Abstract
Background
One of the challenges in insect chemical ecology is to understand how insect pheromones are synthesised, detected and degraded. Genome wide survey by comparative sequencing and gene specific expression profiling provide rich resources for this challenge. A. ipsilon is a destructive pest of many crops and further characterization of the genes involved in pheromone biosynthesis and transport could offer potential targets for disruption of their chemical communication and for crop protection.
Results
Here we report 454 next-generation sequencing of the A. ipsilon pheromone gland transcriptome, identification and expression profiling of genes putatively involved in pheromone production, transport and degradation. A total of 23473 unigenes were obtained from the transcriptome analysis, 86% of which were A. ipsilon specific. 42 transcripts encoded enzymes putatively involved in pheromone biosynthesis, of which 15 were specifically, or mainly, expressed in the pheromone glands at 5 to 120-fold higher levels than in the body. Two transcripts encoding for a fatty acid synthase and a desaturase were highly abundant in the transcriptome and expressed more than 40-fold higher in the glands than in the body. The transcripts encoding for 2 acetyl-CoA carboxylases, 1 fatty acid synthase, 2 desaturases, 3 acyl-CoA reductases, 2 alcohol oxidases, 2 aldehyde reductases and 3 acetyltransferases were expressed at a significantly higher level in the pheromone glands than in the body. 17 esterase transcripts were not gland-specific and 7 of these were expressed highly in the antennae. Seven transcripts encoding odorant binding proteins (OBPs) and 8 encoding chemosensory proteins (CSPs) were identified. Two CSP transcripts (AipsCSP2, AipsCSP8) were highly abundant in the pheromone gland transcriptome and this was confirmed by qRT-PCR. One OBP (AipsOBP6) were pheromone gland-enriched and three OBPs (AipsOBP1, AipsOBP2 and AipsOBP4) were antennal-enriched. Based on these studies we proposed possible A. ipsilon biosynthesis pathways for major and minor sex pheromone components.
Conclusions
Our study identified genes potentially involved in sex pheromone biosynthesis and transport in A. ipsilon. The identified genes are likely to play essential roles in sex pheromone production, transport and degradation and could serve as targets to interfere with pheromone release. The identification of highly expressed CSPs and OBPs in the pheromone gland suggests that they may play a role in the binding, transport and release of sex pheromones during sex pheromone production in A. ipsilon and other Lepidoptera insects.
Background
Lepidoptera sex pheromones are primarily C10-C18 long straight chain unsaturated alcohols, aldehydes or acetate esters [1], biosynthesised and released mainly from pheromone glands located between the 8th and 9th abdominal segments of the female moths. Usually the females use a mixture of compounds in a unique ratio to attract conspecific males [2]. The extremely high specificity and sensitivity of species-specific pheromones make them potential biological control agents for population monitoring, mass trapping and reducing pesticide use in integrated pest management (IPM) programs [3-5]. Further use of pheromones in such strategies would be aided by an understanding of the pathways involved in pheromone biosynthesis and transport.
Most sex pheromone blends of Lepidoptera insects are synthesised de novo via modified fatty acid biosynthesis pathways [2,6,7] and gland-specific enzymes are involved in desaturation, chain shortening, reduction and acetylation [1,2]. Different species use different combinations of these reactions to produce unique species-specific pheromone blends. The first step is the synthesis of saturated fatty acid precursors malonyl-CoA from acetyl-CoA by acetyl-CoA carboxylase (ACC) and fatty acid synthetase (FAS) [8,9]. Labeling studies conducted with acetate indicated that malonyl-CoA and NADPH are used by FAS to produce mainly saturated stearic acid (18:0) and palmitic acid (16:0) with 18 and 16 carbon atoms and no double bonds, respectively, as precursors [10-12]. Modification of the fatty acid chain includes the introduction of a double bond by desaturases specific to pheromone biosynthesis followed by chain shortening using specific β–oxidation enzymes [13,14]. So far, several types of desaturases have been extensively studied through gene characterization and expression analysis, including Δ5 [15], Δ9 [16,17], Δ10 [18], Δ11 [19,20], and Δ14 [21] desaturases. Once unsaturated pheromone precursor with a specific chain-length is produced, the carboxyl carbon is modified to form one of functional groups (aldehyde, alcohol or acetate ester). These modifications require the enzymes fatty acid reductase to produce the alcohols from the fatty acyl precursor [22], which in some species may be oxidized to aldehydes serving as pheromone components [23], and to acetate esters (OAc) by acetyltransferase [24]. Recently, a few members of the reductase gene family have been discovered and functionally characterized in several Lepidoptera species, including Ostrinia scapulalis[25], Heliothis virescens, Heliothis subflexa, Helicoverpa armigera, Helicoverpa assulta[26], Ostrinia nubilalis[27], Yponomeuta evonymellus (L.), Yponomeuta padellus (L.) and Yponomeuta rorellus (Hübner) [28]. A number of pheromone gland-specific enzymes have been identified and their essential functions in pheromone production demonstrated in vitro as well as in vivo. For example, using RNA interference, Matsumoto and colleagues showed that two pheromone gland-specific enzymes (acyl-CoA desaturase and a fatty-acyl reductase) are responsible for pheromone production in the silk moth Bombyx mori[29-31].
After production and release of the sex pheromone components by female moths the males detect the pheromone and respond for mating. It is commonly accepted that pheromone molecules are captured and transported to the pheromone receptors on the dendrites of pheromone-sensitive neurons by olfactory binding proteins, including odorant binding proteins (OBPs) and chemosensory proteins (CSPs) [32-34]. Pheromone binding proteins (PBPs) bind to sex pheromone components and classified into a subclass of OBPs [35]. After activation of the pheromone receptors the olfactory signals must be degraded rapidly to prevent from prolonged neuronal excitation [36]. This may involve pheromone degrading enzymes (PDEs) capable of degrading the pheromone molecules [37].
The black cutworm Agrotis ipsilon is a destructive polyphagous insect pest of many crops and for a strain from China the female sex pheromone blend comprises five main acetate components: (Z)-11-hexadecenyl acetate (Z11-16:OAc), (Z)-9-tetradecenyl acetate (Z9-14:OAc), (Z)-7-dodecenyl acetate (Z7-12:OAc), (Z)-8-dodecenyl acetate (Z8-12:OAc) and (Z)-5-decenyl acetate (Z5-10:OAc) [38]. These components indicate the involvement of different desaturases and ß-oxidases during the sex pheromone biosynthesis. However, the genes/proteins and their specific function in mediating A. ipsilon pheromone production, transport and degradation have not been characterized. Over the last few years, the next generation sequencing such as 454 pyrosequencing technique provides an easy and effective method for the discovery of novel genes. In present study, using the Roche GS FLX Titanium sequencing platform, we report a genetic database of the genes expressed in the pheromone glands of A. ipsilon and the identification of genes with putative roles in pheromone biosynthesis, degradation and transport as well as their tissue expression profiles.
Results and discussion
454 sequencing and unigene assembly
Sequencing of a cDNA library prepared from mRNAs of the pheromone glands of A. ipsilon gave a total of 631,425 raw reads with an average length of 517 base pairs (bp). After trimming adaptor sequences and removing low quality sequences, 629,273 clean reads remained with an average length of 496 bp. The size distribution of the clean reads is shown in Additional file 1. The sequences of all reads have been deposited in the NCBI SRA database with the accession number SRX189143.
The 629,273 clean reads were assembled into 23,473 unigenes, including 20,541 contigs (87.5%) and 2,932 singletons (12.5%), the largest transcriptome dataset so far from moth sex pheromone glands. An overview of the sequencing and assembly results is presented in Table 1. The length of the assembled unigenes ranged from 100 bp to 21842 bp with an average length of 770 bp. Among the unigenes, 22,035 (93.9%) are between 200 bp and 2000 bp long with an average length of 649 bp. These unigenes are in fact transcripts in the A. ipsilon pheromone gland cDNA library. Therefore we refer them as transcripts. All sequences of the unigenes used in the current study are provided in Additional file 2.
Table 1.
Summary of A. ipsilon pheromone gland unigene sequences and assembly
| Sequence number | Average length (bp) | Length distribution(bp) | |
|---|---|---|---|
| Raw reads |
631425 |
517 |
40-1200 bp |
| Clean reads |
629273 |
496 |
40-1200 bp |
| singletons |
2932 |
421 |
100-835 bp |
| contigs |
20541 |
820 |
100-21842 bp |
| unigenes | 23473 | 770 | 100-21842 bp |
Analysis of the transcripts from the A. ipsilon pheromone gland
BLASTx and BLASTn were used to compare each A. ipsilon transcript with a cut-off E-value of 1.0E-5 against GeneBank entries. 12,989 transcripts (55%) had BLASTx hits in the non-redundant protein (nr) databases and 9,392 (40%) had BLASTn hits in the non-redundant nucleotide sequence (nt) databases. This is consistent with a previous report of H. virescens pheromone gland ESTs [39]. Some of the A. ipsilon transcripts were homologous to those from more than one species but in general most were homologous to other Lepidoptera species taking up 2,379 in the 9,392 BLASTn hits, including 1,124 (12%) to B. mori entries. The second highest hits were to Dipteran species with 343 hits to D. melanogaster and 279 and 221 hits to the mosquitoes Anopheles gambiae and Aedes aegypti, respectively. The lowest hits were to the wasp Nasonia vitripennis (190 hits), the beetle Tribolium castaneum (147 hits) and the pea aphid Acyrthosiphon pisum (136 hits). The top 15 insect species that have significant BLASTn hits are shown in Figure 1.
Figure 1.
Top 15 insect species that have significant BLASTn hits. All A. ipsilon pheromone gland unigenes were used in BLASTn searches against the GenBank entries. The significant hits with an E-value >=1.0E-5 for each query were grouped according to species and the number of the unigenes that had significant homology is indicated after the specie name.
Gene Ontology of the genes expressed in the A. ipsilon pheromone gland
The 23,473 assembled transcripts were annotated into different functional groups according to Gene Ontology (GO) analysis. Some transcripts were annotated into more than one GO category. Of the 22,473 transcripts, 7,546 (32%) could be assigned to a GO category (Additional file 3). The “cellular process” and “metabolic process” GO categories were most abundantly represented with 4,056 (17.3%) and 3,361 (14.3%) transcripts, respectively, within the biological process GO ontology. In the “cellular components” GO ontology the transcripts were mainly distributed in cell (18.8%) (4,415 transcripts) and cell part (17.6%) (4,133 transcripts). The GO analysis also showed that in the molecular function ontology 3,271 transcripts (13.9%) were annotated as having binding functions and 3,484 (14.8%) to have catalytic activity.
Comparative analysis of transcripts in Lepidoptera pheromone glands
In order to compare the A. ipsilon pheromone gland transcriptome with those from other Lepidoptera and to identify A. ipsilon transcripts with potential involvement in sex pheromone production and transport we downloaded the pheromone gland ESTs of three other Lepidoptera A. segetum, B. mori and H. virescens from the dbEST database of NCBI and previously published pheromone gland transcriptome of H. virescens[39]. After assembling these ESTs we obtained 925 unigenes from A. segetum, 3943 from B. mori and 8202 from H. virescens with an average length of 384 bp, 692 bp and 474 bp, respectively. These are much lower numbers than that obtained by the current study through the 454 sequencing of the A. ipisilon pheromone gland, demonstrating that our pheromone gland transcriptome is currently the largest transcriptome resource for an insect pheromone gland.
