Abstract
It has long been believed that the DNA-packaging motor of dsDNA viruses utilizes a rotation mechanism. Here we report a revolution rather than rotation mechanism for the bacteriophage phi29 DNA packaging motor. The phi29 motor contains six copies of the ATPase (Schwartz et al., this issue); ATP binding to one ATPase subunit stimulates the ATPase to adopt a conformation with a high affinity for dsDNA. ATP hydrolysis induces a new conformation with a lower affinity, thus transferring the dsDNA to an adjacent subunit by a power stroke. DNA revolves unidirectionally along the hexameric channel wall of the ATPase, but neither the dsDNA nor the ATPase itself rotates along its own axis. One ATP is hydrolyzed in each transitional step, and six ATPs are consumed for one helical turn of 360°. Transition of the same dsDNA chain along the channel wall, but at a location 60° different from the last contact, urges dsDNA to move forward 1.75 base pairs each step (10.5 bp per turn/6ATP=1.75 bp per ATP). Each connector subunit tilts with a left-handed orientation at a 30° angle in relation to its vertical axis that runs anti-parallel to the right-handed dsDNA helix, facilitating the one-way traffic of dsDNA. The connector channel has been shown to cause four steps of transition due to four positively charged lysine rings that make direct contact with the negatively charged DNA phosphate backbone. Translocation of dsDNA into the procapsid by revolution avoids the difficulties during rotation that are associated with DNA supercoiling. Since the revolution mechanism can apply to any stoichiometry, this motor mechanism might reconcile the stoichiometry discrepancy in many phage systems where the ATPase has been found as a tetramer, hexamer, or nonamer.
Keywords: Viral DNA packaging, AAA+ family, Nanobiotechnology, ATPase, Packaging mechanism
Introduction
The AAA+ (ATPases Associated with diverse cellular Activities) superfamily of proteins is a class of motor ATPases with a wide range of functions. Many members of this class of ATPases often fold into hexameric arrangements (Wang et al., 2011; Grainge et al., 2011; Kainov et al., 2008; Mastrangelo et al., 1989; Egelman et al., 1995; Niedenzu et al., 2001; Willows et al., 2004) and are involved in DNA translocation, tracking, and riding (Mueller-Cajar et al., 2011; Lowe et al., 2008; Parsons et al., 1995; Putnam et al., 2001; Iyer et al., 2004a). Despite their functional diversity, the common characteristic of this family is their ability to convert chemical energy obtained from the hydrolysis of the γ-phosphate bond of ATP into mechanical force, a process that usually involves a conformational change in the protein. This change of conformation generates both a loss of affinity for its substrate and a mechanical movement; which in turn is used to either make or break contacts between macromolecules, resulting in local or global protein unfolding, complex assembly or disassembly, or the translocation of DNA, RNA, proteins, or other macromolecules. These activities underlie processes critical to DNA repair, replication, recombination, chromosome segregation, DNA/RNA transportation, membrane sorting, cellular reorganization, and many others (Martin et al., 2005; Ammelburg et al., 2006; Grainge et al., 2007; Grainge, 2008; Lowe et al., 2008). Numerous biochemical and structural aspects of reactions catalyzed by AAA+ proteins have been elucidated, including those occurring during ATP hydrolysis. For instance, the crystal structure of the sliding clamp loader complex has revealed a spiral structure that strikingly correlates with the grooves of helical dsDNA, suggesting a simple explanation for how the loader/DNA helix interaction triggers ATP hydrolysis, and how DNA is released from the sliding clamp (McNally et al., 2010; Guenther et al., 1997).
In both prokaryotic and eukaryotic cells, DNA needs to be transported from one cellular compartment to another. For example, during phage maturation, the genome of dsDNA viruses is translocated into preformed protein shells, termed procapsids (for review, see (Guo and Lee, 2007; Rao and Feiss, 2008; Zhang et al., 2012; Serwer, 2010)). This entropically unfavorable process is accomplished by a nanomotor that uses ATP as an energy source (Guo et al., 1987c; Chemla et al., 2005; Hwang et al., 1996; Sabanayagam et al., 2007; Schwartz et al., 2012; Lee et al., 2008; Shu and Guo, 2003a; Chen and Guo, 1997). The dsDNA packaging motor consists of a proteinaceous channel and two packaging molecules with which it carries out its activities. The larger packaging molecule serves as part of the ATPase complex, and the smaller is responsible for dsDNA binding and cleavage (Guo et al., 1987c, 1998). This model has now become well-established (Guo and Lee, 2007; Rao and Feiss, 2008; Zhang et al., 2012; Serwer, 2010). The bacterial virus phi29 motor involves an ATPase, gp16 (Guo et al., 1987c, 1987b; Huang and Guo, 2003a, 2003b; Lee and Guo, 2006; Lee et al., 2008; Ibarra et al., 2001; Grimes and Anderson, 1990) and a hexameric packaging RNA ring (Guo et al., 1987a, 1998; Shu et al., 2007; Zhang et al., accepted for publication). The connector consists of 12 copies of gp10 that creates a central channel that serves as a pathway for dsDNA translocation (Jimenez et al., 1986; Guasch et al., 2002; Badasso et al., 2000).
The cellular components that show the strongest similarity to the viral DNA packaging motor include FtsK, an AAA+ DNA motor protein that transports DNA and separates intertwined chromosomes during cell division (Iyer et al., 2004b), and SpoIIIE (Demarre et al., 2013), an AAA+ protein responsible for transportation of DNA from a mother cell into the pre-spore during Bacillus subtilis sporulation (Bath et al., 2000). The ATPase of phi29, gp16, is similar to these families in both structure and function (Iyer et al., 2004b; Guo et al., 1998). Both the FtsK and SpoIIE DNA transportation systems rely on assembly of a hexameric machine. FtsK proteins contain three components: one for DNA translocation, one for controlling orientation of movement, and one for anchoring to the substrate (Demarre et al., 2013). Extensive studies suggest that FtsK may employ a “rotary inchworm” mechanism to transport DNA (Massey et al., 2006). The FtsK hexameric motor encircles dsDNA. During each cycle of ATP binding and hydrolysis within each FtsK subunit, one domain tightly binds the helix while another translocates along the DNA. This process causes translational movement, a mechanism that is repeated by the subsequent transfer of the helix to the next adjacent subunit (Massey et al., 2006).
It was suggested that viral DNA packaging motors operate by a rotation mechanism involving a five-/six-fold mismatch structure (Hendrix, 1978). Many subsequent models have been proposed describing the packaging motor of dsDNA (Khan et al., 1995; Serwer, 2003; Astumian, 1997; Guo et al., 1998; Hendrix, 1978; Grimes and Anderson, 1997; Chen and Guo, 1997; Guasch et al., 2002; Hou et al., 2010; Morita et al., 1995b; Sabanayagam et al., 2007; Oram et al., 2008; Serwer, 2010; Shu et al., 2007; Maluf et al., 2006; Yu et al., 2010; Aathavan et al., 2009). The most well-studied bacteriophage phi29 DNA packaging motor was also the first constructed with purified components (Guo et al., 1986) and has been shown to consist of three major components that interact with each other in unison (Fig. 1) (Guo et al., 1987a, 1987c; Lee and Guo, 2006; Ibarra et al., 2001). An RNA component was discovered (Guo et al., 1987a) that was later determined to exist as a hexameric ring (Guo et al., 1998;Zhang et al., 1998). Based on the structure of the hexameric pRNA, it was proposed that the mechanism of the phi29 viral DNA packaging motor is similar to that used by other hexameric DNA tracking motors of the AAA+ family (Guo et al., 1998). The presence of hexameric folds in the motor has been revealed by biochemical analysis (Guo et al., 1998; Zhang et al., 1998; Hendrix, 1998); single molecule photobleaching (Shu et al., 2007); gold labeling imaged by EM (Xiao et al., 2008; Moll and Guo, 2007; Shu et al., 2007); nano-fabrication (Xiao et al., 2010); and RNA crystal structure (Zhang et al., accepted for publication).
Fig. 1.
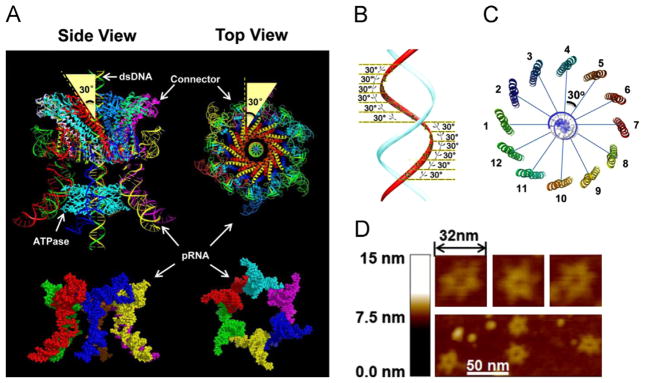
Depiction of structure and function of phi29 DNA-packaging motor. (A) Illustrated model of hexameric pRNA based on a crystal structure (Zhang et al., accepted for publication) and the 30° tilting of the channel subunits, relative to the central axis of the connector (pdb ID: 1H5W); (B) DsDNA showing the shift of 30° angle between two adjacent connector subunits; (C) Connector showing the change of 30° angle between two adjacent connector subunits; (D) AFM images of hexameric pRNA with 7-nt loops (Shu et al., 2013).
