Cytokinin enhances the capacity of plants to tolerate water deficit by improving carbon and nitrogen assimilation processes during stress.
Abstract
The effects of water deficit on carbon and nitrogen metabolism were investigated in flag leaves of wild-type and transgenic rice (Oryza sativa japonica ‘Kitaake’) plants expressing ISOPENTENYLTRANSFERASE (IPT; encoding the enzyme that mediates the rate-limiting step in cytokinin synthesis) under the control of PSARK, a maturation- and stress-induced promoter. While the wild-type plants displayed inhibition of photosynthesis and nitrogen assimilation during water stress, neither carbon nor nitrogen assimilation was affected by stress in the transgenic PSARK::IPT plants. In the transgenic plants, photosynthesis was maintained at control levels during stress and the flag leaf showed increased sucrose (Suc) phosphate synthase activity and reduced Suc synthase and invertase activities, leading to increased Suc contents. The sustained carbon assimilation in the transgenic PSARK::IPT plants was well correlated with enhanced nitrate content, higher nitrate reductase activity, and sustained ammonium contents, indicating that the stress-induced cytokinin synthesis in the transgenic plants played a role in maintaining nitrate acquisition. Protein contents decreased and free amino acids increased in wild-type plants during stress, while protein content was preserved in the transgenic plants. Our results indicate that the stress-induced cytokinin synthesis in the transgenic plants promoted sink strengthening through a cytokinin-dependent coordinated regulation of carbon and nitrogen metabolism that facilitates an enhanced tolerance of the transgenic plants to water deficit.
Plant hormones control many aspects of plant growth and development and the responses of plants to abiotic and biotic stresses. Cytokinins (CKs) have been shown to regulate plant cell differentiation, leaf senescence, and other key developmental processes (Sakakibara, 2006). It has also been shown that CKs regulate assimilate partitioning (Ronzhina and Mokronosov, 1994), sink strength (Kuiper, 1993), and source/sink relationships (Roitsch, 1999). The localized expression in tobacco (Nicotiana tabacum) of a promoterless ISOPENTENYLTRANSFERASE (IPT), a gene encoding the enzyme that catalyzes the rate-limiting step in CK synthesis, enhanced the local sink strength and quickly mobilized nutrients to the tissues with elevated CK (Guivarc’h et al., 2002). Changes in sink/source relationships were also observed in CK-deficient tobacco shoots and roots (Werner et al., 2008). Elevated CK levels enhanced the survival of plants under water-stress conditions (Rivero et al., 2007). The overexpression of IPT under the control of SENESCENCE-ASSOCIATED RECEPTOR KINASE (SARK; a maturation- and stress-induced promoter) improved the drought tolerance of both eudicots (Rivero et al., 2007; Qin et al., 2011) and monocots (Peleg et al., 2011). After a water-stress episode during the reproductive stages (pre and post anthesis), transgenic PSARK::IPT rice (Oryza sativa) plants displayed higher grain yield than the wild type (Peleg et al., 2011). The transgenic PSARK::IPT rice exhibited a differential expression of genes encoding enzymes associated with hormone synthesis and hormone-regulated pathways. These results suggested that changes in hormone homeostasis induced the modification of source/sink relationships in the transgenic plants, resulting in higher grain yields under stress conditions (Peleg et al., 2011).
The maintenance of carbon (C) and nitrogen (N) assimilation is of paramount importance to ensure sink strength and improve stress tolerance without yield penalties. The interactions between C and N metabolism are vital for plant growth and development, and complex mechanisms operate in the plant to coordinate C assimilation with N metabolism (Nunes-Nesi et al., 2010). Thus, plants respond to changes in C and N metabolites through the regulation of translation and posttranslational modification mechanisms. C and N metabolites activate signaling pathways that regulate enzyme and transporter activities that control C and N fluxes, optimizing the plant response to developmental and environmental cues changing source/sink relationships (Coruzzi and Zhou, 2001).