When comparing the pheromone gland transcripts pairwise using best bidirectional hits, we found that there were 461 homologous transcripts between A. ipsilon and A. segetum, 1110 homologous transcripts between A. ipsilon and B. mori, and 2106 homologous transcripts between A. ipsilon and H. virescens (Figure 2). A large portion of A. ipsilon transcripts (86.4%) (20,274 out of 23,473) had no homologous ESTs in the available pheromone gland EST libraries of the other 3 species. This may be due to the larger dataset (23,473 unigenes) for A. ipsilon and lower coverage in the other studies. Nevertheless, it was shown that 309 transcripts, 5,755 transcripts and 2,556 transcripts are only found in A. segetum, H. virescen and B. mori, respectively, in our comparison (Figure 2).
Figure 2.
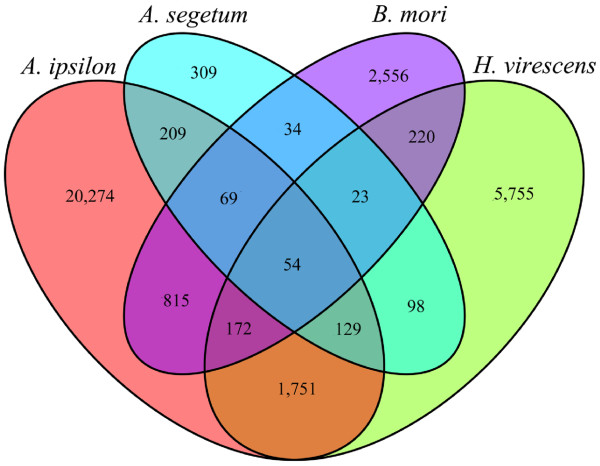
Comparative analysis of A. ipsilon pheromone gland unigenes with other insects. This shows the overlap of blast homology in genes expressed in pheremone glands in four species of Lepidoptera. The comparative analyses of A. ipsilon, H. virescens, B. mori and A. segetum pheromone gland unigenes were performed based on the Best Bidirectional Hits results (reciprocal BLASTn, E-value less than 1.0E-6).
Transcript abundance in the A. ipsilon pheromone gland
The pheromone gland mRNA samples used for constructing the cDNA library were non-normalized and non-amplified by PCR, so the reads in the sequencing dataset most likely represent the relative abundance of each assembled transcript in the pheromone gland as summarized in Table 2. The most abundant transcripts include vitellogenin, a major reproductive protein in insects (2,925 reads per kilobase per million mapped reads (RPKM); 2.2% reads), the precursor of egg yolk proteins for insect egg production [40] and genes involved in PBAN stimulated pheromone production such as lipase 3 [41] (4,731 RPKM; 0.8% reads) and in sex pheromone biosynthesis such as acyl-CoA desaturase (1,206 RPKM; 0.3% reads) and in lipid transport such as apolipophorin III (2894 RPKM 0.4% reads). Another highly abundant transcript (Unigene_721) with 1,365 RPKM encodes a CSP with a 76% protein identity to the H. virescens CSP (Protein ID: ACX53806) and 41% to the ejaculatory bulb-specific protein 3 of D. melanogaster (Protein ID: Q9W1C9).
Table 2.
The most prevalent mRNAs in A. ipsilon sex pheromone gland
| Unigene ID | GenBank annotation | Species | Accession number | Score | E-value | % Identity | No. of Reads |
|---|---|---|---|---|---|---|---|
| Unigene_694 |
vitellogenin |
Spodoptera litura |
ABU68426 |
2195 |
0.0 |
71% |
10106 |
| Unigene_3480 |
lipase 3 |
Danaus plexippus |
EHJ71865 |
344 |
5E-108 |
47% |
5201 |
| Unigene_17140 |
C-type lectin 5 |
Helicoverpa armigera |
AFI47450 |
411 |
4E-141 |
62% |
4131 |
| Unigene_2237 |
vitellogenin |
Spodoptera litura |
ABU68426 |
673 |
0.0 |
66% |
3500 |
| Unigene_688 |
translation elongation factor 2 |
Spodoptera exigua |
AAL83698 |
1565 |
0.0 |
96% |
3122 |
| Unigene_18106 |
apolipophorin precursor |
Bombyx mori |
BAK82317 |
4076 |
0.0 |
61% |
2457 |
| Unigene_692 |
apolipophorin precursor |
Manduca sexta |
AAB53254 |
2536 |
0.0 |
63% |
2274 |
| Unigene_17571 |
calcium ATPase |
Heliothis virescens |
AAD09820 |
1868 |
0.0 |
97% |
2048 |
| Unigene_2742 |
ADP/ATP translocase |
Helicoverpaarmigera |
AAP20934 |
397 |
2E-136 |
92% |
2039 |
| Unigene_17691 |
heat shock e protein 70 |
Agrotis ipsilon |
AEG78288 |
1131 |
0.0 |
100% |
1832 |
| Unigene_728 |
cathepsin |
Helicoverpa armigera |
NP_075125 |
263 |
9E-82 |
43% |
1783 |
| Unigene_7978 |
elongation factor 1 alpha |
Papili opolytes |
BAM18878 |
895 |
0.0 |
99% |
1769 |
| Unigene_178 |
apolipophorin III |
Trichoplusia ni |
ABV68867 |
288 |
2E-96 |
88% |
1709 |
| Unigene_780 |
acyl-CoA desaturase |
Helicoverpa assulta |
AF482909 |
692 |
0.0 |
94% |
1691 |
| Unigene_3446 |
myosin regulatory light chain 2 |
Bombyx mori |
NP_001091813 |
298 |
1E-99 |
98% |
1627 |
| Unigene_18120 |
fatty acid synthase |
Tribolium castaneum |
XP_970417 |
2436 |
0.0 |
57% |
1486 |
| Unigene_721 | Chemosensory protein | Heliothis virescens | ACX53806 | 206 | 3E-66 | 76% | 390 |
Candidate genes in the A. ipsilon pheromone gland with putative functions in pheromone production, transport and degradation
The overall enzymatic steps during pheromone biosynthesis in A. ipsilon are likely to be similar to those in other moth species, which include fatty acid synthesis, desaturation, chain shortening, reduction and acetylation [1,2,6]. By homologous searches we identified members of gene subfamilies in the A. ipsilon pheromone gland transcriptome putatively involved in these biosynthetic processes and pheromone production, including transcripts putatively encoding 3 synthases (2 actyl-CoA carboxylase and 1 fatty acid synthase), 5 desaturases, 13 acyl-CoA reductases, 5 alcohol oxidases and 5 acetyltransferases as well as 11 aldehyde reductases (Table 3); 17 transcripts encoding putative pheromone degradation enzymes (Table 4); 8 transcripts encoding putative CSPs and 7 transcripts encoding putative OBPs (Table 5). Their abundances in the pheromone gland transcriptome are shown in Figures 3 and 4. We further validated and characterized the expression level and the tissue distribution of these genes by RT-PCR and qRT-PCR and summarised below. There is a clear agreement between the transcript abundance estimated by the transcriptome sequencing and transcript expression level in the pheromone gland as measured by RT-PCR and qRT-PCR.
Table 3.
Putative pheromone biosynthesis related genes in the A. ipsilon pheromone gland
| Unigene* | Accession Num. | Length (bp) | No. of Reads | Putative identification | Species | Accession Num.§ | Score (bits) | E-value | % Identity |
|---|---|---|---|---|---|---|---|---|---|
|
Acetyl CoA Carboxylase |
|
|
|
|
|
|
|
|
|
| Unigene_2338-ACC |
JX989149 |
7534 |
305 |
acetyl-CoA carboxylase |
Tribolium castaneum |
XP_969851 |
2644 |
0.0 |
67% |
| Unigene_6244-ACC |
JX989150 |
621 |
17 |
acetyl-CoA carboxylase |
Heliothis virescens |
ACX53705 |
169 |
2E-47 |
56% |
|
Fatty acid synthase |
|
|
|
|
|
|
|
|
|
| Unigene_18120-FAS |
JX989151 |
8703 |
2341 |
fatty acid synthase |
Tribolium castaneum |
XP_970417 |
2436 |
0.0 |
57% |
|
Desaturases |
|
|
|
|
|
|
|
|
|
| Unigene_65-DES |
JX989152 |
1077 |
154 |
acyl-CoA delta 9 desaturase |
Mamestra brassicae |
ABX90048 |
681 |
0.0 |
96% |
| Unigene_741-DES |
JX989153 |
1047 |
630 |
acyl-CoA delta 11 desaturase |
Mamestra brassicae |
ABX90049 |
610 |
0.0 |
87% |
| Unigene_780-DES |
JX989154 |
1077 |
1710 |
acyl-CoA desaturase HassNPVE |
Helicoverpa assulta |
AF482909 |
692 |
0.0 |
94% |
| Unigene_10494-DES |
JX989155 |
254 |
3 |
desaturase |
Spodoptera littoralis |
AAQ74260 |
98.2 |
3E-22 |
64% |
| Unigene_15401-DES |
JX989156 |
489 |
9 |
acyl-CoA desaturaseSexiGATD |
Spodoptera exigua |
AAM28510 |
323 |
1E-110 |
93% |
|
Fatty acyl reductase |
|
|
|
|
|
|
|
|
|
| Unigene_163-FAR |
JX989157 |
1593 |
82 |
fatty-acyl CoA reductase 6 |
Ostrinia nubilalis |
ADI82779 |
536 |
0.0 |
59% |
| Unigene_1098-FAR |
JX989158 |
948 |
43 |
putative fatty acyl-CoA reductase |
Nasonia vitripennis |
XP_001600309 |
261 |
2E-119 |
51% |
| Unigene_2537-FAR |
JX989159 |
1560 |
105 |
putative fatty acyl-CoA reductase |
Bombus terrestris |
XP_003399879 |
585 |
0.0 |
57% |
| Unigene_3905-FAR |
JX989160 |
1593 |
37 |
putative fatty acyl-CoA reductase |
Apis mellifera |
ADI87410 |
541 |
0.0 |
59% |
| Unigene_4078-FAR |
JX989161 |
1017 |
14 |
fatty-acyl CoA reductase 3 |
Ostrinia nubilalis |
ADI82776 |
519 |
2E-178 |
80% |
| Unigene_4302-FAR |
JX989162 |
861 |
56 |
fatty-acyl CoA reductase 6 |
Danaus plexippus |
EHJ76493 |
372 |
8E-174 |
61% |
| Unigene_6708-FAR |
JX989163 |
615 |
7 |
fatty-acyl CoA reductase 4 |
Ostrinia nubilalis |
ADI82777 |
342 |
7E-113 |
77% |
| Unigene_7344-FAR |
JX989164 |
1380 |
43 |
fatty-acyl reductase |
Heliothis virescens |
ACX53790 |
671 |
0.0 |
70% |
| Unigene_8541-FAR |
JX989165 |
375 |
4 |
fatty-acyl CoA reductase 5 |
Ostrinia nubilalis |
ADI82778 |
193 |
4E-63 |
69% |
| Unigene_11561-FAR |
JX989166 |
182 |
2 |
fatty-acyl-CoA reductase |
Heliothis virescens |
ACX53773 |
104 |
2E-26 |
82% |
| Unigene_12329-FAR |
JX989167 |
855 |
|
fatty-acyl CoA reductase 6 |
Ostrinia nubilalis |
ADI82779 |
315 |
3E-101 |
66% |
| Unigene_12652-FAR |
JX989168 |
249 |
2 |
fatty-acyl-CoA reductase |
Heliothis virescens |
ACX53773 |