However, whether the RNA and ATPase are hexamers or pentamers is still debated. Other laboratories have reported the existence of five-fold symmetry (Chistol et al., 2012; Yu et al., 2010; Morais et al., 2008; Simpson et al., 2000; Ding et al., 2011; Harjes et al., 2012). Adherents of the pentamer model have also proposed variations to reconcile the pentamer and hexamer debate. One theory is that a pRNA hexamer is first assembled on the motor, after which one subunit leaves, resulting in a final pentamer state (Morais et al., 2001, 2008; Simpson et al., 2000). An alternative idea suggests that one subunit in the pentameric ring is inactive during each cycle while the other four subunits function sequentially during DNA packaging (Moffitt et al., 2009; Yu et al., 2010).
In previous reports, we have shown that motor intermediates isolated during the active DNA packaging process also contain a hexameric pRNA (Shu et al., 2007). Furthermore, in this issue, we provide data to confirm that the ATPase motor is hexameric (Schwartz et al., this issue) and is a relative of the hexameric AAA+ DNA translocase. In this paper, we show that the motor mechanism of DNA translocation involves revolution, rather than a rotational mechanism that involves a coiling force.
Results
The structure of the hexameric motor
The essential components of the phi29 DNA packaging motor include the dodecameric connector and the ATPase gp16 geared by a ring of RNA. The crystal structure of the three-way junction (3WJ) of the pRNA (Shu et al., 2011), one of the motor components, has recently been solved (Zhang et al., accepted for publication) and the predicted hexameric pRNA ring structure has been confirmed (Zhang et al., 2013; Fig. 1A). AFM images revealed an elaborate, ring-shaped structure consisting of six distinct arms representing the six subunits of pRNA (Fig. 1D).
Sliding of gp16 out of dsDNA verified by addition of steric blocks to the end of dsDNA
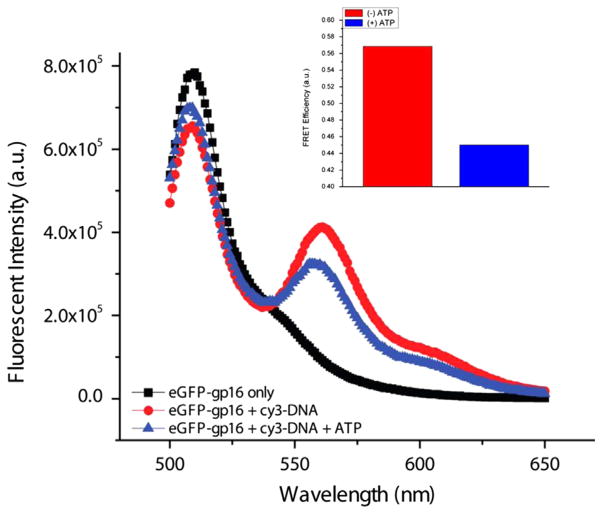
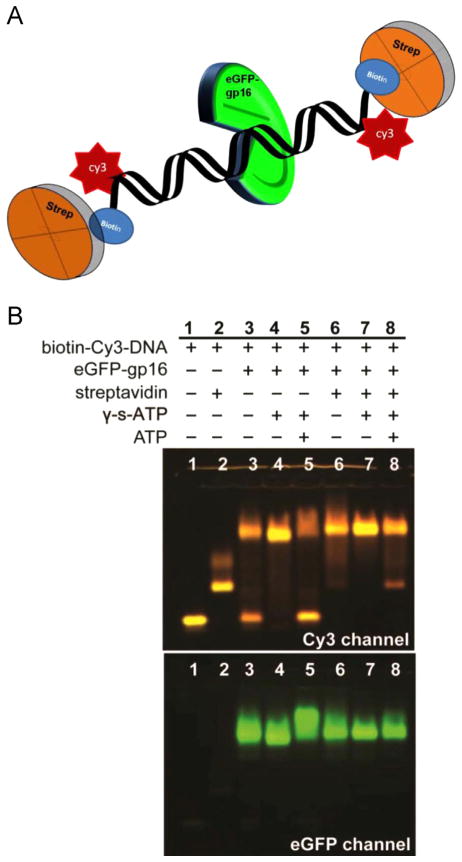
When Cy3-dsDNA is mixed with eGFP-gp16, a transfer of energy from the donor fluorophore (eGFP) to the acceptor fluorophore (Cy3) is observed, indicating that the protein fluorophore is in close proximity to the dsDNA fluorophore. However, after addition of ATP, the Förster Resonance Energy Transfer (FRET) efficiency decreased significantly (Fig. 2), suggesting that the protein had dissociated from the DNA after ATP hydrolysis. We hypothesize that gp16 slides along DNA and then falls off upon reaching the end of the DNA. In contrast, binding of gp16 to dsDNA was significantly enhanced in the presence of non-hydrolyzable ATP analogue, γ-S-ATP, as shown by both gel shift and binding assays. To determine whether the reduced FRET signal is due to dissociation of eGFP-gp16 or a process by the protein moves along DNA, we exploited a streptavidin hindrance test (Fig. 3). The terminally biotinylated DNA was incubated with streptavidin, which should prevent gp16 sliding off the DNA, but not interfere with simple dissociation. Complexes of eGFP-gp16/DNA/γ-S-ATP complexes remain stable in the presence of ATP only when the terminally biotinylated Cy3 DNA was bound to streptavidin (lane 8, Fig. 3).
Fig. 2.
FRET assay of fluorogenic ATPase and short dsDNA. eGFP-gp16 was incubated with Cy3-DNA and without ATP and excited at 480 nm. Energy transfer occurs between the two fluorophores, with light emission at ~560 nm, indicating that gp16 and DNA are in close proximity. Bar graph (top right) showing the FRET efficiency difference between the two samples.
Fig. 3.
EMSA of eGFP-gp16 on terminally blocked short dsDNA. 40 bp Cy3-dsDNA, with biotin attached to each end, was incubated with eGFP-gp16, non-hydrolyzable γ-S-ATP, and streptavidin in different combinations. The complexes that were mixed at approximately a 6:1 molar ratio of protein:DNA were then electrophoresed through an agarose gel and scanned for Cy3 fluorescence of DNA and GFP fluorescence of gp16 (see Materials and Methods).
One defective monomer in the hexameric ATPase blocks function
The Walker A motif of AAA+ proteins is responsible for ATP binding, while the Walker B motif initiates ATP hydrolysis (Story and Steitz, 1992). Both motifs have been identified in phi29 gp16, previously (Guo et. al., 1987c) and (Schwartz et al., this issue). Since other ATPases have been shown to demonstrate cooperativity, the Hill constant for DNA-binding was evaluated using capillary electrophoresis (CE) to distinguish between a sequential or concerted action mechanisms.
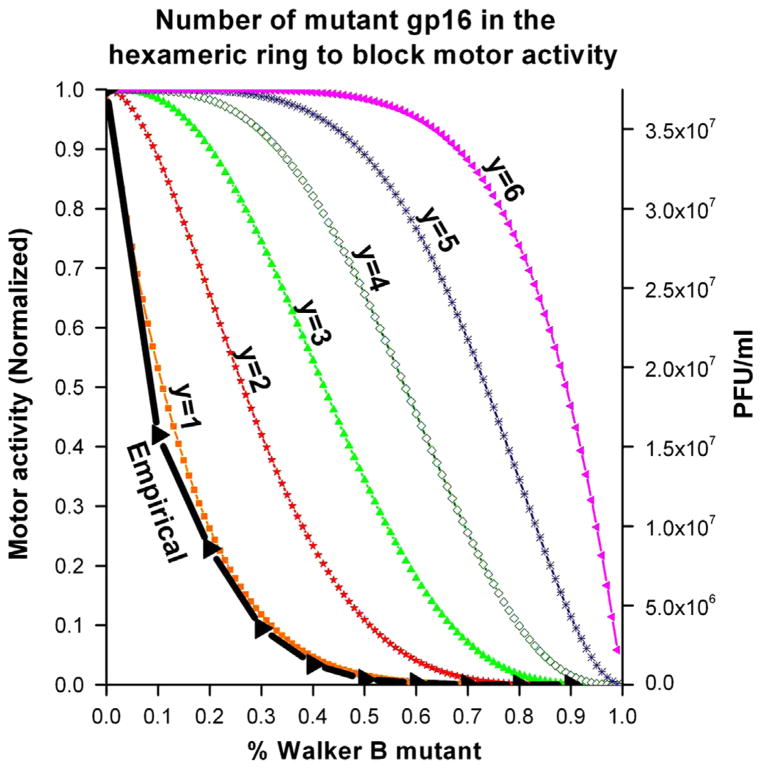
In order to help elucidate the mechanism of the DNA packaging motor, the number of copies of an inactive Walker B mutant within the hexameric ATPase required to block DNA packaging process was determined. The defective mutant was titrated with the active wild-type, were allowed to freely associate, and analyzed for DNA binding (see Materials and Methods). The minimum number (y) of mutant gp16 needed to block the packaging within the hexameric ring was predicted with the equation , where p and q represent the ratio of wildtype and Walker B mutant gp16, respectively, and p+q=1 (Fig. 4). Using this expanded binomial, each term represents a different mixed hexamer where the exponents of p and q, respectively indicate the number of wildtype and mutant monomer in each mixed hexamer. For example, the term indicates that the hexamer contains three wildtype and 3 three Walker B mutant monomers. Our empirical data almost perfectly overlapped with the theoretical curve corresponding to ‘y’ is equal to 1 (Fig. 4), suggesting that one inactive subunit in the hexamer abolishes motor activity.