Plant hormones affect, either directly or indirectly, these pathways and can act antagonistically or synergistically when responding to environmental stress (Wilkinson et al., 2012). The exposure of plants to water-limiting conditions results in abscisic acid (ABA) synthesis that induces ABA-dependent gene expression (Yamaguchi-Shinozaki and Shinozaki, 2006), triggering the closure of stomata and reducing water loss during drought (Wilkinson and Davies, 2010). Other hormones, in particular CK, salicylic acid, ethylene, and jasmonic acid, also play direct or indirect roles in the plant responses to abiotic stress (Peleg and Blumwald, 2011). Under drought stress, plant CK content decreases, and the reduction in CK increases the plant responses to increasing ABA (Davies and Zhang, 1991), inducing stomata closure and inhibiting photosynthesis (Rivero et al., 2010). Our previous results suggested that the stress-induced CK synthesis, driven by a stress-induced promoter, protected against the deleterious effects of water deficit on the photosynthetic apparatus, allowing higher photosynthetic rates and higher yields after water deficit in tobacco (Rivero et al., 2009) and cotton (Gossypium hirsutum; Kuppu et al., 2013) plants grown in the greenhouse and peanut (Arachis hypogaea) plants grown under field conditions (Qin et al., 2011).
Here, we analyzed gene expression profiles, metabolites, and enzymatic and photosynthetic activities of flag leaves of wild-type and transgenic rice expressing PSARK::IPT exposed to water deficit during the reproductive stage and identified metabolic processes associated with the enhanced tolerance of the transgenic plants to water deficit. Our results indicate that the stress-induced CK synthesis in the transgenic plants promoted sink strengthening through the maintenance and coordination of N and C assimilation during water stress.
RESULTS
Effects of Water Stress on C Assimilation in Wild-Type and Transgenic PSARK::IPT Rice Plants
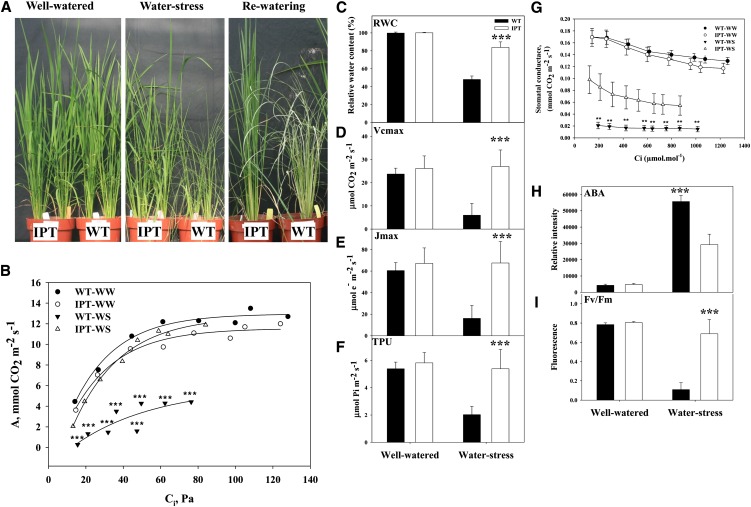
Compared with wild-type plants, which exhibited severe drought stress and reduced growth after 3 d of water stress, transgenic PSARK::IPT rice plants displayed enhanced growth and tolerance to drought stress (Fig. 1A). In addition, PSARK::IPT rice recovered from drought upon rewatering, while wild-type plants did not (Fig. 1A, right panel). Under water-stress conditions, the transgenic plants were able to maintain higher photosynthetic rates, as compared with wild-type plants (Fig. 1B). The relative water content (RWC) of wild-type plants was drastically reduced during water stress, while the decrease in RWC of PSARK::IPT plants was attenuated and remained above 80% (Fig. 1C). The maximum rate of carboxylation (Vcmax), maximum rate of electron transport (Jmax), and triose phosphate utilization (TPU) were calculated in order to evaluate Rubisco activity, the regeneration of ribulose bisphosphate, and phosphate limitations, respectively (Sharkey, 1985; Fig. 1, D–F). Vcmax, Jmax, and TPU remained unchanged in the transgenic plants, while a 75% reduction was noted in the wild-type plants. Transgenic PSARK::IPT plants were able to regulate stomata conductance (Fig. 1G) and displayed a lesser increase in ABA content during water stress (Fig. 1H), which allowed gas exchange and the maintenance of photosynthetic activity during the stress episode (Fig. 1B). In wild-type plants, the stomatal closing after 3 d of water stress correlated well with the decrease in carboxylation efficiency. The maintenance of a constant ratio of variable to maximal chlorophyll fluorescence (Fv/Fm), an indicator of the maximum quantum efficiency of PSII and stress-induced photoinhibition (Krause et al., 1989), further supported the ability of the transgenic plants to maintain C assimilation during the stress episode (Fig. 1I).