117 |
5E-31 |
76% |
| Unigene_15351-FAR |
JX989169 |
714 |
14 |
putative fatty acyl-CoA reductase |
Apis mellifera |
ADI87410 |
283 |
6E-90 |
56% |
| Unigene |
Accession Num. |
Length (bp) |
No. of Reads |
Putative identification |
Species |
Accession Num.§ |
Score (bits) |
E-value |
% Identity |
|
Alcohol oxidase |
|
|
|
|
|
|
|
|
|
| Unigene_195-AOX |
KC007341 |
834 |
25 |
Putative alcohol dehydrogenase |
Danaus plexippus |
EHJ70611 |
221 |
2E-67 |
43% |
| Unigene_307-AOX |
KC007342 |
969 |
56 |
Putative alcohol dehydrogenase |
Danaus plexippus |
EHJ70611 |
327 |
3E-106 |
55% |
| Unigene_397-AOX |
KC007343 |
756 |
253 |
alcohol dehydrogenase |
Heliothis virescens |
ACX53694 |
405 |
6E-139 |
78% |
| Unigene_7733-AOX |
KC007344 |
576 |
6 |
Putative alcohol dehydrogenase |
Danaus plexippus |
EHJ70611 |
219 |
7E-68 |
64% |
| Unigene_10714-AOX |
KC007345 |
501 |
3 |
Putative alcohol dehydrogenase |
Danaus plexippus |
EHJ73729 |
230 |
8E-72 |
68% |
|
Aldehyde reductase |
|
|
|
|
|
|
|
|
|
| Unigene_12563-AR |
KC007346 |
849 |
45 |
putative aldo-ketosereductase 1 |
Papilio dardanus |
CAW30924 |
394 |
2E-134 |
70% |
| Unigene_1274-AR |
KC007347 |
945 |
34 |
aldo-ketoreductase |
Bombyx mori |
ADQ89807 |
491 |
2E-168 |
78% |
| Unigene_17351-AR |
KC007348 |
930 |
43 |
aldo-ketoreductase 2E |
Bombyx mori |
BAL70378 |
342 |
1E-113 |
58% |
| Unigene_1774-AR |
KC007349 |
954 |
37 |
aldo-ketoreductase |
Helicoverpa armigera |
AEB26313 |
577 |
0.0 |
88% |
| Unigene_3134-AR |
KC007350 |
975 |
35 |
putative aldo-ketoreductase |
Danaus plexippus |
EHJ71186 |
410 |
9E-140 |
66% |
| Unigene_4806-AR |
KC007351 |
726 |
9 |
aldehyde reductase 1 |
Culex quinquefasciatus |
XP_001844836 |
302 |
1E-98 |
64% |
| Unigene_5103-AR |
KC007352 |
598 |
8 |
aldo-ketoreductase |
Heliothis virescens |
ACX53715 |
296 |
2E-96 |
72% |
| Unigene_7337-AR |
KC007353 |
456 |
5 |
aldo-ketoreductase |
Heliothis virescens |
ACX53798 |
236 |
2E-74 |
80% |
| Unigene_7554-AR |
KC007354 |
552 |
92 |
aldo-ketoreductase, partial |
Papilio xuthus |
BAM20078 |
183 |
4E-54 |
57% |
| Unigene_9245-AR |
KC007355 |
645 |
8 |
aldo-ketoreductase |
Papilio xuthus |
BAM19656 |
233 |
8E-73 |
53% |
| Unigene_9786-AR |
KC007356 |
963 |
28 |
aldo-ketoreductase |
Danaus plexippus |
EHJ68075 |
407 |
3E-135 |
63% |
|
Acetyltransferase |
|
|
|
|
|
|
|
|
|
| Unigene_173-ATF |
KC007357 |
795 |
43 |
acetyltransferase 1 |
Danaus plexippus |
EHJ65205 |
317 |
1E-145 |
71% |
| Unigene_407-ATF |
KC007358 |
855 |
196 |
acyltransferase |
Heliothis virescens |
ACX53812 |
494 |
5E-172 |
90% |
| Unigene_553-ATF |
KC007359 |
1038 |
141 |
putative acetyl transferase |
Bombyx mori |
NP_001182381 |
528 |
0.0 |
86% |
| Unigene_2015-ATF |
KC007360 |
552 |
67 |
acetyltransferase |
Danaus plexippus |
EHJ65977 |
327 |
6E-110 |
90% |
| Unigene_15362-ATF | KC007361 | 1010 | 69 | putative acetyl-CoA acetyltransferase | Danaus plexippus | EHJ68573 | 444 | 3E-150 | 74% |
*Gene names as deposited in GenBank.§Genbank accession number of the homologous gene. ACC Acetyl-CoA carboxylase, FAS Fatty acid synthase, DES Desaturase, FAR Fatty acyl reductase, AOX alcohol oxidase, AR Aldehyde reductase, ATF Acetyltransferase.
Table 4.
Candidate esterase genes likely involved in A. ipsilon pheromone degradation
| Gene name* | Unigene | Accession num. | Length (bp) | No. of reads | Putative identification | Species | Accession num.§ | Score (bits) | E-value | % Identity |
|---|---|---|---|---|---|---|---|---|---|---|
| AipsCXE1 |
Unigene_8856 |
JX866730 |
459 |
4 |
esterase |
Spodoptera littoralis |
ABH01081 |
219 |
9E-83 |
75% |
| AipsCXE2 |
Unigene_1631 |
JX866731 |
1148 |
22 |
antennal esterase CXE2 |
Spodoptera littoralis |
ACV60229 |
475 |
5E-161 |
67% |
| AipsCXE3 |
Unigene_1377 |
JX866732 |
969 |
20 |
antennal esterase CXE3 |
Spodoptera littoralis |
ACV60230 |
518 |
2E-178 |
82% |
| AipsCXE4 |
Unigene_7378 |
JX866733 |
504 |
6 |
antennal esterase CXE4 |
Spodoptera littoralis |
ACV60231 |
252 |
6E-78 |
77% |
| AipsCXE5 |
Unigene_4213 |
JX866734 |
1047 |
13 |
antennal esterase CXE5 |
Spodoptera littoralis |
ACV60232 |
647 |
0.0 |
87% |
| AipsCXE6 |
Unigene_4686 |
JX866735 |
554 |
7 |
antennal esterase CXE6 |
Spodoptera littoralis |
ACV60233 |
229 |
2E-69 |
75% |
| AipsCXE7 |
Unigene_9837 |
JX866736 |
746 |
5 |
antennal esterase CXE7 |
Spodoptera littoralis |
ACV60234 |
338 |
7E-110 |
70% |
| AipsCXE8 |
Unigene_4231 |
JX866737 |
907 |
11 |
antennal esterase CXE8 |
Spodoptera littoralis |
ACV60235 |
381 |
5E-126 |
69% |
| AipsCXE9 |
Unigene_7661 |
JX866738 |
512 |
6 |
antennal esterase CXE9 |
Spodoptera littoralis |
ACV60236 |
226 |
5E-68 |
64% |
| AipsCXE10 |
Unigene_68 |
JX866739 |
1674 |
54 |
antennal esterase CXE10 |
Spodoptera littoralis |
ACV60237 |
387 |
6E-123 |
41% |
| AipsCXE11 |
Unigene_1628 |
JX866740 |
336 |
9 |
antennal esterase CXE11 |
Spodoptera littoralis |
ACV60238 |
160 |
1E-74 |
83% |
| AipsCXE12 |
Unigene_4126 |
JX866741 |
444 |
16 |
antennal esterase CXE12 |
Spodoptera littoralis |
ACV60239 |
192 |
3E-53 |
50% |
| AipsCXE13 |
Unigene_15183 |
JX866742 |
1843 |
25 |
antennal esterase CXE13 |
Spodoptera littoralis |
ACV60240 |
974 |
0.0 |
83% |
| AipsCXE14 |
Unigene_7537 |
JX866743 |
451 |
5 |
antennal esterase CXE14 |
Spodoptera littoralis |
ACV60241 |
256 |
5E-80 |
85% |
| AipsCXE15 |
Unigene_5546 |
JX866744 |
1380 |
19 |
antennal esterase CXE15 |
Spodoptera littoralis |
ACV60242 |
498 |
5E-167 |
52% |
| AipsCXE16 |
Unigene_7483 |
JX866745 |
474 |
6 |
antennal esterase CXE16 |
Spodoptera littoralis |
ACV60243 |
268 |
3E-84 |
77% |
| AipsCXE20 | Unigene_5325 | JX866746 | 622 | 8 | antennal esterase CXE20 | Spodoptera littoralis | ACV60247 | 343 | 2E-112 | 81% |
*Gene names as deposited in GenBank. §Genbank accession number of the homologous gene. CXE carboxylesterase.
Table 5.
Candidate olfactory genes involved in A. ipsilon pheromone reception
| Gene Name* | Unigene | Accession num. | Length (AA) | Signal peptide | No. of reads | Putative identification | Species | Accession num.§ | Score (bits) | E-value | % Identity |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Chemosensory proteins | |||||||||||
| AipsCSP1 |
Unigene_468 |
JX863696 |
124 |
1-16 aa |
21 |
chemosensory protein |
Heliothis virescens |
ACX53825 |
130 |
5E-36 |
59% |
| AipsCSP2 |
Unigene_1704 |
JX863697 |
119 |
1-16 aa |
117 |
chemosensory protein |
Papili oxuthus |
BAF91716 |
159 |
8E-48 |
66% |
| AipsCSP3 |
Unigene_1767 |
JX863698 |
128 |
1-18 aa |
19 |
chemosensory protein |
Mamestra brassicae |
AAF71290 |
223 |
2E-72 |
83% |
| AipsCSP4 |
Unigene_2047 |
JX863699 |
120 |
1-16 aa |
46 |
chemosensory protein 2 |
Helicoverpa armigera |
AEX07265 |
222 |
2E-72 |
86% |
| AipsCSP5 |
Unigene_6052 |
JX863700 |
107 |
1-18 aa |
16 |
chemosensory protein |
Danaus plexippus |
EHJ67380 |
186 |
3E-55 |
84% |
| AipsCSP6 |
Unigene_15000 |
JX863701 |
127 |
1-18 aa |
13 |
chemosensory protein 2 |
Heliothis virescens |
AAM77040 |
227 |
3E-74 |
87% |
| AipsCSP7 |
Unigene_15308 |
JX863702 |
128 |
1-16 aa |
18 |
chemosensory protein |
Heliothis virescens |
ACX53804 |
211 |
5E-68 |
75% |
| AipsCSP8 |
Unigene_721 |
JX863703 |
123 |
1-18 aa |
390 |
chemosensory protein |
Heliothis virescens |
ACX53806 |
206 |
3E-66 |
76% |
|
Odorant binding proteins | |||||||||||
| AipsOBP1 |
Unigene_520 |
JX863689 |
183 |
ND |
44 |
odorant binding protein |
Heliothis virescens |
ACX53761 |
197 |
2E-60 |
52% |
| AipsOBP2 |
Unigene_2120 |
JX863690 |
148 |
1-21 aa |
44 |
pheromone binding protein 4 |
Mamestra brassicae |
AAL66739 |
241 |
6E-77 |
79% |
| AipsOBP3 |
Unigene_6517 |
JX863691 |
108 |
ND |
10 |
odorant-binding protein 19 |
Helicoverpa armigera |
AFM93773 |
127 |
4E-34 |
53% |
| AipsOBP4 |
Unigene_8860 |
JX863692 |
121 |
ND |
4 |
antennal binding protein |
Heliothis virescens |
CAC33574 |
173 |
7E-53 |
65% |
| AipsOBP5 |
Unigene_15218 |
JX863693 |
137 |
1-16 aa |
20 |
odorant binding protein |
Heliothis virescens |
ACX53795 |
193 |
2E-60 |
70% |
| AipsOBP6 |
Unigene_15711 |
JX863694 |
125 |
ND |
5 |
odorant binding protein |
Heliothis virescens |
ACX53743 |
243 |
7E-80 |
86% |
| AipsOBP7 | Unigene_15861 | JX863695 | 145 | 1-23 aa | 4 | odorant binding protein 3 | Helicoverpa armigera | AEB54582 | 97.8 | 9E-23 | 38% |
*Gene names as deposited in GenBank. §Genbank accession number of the homologous gene. ND, not detected signal peptide because the N-terminus is missing. CSP chemosensory protein, OBP odorant binding protein.