Fig. 4.
Binomial distribution assay to determine the minimum number (y) of defective eGFP-gp16 in the hexameric ring to block motor activity. Theoretical plot of percent Walker B mutant gp16 versus yield of infectious virions in in vitro phage assembly assays. Predictions were made with the equation as seen in the Materials and Methods.
Motor ATPase tightly clinches dsDNA after binding to ATP and subsequently pushes the dsDNA away after ATP hydrolysis
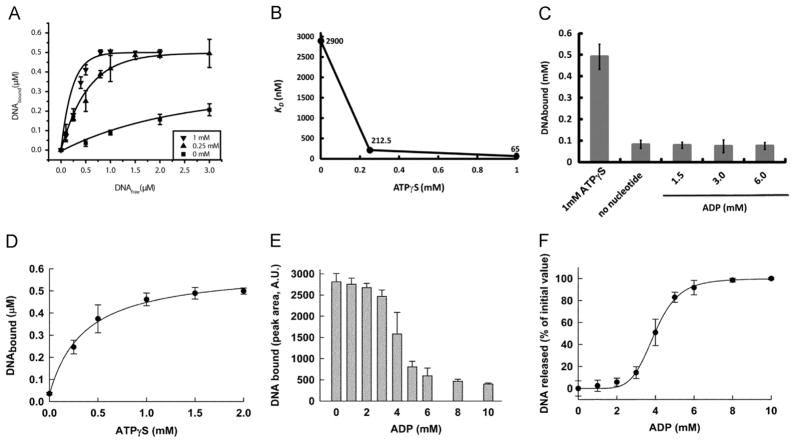
Similar to the AAA+ motor proteins that undergo conformational changes during their interaction with ATP and adopt two distinct states, the phi29 motor ATPase also exhibits a high or low affinity state for DNA. EMSA revealed that the affinity of gp16 for dsDNA increases in the presence of γ-S-ATP (Schwartz et al., 2012). We utilized a CE assay that allowed for direct quantification of the amount of DNA bound to gp16. At increasing concentrations of γ-S-ATP, the amount of bound DNA increased progressively, indicating that gp16 transitioned from a state in which binding to DNA was unfavorable to one in which binding was preferred (Fig. 5A). A regression plot of dissociation constant (Kd) for dsDNA versus concentration of γ-S-ATP indicated that the affinity of gp16 for substrate increased 40-fold with saturating amounts of γ-S-ATP (Fig. 5B). This significant increase strongly suggests that the species that binds to DNA is the gp16-ATP complex and the gp16 binds first to ATP and secondly to DNA, as also suggested previously (Schwartz et al., 2012). However, adding ADP, even at non-physiological conditions (up to 6 mM), failed to promote an increase in dsDNA-binding affinity (Fig. 5C). Furthermore, the amount of DNA bound to g16 was comparable to a condition where no nucleotide was added. These observations indicate that gp16 cycles through states of ATP binding/DNA loading and ATP hydrolysis/DNA release or pushing. This conclusion was further supported by the finding that addition of normal ATP to the gp16/DNA/γ-S-ATP complex promoted the departure of the dsDNA from the complex (Schwartz et al., this issue).
Fig. 5.
One γ-S-ATP is sufficient to bind to one subunit of the gp16 hexamer and promote a high affinity state for dsDNA. Sequential binding of gp16 for dsDNA substrate involves γ-S-ATP substep. (A) The Kd for dsDNA at varying concentrations of γ-S-ATP. (B) The relative Kd of gp16 decreased 40-fold as the concentration of γ-S-ATP increased from 0 mM to 1 mM. (C) ADP, a derivative of ATP hydrolysis, was unable to promote binding and had the similar effect as no nucleotide addition. The hyperbolic curve (D) suggests a cooperativity factor of 1, indicating that one γ-S-ATP is sufficient to produce the high affinity state of gp16 for DNA. DNA releases from the complex DNA-gp16-γ-S-ATP mediated by ADP (E), forming a sigmoidal curve (F) with a cooperativity factor of 6 indicating that all six subunits of gp16 need to be bound to ADP to release DNA from the protein.
Only one molecule of ATP is sufficient to generate the high affinity state for DNA in the ring of the motor ATPase
Next, we sought the answer to how many nucleotides were required for gp16 to generate the high affinity state for dsDNA; in other words, how many subunits need to bind to ATP in order for the gp16 hexamer to stably associate to dsDNA. This information is useful in understanding how the hexameric complex of gp16 utilizes the substrate in order to generate unidirectional DNA translocation. AAA+ proteins are typically organized into a homo-oligomeric assembly where each component contains the recognition motifs required for binding of the substrate. In principle, one can imagine that each subunit can bind to the substrate independently from the others; however, such an arrangement can lead to futile cycles of ATP consumption. Two major configurations can be hypothesized to avoid the above described scenario. First, it may be possible that the binding sites for the substrate consist of the same recognition motifs in all the subunits, and in this case, all subunits can bind at the same time to the substrate. In this hypothetical situation, it is intuitive to imagine that a form of coordination among the subunits must also exist at the level of ATP hydrolysis, since the most effective mechanism of translocation would allow all subunits to hydrolyze at the same time corresponding to an exodus of the dsDNA substrate. The second possibility is that DNA is bound at any given time to only one subunit of the oligomer, and after the cycle of ATP hydrolysis is terminated in the specific subunit that binds DNA, the substrate is then passed to the next subunit in the high ATP affinity state in order to initiate another cycle of hydrolysis. To distinguish between these two scenarios, we analyzed the amount of DNA bound to gp16 by keeping the concentration of gp16 and DNA constant and varying the concentration of γ-S-ATP in the reaction mixture (Fig. 5D). If more than one γ-S-ATP per oligomer of gp16 is required to generate the high affinity state for DNA in the protein, the plot would show a cooperativity profile, with a Hill coefficient representing the amount of γ-S-ATP required to be bound to gp16. Our data exhibit no cooperativity in binding (Hill coefficient=1.5) indicating that not all of the subunits of gp16 are required to be bound to γ-S-ATP to stabilize binding to DNA.
In principle, a Hill coefficient close to one indicates that only one γ-S-ATP-activated subunit in the oligomer is required for DNA binding or that the binding of DNA is progressively increased with the number of subunits that are bound to γ-S-ATP. To address this question, we performed an experiment similar to the CE assay described above. A complex of gp16-DNA was assembled in the presence of saturating γ-S-ATP. After the complex formed, increasing amounts of ADP were added in order to compete with γ-S-ATP for the active sites of gp16 and to ultimately promote DNA release. The reaction is remarkably cooperative (Fig. 5E, F). From the fractional inhibition plot we extrapolated a Hill coefficient close to 6, indicating that six molecules of ADP must be bound to gp16 before dsDNA is released from the protein. This indicates that only one ATP-bound subunit stably binds DNA and prevents ADP-mediated release. Furthermore, the data indicate that gp16 most likely binds to dsDNA at only one subunit per round of ATP hydrolysis. As mentioned above, a Hill coefficient close to one indicates that binding of DNA progressively increases with the number of subunits that are bound to γ-S-ATP. However, the 3.6-nm diameter of the motor channel, as measured from the crystal structure (Guasch et al., 2002; Badasso et al., 2000), suggests that only one dsDNA can be bound within the channel; indicating that dsDNA shifts to a neighboring gp16 subunit upon release of the first subunit. In combination with the finding that one Walker B mutant gp16 was found to be sufficient to block the motor for DNA packaging, these results support a model where the motor ATPase works sequentially, and upon ATP hydrolysis the subunit of the ATPase gp16 assumes a new conformation and pushes dsDNA away from the subunit and transfers it to an adjacent subunit (Fig. 7).
Fig. 7.