Figure 1.
Analysis of carbon assimilation in flag leaves of wild-type (WT) and transgenic PSARK::IPT rice plants subjected to 3 d of water stress. A, Plants grown under well-watered conditions, 3 d of water stress, and 3 d after rewatering at pre anthesis (booting) stage. B, A/Ci curves of wild-type well-watered (WW; black circles), wild-type stressed (WS; black triangles), PSARK::IPT well-watered (white circles), and PSARK::IPT stressed (white triangles) plants. C, RWC. D, Vcmax. E, Jmax. F, TPU. G, Stomatal conductance (mmol water m−2 s−1). H, ABA content. I, Fv/Fm. Values are means ± sd (n = 7). Seven individual plants per genotype/treatment were used in each of the measurements, and the data were analyzed using one-way ANOVA followed by Student’s t test. Asterisks indicate significant differences between genotypes per treatment (**P ≤ 0.01, ***P ≤ 0.001).
Impact of IPT Expression on the Regulation of C Metabolism Under Water Stress
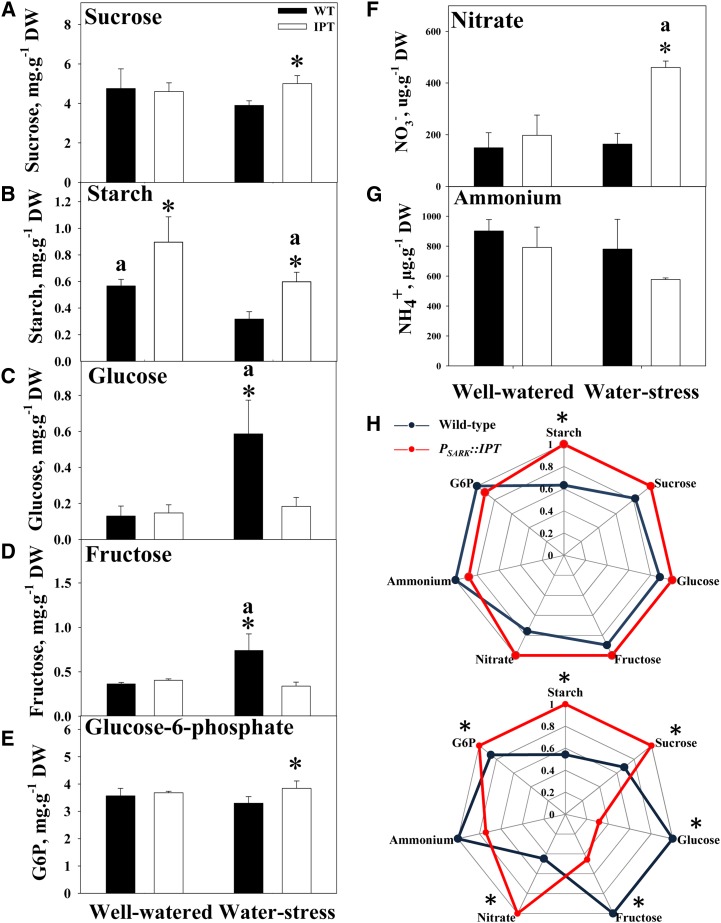
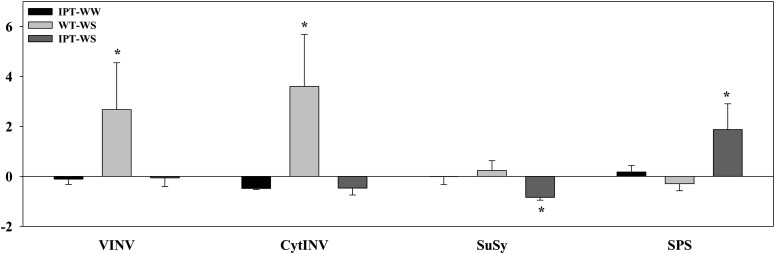
Changes in C metabolism in PSARK::IPT rice plants were associated with the maintenance of photosynthetic activity during water stress. Although both wild-type and PSARK::IPT plants showed reductions in starch content under stress (Fig. 2) correlated with the increase in amylolytic activity with the stress (Table I), PSARK::IPT rice plants displayed higher starch content than wild-type plants grown under both well-watered and water-stress conditions (Fig. 2B; Peleg et al., 2011). Despite the increase in amylolytic activity found in wild-type plants with the stress, no significant differences were found in amylolytic activity during stress between transgenic and wild-type plants (Table I). Suc, Glc, Fru, and Glc-6-P contents were similar in wild-type and PSARK::IPT plants grown under well-watered conditions (Fig. 2). During water stress, Suc contents increased in PSARK::IPT plants (Fig. 2), and this increase was correlated with higher Suc synthesis (sucrose phosphate synthase [SPS] activity) and lower cytosolic Suc degradation (sucrose synthase [SuSy] and cytosolic invertase activities; Table I). The reduction in Suc synthesis seen in wild-type plants during water stress was accompanied by increased Suc degradation (Table I) and the concomitant increase in Glc and Fru (Fig. 2). The flow of C from starch and/or Suc to Glc and Fru indicated a typical osmotic adjustment response of the wild-type plants to water stress (Merewitz et al., 2011; Nio et al., 2011). This osmotic stress response was not seen in the transgenic PSARK::IPT plants. The increase in cytosolic invertase and SuSy activities in wild-type plants during water stress was