processing sites (numbered 1–4; Fig. 3A), exceeding background counts by ~10–100-fold. Additional minor termini locate to the helical region that corresponds to the anticodon arm. The mapped transcript termini provide evidence that maturation of the mt-tmRNA precursor proceeds in three principal steps, notably removal of the flanking tRNA gene sequences (presumably catalyzed by RNase P and a tmRNA-specific processing activity, respectively) and two cleavage events releasing the intervening sequence (by RNase P and a tRNA 3′ processing endonuclease). The resulting mature product is a two-piece RNA (Fig. 3B). The few additional fragments probably result from non-specific degradation.
Figure 3. Primary transcript and maturation of P. sojae mt-tmRNA. The precursor mt-tmRNA of P. sojae is located between two glycine tRNA genes [note that due to the particular two-piece tmRNA configuration, the RNA precursor sequence is shown in 3′ (left) to 5′ (right) direction]. Short lines and solid circles indicate Watson-Crick and G-U pairs, respectively. Major RNA processing sites are inferred from RNA-Seq data and numbered and indicated by arrows; small arrows point to minor sites. Processing of sites 1 and 2 is likely catalyzed by RNase P, site 3 by a 3′ tRNA endonuclease, and site 4 by a tmRNA-specific activity. Nucleotides cleaved off from the precursor are shown in lower case, and the post-transcriptionally added CCA is boxed.
About 85% of the tm-RNA transcripts in RNA-Seq data carry the post-transcriptionally added nucleotides CCA at processing site 3, which are attached to the 3′ terminal A that corresponds to the discriminator position of tRNAs. The CCA addition is a molecular hallmark of mitochondrial tRNAs and tmRNAs (Fig. 3B).
Sequence and secondary structure comparison of mt-tmRNAs
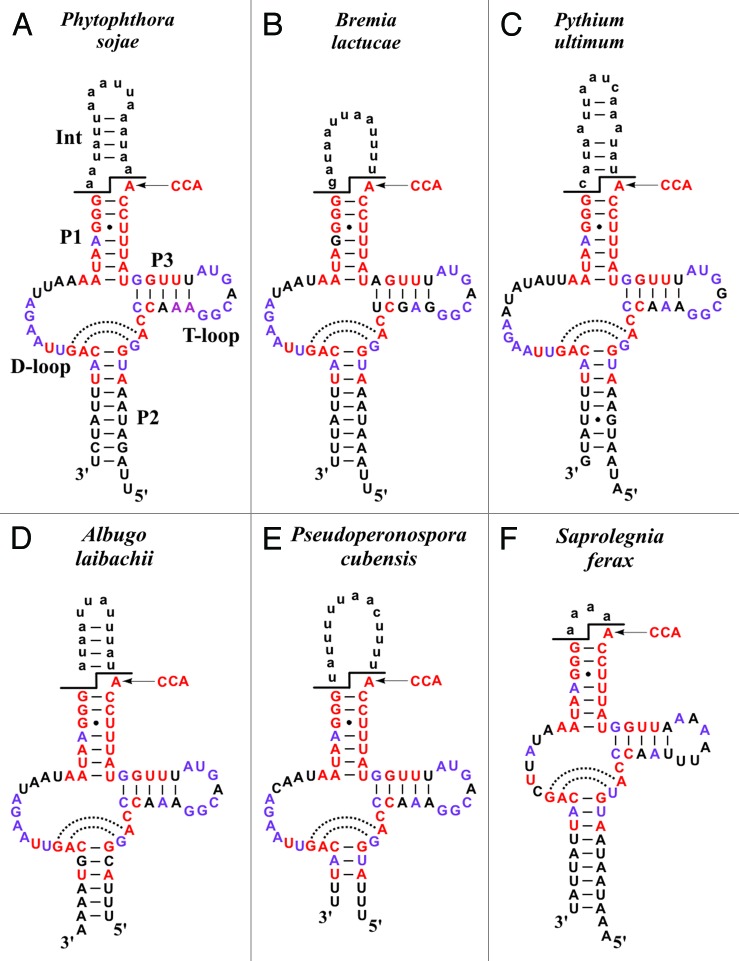
Based on the nucleotide-precise mapping of mt-tmRNA termini in P. sojae, we built secondary structure models for this molecule and for the inferred mt-tmRNAs from five other, representative oomycetes (Fig. 4). Most highly conserved is the functionally important seven bp-long amino acid acceptor domain (termed P1, Fig. 4A) with an invariant A in the discriminator position and a universal G-U pair at position 3. This base pair is the recognition site for alanyl-tRNA synthetase.25
Figure 4. Representative oomycete mt-tmRNA secondary structures. All oomycete mt-tmRNAs feature the two-piece configuration, and lack (as in jakobids) an mRNA-like domain. Lower-case nucleotides represent portions of the precursor transcript that are removed by RNA processing. Nucleotides identical across oomycete mt-tmRNAs are highlighted by color (red, invariant; purple, identical in five out of the six shown examples). Black lines indicate the two processing sites shared with tRNAs (5′ processing by RNase P and 3′ processing by a specific endonuclease); for an experimental confirmation of these processing sites in P. sojae tmRNA, see Figure 3. Black arrows indicate post-transcriptional addition of CCA at the 3′ discriminator nucleotide, which is invariantly A.