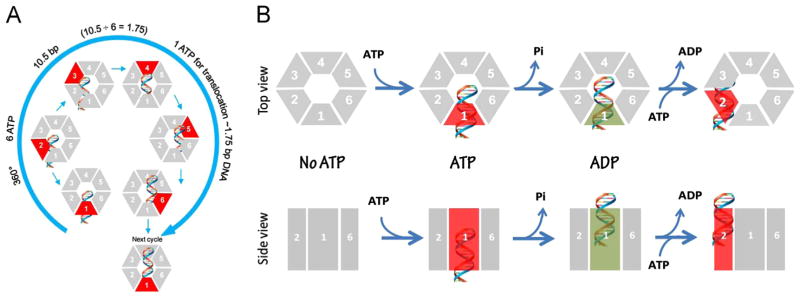
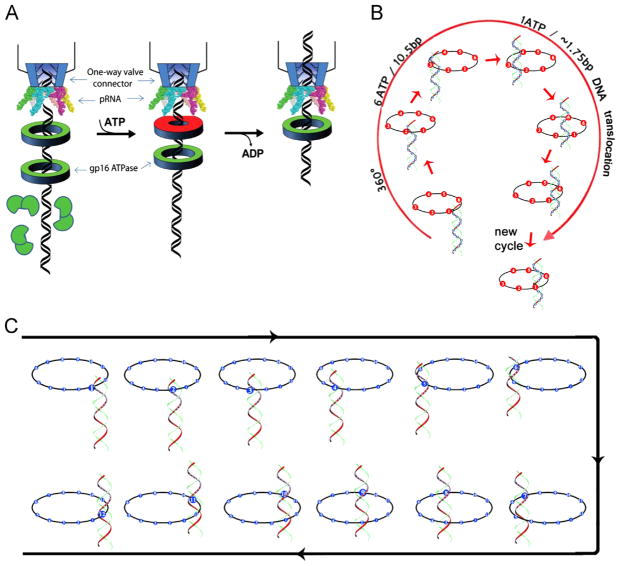
Schematic of gp16 binding to DNA and mechanism of sequential revolution in translocating genomic DNA. The connector is a one way valve that allows dsDNA to move into the procapsid, but does not allow movement in the opposite direction. Gp16, which is bridged by pRNA to associate with the connector, provides the pushing force. The binding of ATP to one subunit stimulates gp16 to adopt a conformation with a higher affinity for dsDNA. ATP hydrolysis forces gp16 to assume a new conformation with a lower affinity for dsDNA, thus pushing dsDNA away from the subunit and transferring it to an adjacent subunit. DsDNA is translocated at a pace of 1.75 base pairs per transfer to the neighboring subunit and is bound at a location 60° different from the first subunit on the same phosphate backbone chain. Rotation of neither the hexameric ring nor the dsDNA is required since the dsDNA revolves around the diameter of the ATPase. In each transitional step, one ATP is hydrolyzed, and in one cycle, six ATPs are required to translocate dsDNA one helical turn of 360° (10.5 base pairs). An animation is available at http://nanobio.uky.edu/movie.html.
Mixed oligomer between wildtype and mutants display negative cooperativity and communication between the subunits of gp16 oligomer
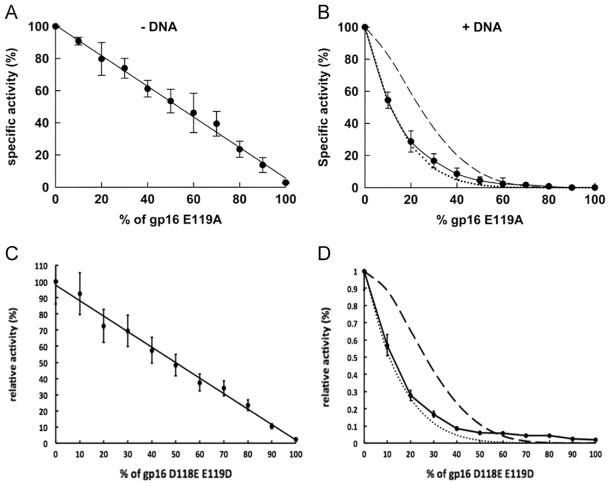
The fact that dsDNA only binds to one gp16 subunit at a time suggests that gp16 undergoes cooperativity during translocation. To verify this hypothesis we analyzed ATPase activity by studying the effect on the oligomerization of gp16 when mutant subunits were introduced (Trottier and Guo, 1997; Chen et al., 1997). If we assume communication between the subunits of the ATPase, the effect on the ATPase activity mediated by one inactive subunit should be higher than the simple sum of the ATPase activity of the single subunit. When the ATPase activity was measured in the absence of dsDNA, increasing amounts of Walker B mutants added to the overall oligomer of gp16 failed to provide any significant effect on the rate of hydrolysis (Fig. 6A, C), suggesting that each subunit of gp16 is able to hydrolyze ATP independently. However, when saturating amounts of dsDNA were added to the reaction, we observed a strong negative cooperative effect with a profile that mostly overlapped with the one predicted for the case in which one single inactive subunit is able to inactivate a whole oligomer (Fig. 6B, D) using an equation derived from a binomial distribution inhibition assay (Trottier and Guo, 1997; Chen et al., 1997) (see also Fig. 4). The results suggest that in the presence of dsDNA, a rearrangement occurs within the subunits of gp16, enabling them to communicate and “sense” the nucleotide state of the neighboring subunit. The fact that dsDNA needs to be present in the reaction indicates that dsDNA binds to the inactive subunit during the catalytic cycle and remains bound to it, which generates a stalled ATP hydrolysis cycle. This observation supports the idea that only the subunit that is binding to the substrate at any given time is permitted to hydrolyze ATP, thus performing translocation while the other subunits are in a type of ‘stalled’ or ‘loaded’ state. The scenario suggests an extremely high level of coordination on protein function, which is likely the most efficient process to couple energy production with DNA translocation via ATP hydrolysis.
Fig. 6.
ATPase inhibition assay by Walker B mutants reveals complete negative cooperativity. The inhibition ability of the Walker B mutants E119A and D118E/E119D was assayed by ATPase activity in the absence (left) and presence (right) of dsDNA. In the presence of DNA (right), the experimental data (solid line) overlapped with a theoretical curve indicating that one inactive subunit (dotted line) within the hexamer is able to completely block the activity of the hexameric gp16 and abolish ATPase activity, demonstrating negative cooperativity (see also Fig. 4). The dashed line is the theoretical curve where two inactive subunits are necessary for inhibition.
Direct observation of multiple ATPase gp16s lining up in queue along dsDNA as the initiation step in DNA packaging
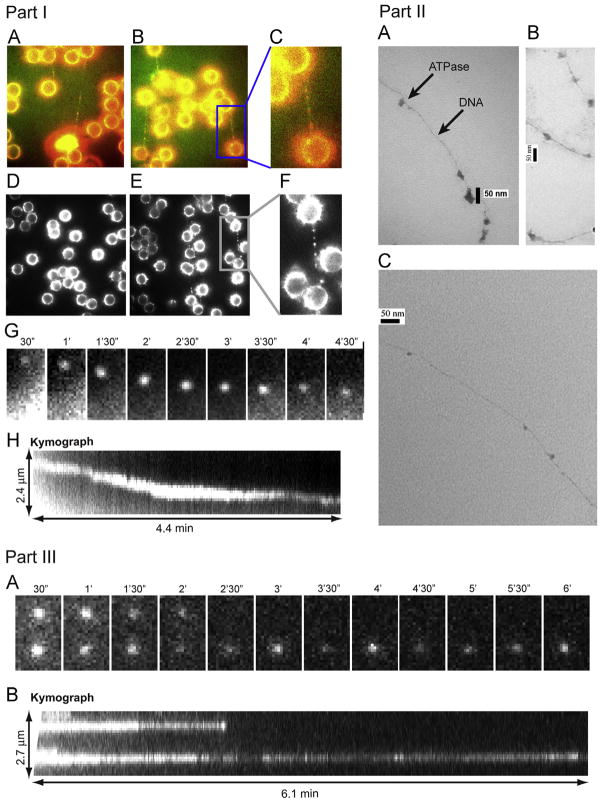
The consensus idea from extensive investigation of viral packaging motors is that the ATPase binds to the procapsid to form a procapsid/ATPase complex as the first step of motor action (Fujisawa et al., 1991; Guo et al., 1987b; Koti et. al., 2008). To investigate the sequence of interaction between motor components during DNA packaging, a fluorescent Cy3-conjugated gp16 was used to visualize the protein. Interestingly, we found that the first step in DNA packaging was the binding of multiple gp16 queued along the dsDNA, as observed by both single molecule imaging (Fig. 8 Part I) and by binding affinity studies. Moreover, negatively stained electron microscopy images have been taken of a multimeric gp16 complex along long genomic DNA (Fig. 8 Part II), lending further support to our conclusions.
Fig. 8.
Part I. Direct observation of ATPase complex queued and moving along dsDNA. Cy3 conjugated gp16 was incubated with (A, B, E) and without (D) dsDNA, tethered between two polylysine beads where (C, F) are magnified images of the framed regions of (B, E), respectively. (A–C) are overlapped pseudocolor images indicating the binding of Cy3-labeled gp16 along the To-Pro-3 stained dsDNA chain (Red: Cy3-gp16; Green: To-Pro-3 DNA). (G, H) The motion of the Cy3-gp16 spot was analyzed and a kymograph was produced to characterize the ATPase walking. (Actual motion videos can be found in the supplementary information and at http://nanobio.uky.edu/movie.html). Part II. Negatively stained transmission electron microscopy images of ATPase queued along dsDNA. gp16 was bound to non-specific dsDNA in queue. Part III. Recording of two Cy3-gp16/dsDNA complexes showing motionless gp16 spots in a buffer containing no ATP. (A) Sequential images of the recording. (B) Kymograph of the two spots.