Figure 3.
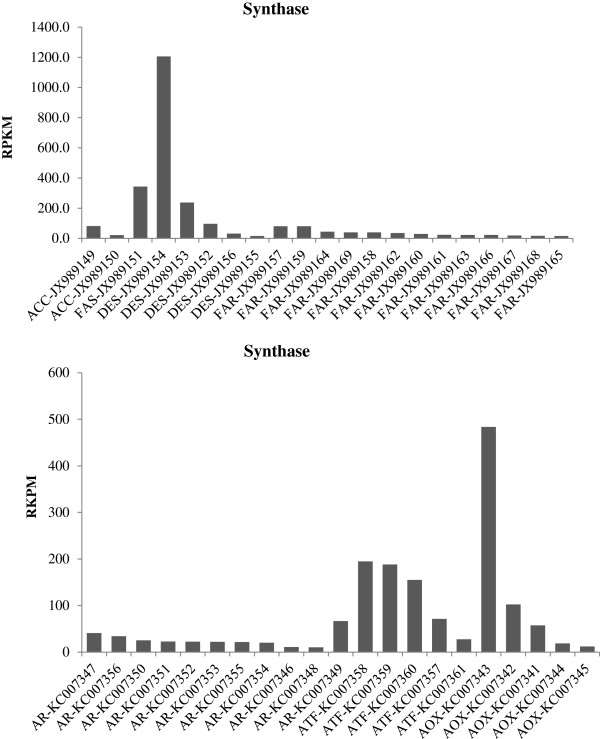
The abundance of the unigenes encoding the sex pheromone synthase in the A. ipsilon transcriptome dataset presented as normalized read count in reads per kilobase per million mapped reads (RPKM). The putative enzyme names are indicated as gene abbreviations followed by Genbank accession numbers. ACC Acetyl-CoA carboxylase, AOX Alcohol oxidase, AR Aldehyde reductase, ATF Acetyltransferase, DES Desaturase, FAR Fatty acyl reductase, FAS Fatty acid synthase.
Figure 4.
The abundance of unigenes encoding chemosensory proteins (CSPs), odorant-binding proteins (OBPs) and esterase (EST) in the A. ipsilon transcriptome dataset presented as normalized reads in reads per kilobase per million mapped reads (RPKM).
Receptor for the pheromone biosynthesis activating neuropeptide (PBAN)
PBAN is released from the suboesophagal ganglion in the brain and goes to the hemolymph, where it binds to the PBAN receptor in the membrane of the pheromone gland and triggers the pheromone production [42,43]. Although there was no PBAN receptor found in the pheromone gland transcriptome of H. virescens[39] we found one transcript (Unigene_3821) encoding a protein highly homologous to PBAN receptor isoform B. It has very low abundance in the A. ipsilon transcriptome (31 RPKM) but high amino acid identity of 97% to H. virescens PBAN receptor in GenBank (Protein IDs: ABU93813) [44].
Acetyl-CoA carboxylase (ACC)
Saturated long chain fatty acids are the precursors of sex pheromones in most moth species. Their biosynthesis is started by ACC catalysing the production of malonyl-CoA from acetyl-CoA in the first committed biosynthesis step [8,9]. In the A. ipsilon pheromone gland we found two transcripts (ACC-JX989149 and ACC-JX989150) encoding ACCs. ACC-JX989149 with an open reading frame (ORF) of 5841 bp encodes for a ACC with 67% amino acid identity with the ACC of T. castaneum (Protein ID: XP_969851) and ACC-JX989150 encodes a protein with 56% amino acid identity with the ACC of H. virescens (Protein ID: ACX53705) (Table 3). The RT-PCR and qRT-PCR revealed that both ACC-JX989149 and ACC-JX989150 are highly expressed in the pheromone gland as compared to the body (Figure 5 and Figure 6). However, they have very low abundance (81 and 21 RPKM) in the transcriptome (Figure 3).
Figure 5.
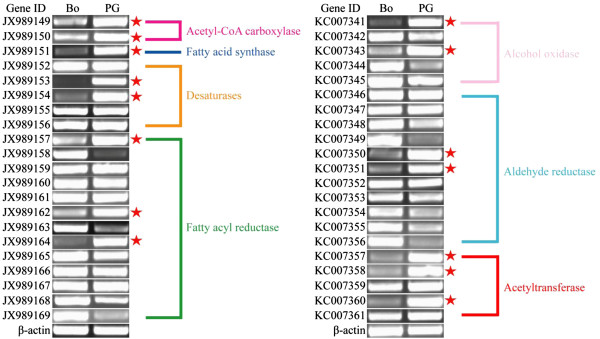
RT-PCR results showing the relative expression of the A. ipsilon pheromone biosynthesis-related genes in pheromone gland (PG) and the body (BO). The genes that are more highly expressed in the pheromone gland are labeled with red pentagram. β-actin was used as internal reference gene to test the integrity of each cDNA templates; the similar intensity of β-actin bands between the pheromone gland and the body part indicate the use of equal template concentrations.
Figure 6.
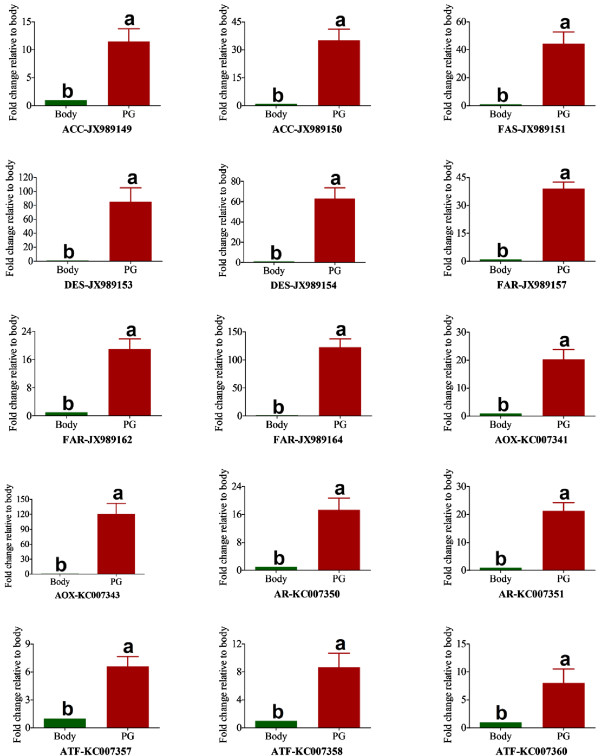
qRT-PCR results showing the relative expression levels of the A. ipsilon pheromone biosynthesis related genes between the pheromone gland (PG) and the body (BO). The putative enzyme names are indicated as gene abbreviations followed by Genbank accession numbers. ACC Acetyl-CoA carboxylase, FAS Fatty acid synthase, DES Desaturase, FAR Fatty acyl reductase, AOX alcohol oxidase, AR Aldehyde reductase, ATF Acetyltransferase. The internal control β-actin and ribosomal protein S3 were used to normalize transcript levels in each sample. This figure was presented using β-actin as reference gene to normalize the target gene expression and correct sample-to-sample variation; similar results were also obtained with ribosomal protein S3 as reference gene. The standard error is represented by the error bar, and the different letters (a, b) above each bar denote significant differences (p >0.05).
Fatty acid synthase (FAS)
FAS has been shown to catalyse the conversion of malonyl-CoA and NADPH to produce saturated fatty acids [8]. We identified one putative FAS transcript (FAS-JX989151) in the A. ipsilon pheromone gland (Table 3), containing an ORF of 7176 bp and encoding a FAS with 57% amino acid identity to the FAS of T. castaneum (Protein ID: XP_970417). The RT-PCR and qRT-PCR revealed that FAS-JX989151 is highly expressed in the pheromone gland (40-fold higher than in the body, Figure 5 and Figure 6) and also has a high abundance (343 RPKM) in the transcriptome (Figure 3).
Desaturase (DES)
Pheromone-specific desaturases introduce double bond(s) into the fatty acids at specific positions along the chain. Five putative sex pheromone components extracted from A. ipsilon sex pheromone gland are unsaturated fatty acids with acetate as the functional group and 16 or less carbons [38]. At least three active pheromone components (Z7-12:OAc, Z9-14:OAc and Z11-16:OAc) have been identified in A. ipsilon strains from China [38], North America [45], France [46] and Japan [47]. It is reasonable to propose that the saturated fatty acid precursor of A. ipsilon sex pheromones would be palmitic acid (16:0) which is desaturated by ∆11-desaturase to form the precursor Z11-16:acyl-CoA for the production of two major (Z7-12:OAc and Z9-14:OAc) and two minor (Z11-16:OAc and Z5-10:Ac) pheromone components (Figure 7). It is not clear how the minor pheromone component (Z8-12:OAc) is synthesized in A. ipsilon, which should involve a ∆12-desaturase. Other studies in Lepidoptera species support a ∆11-desaturase acting on palmitic acid and leading to the production of the sex pheromone components [19,20,48]. In the A. ipsilon pheromone gland transctiptome 5 transcripts have high homology to genes encoding desaturases (Table 3). DES-JX989152 is homologous to a gene encoding an acyl-CoA ∆9-desaturase in M. brassicae (Protein ID: ABX90048) with an amino acid identity of 96%. ∆9-desaturase makes oleic acid from stearic acid (18:0) and possibly palmitoleic acid from palmitic acid [16,17,49]. It would not participate in the biosynthesis of A. ipsilon sex pheromones. DES-JX989153 encodes a protein with 87% amino acid identity with the acyl-CoA ∆11 desaturase of M. brassicae (Protein ID: ABX90049). DES-JX989154, DES-JX989155 and DES-JX989156 encode proteins, respectively, with 94% amino acid identity to the acyl-CoA desaturase from H. assulta (Protein ID: AF482909), 64% amino acid identity to a S. littoralis desaturase (Protein ID: AAQ74260) and 93% amino acid identity to an acyl-CoA desaturase of S. exigua (Protein ID: AAM28510). These transcripts could possibly encode ∆12-desaturases in A. ipsilon in formation of the minor pheromone component Z8-12:OAc from the precursor Z12-16:acyl-CoA. However, they could also function as ∆9-desaturase. Further study on their enzyme activity could confirm their role in the sex pheromone biosynthesis. The RT-PCR and qRT-PCR results indicated that DES-JX989153 and DES-JX989154 are highly expressed in the A. ipsilon pheromone gland compared with the body (85 and 63 fold higher, respectively) (Figure 5 and Figure 6). One of the transcripts (DES-JX989154) is also highly abundant (1206 RPKM) in the pheromone gland transcriptome (Figure 3), suggesting a possible role in A. ipsilon sex pheromone biosynthesis.
Figure 7.
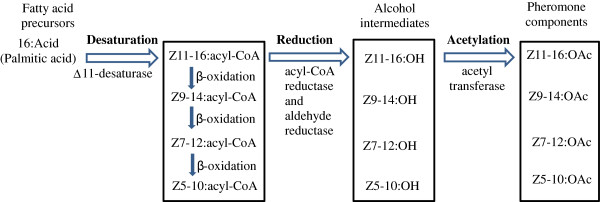
Putative biosynthesis pathways of the sex pheromones in Agrotis ipsilon. The saturated fatty acid precursor palmitic acid (16:0) is desaturated by ∆11-desaturase to form the precursor Z11-16:acyl-CoA for the production of three major and one minor pheromone components (adapted from [2,6,12,13,50]).