Juxtaposed to P1 is a 3–10 bp long helix corresponding to the tRNA anticodon stem, referred here to as P2 (Fig. 4A). The role of P2 is likely in stabilizing the secondary structure and protecting the molecule against exonucleases. An anticodon loop is not present (and not required for tmRNA function). Four nucleotide positions in P2 are invariant across oomycetes and jakobids suggesting that they play an additional, currently unknown role; the helix can be extended by two non-Watson-Crick base pairs as discussed below. The third stem, P3, located between P1 and P2, has five base pairs and corresponds to the T-arm of tRNAs. The T-loop-like sequence of tmRNA is highly conserved in oomycetes and jakobids (Fig. 2), with only a few species deviating from the common pattern (e.g., S. ferax, Fig. 4F, and Andalucia godoyi15). Finally, mitochondrial tmRNAs have a 9–18 nt long D-loop region without D-stem, which is also characteristic of bacterial tmRNAs26,27 and reminiscent of certain, non-canonical animal mitochondrial tRNAs (for a review, see ref. 28).
Precursor transcripts of two-piece mt-tmRNAs include the intervening sequence (labeled “Int” in Fig. 4), looping out of P1. This sequence is A+U-rich and not only highly variable in sequence but also in length, which ranges in jakobids from 12‒34 nt12,15 and in oomycetes from as little as four (S. ferax; Fig. 4F) to 35 nt (Phytophthora karsurae, not shown). Among all known mt-tmRNAs, that of the oomycete S. ferax is the most divergent one, with all loops shortened and less well conserved at the sequence level.
Tertiary interactions in mt-tmRNAs
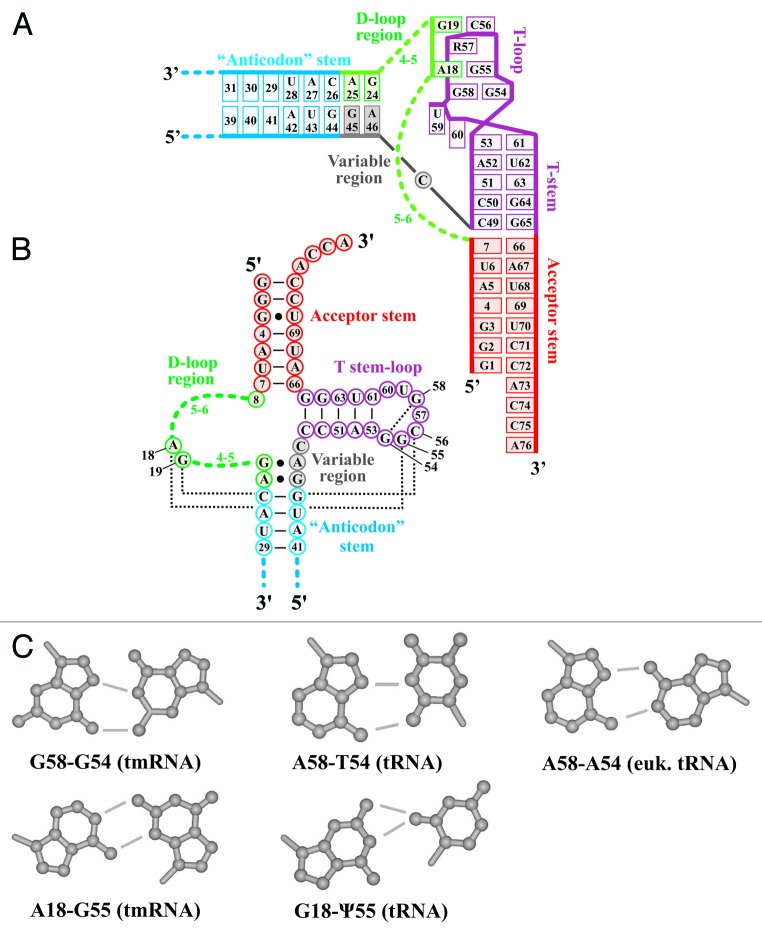
With mt-tmRNA sequences now available from diverse eukaryotes, we conducted manual/comparative three-dimensional structure modeling of this molecule. We observed that the T-loop sequence of mt-tmRNAs differs conspicuously from that of canonical tRNAs (see Fig. S1), with the first and fifth nucleotides (positions 54 and 58 according to tRNA nomenclature) predominantly occupied by guanosine (Figs. 2 and 515). However, at the tertiary structure level, these two nucleotides in mt-tmRNAs form a reverse-Hoogsteen G-G base pair (Fig. 5C) that is practically isosteric with the base pairs A58-A54 or A58-T54 (T in RNA is the 5-methylated derivative of uridine) of eukaryotic initiator and other cytosolic tRNAs, respectively.