paralleled by the increase in transcripts encoding for vacuolar and cytosolic invertases (Fig. 3) that resulted in the increase of Glc and Fru contents. Also, the increase in SPS activity in the transgenic PSARK::IPT plants was matched by an increase in transcripts encoding SPS (Fig. 3). Trehalase activity displayed similar patterns in response to stress between wild-type and transgenic plants under both conditions (Table I). Interestingly, PSARK::IPT plants displayed higher hexokinase (HK) activity than wild-type plants under both experimental conditions, and the high HK activity correlated well with the Glc-6-P contents of the transgenic plants (Fig. 2). These results appear to be consistent with the role of Glc-6-P as a substrate for Suc synthesis via SPS, in contrast to the wild-type plants, where Suc was degraded. Our results show that increased HK activity in PSARK::IPT plants is not associated with detrimental effects on photosynthesis and growth or in accelerating senescence, indicating that the differences between the regulation driven by changes in gene expression and protein activity might involve different signaling pathways, as suggested previously (Cho et al., 2009).
Figure 2.
Sugar and N-related metabolite contents in flag leaf tissue at the pre anthesis stage of wild-type (WT) and transgenic PSARK::IPT plants. A, Suc. B, Starch. C, Glc. D, Fru. E, Glc-6-P. F, Nitrate. G, Ammonium. H, Radar charts comparing the metabolic profile of wild-type (blue) and transgenic PSARK::IPT (red) plants under control (well-watered) conditions (top) and water-stress conditions (bottom). Values are means ± sd (n = 6). Six individual plants per genotype/treatment were used in each of the measurements, and the data were analyzed using one-way ANOVA followed by Student’s t test. Asterisks indicate significant differences by genotypes (*P ≤ 0.05); a indicates significant differences between treatments (P ≤ 0.05). DW, Dry weight.
Table I. C-related enzyme activities during pre anthesis in flag leaves.
C-related enzyme activities were determined in flag leaves of wild-type and transgenic PSARK::IPT plants subjected to 3 d of water stress. Enzyme activities are expressed as moles of metabolite generated/consumed per milligram of protein per unit of time. Additional details are provided in “Materials and Methods.” Values are presented as means ± sd of six individual plants per genotype/treatment. The data were analyzed using one-way ANOVA followed by Student’s t test. Boldface values indicate significant differences by genotype comparisons (P ≤ 0.05); asterisks indicate significant differences by treatment comparisons (P ≤ 0.05); CWInv, Cell wall invertase.