Fatty acyl-CoA reductase (FAR)
Once a specific Δ11 and possibly Δ12 double bond is introduced into fatty acid precursors to form a fatty acyl-CoA precursor, the chain of the precursors is then shortened sequentially by ß–oxidation to form different shorter chain fatty acyl-CoA precursors [6]. These precursors are further reduced individually by fatty acyl reductase (FAR) to form corresponding fatty alcohols [26,28,51]. In the A. ipsilon pheromone gland transcriptome there are 13 transcripts homologous to putative FAR genes (Table 3). Among them, 5 transcripts encode proteins with 59%-80% amino acid identity to the fatty-acyl CoA reductases of Ostrinia nubilalis (Protein IDs: ADI82776, ADI82777, ADI82778 and ADI82779). Other FAR transcripts are homologous to the fatty acyl-CoA reductase from a wide range of insect species including H. virescens, N. vitripennis, Danaus plexippus, Bombus terrestris and Apis mellifera with amino acid identities of about 60% (Table 3). The RT-PCR and qRT-PCR results indicated that three transcripts (FAR-JX989157, FAR-JX989162 and FAR-JX989164) are highly expressed in the pheromone gland (Figure 5 and Figure 6). The other ten transcripts seem equally expressed in the pheromone gland and the body or highly expressed in the body. All FAR transcripts except two (FAR-JX989157 and FAR-JX989159) have low abundance (from 81 and 16 RPKM) in the pheromone gland transcriptome (Figure 3).
Alcohol oxidase/dehydrogenase (AOX)
Fatty alcohols can be used as pheromone components in many moth species, and they are also pheromone intermediates to produce aldehyde pheromones by the alcohol oxidases [52,53]. In the A. ipsilon PG 5 homologous genes of alcohol oxidase/dehydrogenase were identified, the BLASTx results revealed three unigenes (AOX-KC007341, AOX-KC007342 and AOX-KC007344) are with the amino acid identity of 43%, 55% and 64%, respectively, to a putative alcohol dehydrogenase of D. plexippus (Protein ID: EHJ70611), and one unigene (AOX-KC007345) are homologous to another putative alcohol dehydrogenase of D. plexippus (Protein ID: EHJ73729 ) with the amino acid identity of 68%. AOX-KC007343 showed 78% amino acid identity with the alcohol dehydrogenase of H. virescens (Protein ID: ACX53694). The RT-PCR and qRT-PCR results indicated that AOX-KC007341 and AOX-KC007343 showed a higher expressed level in the PG than in the body (Figure 5 and Figure 6).
Aldehyde reductase (AR)
Aldehyde reductases are members of the aldo-ketoreductase superfamily and could be used to reduce long-chain acyl-CoA to form alcohol intermediates [13]. In the A. ipsilon pheromone gland we identified 11 transcripts with homology to the aldo-ketoreductases of Papilio dardanus, B. mori, H. armigera, D. plexippus, Culex quinquefasciatus, H. virescens and Papilio xuthus (Table 3). The derived protein sequences of these 11 transcripts show 53%-88% amino acid identity with their homologs in other insects. The RT-PCR and qRT-PCR results indicated that AR-KC007350 and AR-KC007351 are mainly expressed in the pheromone gland, while the other 9 putative aldehyde reductase transcripts have equal expression levels between the pheromone gland and the body or a higher expression level in the body (Figure 5 and Figure 6). All aldehyde reductase transcripts are present at low abundance (from 67 to 10 RPKM) in the pheromone gland transcriptome (Figure 3). The involvement of aldehyde reductase in sex pheromone biosynthesis has not been demonstrated in moth species.
Acetyltransferase (ATF)
The fatty acid alcohols are used as pheromone components in many moth species. In A. ipsilon whose sex pheromone blends comprise only acetates, they are intermediates and acetylated to pheromone components as acetate esters by actyltransferases [13]. In the A. ipsilon pheromone gland transcriptome 5 acetyltransferase homologous transcripts were identified (Table 3), 3 of them (ATF-KC007357, ATF-KC007360 and ATF-KC007361) encode proteins that are homologous to the acetyltransferase of D. plexippus (Protein IDs: EHJ65205, EHJ65977 and EHJ68573) with relatively high amino acid identities (<70%), one (ATF-KC007358) encodes a protein with 90% amino acid identity to H. virescens acetyltransferase (Protein ID: ACX53812) and one (ATF-KC007359) encodes a protein with 86% amino acid identity with the acetyltransferase of B. mori (Protein ID: NP_001182381). The RT-PCR and qRT-PCR revealed that three transcripts (ATF-KC007358, ATF-KC007360 and ATF-KC007357) are mainly expressed in the pheromone gland (Figure 5 and Figure 6) and have a relative high abundance of 195, 155 and 71 RPKM, respectively in the pheromone gland transcriptome (Figure 3).
Genes encoding candidate pheromone degrading enzymes in the A. ipsilon pheromone gland
It would be potentially harmful to insects if pheromone molecules and other odorants remained on the olfactory receptors after they had stimulated the olfactory receptor neurons (ORNs). It is therefore thought that there are mechanisms to protect the ORNs by odorant degrading enzymes (ODEs) [37] including esterases [54,55], aldehyde oxidases [56-58], cytochromes P450 [59-61], carboxyl esterase [62], and glutathione S-transferase (GST) [63]. In this study, we identified 17 transcripts predicted to encode esterases in the A. ipsilon pheromone gland, and the BLASTx results showed that all have very high amino acid identities with the antennal esterases of S. littoralis (Table 4), we named them as AipsCXE1-AipsCXE16 and AipsCXE20 following the nomenclature in S. littoralis. Our qRT-PCR results revealed that 7 of the transcripts (AipsCXE3, AipsCXE7, AipsCXE8, AipsCXE9, AipsCXE11, AipsCXE14 and AipsCXE20) are antennal-enriched, 3 (AipsCXE5, AipsCXE10 and AipsCXE15) are both antennal- and pheromone gland-enriched and the remaining 7 (AipsCXE1, AipsCXE2, AipsCXE4, AipsCXE6, AipsCXE12, AipsCXE13 and AipsCXE16) have similar expression levels in antennae, body and pheromone gland, suggesting they are not pheromone specific (Figure 8).
Figure 8.
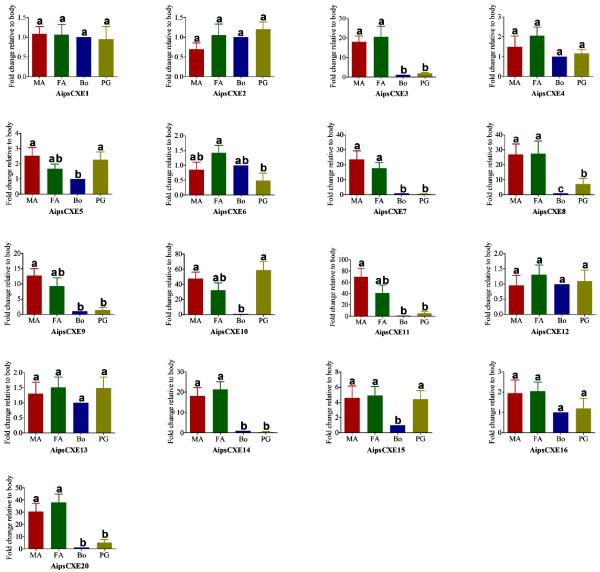
qRT-PCR results showing the expression of A. ipsilon unigenes encoding the putative esterase (CXE) identified in the pheromone gland in the male antennae (MA), the female antennae (FA), the body (BO) and the pheromone gland (PG). The standard error is represented by the error bar, and the different letters (a, b, c) above each bar denote significant differences (p > 0.05).
Genes encoding candidate pheromone carrier proteins in the A. ipsilon pheromone gland
Moth sex pheromones are synthesised and protected from degradation until being released from the female pheromone gland and it has been proposed that OBPs and CSPs could participate in this process. In this study we have identified transcripts of 7 OBPs and 8 CSPs from the A. ipsilon pheromone gland (Table 5), all of these have the typical insect OBP sequence motif C1-X15-39-C2-X3-C3-X21-44-C4-X7-12-C5-X8-C6 [35,64] or CSP sequence motif C1-X6-8-C2-X16-21-C3-X2-C4[65]. One CSP transcript, AipsCSP2 seems to be gland-specific and has an extremely high expression level (<100 folds) in the pheromone glands compared with the antennae and body and a relative high abundance in the pheromone gland transcriptome. AipsCSP8 shows a higher expression level in the pheromone gland (10-fold higher than in body) (Figure 9) and is extremely abundant with 1,364 RPKM in the pheromone gland transcriptome (Figure 4).
Figure 9.
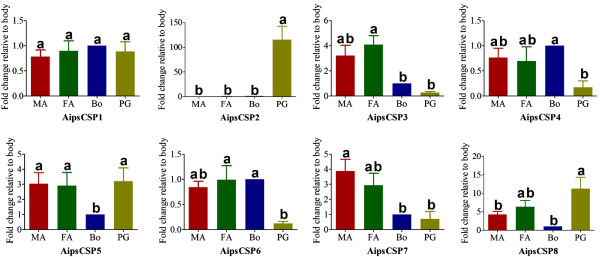
qRT-PCR results showing the relative expression of the A. ipsilon unigenes encoding putative chemosensory proteins (CSP) identified in the pheromone gland in the male antennae (MA), the female antennae (FA), the body (BO) and the pheromone gland (PG). The standard error is represented by the error bar, and the different letters (a, b) above each bar denote significant differences (p >0.05).
There is one OBP transcript (AipsOBP6) which is highly expressed in the pheromone gland (more than 3-fold higher than in the antennae), and 3 OBPs (AipsOBP1, AipsOBP2 and AipsOBP4) are highly expressed in the antennae (Figure 10). This high expression of OBPs and CSPs in the pheromone gland is interesting because it suggests a possible involvement in carrying and releasing sex pheromones as demonstrated for the antennal OBPs and CSPs. However, the molecular mechanisms that connect these proteins with the involvement of pheromone production needs further investigation. No ORs, IRs and SNMPs are identified in the A. ipsilon pheromone gland.
Figure 10.
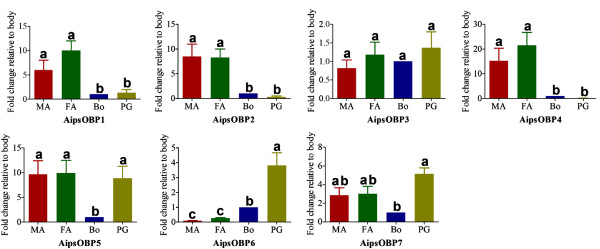
qRT-PCR results showing the relative expression of the A. ipsilon unigenes encoding putative odorant binding proteins (OBP) identified in the pheromone gland in the male antennae (MA), the female antennae (FA), the body (BO) and the pheromone gland (PG). The standard error is represented by the error bar, and the different letters (a, b, c) above each bar denote significant differences (p > 0.05).
Conclusions
The black cutworm A. ipsilon is a destructive pest of many crops [66,67] and mainly controlled by chemical pesticides, which has led to the development of resistance to various compounds [68]. Our study provides information and resource to identify and facilitate functional studies of genes responsible for pheromone production, transport and degradation at the molecular level both in vivo and in vitro.