Figure 5. Tertiary structural model of mitochondrial two-piece tmRNAs. Nucleotides are numbered in accordance with the standard tRNA nomenclature, which is based on the yeast Phe-tRNA.42 (A) L-form tertiary structure of tmRNA. Rectangles represent individual nucleotides. The various tmRNA structural domains are marked with distinct colors. Within the T-loop, base pair G58-G54 is equivalent to base pairs A58-T54 and A58-A54 found in tRNAs. Two inter-loop base pairs are present, A18-G55 and G19-C56, separated by a purine at position 57. The dinucleotide at positions 59–60 bulges between base pairs 53–61 and 54–58. The green-dashed lines connect the T-loop with the anticodon stem and acceptor stem. (B) The tmRNA cloverleaf. Nucleotides of the stem-loops are shown in the same color and with the same numbering as in (A). The most frequent number of nucleotides in the variable D-loop region are shown in green; positions of nucleotides involved in tertiary interactions are in black. Short lines connect nucleotides forming Watson-Crick base pairs within stems. Dotted lines connect nucleotides involved in conserved tertiary interactions. Identities of these nucleotides (above 90%) are indicated. (C) Structures of base pairs at positions 58–54 (upper three) and 18–55 (lower two) in tmRNAs compared with tRNAs. The identities of bases and the inter-base hydrogen bonds are indicated.
While nucleotide 55 in the T-loop of tRNAs is a pseudouridine (ψ, ribosyluracil) engaging in tertiary interaction with G18 (G18-ψ5529), position 55 in mt-tmRNAs is predominantly a G. G55 is also known from some mitochondrial tRNAs and T-loop-like structures in RNase P and correlates with a substitution of G by A at position 18 in the D-loop (reviewed in ref. 30) A18-G55 pairing was demonstrated experimentally for the T-loop-like domains of RNase P,31 a pairing almost isosteric to the standard G18-ψ55 of tRNAs (Fig. 5C). We assume that A18-T55 interactions also exist in mt-tmRNAs, as the corresponding D-loop region contains several candidate adenines (Fig. 2).
In agreement with tRNA interactions, the nucleotide at position 56 of mt-tmRNAs is predicted to interact with position 19 in the D-loop region typically generating a G19-C56 Watson-Crick pair (Figs. 2 and 5A). Like in bacterial tRNA structures, the dinucleotide 18–19 of the mt-tmRNA D-loop is flanked by unpaired regions of 3–8 nt containing predominantly adenosines and uridines whose tertiary arrangement is difficult to model.
Further, in standard tRNAs, nucleotide 57 of the T-loop intercalates between the two tertiary base pairs 18–55 and 19–56. In tRNAs this arrangement is stabilized by a purine in position 57, which is also found in virtually all mt-tmRNA sequences. In summary, tmRNA regions corresponding to the D- and T-loops appear to form a stable, tRNA-like three-dimensional structure. Differences between mt-tmRNAs and tRNAs at the sequence level do obviously not affect charging with alanine as shown for the J. libera molecule,12 and most likely also preserve the mt-tmRNA interaction with the translational elongation factor EF-Tu.
Finally, in most mt-tmRNA sequences including those of jakobids, the anticodon stem is flanked on both sides by GA dinucleotides, which also exist in the bacterial tmRNA and are known to form tertiary base pairs (G25-A45 and A26-G44, according to the E. coli tmRNA nomenclature26,27).