| Enzyme Activities | Flag Leaf |
|||
|---|---|---|---|---|
| Well Watered |
Water-Stress |
|||
| Wild Type | PSARK::IPT | Wild Type | PSARK::IPT | |
| SPS (µmol Suc mg−1 protein h−1) | 24.9 ± 6.5* | 18.5 ± 2.7 | 16.3 ± 3.4 | 24 ± 2.6* |
| SuSy (synthesis) [µmol Suc mg−1 protein h−1] | 5.9 ± 1.3 | 5.8 ± 2.6 | 5 ± 2.4 | 5.8 ± 2.72 |
| SuSy (degradation) [µmol Fru mg−1 protein h−1] | 4.7 ± 0.5 | 3.3 ± 0.7 | 5.7 ± 0.4 | 4. ± 0.8 |
| CytINV (µmol Glc mg−1 protein h−1) | 2.9 ± 0.6 | 2.9 ± 0.2 | 4.4 ± 0.5 | 2.3 ± 0.6 |
| VINV (µmol Glc mg−1 protein h−1) | 7.7 ± 1.7 | 9.1 ± 1.2 | 11.6 ± 1.7* | 9.5 ± 0.6 |
| CWInv (µmol Glc mg−1 protein h−1) | 9 ± 3.2 | 11 ± 3.4 | 11.8 ± 3 | 9.5 ± 3.9 |
| Amylolytic activity (µmol Glc mg−1 protein h−1) | 6 ± 0.8 | 5.8 ± 0.8 | 7.9 ± 1.6* | 6.7 ± 0.7 |
| Trehalase (µmol Glc mg−1 protein h−1) | 0.6 ± 0.1 | 0.5 ± 0.2 | 1 ± 0.06* | 1.2 ± 0.1* |
| HK (nmol NADH mg−1 protein min−1) | 6.1 ± 0.6 | 9.4 ± 0.4 | 8.8 ± 0.9* | 12 ± 1.3* |
Figure 3.
Relative expression of C-related genes in flag leaves of wild-type (WT) and transgenic PSARK::IPT plants grown under well-watered (WW) and water-stress (WS) conditions. Values were calculated and normalized using the transcription elongation factor (LOC_Os03g08060) as an internal control. Expression values were calculated relative to the expression of wild-type plants grown under well-watered conditions (2−ΔΔCt = 0). Values represent means ± sd (n = 6). C-related genes are as follows: VINV, vacuolar invertase (LOC_Os02g01590); CytINV, cytosolic neutral invertase (LOC_Os01g22900); SuSy (LOC_Os07g42490); SPS (LOC_Os06g43630). Asterisks indicate significant differences between genotypes (*P ≤ 0.05).
Effects of Water Stress on the Interaction between C and N Metabolism
No difference in NH4+ contents were seen in wild-type and PSARK::IPT transgenic plants under well-watered and water-stress conditions (Fig. 2). However, equal NH4+ contents could result from different NH4+ generation processes. Under drought stress, wild-type plants displayed increased glutamate dehydrogenase (GDH) deamination activity (Table II). This process has been shown to be active under stress conditions and serves to direct amino acid synthesis in conditions where N assimilation is decreased (Masclaux-Daubresse et al., 2010), as reflected by the decrease in nitrate reductase (NR) activity in wild-type plants under stress (Table II). PSARK::IPT plants were able to maintain primary N assimilation without the activation of reassimilation processes, increasing NO3− uptake together with higher NR and nitrite reductase (NiR) activities (Table II).
Table II. N-related enzyme activities during pre anthesis in flag leaves.
N-related enzyme activities were determined in flag leaves of wild-type and transgenic PSARK::IPT plants subjected to 3 d of water stress. Enzyme activities are expressed as moles of metabolite generated/consumed per milligram of protein per unit of time. Additional details are provided in “Materials and Methods.” Values are presented as means ± sd of six individual plants per genotype/treatment. The data were analyzed using one-way ANOVA followed by Student’s t test. Boldface values indicate significant differences by genotype comparisons (P ≤ 0.05); asterisks indicate significant differences by treatment comparisons (P ≤ 0.05).