By deep sequencing of the A. ipsilon sex pheromone gland transcriptome, we have identified 42 transcripts encoding enzymes putative involved in pheromone production. This is the first study reporting the key enzyme ∆11-desaturase involved in A. ipsilon sex pheromone biosynthesis. One new transcript (DES-JX989154) encoding a desaturase is highly abundant in the transcriptome and highly expressed in the pheromone gland, suggesting this desaturase encoded by DES-JX989154 or other newly identified transcripts (DES-JX989155 and DES-JX989156) may play important roles in A. ipsilon sex pheromone biosynthesis. They may contribute in the introducing a double bond at C11 and C12 positions of the saturated fatty acid precursor palmitic acid for the production of pheromone precursors. Further studies are needed to confirm the substrates and the products thus the involvement of these desaturases and other newly identified genes such as those encoding for aldehyde reductases and acetyltransferases in A. ipsilon sex pheromone biosynthesis. Two of the CSPs are highly abundant transcripts (AipsCSP2 and AipsCSP8) with 100- and 10-fold higher transcription level, respectively than in the body. Furthermore AipsCSP2 and AipsOBP6 are pheromone gland-specific and –enriched, respectively (Figure 9 and Figure 10). This suggests a functional role of the PG-enriched CSPs and OBPs in sex pheromone transport and release. It is clear that during perireceptor event after pheromones and odorants enter the sensillun lymph that the antennae-specific odorant binding proteins (OBPs) capture these hydrophobic pheromone and odorant and deliver them to the membrane-bound olfactory receptors (ORs) [35]. Further study of these PG-expressed OBPs, especially their binding to sex pheromone components is needed to confirm its function.
Methods
Insect material
The A. ipsilon colony has been reared in our laboratory (State Key Laboratory for Biology of Plant Diseases and Insect Pests, Chinese Academy of Agricultural Sciences, Beijing, China) since 2006 with field-collected moths introduced each summer to prevent inbreeding effects. The larvae were reared on an artificial diet comprising wheat germ, casein and sucrose as the main components. The colony was kept at 24°C with 75% relative humidity and a 14h:10h light:dark photoperiod. Pupae were sexed and kept separately in hyaline plastic cups before emergence. Adult moths were given 20% honey solution after emergence.
Pheromone gland dissection
The pheromone gland plus associated ovipositor valves and parts of the terminal abdominal segments were dissected with fine scissors [39] from the rest of the body parts refereed as ‘body’ which comprises of heads, thoraxes, legs, wings and abdomens (without the pheromone glands). The calling behavior of female A. ipsilon moths begins on the first night after eclosion and increases sharply, peaking on the third night [38]. So in order to cover all genes involved in pheromone biosynthesis, four glands of 1-day-old females, four glands of 2-day-old females and ten glands of 3-day-old females were dissected during the second half of the scotophase, which is reported to be the calling period of this moth [69-71]. The eighteen glands were mixed in one RNase-free centrifuge tube for total RNA extraction and frozen in liquid nitrogen until further processing.
RNA extraction and cDNA library construction
Total RNA was extracted using TRIzol regent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The quantity of RNA was determined using a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and 1.1% agarose gel electrophoresis. About 500 ng mRNA was further purified from 50 μg total RNA using the polyATtract mRNA isolation system III (Promega, Madison, WI, USA). The mRNA was then sheared into about 800 nucleotides using a RNA fragmentation solution (Autolab, Beijing, China) at 70°C for 30 sec, and then cleaned and condensed using RNeasyMinElute RNA Cleaning Up kit (Qiagen, Valencia, CA, USA). The mRNA was used as a template for first-strand cDNA synthesis using N6 random primers and MMLV reverse transcriptase (TaKaRa, Dalian, China) and the second strands were synthesized using Secondary Strand cDNA synthesis enzyme mixtures (Autolab, Beijing, China). cDNAs with appropriate length were purified with the QIAquick PCR Purification kit (Qiagen, Valencia, CA, USA) and eluted with 10 μl Elution Buffer. After blunt ending and the addition of a poly-A tail at the 3’ end according to the Roche’s Rapid Library Preparing protocols (Roche, USA), the purified cDNAs were linked to GS-FLX sequencing Adaptors (Roche, USA). Finally, the cDNAs shorter than 500 bp were removed using Ampure Beads according to the manufactures’ instruction (Beckman, USA) before the preparation of the cDNA library.
454 sequencing
Pyrosequencing of the cDNA library was performed by Beijing Autolab Biotechnology Company using a 454 GS-FLX sequencer (Roche, IN, USA). All sequencing reads were deposited into the Short Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under the accession number SRX189143.
Sequence analysis and assembly
Base calling of the raw 454 reads in SFF files was carried out using the python script sff_extract.py developed by COMAV (http://bioinf.comav.upv.es). All raw reads were then processed to remove low quality and adaptor sequences using programs tagdust [72], LUCY [73] and SeqClean [74] with default parameters. The resulting sequences were then screened against the NCBI UniVec database (http://www.ncbi.nlm.nih.gov/VecScreen/UniVec.html) to remove possible vector sequence contamination. Cleaned reads shorter than 60 bases were discarded because they are likely to be sequencing artifacts [75].
Two steps were taken to assemble the clean reads. First MIRA3 [76] was used with the assembly settings of minimum sequence overlap of 30 bp and minimum percentage overlap identity of 80%. Then CAP3 was used with assembly parameters of overlap length cutoff <30 and overlap percent identity cutoff <90% [77]. The resulting contigs and singletons of more than 100 bases were retained as unigenes and annotated as described below.
Homology searches and functional classification
Following the assembly, homology searches of all unigenes were performed using BLASTx and BLASTn programs against the GenBank non-redundant protein (nr) and nucleotide sequence (nt) database at NCBI [78]. Matches with an E-value less than 1.0E-5 were considered significant [79]. Gene names were assigned to each unigene based on the best BLASTx hit with the highest score value.
Gene Ontology terms were assigned by Blast2GO [80] through BLASTx program with an E-value less than 1.0E-5. Then, WEGO [81] software was used for assignment of each GO ID to the related ontology entries. The longest open reading frame (ORF) of each unigene was determined by an ORF finder tool (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
Pheromone gland ESTs from other insects
The H. virescens pheromone gland ESTs (14112 with accession number: GR958232-GR972305, GT067784-GT067747) [39], the A. segetum pheromone gland ESTs (2286 with accession number: ES582156-ES584441) [82] and the B. mori pheromone gland ESTs (10501 with accession number: BP184340-BP182009; AV404455-AV403746; DC552314-DC544856) were downloaded from the dbEST database at NCBI (http://www.ncbi.nlm.nih.gov/nucest) and saved as fasta files. All the EST sequences were assembled using the CAP3 program with the same parameters as used in the A. ipsilon assembly. The comparative analyses of A. ipsilon, H. virescens, B. mori and A. segetum pheromone gland unigenes were performed based on the Best Bidirectional Hits results (reciprocal BLASTn, E-value less than 1.0E-6).
Identification of candidate genes associated with moth pheromone biosynthesis
Some putative genes and enzymes have been reported previously as being involved in moth sex pheromone production. We focused our research on the target genes: (1) Acetyl-CoA carboxylase; (2) Fatty acid synthase; (3) Desaturase; (4) Fatty acyl reductase; (5) Alcohol oxidase; (6) Aldehyde reductase; (7) Acetyltransferase.
Identification of putative genes involved in pheromone degradation
Since the sex pheromone blend of A. Ipsilon is comprised of acetate esters (Z)-7-dodecenyl acetate (Z7-12:Ac) (40.5%), (Z)-9-tetradecenyl acetate (Z9-14:Ac) (13.2%), (Z)-11-hexadecenyl acetate (Z11-16:Ac) (14.9%), (Z)-8-dodecenyl acetate (Z8-12:Ac) (17.2%) and (Z)-5-decenyl acetate (Z5-10:Ac) (14.3%) [38], esterases may play a major role in pheromone degradation. Therefore, we performed BLASTx and BLASTn searches to identify candidate esterase genes in the A. ipsilon pheromone gland NGS dataset.
Identification of putative genes involved in pheromone transport
Genes encoding odorant binding proteins (OBPs) and chemosensory proteins (CSPs) were identified using the “OBP sequence motif” C1-X15-39-C2-X3-C3-X21-44-C4-X7-12-C5-X8-C6 [64] and the “CSP sequence motif” C1-X6-8-C2-X16-21-C3-X2-C4, [65]. Candidate olfactory receptors (ORs), ionotropic receptors (IRs), sensory neuron membrane proteins (SNMPs) genes were identified by BLASTx and BLASTn searches.
Sequence analyses
The putative N-terminal signal peptides and most likely cleavage sites were predicted by the SignalP V3.0 program [83] (http://www.cbs.dtu.dk/services/SignalP/). Sequence alignments were done with ClustalX 1.83 [84] with default gap penalty parameters of gap opening 10 and extension 0.2.
RT-PCR and qRT-PCR
The cDNAs from female pheromone glands and other body parts (mixture of heads, thoraxes, legs, wings and abdomens (without the pheromone glands)) were synthesized using PrimeScript RT Reagent with gDNA Eraser (TaKaRa, Dalian, China). 200 ng cDNA was used as RT-PCR and qRT-PCR templates. Specific primer pairs for RT-PCR analysis were designed with Primer 3 (http://frodo.wi.mit.edu/) or Primer Premier 5 (see Additional file 4). To test the integrity of the cDNA templates, a pair of control primers for the β-actin (GenBank Acc. JQ822245) of A. ipsilon was used. The PCR cycling profile was: 95°C for 2 min, followed by 35 cycles of 95°C for 30 sec, 60°C for 30 sec, 72°C for 1 min and a final extension for 10 min at 72°C. PCR products were separated in 1.2% agarose gels and stained with ethidium bromide. Each reaction was done at least six times with three biological replicates.
qRT-PCR analysis was conducted using the ABI 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA). The primers were designed by Beacon Designer 7.90 (PREMIER Biosoft International) (see Additional file 5). Two reference genes, β-actin (GenBank Acc. JQ822245) and ribosomal protein S3 (GenBank Acc. JQ822246) were used for normalizing expression of the target gene and correcting for sample-to-sample variation. qRT-PCRs were done in a 25 μl reaction containing 12.5 μl of Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, Shanghai, China), 0.5 μl of each primer (10 pmol/ μl), 0.5 μl of Rox Reference Dye, 1 μl of sample cDNA (200 ng/μl), 10 μl of sterilized H2O. The cycling parameters were: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec. Then, the PCR products were heated to 95°C for 15 sec, cooled to 60°C for 1 min and heated to 95°C for 30 sec and cooled to 60°C for 15 sec to measure the dissociation curves. Negative controls, without either template or transcriptase, were included in each experiment. To check reproducibility, each qRT-PCR reaction for each sample was carried out in three technical replicates and three biological replicates.
qRT-PCR data analysis
Relative quantification was performed using the comparative 2-ΔΔCt method [85]. All data were normalized to endogenous β-actin or ribosomal protein S3 levels from the same individual samples. In the analysis of the relative fold change in different tissues, the body sample was taken as the calibrator. Thus, the relative fold change in different tissues was assessed by comparing the expression level of each target gene in other tissues to that in the body part. The results are presented as the mean of the fold change in three biological samples. The comparative analyses of each OBP, CSP and CXE gene among different tissues were determined with one-way nested analysis of variance (ANOVA), followed by a Tukey’s honestly significance difference (HSD) test using SPSS Statistics 18.0 (SPSS Inc., Chicago, IL, USA). The comparative analyses of each putative pheromone biosynthesis gene between pheromone gland (PG) and body part were determined with paired t-test. When applicable, values were presented as mean ± SE.
Abbreviations
CSP: Chemosensory protein; OBP: Odorant binding protein; CXE: Carboxylesterase; EST: Expressed sequenced tag; OR: Olfactory receptor; IR: Ionotropic receptor; SNMP: Sensory neuron membrane protein; ODE: Odorant-degrading enzyme; NGS: Next generation sequencing; PCR: Polymerase chain reaction; PDE: Pheromone degrading enzyme; PBP: Pheromone binding protein; PBAN: Pheromone biosynthesis activating neuropeptide; ACC: Acetyl-CoA carboxylase; FAS: Fatty acid synthetase; DES: Desaturase; FAR: Fatty acyl-CoA reductase; AR: Aldehyde reductase; ATF: Acetyltransferase; AOX: Aldehyde oxidase; GST: Glutathione S-transferase; Z7-12:OAc: (Z)-7-dodecenyl acetate; Z9-14:OAc: (Z)-9-tetradecenyl acetate; Z11-16:OAc: (Z)-11-hexadecenyl acetate; Z5-10:OAc: (Z)-5-decenyl acetate; Z8-12:OAc: (Z)-8-dodecenyl acetate; ORN: Olfactory receptor neuron.