Discussion
Additional mt-tmRNA genes outside jakobids and oomycetes?
With the realization that tmRNAs may be encoded in either a contiguous or permuted configuration, ssrA genes have been detected in virtually all bacterial genomes.16,17 In contrast, mitochondrial ssrA genes are currently only known from two unrelated protist groups (for a phylogenetic tree, see Fig. S2). One possible explanation for this apparently punctate distribution is that mt-tmRNAs are generally nucleus-encoded and imported into the organelle. It is also conceivable that eukaryotes outside jakobids and oomycetes do possess mtDNA-encoded tmRNAs, but that the gene has diverged even more than that of S. ferax, which is the most highly derived sequence in our study (Fig. 2F). Indeed, highly divergent small structural RNAs are often unrecognizable based on sequence alone, as is the case of the long-elusive mitochondrial RNase P-RNA of fission yeast.32
Note that our mt-tmRNA-search model reports bona fide permuted tmRNA genes with high confidence (within an e-value range from 2.4e-8‒1.7e-16). Although based on two-piece molecules with permuted gene sequence, the model also detects the 3′ portion of the single-piece ssrA (e-value 8.7e-3) encoded in the mitochondrial genome of J. libera,15 providing a valuable lead for modeling its full structure manually.
However, the model does not find a match in mtDNA of the close relative Jakoba bahamiensis, and this may have several reasons. In addition to extreme sequence divergence, detection of mitochondrial ssrA might be hampered by long Int sequences (Fig. 2). Our in silico experiments indicate (not shown) that if Int regions are longer than ~500 nt or if the gene is split with fragments located on different strands of mtDNA, current covariance methods will fail to recognize the gene of two-piece mt-tmRNAs in genomic sequence. Descriptor-based search engines such as Erpin33 do not have this limitation as we demonstrated for large, poorly conserved group I intron sequences.34 However, descriptor models are much less sensitive than covariance models. Therefore, annotation of ssrA genes that carry long inserts or are fragmented will require the development of a novel modular covariance search algorithm.
In silico methods for identifying mt-tmRNAs would be probably greatly assisted by RNA-Seq data analysis. Most promising targets would be the above-mentioned jakobid J. bahamiensis, stramenopile sister groups of oomycetes and any other protist with moderately derived mtDNAs (e.g., Malawimonada, Heterolobosea, Cryptophyta and Prasinophyta).
Are mitochondrial tmRNAs indeed lacking the mRNA-like domain?
Without exception, bacterial ssrA genes encode both the tRNAAla-like domain and the mRNA domain coding for the protein tag. In contrast, mitochondrial ssrA, now known from two unrelated protist groups, only include one portion, the tRNAAla-like domain. Apparently, the mRNA-like domain was lost very early in mtDNA evolution.12,15
Whether the terminally added alanine alone is sufficient to signal degradation of incomplete polypeptides has been discussed previously.12 This question is difficult to address experimentally given the low concentration of such aberrant molecules. Alternative protein quality control pathways are known from baker’s yeast and Neurospora crassa, where non-assembled or misfolded mitochondrial polypeptides are selectively degraded by eukaryotic AAA proteases (membrane-embedded ATP-dependent proteases35-37). In mitochondria, this pathway may substitute for the bacterial proteolysis tagging mode of incomplete polypeptides.
A final possibility is that an unrecognized mRNA-like domain is encoded separately in jakobid and oomycete mtDNAs, or even in the nucleus, to assemble with the known mt-tmRNA into a larger unit. This scenario, as well as the potential import of AAA proteases into jakobid and oomycete mitochondria, could be tested experimentally by purifying tmRNA-containing mitochondrial RNP particles and identifying their RNA and protein subunits. This would be not a trivial undertaking, because there are currently no established protocols for the isolation of mitochondria in sufficient quantity and purity from these organisms.