DNA tightropes were constructed (Kad et al., 2010), that not only generated a straight DNA chain, but also lifted the DNA a few microns away from the surface of the slide within the sample chamber. Background fluorescence from non-specific binding of Cy3-gp16 to the surface of the slide is therefore eliminated when the focus of the imaging plane is on the DNA-bound Cy3-gp16 molecules. A string of multiple Cy3 spots representing Cy3-gp16 complexes are observed along the DNA chains (Fig. 8 Part I A–C, E, F). In the absence of DNA, a Cy3 signal was not observed between the polylysine beads (Fig. 8 Part I D), indicating that the queued Cy3 signals were truly from the multiple Cy3-gp16 bound to DNA. The results suggest that ATPase gp16 lines up in a queue along dsDNA at the initiation step in DNA packaging. These data are in accordance with another study where complexes of procapsid containing partially packaged dsDNA were isolated by sucrose sedimentation; conversion of the complexes to complete the DNA packaging process required the addition of ATPase gp16, but not pRNA (Shu and Guo, 2003b). The same publication also indicated that multiple gp16 molecules, but only a single hexameric pRNA, were required for packaging (Shu and Guo, 2003b).
The motion of gp16 along the lifted dsDNA tightrope was observed by single molecule fluorescence imaging. Sequential images were taken after washing with different buffers to illustrate the displacement of Cy3-gp16 over time. When the sample was washed with a buffer, a total of 195 Cy3-gp16 spots were studied. In the absence of ATP, the vast majority of these Cy3-gp16 spots did not show any motion along the DNA chain. After 20 mM ATP was added to the washing buffer, active motion of eGFP-gp16 along the dsDNA was observed, as shown by the sequential images (Fig. 8 Part III A) and kymographs (Fig. 8 Part III B). Actual motion videos can be found in the supplementary information and at http://nanobio.uky.edu/movie.html.
Terminases of viral DNA packaging motors bind to procapsids, although with an extremely low affinity and at low efficiency (Shibata et al., 1987; Morita et al., 1995b; Morita et al., 1995a; Guo et al., 1987b; Fujisawa et al., 1991; Lee and Guo, 2006). Our finding that gp16 first binds to dsDNA and then moves along dsDNA before reaching and binding to the procapsid is not in contradiction, rather a further refinement of the previous understanding. We hypothesize that gp16 contains two domains, one for dsDNA binding and one for connector/procapsid binding. In the absence of genomic DNA, gp16 binds to the procapsid, albeit with lower affinity. The key to understanding the sequence of interactions is based on the relative affinity of the protein for its substrate. Gp16 has a higher binding affinity for genomic DNA compared to that of the procapsid (Fig. 9). In the absence of dsDNA, gp16 and other terminases bind to the procapsid (Guo et al., 1987b). However, it is hypothesized that in the presence of genomic DNA, gp16 and other terminases prefer to bind to genomic DNA and track along it until reaching the packaging RNA and other motor components of the procapsid.
Fig. 9.
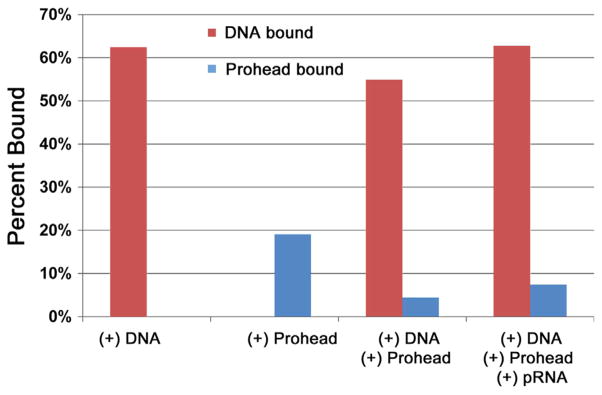
Binding affinity of gp16 to dsDNA and procapsid/pRNA complex measured using sucrose sedimentation. Ratio of procapsid-bound and DNA-bound gp16 under different treatments where the percent of bound gp16 to total gp16 is expressed, showing gp16’s affinity to DNA is much greater than to procapsid/pRNA complex.
To test this hypothesis, we measured the interaction of ATPase gp16 with the procapsid (Fig. 9). Gp16 is sticky and binds to all kinds of substrate, including nonspecifically to the procapsid. No significant difference was observed during the formation of the procapsid/gp16 complex in the presence or absence of pRNA (Fig. 9), which has been reported to serve as the bridge for gp16 binding to procapsid (Lee and Guo, 2006), but gp16 exhibited substantially higher affinity for dsDNA than for procapsid/pRNA complex. Although the ATPase may contain both dsDNA and procapsid binding domains, we suggest it prefers to bind to the procapsid only after tracking along the genomic DNA; that is, gp16 prefers to bind to genomic DNA first before reaching the procapsid.
Translocation of dsDNA helix by revolution without involvement of coiling or tension force
It has been demonstrated that the connector acts as a one way valve (Schwartz et al., 2012; Fang et al., 2012; Jing et al., 2010), only allowing dsDNA to move into the procapsid, but not in the opposite direction. Gp16, which is bridged by hexameric pRNA to associate with the connector, is expected to be the pushing force (Fig. 10A). The binding of ATP to one subunit stimulates gp16 to adopt a conformation with high affinity for dsDNA, while ATP hydrolysis forces gp16 to assume a new conformation with lower affinity, thus shifting dsDNA away from the first subunit and transferring it to an adjacent subunit due to the higher affinity for the next subunit (Fig. 10). Since the contact of the connector with dsDNA chain is transferred from one point on the phosphate backbone to another, rotation of neither the hexameric ring nor the dsDNA is required. One ATP is hydrolyzed in each step, and six ATPs are consumed for one cycle to translocate dsDNA one helical turn of 360° (10.5 base pairs). The binding of gp16 to the same phosphate backbone chain, but at a location 60° different from the last subunit, causes dsDNA to move forward 1.75 base pairs (10.5 bp per turn/6 ATP=1.75 bp per ATP), in good agreement with the 2 bp/ATP(Guo et al., 1987c) or 1.8 bp/ATP previously quantified empirically (Morita et al., 1993).
Fig. 10.
Mechanism of sequential revolution in translocating genomic DNA. Connector is a one way valve (Jing et. al., 2010) that allows dsDNA to move into the procapsid, but does not allow movement in the opposite direction. (A) Binding of ATP to one gp16 subunit stimulates it to adapt a conformation with higher affinity for dsDNA. ATP hydrolysis forces gp16 to assume a new conformation with lower affinity for dsDNA, thus pushing dsDNA away from this subunit and transferring it to an adjacent subunit. (B) Binding of gp16 to the same phosphate backbone chain, but at a location 60° different from last subunit urges dsDNA to move forward 1.75 base pairs. Since the dsDNA chain is transferred from one point on the phosphate backbone to another point, the rotation of the hexameric ring or the dsDNA is not required. (C) The revolution of dsDNA along the 12 subunits of the connector channel.
Translocation of dsDNA helix by revolution through the 30°-tilted connector subunits facilitated by anti-parallel displacement between the right-handed dsDNA helix and the left-handed connector channel subunits
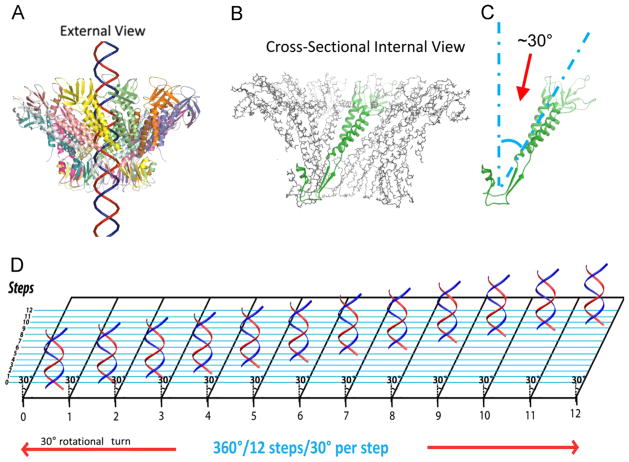
The crystal structure of the connector revealed that all 12 subunits of the connector protein are tilted at a 30° relative to the DNA axis in a configuration opposite in handedness to the dsDNA helix during packaging, to form the channel (Guasch et al., 2002; Badasso et al., 2000). The structural relationship for the left-handed/right-handed anti-parallel displacement between the connector whirl and dsDNA helix can be visualized from an external viewpoint, in which dsDNA propels through the connector potentially making contact at every 30° subunit (Fig. 11). The left-handed/right-handed anti-parallel displacement argues against the rotation threading mechanism that requires the threads to be in the same direction to move. On the contrary, this arrangement greatly facilitates controlled single directional revolution motion; supporting the model that dsDNA revolves through the connector channel without producing a coiling or torsion force, and touching each of the 12 connector subunits in 12 discrete steps of 30° transitions. Since each change of 30° (360°/12=30°) during the advancement of the DNA helical pitch is compensated by the connector subunit 30° tilting (Fig. 11D), rotation of dsDNA is not necessary during translocation.