Competing interests
The author(s) declare that they have no competing interests.
Authors’ contributions
SHG, YJZ and JJZ initiated the project, conceived and design the study. SHG, YJZ and JJZ wrote the manuscript. SHG carried out sample collection, library construction, the data processing, bioinformatics analysis, RT-PCR and qRT-PCR. YYG and KMW coordinated the study. JAP and LMF contributed data analyses, data interpretation and extensively revised the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Size distribution of the clean reads (A) and the assembled unigenes (B).
The identified transcripts with putative roles in Agrotis ipsilon sex pheromone biosynthesis and transport (Tables 3,4,5).
Gene Ontology (GO) classifications of the 23473 A. ipsilon pheromone gland unigenes according to their involvement in biological processes, cellular component and molecular function.
Primers used for RT-PCR analysis of enzyme genes of the A. ipsilon PG.
Primers used in real-time PCR for determination expression level of target genes.
Contributor Information
Shao-Hua Gu, Email: shgu@ippcaas.cn.
Kong-Ming Wu, Email: kmwu@ippcaas.cn.
Yu-Yuan Guo, Email: yyguo@ippcaas.cn.
John A Pickett, Email: john.pickett@rothamsted.ac.uk.
Linda M Field, Email: lin.field@rothamsted.ac.uk.
Jing-Jiang Zhou, Email: jing-jiang.zhou@rothamsted.ac.uk.
Yong-Jun Zhang, Email: yjzhang@ippcaas.cn.
Acknowledgements
This work was supported by the China National “973” Basic Research Program (2012CB114104) and the National Natural Science Foundation of China (31071694, 31171858 and 31272048). JJ Zhou and YJ Zhang acknowledge financial support from the Royal Society, UK for the international joint project between China and UK (31111130203; JP100849) and BBSRC International Partnering Award (BB/J020281). Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom. The authors thank Ms. RN Yang, XY Qi and W Wang for their contributions in the insect rearing and the RT-PCR experiment.
References
- Ando T, Inomata S, Yamamoto M. Lepidopteran sex pheromones. Top Curr Chem. 2004;239:51–96. doi: 10.1007/b95449. [DOI] [PubMed] [Google Scholar]
- Tillman JA, Seybold SJ, Jurenka RA, Blomquist GJ. Insect pheromones—an overview of biosynthesis and endocrine regulation. Insect Biochem Mol Biol. 1999;29:481–514. doi: 10.1016/S0965-1748(99)00016-8. [DOI] [PubMed] [Google Scholar]
- McNeil JN. Behavioral ecology of pheromone-mediated communication in moth and its importance in the use of pheromone traps. Annu Rev Entomol. 1991;36:407–430. doi: 10.1146/annurev.en.36.010191.002203. [DOI] [Google Scholar]
- Witzgall P, Stelinski L, Gut L, Thomson D. Codling moth management and chemical ecology. Annu Rev Entomol. 2008;53:503–522. doi: 10.1146/annurev.ento.53.103106.093323. [DOI] [PubMed] [Google Scholar]
- Witzgall P, Kirsch P, Cork A. Sex pheromones and their impact on pest management. J Chem Ecol. 2010;36:80–100. doi: 10.1007/s10886-009-9737-y. [DOI] [PubMed] [Google Scholar]
- Jurenka R. Insect pheromone biosynthesis. Top Curr Chem. 2004;239:97–132. doi: 10.1007/b95450. [DOI] [PubMed] [Google Scholar]
- Matsumoto S. Molecular mechanisms underlying sex pheromone production in moths. Biosci Biotechnol Biochem. 2010;74:223–231. doi: 10.1271/bbb.90756. [DOI] [PubMed] [Google Scholar]
- Volpe JJ, Vagelos PR. Saturated fatty acid biosynthesis and its regulation. Annu Rev Biochem. 1973;42:21–60. doi: 10.1146/annurev.bi.42.070173.000321. [DOI] [PubMed] [Google Scholar]
- Pape ME, Lopez-Casillas F, Kim KH. Physiological regulation of acetyl-CoA carboxylase gene expression: effects of diet, diabetes, and lactation on acetyl-CoA carboxylase mRNA. Arch Biochem Biophys. 1988;267:104–109. doi: 10.1016/0003-9861(88)90013-6. [DOI] [PubMed] [Google Scholar]
- Bjostad LB, Roelofs WL. Biosynthesis of sex pheromone components and glycerolipid precursors from sodium [1–14C] acetate in redbanded leafroller moth. J Chem Ecol. 1984;10:681–691. doi: 10.1007/BF00994228. [DOI] [PubMed] [Google Scholar]
- Tang JD, Charlton RE, Jurenka RA, Wolf WA, Phelan PL, Sreng L, Roelofs WL. Regulation of pheromone biosynthesis by a brain hormone in two moth species. Proc Natl Acad Sci USA. 1989;86:1806–1810. doi: 10.1073/pnas.86.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurenka RA, Jacquin E, Roelofs WL. Control of the pheromone biosynthetic pathway in Helicoverpa zea by the pheromone biosynthesis activating neuropeptide. Arch Insect Biochem Physiol. 1991;17:81–91. doi: 10.1002/arch.940170203. [DOI] [Google Scholar]
- Morse D, Meighen EA. In: Pheromone Biochemistry. Prestwich GD, Bloriiquist GJ, editor. Orlando, FL: Academic Press; 1987. Pheromone biosynthesis: enzymatic studies in lepidoptera; pp. 121–158. [Google Scholar]
- Hoskovec MA, Luxová A, Svatoš A, Boland W. Biosynthesis of sex pheromones in moths: stereochemistry of fatty alcohol oxidation in Manduca sexta. Tetrahedron. 2002;58:9193–9201. doi: 10.1016/S0040-4020(02)01199-7. [DOI] [Google Scholar]
- Foster SP, Roelofs WL. Sex pheromone biosynthesis in the tortricid moth, Ctenopseustis herana (Felder & Rogenhofer) Arch Insect Biochem Physiol. 1996;33:135–147. doi: 10.1002/(SICI)1520-6327(1996)33:2<135::AID-ARCH4>3.0.CO;2-X. [DOI] [Google Scholar]
- Löfstedt C, Bengtsson M. Sex pheromone biosynthesis of (E, E)-8,10-dodecadienol in codling moth Cydia pomonella involves E9 desaturation. J Chem Ecol. 1988;14:903–915. doi: 10.1007/BF01018782. [DOI] [PubMed] [Google Scholar]
- Martinez T, Fabriás G, Camps F. Sex pheromone biosynthetic pathway in Spodoptera littoralis and its activation by a neurohormone. J Biol Chem. 1990;265:1381–1387. [PubMed] [Google Scholar]
- Foster SP, Roelofs WL. Sex pheromone biosynthesis in the leafroller moth Planotortix excessana by Δ10 desaturation. Arch Insect Biochem Physiol. 1988;8:1–9. doi: 10.1002/arch.940080102. [DOI] [Google Scholar]
- Bjostad LB, Roelofs WL. Sex pheromone biosynthesis from radiolabeled fatty acids in the redbanded leafroller moth. J Biol Chem. 1981;256:7936–7940. [PubMed] [Google Scholar]
- Bjostad LB, Roelofs WL. Sex pheromone biosynthesis in Trichoplusia ni: key steps involve delta-11 desaturation and chain-shortening. Science. 1983;220:1387–1389. doi: 10.1126/science.220.4604.1387. [DOI] [PubMed] [Google Scholar]
- Zhao C, Löfstedt C, Wang X. Sex pheromone biosynthesis in the Asian corn borer Ostrinia furnacalis (II): biosynthesis of (E)- and (Z)-12-tetradecenyl acetate involves Δ14 desaturation. Arch Biochem Physiol. 1990;15:57–65. doi: 10.1002/arch.940150106. [DOI] [Google Scholar]
- Lassance JM, Liénard MA, Antony B, Qian S, Fujii T, Tabata J, Ishikawa Y, Löfstedt C. Functional consequences of sequence variation in the pheromone biosynthetic gene pgFAR for Ostrinia moths. Proc Natl Acad Sci USA. 2013;110:3967–3972. doi: 10.1073/pnas.1208706110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teal PEA, Tumlinson JH. The role of alcohols in pheromone biosynthesis by 2 noctuid moths that use acetate pheromone components. Arch Insect Biochem Physiol. 1987;4:261–269. doi: 10.1002/arch.940040404. [DOI] [Google Scholar]
- Roelofs WL, Wolf WA. Pheromone biosynthesis in Lepidoptera. J Chem Ecol. 1998;14:2019–2031. doi: 10.1007/BF01014247. [DOI] [PubMed] [Google Scholar]
- Antony B, Fujii T, Moto K, Matsumoto S, Fukuzawa M, Nakano R, Tatsuki S, Ishikawa Y. Pheromone-gland-specific fatty-acyl reductase in the adzuki bean borer, Ostrinia scapulalis (Lepidoptera: Crambidae) Insect Biochem Mol Biol. 2009;39:90–95. doi: 10.1016/j.ibmb.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Hagström AK, Lienard MA, Groot AT, Hedenström E, Löfstedt C. Semi-selective fatty acyl reductases from four heliothine moths influence the specific pheromone composition. PLoS ONE. 2012;7(5):e37230. doi: 10.1371/journal.pone.0037230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassance JM, Groot AT, Liénard MA, Antony B, Borgwardt C, Andersson F, Hedenström E, Heckel DG, Löfstedt C. Allelic variation in a fatty-acyl reductase gene causes divergence in moth sex pheromones. Nature. 2010;466:486–489. doi: 10.1038/nature09058. [DOI] [PubMed] [Google Scholar]
- Lienard MA, Hagström AK, Lassance JM, Löfstedt C. Evolution of multicomponent pheromone signals in small ermine moths involves a single fatty-acyl reductase gene. Proc Natl Acad Sci USA. 2010;107:10955–10960. doi: 10.1073/pnas.1000823107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moto K, Kojima H, Kurihara M, Iwami M, Matsumoto S. Cell-specific expression of enhanced green fluorescence protein under the control of neuropeptide gene promoters in the brain of the silkworm, Bombyx mori, using Bombyx mori nucleopolyhedrovirus-derived vectors. Insect Biochem Mol Biol. 2003;33:7–12. doi: 10.1016/S0965-1748(02)00185-6. [DOI] [PubMed] [Google Scholar]
- Moto K, Suzuki MG, Hull JJ, Kurata R, Takahashi S, Yamamoto M, Okano K, Imai K, Ando T, Matsumoto S. Involvement of a bifunctional fatty-acyl desaturase in the biosynthesis of the silkmoth, Bombyx mori, sex pheromone. Proc Natl Acad Sci USA. 2004;101:8631–8636. doi: 10.1073/pnas.0402056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi A, Hull JJ, Matsumoto S. Targeted disruption of genes in the Bombyx mori sex pheromone biosynthetic pathway. Proc Natl Acad Sci USA. 2006;103:4398–4403. doi: 10.1073/pnas.0511270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honson NS, Gong Y, Plettner E. Structure and function of insect odorant and pheromone-binding proteins (OBPs and PBPs) and chemosensory-specific proteins (CSPs) Recent Ad Phytochem. 2005;39:227–268. [Google Scholar]
- Pelosi P, Zhou JJ, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 2006;63:1658–1676. doi: 10.1007/s00018-005-5607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- Zhou JJ. Odorant-binding proteins in insects. Vitam Horm. 2010;83:241–272. doi: 10.1016/S0083-6729(10)83010-9. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Riddiford LM. Scale esterase a pheromone degrading enzyme from the wing scales of the silk moth Antheraea polyphemus. J Chem Ecol. 1986;12:469–482. doi: 10.1007/BF01020567. [DOI] [PubMed] [Google Scholar]
- Prestwich GD. Chemistry of pheromone and hormone metabolism in insects. Science. 1987;237:999–1006. doi: 10.1126/science.3616631. [DOI] [PubMed] [Google Scholar]
- Xiang YY, Yang MF, Li ZZ. Sex pheromone components of the female black cutworm moth in China: identification and field trials. Zool Res. 2009;30:59–64. doi: 10.3724/SP.J.1141.2009.01059. [DOI] [Google Scholar]