Materials and Methods
Identification and secondary structure analysis of mt-tmRNAs
Previously identified mitochondrial ssrA gene sequences from jakobids were taken from GenBank accession nos. KC_353352–59.15 The two-piece jakobid mt-tmRNA sequences were aligned with Muscle version 3.6.38 The resulting alignment was manually curated for an improved fit of primary sequence plus secondary structure, and the minimum-free energy consensus secondary structure was predicted with RNAalifold.39 The final alignment was processed using components of the Infernal package v1.1rc2,21,23 notably cmbuild and cmcalibrate to build a covariance search model, followed by cmsearch to screen all publicly available complete mtDNAs. Sequence-conserved tmRNAs were identified by searching potential homologs of the P. sojae ssrA in GenBank nr by BLAST (www.ncbi.nlm.nih.gov/BLAST).40 Candidate genes were compiled with GeneDoc v2.5.010.41 See Supplemental Materials, Data set 1 for the alignment of representative jakobid plus oomycete mt-tmRNAs that was used for generating a comprehensive mt-tmRNA model.
Mapping of P. sojae tmRNA termini based on RNA-Seq data
RNA-Seq sequences of P. sojae were generated by others from small total RNAs.24 Sequences in fastq format were quality-trimmed (phred 20, minimum sequence length 20 nt) and adaptor-clipped using Seqtrimnext (rubygems.org/gems/seqtrimnext). Termini of tmRNA transcripts were identified by scanning the trimmed sequences with a 20-nt long window (using the basic Linux tools grep and wc).
Access to the mt-tmRNA model in Rfam
The covariance model is based on mtDNA- encoded sequences from jakobids and oomycetes. The model plus sequence alignment will be made available at Rfam under the name mt-tmRNA.
Supplementary Material
Acknowledgments
We thank Dr Mark Gijzen (Agriculture Canada) for kindly providing RNA-Seq data from the P. sojae type strain prior to publication and Michael W. Gray (Dalhousie University) for critically reading the manuscript. We also acknowledge Lise Forget for assistance with RNA experiments, and Sandrine Rousseau (Université de Montréal) for help with filtering Illumina readings.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Financial support including salary support (M.H.) was provided by Genome Quebec/Canada (B.F.L., G.B.), by the Canadian Institute of Health Research [CIHR; MOP-79309 (G.B.) and MOP-89923 (S.V.S.)] and by the National Research Council (NSERC; B.F.L.).
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/25376
References
- 1.Barends S, Kraal B, van Wezel GP. The tmRNA-tagging mechanism and the control of gene expression: a review. Wiley Interdiscip Rev RNA. 2011;2:233–46. doi: 10.1002/wrna.48. [DOI] [PubMed] [Google Scholar]
- 2.Himeno H, Nameki N, Tadaki T, Sato M, Hanawa K, Fukushima M, et al. Escherichia coli tmRNA (10Sa RNA) in trans-translation. Nucleic Acids Symp Ser. 1997;37:185–6. [PubMed] [Google Scholar]
- 3.Felden B, Gesteland RF, Atkins JF. Eubacterial tmRNAs: everywhere except the alpha-proteobacteria? Biochim Biophys Acta. 1999;1446:145–8. doi: 10.1016/S0167-4781(99)00085-8. [DOI] [PubMed] [Google Scholar]
- 4.Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J. Visualizing tmRNA entry into a stalled ribosome. Science. 2003;300:127–30. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- 5.Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol. 2000;7:449–55. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 6.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–3. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 7.Mehta P, Richards J, Karzai AW. tmRNA determinants required for facilitating nonstop mRNA decay. RNA. 2006;12:2187–98. doi: 10.1261/rna.247706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barends S, Karzai AW, Sauer RT, Wower J, Kraal B. Simultaneous and functional binding of SmpB and EF-Tu-TP to the alanyl acceptor arm of tmRNA. J Mol Biol. 2001;314:9–21. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- 9.Ramrath DJ, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, et al. The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Nature. 2012;485:526–9. doi: 10.1038/nature11006. [DOI] [PubMed] [Google Scholar]
- 10.Rudinger-Thirion J, Giegé R, Felden B. Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA. 1999;5:989–92. doi: 10.1017/S135583829999101X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gueneau de Novoa P, Williams KP. The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 2004;32(Database issue):D104–8. doi: 10.1093/nar/gkh102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacob Y, Seif E, Paquet PO, Lang BF. Loss of the mRNA-like region in mitochondrial tmRNAs of jakobids. RNA. 2004;10:605–14. doi: 10.1261/rna.5227904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Kelly CJ. The jakobid flagellates: structural features of Jakoba, Reclinomonas and Histiona and implications for the early diversification of eukaryotes. J Eukaryot Microbiol. 1993;40:627–36. doi: 10.1111/j.1550-7408.1993.tb06120.x. [DOI] [Google Scholar]