| Enzyme Activities | Flag Leaf |
|||
|---|---|---|---|---|
| Well Watered |
Water-Stress |
|||
| Wild Type | PSARK::IPT | Wild Type | PSARK::IPT | |
| NR (nmol NO2 mg−1 protein min−1) | 3.2 ± 0.4* | 3.4 ± 0.3 | 1.7 ± 0.5 | 5.1 ± 0.2* |
| NiR (nmol NO2 mg−1 protein min−1) | 98.9 ± 26.1* | 138.1 ± 38.4 | 14.7 ± 8.3 | 118 ± 8 |
| NADH-GOGAT (nmol NADH mg−1 protein min−1) | 7.7 ± 1.2 | 7.3 ± 1.2 | 8. 5 ± 1 | 6.9 ± 0.1 |
| GS (µmol Gln mg−1 protein h−1) | 198.3 ± 53.5 | 191.6 ± 23.1 | 215.4 ± 51.7 | 215.4 ± 26.1 |
| GDH (amination) [nmol NADH mg protein−1 min−1] | 8.1 ± 1.5 | 7.1 ± 1.1 | 6.6 ± 0.6 | 6.2 ± 1 |
| GDH (deamination) [nmol NADH mg−1 protein min−1] | 3.5 ± 0.3 | 5.3 ± 0.7 | 8.4 ± 2.4* | 4.7 ± 1.5 |
| Ala aminotransferase (nmol NADH mg−1 protein min−1) | 45 ± 6.5 | 41.5 ± 3 | 35.9 ± 5.6 | 43.5 ± 11.6 |
| Asp aminotransferase (nmol NADH mg−1 protein min−1) | 13.4 ± 0.9 | 12.9 ± 1.1 | 13.13 ± 1.6 | 13.9 ± 1.5 |
The water stress-induced inhibition of photosynthesis (Fig. 1) and the reduction in C and N assimilation in the wild-type plants were accompanied by a dramatic accumulation of amino acids (Table III). The increase in amino acid contents, together with the accumulation of Glc and Fru (Fig. 2), are characteristic hallmarks of plant osmotic adjustments to water deficit, providing protection against structural and functional damage caused by dehydration (Chen and Jiang, 2010).
Table III. Free amino acid content and relative TCA- and glycolysis-related metabolite contents in flag leaf of wild-type and transgenic PSARK::IPT plants grown under control and water-stress conditions.
Values are means ± sd for each compound and were calculated from semiquantitative data that were scaled with respect to the median response observed for each metabolite and in which null values were imputed with the minimum observed value. Six individual plants were used per genotype/treatment. Data were analyzed using one-way ANOVA followed by Student’s t test. Boldface values indicate significant differences by genotype (P ≤ 0.05); asterisks indicate significant differences by treatment (P ≤ 0.05). BDL, Below detection limits; DW, dry weight.
| Amino Acid/Metabolite |
Well Watered |
Water-Stress |
||
|---|---|---|---|---|
| Wild Type |
PSARK::IPT | Wild Type | PSARK::IPT | |
| Amino acid composition (nmol g−1 DW) | ||||
| Ala | 1.4 ± 0.1 | 0.8 ± 0.2 | 2.1 ± 0.2* | 0.8 ± 0.2 |
| Arg | 0.2 ± 0.07 | 0.2 ± 0.01* | 0.8 ± 0.1* | 0.07 ± 0.02 |
| 4-Guanidinobutanoate | 0.06 ± 0.01* | 0.06 ± 0.01 | 0.03 ± 0.003 | 0.05 ± 0.01 |
| Asn | 2.8 ± 0.4 | 3.3 ± 0.4* | 3.9 ± 0.03* | 2.4 ± 0.2 |
| Asp | 1.5 ± 0.2 | 1.8 ± 0.3 | 1.6 ± 0.21 | 1.5 ± 0.1 |
| Creatine | 0.2 ± 0.04 | 0.2 ± 0.04 | 0.3 ± 0.2 | 0.1 ± 0.05 |
| Ethanolamine | 0.1 ± 0.01 | 0.1 ± 0.02 | 0.2 ± 0.02 | 0.13 ± 0.02 |