Fig. 11.
DNA revolves and transports through 30° tilted connector subunits facilitated by anti-parallel helices between dsDNA helix and connector protein subunits. The anti-parallel configuration can be visualized in an external view (A) in which DNA revolves through the connector making contacts at every 30° subunit (B,C). A planar view is suggested (D) in which DNA is advanced and travels along the circular wall of the connector channel with no torsion or coiling force, through the connector channel, touching each subunit translating to 12 discrete steps of 30° revolving turns for each step.
Discussion
A rotation mechanism for viral DNA packaging has long been proposed (Hendrix, 1978) and has been well-regarded by the scientific community. However, studies in which the connector was covalently linked to the capsid protein have suggested that the connector does not rotate (Baumann et al., 2006; Maluf and Feiss, 2006). When the connector and the procapsid protein were fused to each other, rotation of the connector within the procapsid was not possible since motors were still active in packaging, implying that connector rotation is not necessary for DNA packaging. The lack of connector rotation was also shown by experiments combining the methods of single-molecule force spectroscopy with a polarization-sensitive single-molecule fluorescence trap (Hugel et al., 2007). Because the connector does not rotate, there is no reason to expect that gp16 would rotate since the gp16 ring is tightly bound to the pRNA ring (Lee and Guo, 2006) that is immobilized to the stationary connector. Single molecule studies by three separate labs using beads tethered to the end of the phi29 dsDNA have revealed that dsDNA still translocates into the procapsid even with such tethering (Shu et al., 2007; Chang et al., 2008; Moffitt et al., 2009; Yu et al., 2010). This data led to a mystery regarding the operation of the phi29 DNA translocation motor since it does not follow the classical rotational model. The finding that phi29 DNA packaging motor utilizes a revolution rather than a rotation mechanism is in good agreement with all data reported in the literature. The connector was recently shown to only allow unidirectional movement of dsDNA (Jing et al., 2010), and a model using a “push through a one-way valve” mechanism has been described (Schwartz et al., 2012; Fang et al., 2012) that is in accordance with the proposed ratchet (Serwer, 2003) and compression (Ray et al., 2010a, 2010b) models that explain how DNA is prevented from reversing out of the capsid during packaging (Black, 1989; Feiss and Rao, 2012; Casjens, 2011; Guo and Lee, 2007). The revolution mechanism is independent of any specific stoichiometry, thus motors with different oligomeric states can act similarly. This idea alleviates a current question that different phage packaging ATPases are found to be tetramers (Maluf et al., 2006; Feiss et al., 2010), hexamers (Guo et al., 1998; Schwartz et al., this issue; Shu et al., 2007), and nonamers (Roy et al., 2011).
Materials and methods
Cloning, mutagenesis and protein purification
The engineering of eGFP-gp16 and the purification of gp16 fusion protein have been reported previously (Lee et al., 2009). eGFP-gp16 mutants G27D, E119A, R146A, and D118E/E119D were cloned previously (Schwartz et al., this issue).
Measurement of gp16 ATPase activity
Enzymatic activity via fluorescence was described previously (Lee et al., 2008).
In vitro virion assembly assay
Purified in vitro components were mixed and subjected to virion assembly assay, as previously described (Lee and Guo, 1994).
Statistical analysis and data plotting
Most statistical analysis was performed using Sigmaplot 11. Determination of the Hill coefficient was obtained by nonlinear regression fitting of the experimental data to the following equation: , where E and Emax refer to the concentration of gp16/DNA complex, X is the concentration of ATP or ADP, Kapp is the apparent binding constant, and n is the Hill coefficient.
CE experiments to determine ratio of gp16 to bound dsDNA
CE (Capillary electrophoresis) experiments were performed on a Beckman MDQ system equipped with double fluorescent detectors (488 nm and 635 nm excitation). The capillary used was a bare borosilicate capillary 60 cm in total length with a 50 μm inner section. The method consisted of a 20 min separation at 30 KV normal polarity. Typical assay conditions contained an optimized buffer (Huang and Guo, 2003a, 2003b) of 50 mM Tris–HCl, 100 mM borate at pH 8.00, 5 mM MgCl2, 10% PEG 8000 (w/v), 0.5% acetone (v/v), 3 μM eGFP-gp16 monomer and variable amounts of ATP/ADP and DNA (Schwartz et al., this issue). Peaks were quantified and analyzed by Sigma Plot for DNA binding.
Sucrose gradient sedimentation of gp16/procapsid
Procapsids (1.6 mg/mL) were purified by opti-prep sedimentation, a sterile density gradient solution used in isolation of virus purification, and incubated with eGFP-gp16 (3 μM) and pRNA (62.5 ng/μL) at room temperature for 30 min. Samples were then loaded on top of a 5–20% sucrose gradient containing 50 mM NaCl, 25 mM Tris pH 8.0, 2% glycerol, 0.01% tween-20, 5 mM MgCl2, and 0.25 mM γ-S-ATP; 200 μL 60% sucrose was used as a cushion and were then sedimented at 35,000 rpm with a SW55 rotor for 4 h. After fractionation, the fluorescent signal was captured using a Synergy IV microplate reader.
Electrophoretic mobility shift assay (EMSA)
The engineering of eGFP-gp16 and the purification of gp16 fusion protein (Lee et al., 2009), as well as the gp16 and dsDNA binding assay (Schwartz et al., 2012), have been reported previously. Cy3- or Cy5-dsDNA (40 bp) was prepared by annealing two complementary DNA oligos containing Cy3 or Cy5 labels at their 5′ ends (IDT). The annealed product was purified from 10% polyacrylamide gel. The samples for EMSA assay were prepared in 20μl buffer A (20 mM Tris–HCl, 50 mM NaCl, 1.5% glycerol, 0.1 mM Mg2+). 1.78 μM eGFP-gp16 was mixed with 0.3 μM 40 bp Cy3-DNA at various conditions in the typical fashion. The samples were incubated at ambient temperature for 20 min and then loaded onto a 1% agarose gel (44.5 mM Tris borate) and electrophoresed at 8 V/cm for 1 h at 4 °C. The eGFP-gp16 and Cy3-DNA in the gel were analyzed by a fluorescent LightTools Whole Body Imager using 488 nm and 540 nm excitation wavelengths for GFP and Cy3, respectively.
Observation of gp16 motion
Double-stranded lambda DNA (48 kbp) was tethered between two polylysine coated 4 μm silica beads (Kad et al., 2010). The dsDNA was bound between beads by back-and-forth infusion of DNA over the beads for 10 min; binding occurred as a result of charge–charge interactions. The back and forth motion of DNA over the polylysine beads stretched the DNA taut and lifted the chain above the surface by the 4 μm silica beads as visualized under the microscope. The incident angle of the excitation beam in objective-type TIRF (total internal reflection fluorescence) was adjusted to a sub-critical angle in order to image the samples a few microns above the surface; this yields a low fluorescence background (Kad et al., 2010). To-Pro-3 was used to confirm the formation of the DNA tightropes. After the DNA tightrope was formed, a 30 μL mixture with a final concentration of 1 nM Cy3-gp16 with 100 nM unlabeled gp16 in buffer B (25 mM Tris, pH 6.1, 25 mM NaCl, 0.25 mM MgCl2) was infused into the sample chamber for binding to the stretched DNA. After 30 min incubation, 30 μL of a solution containing anti-photobleaching reagents (Shu et al., 2007) was infused into the chamber in order to prevent photobleaching of less photostable fluorophores and to detect binding. Movies were taken after the chamber was washed with buffer C (25 mM Tris, pH 8, 25 mM NaCl, 0.25 mM MgCl2). A comparison was made of washings with buffer C, with and without 20 mM ATP. Since the DNA has been fixed by charge interactions and the protein fixed by binding affinity to the tethered DNA, washing does not remove pertinent material. Sequential images were acquired with a 0.2 s exposure time at an interval of 0.22 s, with a laser of 532 nm for excitation. The movies were taken for about 8 min, or until the Cy3 fluorophores lost their fluorescence due to photobleaching. Image J software was utilized to generate kymographs to show the displacement of the Cy3-gp16 spots along the DNA chains.