- Vogel H, Heidel AJ, Heckel DG, Groot AT. Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens. BMC Genomics. 2010;11:29. doi: 10.1186/1471-2164-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann F. Insect vitellogenin: identification, biosynthesis, and role in vitellogenesis. Adv Insect Physiol. 1979;14:49–108. [Google Scholar]
- Du M, Yin X, Zhang S, Zhu B, Song Q, An S. Identification of lipases involved in PBAN stimulated pheromone production in Bombyx mori using the DGE and RNAi approaches. PLoS ONE. 2012;7:e31045. doi: 10.1371/journal.pone.0031045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull JJ, Ohnishi A, Moto K, Kawasaki Y, Kurata R, Suzuki MG, Matsumoto S. Cloning and characterization of the pheromone biosynthesis activating neuropeptide receptor from the silkmoth, Bombyx mori: significance of the carboxyl terminus in receptor internalization. J Biol Chem. 2004;279:51500–51507. doi: 10.1074/jbc.M408142200. [DOI] [PubMed] [Google Scholar]
- Rafaeli A, Bober R, Becker L, Choi MY, Fuerst EJ, Jurenka R. Spatial distribution and differential expression of the PBAN receptor in tissues of adult Helicoverpa spp. (Lepidoptera: Noctuidae) Insect Mol Biol. 2007;16:287–293. doi: 10.1111/j.1365-2583.2007.00725.x. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Nachman RJ, Aimanova K, Gill S, Adams ME. The pheromone biosynthesis activating neuropeptide (PBAN) receptor of Heliothis virescens: identification, functional expression, and structure-activity relationships of ligand analogs. Peptides. 2008;29:268–275. doi: 10.1016/j.peptides.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemeno C, Haynes KF. Chemical and behavioral evidence for a third pheromone component in a North American population of the black cutworm moth, Agrotis ipsilon. J Chem Ecol. 1998;24:999–1011. doi: 10.1023/A:1022398318465. [DOI] [Google Scholar]
- Picimbon JF, Gadenne C, Bécard JM, Clément JL, Sreng L. Sex pheromone of the French black cutworm moth, Agrotis ipsilon (Lepidoptera: Noctuidae): identification and regulation of a multicomponent blend. J Chem Ecol. 1997;23:211–230. [Google Scholar]
- Wakamura S, Struble DL, Matsuura H, Sato M, Kegasawa K. Sex pheromone of the black cutworm moth, Agrotis ipsilon Hufnagel (Lepidoptera: Noctuidae): attractant synergist and improved formulation. Appl Entomol Zool. 1986;21:299–304. [Google Scholar]
- Lienard MA, Strandh M, Hedenström E, Johansson T, Löfstedt C. Key biosynthetic gene subfamily recruited for pheromone production prior to the extensive radiation of Lepidoptera. BMC Evol Biol. 2008;8:270. doi: 10.1186/1471-2148-8-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallerac R, Labeur C, Jallon JM, Knipple DC, Roelofs WL, Wicker-Thomas C. A Δ9 desaturase gene with a different substrate specificity is responsible for the cuticular diene hydrocarbon polymorphism in Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:9449–9454. doi: 10.1073/pnas.150243997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafaeli A. Mechanisms involved in the control of pheromone production in female moths: recent developments. Entomol Exp Appl. 2005;115:7–15. doi: 10.1111/j.1570-7458.2005.00292.x. [DOI] [Google Scholar]
- Lienard MA, Löfstedt C. Functional flexibility as a prelude to signal diversity?: role of a fatty acyl reductase in moth pheromone evolution. Commun Integr Biol. 2010;3(6):586–588. doi: 10.4161/cib.3.6.13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang NB, Teal PEA, Tumlinson JH. PBAN regulation of pheromone biosynthesis in female tobacco hornworm moths, Manduca sexta (L.) Arch Insect Biochem Physiol. 1995;29:35–44. doi: 10.1002/arch.940290105. [DOI] [Google Scholar]
- Luxová A, Svatoš A. Substrate specificity of membrane-bound alcohol oxidase from the tobacco hornworm moth (Manduca sexta) female pheromone glands. J Mol Catal B-Enzym. 2006;38:37–42. doi: 10.1016/j.molcatb.2005.10.006. [DOI] [Google Scholar]
- Ishida Y, Leal WS. Rapid inactivation of a moth pheromone. Proc Natl Acad Sci USA. 2005;102:14075–14079. doi: 10.1073/pnas.0505340102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlin C, Rosell G, Carot-Sans G, François MC, Bozzolan F, Pelletier J, Jacquin-Joly E, Guerrero A, Maïbèche-Coisne M. Antennal esterase cDNAs from two pest moths, Spodoptera littoralis and Sesamia nonagrioides, potentially involved in odourant degradation. Insect Mol Biol. 2007;16:73–81. doi: 10.1111/j.1365-2583.2006.00702.x. [DOI] [PubMed] [Google Scholar]
- Merlin C, François MC, Bozzolan F, Pelletier J, Jacquin-Joly E, Maïbèche-Coisne M. A new aldehyde oxidase selectively expressed in chemosensory organs of insects. Biochem Biophys Res Commun. 2005;332:4–10. doi: 10.1016/j.bbrc.2005.04.084. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Bozzolan F, Solvar M, François MC, Jacquin-Joly E, Maïbèche-Coisne M. Identification of candidate aldehyde oxidases from the silkworm Bombyx mori potentially involved in antennal pheromone degradation. Gene. 2007;404:31–40. doi: 10.1016/j.gene.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Rybczynski R, Reagan J, Lerner MR. A pheromone-degrading aldehyde oxidase in the antennae of the moth Manduca sexta. J Neurosci. 1989;9:1341–1353. doi: 10.1523/JNEUROSCI.09-04-01341.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maïbèche-Coisne M, Nikonov AA, Ishida Y, Jacquin-Joly E, Leal WS. Pheromone anosmia in a scarab beetle induced by in vivo inhibition of a pheromone-degrading enzyme. Proc Natl Acad Sci USA. 2004;101:11459–11464. doi: 10.1073/pnas.0403537101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maïbèche-Coisne M, Merlin C, François MC, Porcheron P, Jacquin-Joly E. P450 and P450 reductase cDNAs from the moth Mamestra brassicae: cloning and expression patterns in male antennae. Gene. 2005;346:195–203. doi: 10.1016/j.gene.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Wojtasek H, Leal WS. Degradation of an alkaloid pheromone from the pale-brown chafer, Phyllopertha diversa (Coleoptera: Scarabaeidae), by an insect olfactory cytochrome P450. FEBS Lett. 1999;458:333–336. doi: 10.1016/S0014-5793(99)01178-3. [DOI] [PubMed] [Google Scholar]
- Durand N, Carot-Sans G, Chertemps T, Montagné N, Jacquin-Joly E, Debernard S, Maïbèche-Coisne M. A diversity of putative carboxylesterases are expressed in the antennae of the noctuid moth Spodoptera littoralis. Insect Mol Biol. 2010;19:87–97. doi: 10.1111/j.1365-2583.2009.00939.x. [DOI] [PubMed] [Google Scholar]
- Rogers ME, Jani MK, Vogt RG. An olfactory-specific glutathione-S-transferase in the sphinx moth Manduca sexta. J Exp Biol. 1999;202:1625–1637. doi: 10.1242/jeb.202.12.1625. [DOI] [PubMed] [Google Scholar]
- Zhou JJ, He XL, Pickett JA, Field LM. Identification of odorant-binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. Insect Mol Biol. 2008;17:147–163. doi: 10.1111/j.1365-2583.2007.00789.x. [DOI] [PubMed] [Google Scholar]
- Zhou JJ, Kan YC, Antoniw J, Pickett JA, Field LM. Genome and EST analyses and expression of a gene family with putative functions in insect chemoreception. Chem Senses. 2006;31:453–465. doi: 10.1093/chemse/bjj050. [DOI] [PubMed] [Google Scholar]
- Clement SL, Show ED, Way MO. Black cutworm pheromone trapping in strawberries. Calif Agric. 1982;36:20–21. [Google Scholar]
- Rings RW, Arnold FJ, Keaster AJ, Musick GJ. A worldwide annotated bibliography of the black cutworm, Agrotis ipsilon (Hufnagel) Ohio Agric Res Dev Cent Res Circ. 1974;198:1–106. [Google Scholar]
- Han ZJ. Toxicological responses and resistances of the black cutworm, Agrotis ypsilon (Rottemberg), to several groups of insecticides. Acta Phytophylacica Sinica. 1986;13:125–130. [Google Scholar]
- Gemeno C, Haynes KF. Periodical and age-related variation in chemical communication system of black cutworm moth, Agrotis ipsilon. J Chem Ecol. 2000;26:329–342. doi: 10.1023/A:1005468203045. [DOI] [Google Scholar]
- Swier SR, Rings RW, Musick GJ. Age-related calling behavior of the black cutworm, Agrotis ipsilon. Ann Entomol Soc Am. 1977;70:919–924. [Google Scholar]
- Xiang YY, Yang MF, Li ZZ. Calling behavior and rhythms of sex pheromone production in the black cutworm moth in China. J Insect Behav. 2010;23:35–44. doi: 10.1007/s10905-009-9193-0. [DOI] [Google Scholar]
- Lassmann T, Hayashizaki Y, Daub CO. TagDust-a program to eliminate artifacts from next generation sequencing data. Bioinformatics. 2009;25:2839–2840. doi: 10.1093/bioinformatics/btp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- Chen YA, Lin CC, Wang CD, Wu HB, Hwang PI. An optimized procedure greatly improves EST vector contamination removal. BMC Genomics. 2007;8:416. doi: 10.1186/1471-2164-8-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Aglyamova GV, Wang S, Buchanan-Carter J, Abrego D, Colbourne JK, Willis BL, Matz MV. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics. 2009;10:219. doi: 10.1186/1471-2164-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WEG, Wetter T, Suhai S. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XQ, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson I, Brass A. Searching DNA databases for similarities to DNA sequences: when is a match significant? Bioinformatics. 1998;14:349–356. doi: 10.1093/bioinformatics/14.4.349. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, Wang J, Li ST, Li RQ, Bolund L, Wang J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandh M, Johansson T, Ahrén D, Löfstedt C. Transcriptional analysis of the pheromone gland of the turnip moth, Agrotis segetum (Noctuidae), reveals candidate genes involved in pheromone production. Insect Mol Biol. 2008;17:73–85. doi: 10.1111/j.1365-2583.2008.00782.x. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: signalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Size distribution of the clean reads (A) and the assembled unigenes (B).
The identified transcripts with putative roles in Agrotis ipsilon sex pheromone biosynthesis and transport (Tables 3,4,5).
Gene Ontology (GO) classifications of the 23473 A. ipsilon pheromone gland unigenes according to their involvement in biological processes, cellular component and molecular function.
Primers used for RT-PCR analysis of enzyme genes of the A. ipsilon PG.
Primers used in real-time PCR for determination expression level of target genes.