- 14.Lang BF, Burger G, O’Kelly CJ, Cedergren R, Golding GB, Lemieux C, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–7. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 15.Burger G, Gray MW, Forget L, Lang BF. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 2013;5:418–38. doi: 10.1093/gbe/evt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keiler KC, Shapiro L, Williams KP. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: A two-piece tmRNA functions in Caulobacter. Proc Natl Acad Sci USA. 2000;97:7778–83. doi: 10.1073/pnas.97.14.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao C, Bhardwaj K, Sharkady SM, Fish RI, Driscoll T, Wower J, et al. Variations on the tmRNA gene. RNA Biol. 2009;6:355–61. doi: 10.4161/rna.6.4.9172. [DOI] [PubMed] [Google Scholar]
- 18.Lang BF, Lavrov D, Beck N, Steinberg V. Mitochondrial tRNA structure, identity and evolution of the genetic code. In: Bullerwell CE, ed. Organelle Genetics. Berlin, Heidelberg, 2012: Springer, 2011. [Google Scholar]
- 19.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41(Database issue):D226–32. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37(Database issue):D136–40. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eddy S. Infernal website. 2008; http://infernal.janelia.org
- 22.Eddy SR, Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–88. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–7. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qutob D, Chapman BP, Gijzen M. Transgenerational gene silencing causes gain of virulence in a plant pathogen. Nat Commun. 2013;4:1349. doi: 10.1038/ncomms2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naganuma M, Sekine S, Fukunaga R, Yokoyama S. Unique protein architecture of alanyl-tRNA synthetase for aminoacylation, editing, and dimerization. Proc Natl Acad Sci USA. 2009;106:8489–94. doi: 10.1073/pnas.0901572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burks J, Zwieb C, Müller F, Wower I, Wower J. Comparative 3-D modeling of tmRNA. BMC Mol Biol. 2005;6:14. doi: 10.1186/1471-2199-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanawa-Suetsugu K, Bordeau V, Himeno H, Muto A, Felden B. Importance of the conserved nucleotides around the tRNA-like structure of Escherichia coli transfer-messenger RNA for protein tagging. Nucleic Acids Res. 2001;29:4663–73. doi: 10.1093/nar/29.22.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang BF, Lavrov D, Beck N, Steinberg S. Mitochondrial tRNA structure, identity and evolution of the genetic code. In: Bullerwell CE, ed. Organelle Genetics - Evolution of organelle genomes and gene expression, 2012:431-74. [Google Scholar]
- 29.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37(Database issue):D159–62. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doyon FR, Zagryadskaya EI, Chen J, Steinberg SV. Specific and non-specific purine trap in the T-loop of normal and suppressor tRNAs. J Mol Biol. 2004;343:55–69. doi: 10.1016/j.jmb.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 31.Krasilnikov AS, Yang X, Pan T, Mondragón A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–4. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 32.Seif ER, Forget L, Martin NC, Lang BF. Mitochondrial RNase P RNAs in ascomycete fungi: lineage-specific variations in RNA secondary structure. RNA. 2003;9:1073–83. doi: 10.1261/rna.5880403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gautheret D, Lambert A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J Mol Biol. 2001;313:1003–11. doi: 10.1006/jmbi.2001.5102. [DOI] [PubMed] [Google Scholar]
- 34.Lang BF, Laforest MJ, Burger G. Mitochondrial introns: a critical view. Trends Genet. 2007;23:119–25. doi: 10.1016/j.tig.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Käser M, Langer T. Protein degradation in mitochondria. Semin Cell Dev Biol. 2000;11:181–90. doi: 10.1006/scdb.2000.0166. [DOI] [PubMed] [Google Scholar]
- 36.Pajic A, Tauer R, Feldmann H, Neupert W, Langer T. Yta10p is required for the ATP-dependent degradation of polypeptides in the inner membrane of mitochondria. FEBS Lett. 1994;353:201–6. doi: 10.1016/0014-5793(94)01046-3. [DOI] [PubMed] [Google Scholar]
- 37.Klanner C, Prokisch H, Langer T. MAP-1 and IAP-1, two novel AAA proteases with catalytic sites on opposite membrane surfaces in mitochondrial inner membrane of Neurospora crassa. Mol Biol Cell. 2001;12:2858–69. doi: 10.1091/mbc.12.9.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholas KB, Nicholas HB, Deerfield DW. GeneDoc: analysis and visualization of genetic variation. EMB News. 1997;4:14. [Google Scholar]
- 42.Rich A, RajBhandary UL. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–60. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.