Supplementary Material
Acknowledgments
We would like to thank Dr. Guo-Min Li for his valuable comments, Zhengyi Zhao, Emil Khisamutdinov, and Hui Li for their diligent work on the animation figures, Drs. Bruce Maley and Mary Gail Engle for EM images, and Jeannie Haak for modifying this manuscript. The work was supported by NIH grants R01 EB012135, U01 CA151648 and R01 EB003730 to PG, who is a co-founder of Kylin Therapeutics, Inc, and Biomotor and Nucleic Acids Nanotech Development, Ltd.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-Non Commercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–673. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J Struct Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Astumian RD. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917–922. doi: 10.1126/science.276.5314.917. [DOI] [PubMed] [Google Scholar]
- Badasso MO, Leiman PG, Tao Y, He Y, Ohlendorf DH, Rossmann MG, Anderson D. Purification, crystallization and initial X-ray analysis of the head- tail connector of bacteriophage phi29. Acta Crystallogr D Biol Crystallogr. 2000;56 (Pt 9):1187–1190. doi: 10.1107/s0907444900009239. [DOI] [PubMed] [Google Scholar]
- Bath J, Wu LJ, Errington J, Wang JC. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science. 2000;290:995–997. doi: 10.1126/science.290.5493.995. [DOI] [PubMed] [Google Scholar]
- Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- Black LW. DNA Packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- Chang C, Zhang H, Shu D, Guo P, Savran C. Bright-field analysis of phi29 DNA packaging motor using a magnetomechanical system. Appl Phys Lett. 2008;93:153902–153902-3. doi: 10.1063/1.3000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Chen C, Guo P. Sequential action of six virus-encoded DNA-packaging RNAs during phage phi29 genomic DNA translocation. J Virol. 1997;71:3864–3871. doi: 10.1128/jvi.71.5.3864-3871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Trottier M, Guo P. New approaches to stoichiometry determination and mechanism investigation on RNA involved in intermediate reactions. Nucleic Acids Symp Ser. 1997;36:190–193. [Google Scholar]
- Chistol G, Liu S, Hetherington CL, Moffitt JR, Grimes S, Jardine PJ, Bustamante C. High Degree of Coordination and Division of Labor among Subunits in a Homomeric Ring ATPase. Cell. 2012;151:1017–1028. doi: 10.1016/j.cell.2012.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarre G, Galli E, Barre FX. The FtsK family of DNA pumps. Adv Exp Med Biol. 2013;767:245–262. doi: 10.1007/978-1-4614-5037-5_12. [DOI] [PubMed] [Google Scholar]
- Ding F, Lu C, Zhao W, Rajashankar KR, Anderson DL, Jardine PJ, Grimes S, Ke A. Structure and assembly of the essential RNA ring component of a viral DNA packaging motor. Proc Natl Acad Sci U S A. 2011;108:7357–7362. doi: 10.1073/pnas.1016690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman HH, Yu X, Wild R, Hingorani MM, Patel SS. Bacteriophage T7 helicase/primase proteins form rings around single- stranded DNA that suggest a general structure for hexameric helicases. Proc Natl Acad Sci U S A. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Jing P, Haque F, Guo P. Role of channel lysines and “Push Through a One-way Valve” mechanism of viral DNA packaging motor. Biophys J. 2012;102:127–135. doi: 10.1016/j.bpj.2011.11.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiss M, Rao VB. The bacteriophage DNA packaging machine. Adv Exp Med Biol. 2012;726:489–509. doi: 10.1007/978-1-4614-0980-9_22. [DOI] [PubMed] [Google Scholar]
- Feiss M, Reynolds E, Schrock M, Sippy J. DNA packaging by lambda-like bacteriophages: mutations broadening the packaging specificity of terminase, the lambda-packaging enzyme 7. Genetics. 2010;184:43–52. doi: 10.1534/genetics.109.108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa H, Shibata H, Kato H. Analysis of interactions among factors involved in the bacteriophage T3 DNA packaging reaction in a defined in vitro system. Virology. 1991;185:788–794. doi: 10.1016/0042-6822(91)90550-u. [DOI] [PubMed] [Google Scholar]
- Grainge I. Sporulation: SpoIIIE is the key to cell differentiation. Curr Biol. 2008;18:R871–R872. doi: 10.1016/j.cub.2008.07.047. [DOI] [PubMed] [Google Scholar]
- Grainge I, Bregu M, Vazquez M, Sivanathan V, Ip SC, Sherratt DJ. Unlinking chromosome catenanes in vivo by site-specific recombination. EMBO J. 2007;26:4228–4238. doi: 10.1038/sj.emboj.7601849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainge I, Lesterlin C, Sherratt DJ. Activation of XerCD-dif recombination by the FtsK DNA translocase. Nucleic Acids Res. 2011;39:5140–5148. doi: 10.1093/nar/gkr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes S, Anderson D. RNA Dependence of the Bateriophage phi29 DNA Packaging ATPase. J Mol Biol. 1990;215:559–566. doi: 10.1016/s0022-2836(05)80168-8. [DOI] [PubMed] [Google Scholar]
- Grimes S, Anderson D. The bacteriophage phi29 packaging proteins super-coil the DNA ends. J Mol Biol. 1997;266:901–914. doi: 10.1006/jmbi.1996.0843. [DOI] [PubMed] [Google Scholar]
- Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol. 2002;315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- Guenther B, Onrust R, Sali A, O’Donnell M, Kuriyan J. Crystal structure of the delta’ subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi29 DNA. Science. 1987a;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- Guo P, Grimes S, Anderson D. A defined system for in vitro packaging of DNA-gp3 of the Bacillus subtilis bacteriophage phi29. Proc Natl Acad Sci U S A. 1986;83:3505–3509. doi: 10.1073/pnas.83.10.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Peterson C, Anderson D. Initiation events in in vitro packaging of bacteriophage φ29 DNA-gp3. J Mol Biol. 1987b;197:219–228. doi: 10.1016/0022-2836(87)90120-3. [DOI] [PubMed] [Google Scholar]
- Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage φ29. J Mol Biol. 1987c;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- Guo P, Zhang C, Chen C, Trottier M, Garver K. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- Guo PX, Lee TJ. Viral nanomotors for packaging of dsDNA and dsRNA. Mol Microbiol. 2007;64:886–903. doi: 10.1111/j.1365-2958.2007.05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjes E, Kitamura A, Zhao W, Morais MC, Jardine PJ, Grimes S, Matsuo H. Structure of the RNA claw of the DNA packaging motor of bacteriophage Phi29. Nucleic Acids Res. 2012;40:9953–9963. doi: 10.1093/nar/gks724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage DNA packaging: RNA gears in a DNA transport machine (Minireview) Cell. 1998;94:147–150. doi: 10.1016/s0092-8674(00)81413-0. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci U S A. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Yang F, Li L, Song Y, Jiang L, Zhu D. A biomimetic asymmetric responsive single nanochannel. J Am Chem Soc. 2010;132:11736–11742. doi: 10.1021/ja1045082. [DOI] [PubMed] [Google Scholar]
- Huang LP, Guo P. Use of acetone to attain highly active and soluble DNA packaging protein gp16 of phi29 for ATPase assay. Virology. 2003a;312:449–457. doi: 10.1016/s0042-6822(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Huang LP, Guo P. Use of PEG to acquire highly soluble DNA-packaging enzyme gp16 of bacterial virus phi29 for stoichiometry quantification. J Virol Methods. 2003b;109:235–244. doi: 10.1016/s0166-0934(03)00077-6. [DOI] [PubMed] [Google Scholar]
- Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Faik W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol. 2007;5:558–567. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y, Catalano CE, Feiss M. Kinetic and mutational dissection of the two ATPase activities of terminase, the DNA packaging enzyme of bacteriophage lambda. Biochemistry. 1996;35:2796–2803. doi: 10.1021/bi952322z. [DOI] [PubMed] [Google Scholar]
- Ibarra B, Valpuesta JM, Carrascosa JL. Purification and functional characterization of p16, the ATPase of the bacteriophage phi29 packaging machinary. Nucleic Acids Res. 2001;29:4264–4273. doi: 10.1093/nar/29.21.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA plus ATPases. J Struct Biol. 2004a;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004b;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J, Santisteban A, Carazo JM, Carrascosa JL. Computer graphic display method for visualizing three-dimensional biological structures. Science. 1986;232:1113–1115. doi: 10.1126/science.3754654. [DOI] [PubMed] [Google Scholar]
- Jing P, Haque F, Shu D, Montemagno C, Guo P. One-way traffic of a viral motor channel for double-stranded DNA translocation. Nano Lett. 2010;10:3620–3627. doi: 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kad NM, Wang H, Kennedy GG, Warshaw DM, Van HB. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Mol Cell. 2010;37:702–713. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainov DE, Mancini EJ, Telenius J, Lisai J, Grimes JM, Bamford DH, Stuart DI, Tuma R. Structural basis of mechanochemical coupling in a hexameric molecular motor. J Biol Chem. 2008;283:3607–3617. doi: 10.1074/jbc.M706366200. [DOI] [PubMed] [Google Scholar]
- Koti JS, Morais MC, Rajagopal R, Owen BA, McMurray CT, Anderson D. DNA packaging motor assembly intermediate of bacteriophage phi29. J Mol Biol. 2008;381:1114–1132. doi: 10.1016/j.jmb.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA, Hayes SJ, Wright ET, Watson RH, Serwer P. Specific single-stranded breaks in mature bacteriophage T7 DNA. Virology. 1995;211:329–331. doi: 10.1006/viro.1995.1411. [DOI] [PubMed] [Google Scholar]
- Lee CS, Guo P. A highly sensitive system for the assay of in vitro viral assembly of bacteriophage phi29 of Bacillus subtilis. Virology. 1994;202:1039–1042. doi: 10.1006/viro.1994.1434. [DOI] [PubMed] [Google Scholar]
- Lee TJ, Guo P. Interaction of gp16 with pRNA and DNA for genome packaging by the motor of bacterial virus phi29. J Mol Biol. 2006;356:589–599. doi: 10.1016/j.jmb.2005.10.045. [DOI] [PubMed] [Google Scholar]
- Lee TJ, Zhang H, Chang CL, Savran C, Guo P. Engineering of the fluorescent-energy-conversion arm of phi29 DNA packaging motor for single-molecule studies. Small. 2009;5:2453–2459. doi: 10.1002/smll.200900467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, Zhang H, Liang D, Guo P. Strand and nucleotide-dependent ATPase activity of gp16 of bacterial virus phi29 DNA packaging motor. Virology. 2008;380:69–74. doi: 10.1016/j.virol.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Ellonen A, Allen MD, Atkinson C, Sherratt DJ, Grainge I. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol Cell. 2008;31:498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluf NK, Feiss M. Virus DNA translocation: progress towards a first ascent of mount pretty difficult. Mol Microbiol. 2006;61:1–4. doi: 10.1111/j.1365-2958.2006.05214.x. [DOI] [PubMed] [Google Scholar]
- Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45:15259–15268. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA+motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Lowe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Mastrangelo IA, Hough PV, Wall JS, Dodson M, Dean FB, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- McNally R, Bowman GD, Goedken ER, O’Donnell M, Kuriyan J. Analysis of the role of PCNA-DNA contacts during clamp loading. BMC Struct Biol. 2010;10:3. doi: 10.1186/1472-6807-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll D, Guo P. Grouping of Ferritin and Gold Nanoparticles Conjugated to pRNA of the Phage phi29 DNA-packaging motor. J Nanosci Nanotechnol. 2007;7:3257–3267. doi: 10.1166/jnn.2007.914. [DOI] [PubMed] [Google Scholar]
- Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson D, Rossman MG. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure. 2008;16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais MC, Tao Y, Olsen NH, Grimes S, Jardine PJ, Anderson D, Baker TS, Rossmann MG. Cryoelectron-Microscopy Image Reconstruction of Symmetry Mismatches in Bacteriophage phi29. J Struct Biol. 2001;135:38–46. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Tasaka M, Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: importance of the C-terminal region in prohead binding. J Mol Biol. 1995b;245:635–644. doi: 10.1006/jmbi.1994.0052. [DOI] [PubMed] [Google Scholar]
- Morita M, Tasaka M, Fujisawa H. DNA packaging ATPase of bacteriophage T3. Virology. 1993;193:748–752. doi: 10.1006/viro.1993.1183. [DOI] [PubMed] [Google Scholar]
- Morita M, Tasaka M, Fujisawa H. Analysis of the fine structure of the prohead binding domain of the packaging protein of bacteriophage T3 using a hexapeptide, an analog of a prohead binding site. Virology. 1995a;211:516–524. doi: 10.1006/viro.1995.1433. [DOI] [PubMed] [Google Scholar]
- Mueller-Cajar O, Stotz M, Wendler P, Hartl FU, Bracher A, Hayer-Hartl M. Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature. 2011;479:194–199. doi: 10.1038/nature10568. [DOI] [PubMed] [Google Scholar]
- Niedenzu T, Roleke D, Bains G, Scherzinger E, Saenger W. Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. J Mol Biol. 2001;306:479–487. doi: 10.1006/jmbi.2000.4398. [DOI] [PubMed] [Google Scholar]
- Oram M, Sabanayagam C, Black LW. Modulation of the Packaging Reaction of Bacteriophage T4 Terminase by DNA Structure. J Mol Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons CA, Stasiak A, Bennett RJ, West SC. Structure of a multisubunit complex that promotes DNA branch migration. Nature. 1995;374:375–378. doi: 10.1038/374375a0. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Clancy SB, Tsuruta H, Gonzalez S, Wetmur JG, Tainer JA. Structure and mechanism of the RuvB Holliday junction branch migration motor. J Mol Biol. 2001;311:297–310. doi: 10.1006/jmbi.2001.4852. [DOI] [PubMed] [Google Scholar]
- Rao VB, Feiss M. The Bacteriophage DNA Packaging Motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- Ray K, Ma J, Oram M, Lakowicz JR, Black LW. Single-molecule and FRET fluorescence correlation spectroscopy analyses of phage DNA packaging: colocalization of packaged phage T4 DNA ends within the capsid. J Mol Biol. 2010a;395:1102–1113. doi: 10.1016/j.jmb.2009.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Sabanayagam CR, Lakowicz JR, Black LW. DNA crunching by a viral packaging motor: compression of a procapsid-portal stalled Y-DNA substrate. Virology. 2010b;398:224–232. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Bhardwaj A, Cingolani G. Crystallization of the nonameric small terminase subunit of bacteriophage P22. Acta Crystallogr F-Struct Biol Cryst Commun. 2011;67:104–110. doi: 10.1107/S174430911004697X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabanayagam CR, Oram M, Lakowicz JR, Black LW. Viral DNA packaging studied by fluorescence correlation spectroscopy. Biophys J. 2007;93:L17–L19. doi: 10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, et al. The ATPase of the phi29 DNA packaging motor is a member of the hexameric AAA+ superfamily. Virology. doi: 10.1016/j.virol.2013.04.004. http://dx.doi.org/10.1016/j.virol.2013.04.004i, this issue. [DOI] [PMC free article] [PubMed]
- Schwartz C, Fang H, Huang L, Guo P. Sequential action of ATPase, ATP, ADP, Pi and dsDNA in procapsid-free system to enlighten mechanism in viral dsDNA packaging. Nucleic Acids Res. 2012;40:2577–2586. doi: 10.1093/nar/gkr841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwer P. Models of bacteriophage DNA packaging motors. J Struct Biol. 2003;141:179–188. doi: 10.1016/s1047-8477(02)00628-7. [DOI] [PubMed] [Google Scholar]
- Serwer P. A hypothesis for bacteriophage DNA packaging motors. Viruses. 2010;2:1821–1843. doi: 10.3390/v2091821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Fujisawa H, Minagawa T. Early events in a defined in vitro system for packaging of bacteriophage T3 DNA. Virology. 1987;159:250–258. doi: 10.1016/0042-6822(87)90462-4. [DOI] [PubMed] [Google Scholar]
- Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junctions for constructing multifunctional nanoparticles for delivery of therapeutics. Nat Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Guo P. A Viral RNA that binds ATP and contains an motif similar to an ATP-binding aptamer from SELEX. J Biol Chem. 2003a;278 (9):7119–7125. doi: 10.1074/jbc.M209895200. [DOI] [PubMed] [Google Scholar]
- Shu D, Guo P. Only one pRNA hexamer but multiple copies of the DNA-packaging protein gp16 are needed for the motor to package bacterial virus phi29 genomic DNA. Virology. 2003b;309 (1):108–113. doi: 10.1016/s0042-6822(03)00011-4. [DOI] [PubMed] [Google Scholar]
- Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Haque F, Shu D, Li W, Zhu Z, Kotb M, Lyubchenko Y, Guo P. Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA. 2013;19(6):767–777. doi: 10.1261/rna.037002.112. http://dx.doi.org/10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story RM, Steitz TA. Structure of the rec-A protein-ADP complex. Nature. 1992:355. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- Trottier M, Guo P. Approaches to determine stoichiometry of viral assembly components. J Virol. 1997;71:487–494. doi: 10.1128/jvi.71.1.487-494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Mei Z, Qi Y, Yan C, Hu Q, Wang J, Shi Y. Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature. 2011;471:331–335. doi: 10.1038/nature09780. [DOI] [PubMed] [Google Scholar]
- Willows RD, Hansson A, Birch D, Al-Karadaghi S, Hansson M. EM single particle analysis of the ATP-dependent BchI complex of magnesium chelatase: an AAA(+) hexamer. J Struct Biol. 2004;146:227–233. doi: 10.1016/j.jsb.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Xiao F, Zhang H, Guo P. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res. 2008;36:6620–6632. doi: 10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Demeler B, Guo P. Assembly mechanism of the sixty-subunit nanoparticles via interaction of RNA with the reengineered protein connector of phi29 DNA-packaging motor. ACS Nano. 2010;4:3293–3301. doi: 10.1021/nn100158k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Moffitt J, Hetherington CL, Bustamante C, Oster G. Mechanochem-istry of a viral DNA packaging motor. J Mol Biol. 2010;400:186–203. doi: 10.1016/j.jmb.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Zhang F, Lemieux S, Wu X, St-Arnaud S, McMurray CT, Major F, Anderson D. Function of hexameric RNA in packaging of bacteriophage phi29 DNA in vitro. Mol Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- Zhang H, Schwartz C, De Donatis GM, Guo P. “Push Through One-Way Valve” mechanism of viral DNA packaging. Adv Virus Res. 2012;83:415–465. doi: 10.1016/B978-0-12-394438-2.00009-8. [DOI] [PubMed] [Google Scholar]
- Zhang H, Endrizzi JA, Shu Y, Haque F, Guo P, Chi YI. Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA. doi: 10.1261/rna.037077.112. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.