| γ-Aminobutyric acid | 0.2 ± 0.01 | 0.1 ± 0.01 | 0.2 ± 0.07 | 0.1 ± 0.02 |
| Glu | 3.8 ± 0.5 | 4.5 ± 0.01* | 5.4 ± 0.03* | 3.3 ± 0.3 |
| Gln | 2.6 ± 0.27 | 2.1 ± 0.3* | 5.8 ± 1.2* | 1.5 ± 0.04 |
| Gly | 0.1 ± 0.01 | 0.07 ± 0.01* | 0.2 ± 0.05 | 0.04 ± 0.01 |
| His | 0.06 ± 0.01 | 0.05 ± 0.01 | 0.6 ± 0.1* | 0.04 ± 0.01 |
| Ile | 0.04 ± 0.002 | 0.04 ± 0.01 | 1.2 ± 0.1* | 0.03 ± 0.01 |
| Leu | 0.04 ± 0.001 | 0.03 ± 0.01 | 1.7 ± 0.2* | 0.03 ± 0.01 |
| Lys | 0.05 ± 0.004 | 0.05 ± 0.01 | 0.5 ± 0.06* | 0.03 ± 0.01 |
| Pipecolate | 0.04 ± 0.01 | 0.06 ± 0.02 | 0.04 ± 0.01 | 0.1 ± 0.03* |
| Met | 0.04 ± 0.02 | 0.03 ± 0.003 | 0.2 ± 0.01* | 0.03 ± 0.01 |
| Pro | BDL | 0.04 ± 0.01 | 1.1 ± 0.1* | BDL |
| Phe | 0.05 ± 0.01 | 0.05 ± 0.01 | 1.2 ± 0.2* | 0.04 ± 0.01 |
| Sarcosine | 1.4 ± 0.5 | 1.2 ± 0.2 | 1.1 ± 0.3 | 1.4 ± 0.2 |
| Ser | 1 ± 0.07 | 0.96 ± 0.1* | 1.2 ± 0.1 | 0.6 ± 0.1 |
| Thr | 0.4 ± 0.07 | 0.47 ± 0.03* | 1.6 ± 0.1 | 0.3 ± 0.04 |
| Trp | BDL | BDL | 0.6 ± 0.2 | BDL |
| Tyr | 0.05 ± 0.01 | 0.04 ± 0.003 | 0.7 ± 0.1 | 0.04 ± 0.003 |
| Val | 0.07 ± 0.005 | 0.08 ± 0.02 | 1.8 ± 0.2 | 0.05 ± 0.01 |
| TCA cycle metabolites (relative intensity) | ||||
| 2-Oxoglutarate | 1 ± 0.2* | 1.2 ± 0.2 | 0.8 ± 0.3 | 1 ± 0.3 |
| cis-Aconitate | 1.1 ± 0.2* | 1.1 ± 0.2 | 0.8 ± 0.1 | 1 ± 0.07 |
| Citrate | 1.2 ± 0.2* | 1.4 ± 0.2* | 0.8 ± 0.08 | 0.9 ± 0.08 |
| Fumarate | 1 ± 0.06 | 1.1 ± 0.1 | 1 ± 0.1 | 1.1 ± 0.3 |
| Malate | 1 ± 0.08 | 1.1 ± 0.2 | 1 ± 0.2 | 0.9 ± 0.08 |
| Succinate | 1.2 ± 0.1 | 1.1 ± 0.2 | 0.8 ± 0.05 | 1 ± 0.2 |
| Glycolysis (relative intensity) | ||||
| 1,3-Dihydroxyacetone | 0.9 ± 0.05 | 1 ± 0.1 | 1 ± 0.1 | 1.1 ± 0.07 |
| Fru-6-P | 1.1 ± 0.1 | 1.1 ± 0.3 | 0.8 ± 0.26 | 0.8 ± 0.4 |
| Glc | 0.07 ± 0.01 | 0.07 ± 0.02 | 0.2 ± 0.06* | 0.1 ± 0.02 |
| Glc-6-P | 1 ± 0.08 | 1 ± 0.02 | 0.9 ± 0.07 | 1.1 ± 0.06 |
| Glycerate | 1 ± 0.08 | 1.1 ± 0.2 | 0.8 ± 0.1 | 1.1 ± 0.3 |
| Pyruvate | 1.2 ± 0.1* | 1.3 ± 0.3* | 0.5 ± 0.1 | 0.7 ± 0.2 |
| Fru | 0.1 ± 0.05 | 0.1 ± 0.01 | 0.2 ± 0.06* | 0.07 ± 0.03 |
| Lipid metabolism (relative intensity) | ||||
| Glycerol | 0.9 ± 0.1 | 1 ± 0.3 | 1.5 ± 0.3* | 1 ± 0.2 |
| Glycerol 3-phosphate | 0.9 ± 0.08 | 1 ± 0.1 | 1.2 ± 0.2* | 1 ± 0.1 |
The accumulation of pipecolate and 4-guanidine butanoate during stress in the PSARK::IPT plants is noteworthy (Table III). Pipecolate and 4-guanidine butanoate are intermediate components of Lys and Arg metabolism, respectively. In plants, these compounds act as N reservoirs and control the catabolism of Lys and Arg when stress conditions recede (Moulin et al., 2006). These results are consistent with the N remobilization patterns shown by PSARK::IPT plants, which were able to maintain N metabolism levels similar to those under control growth conditions (Fig. 4) and recovered after rewatering.
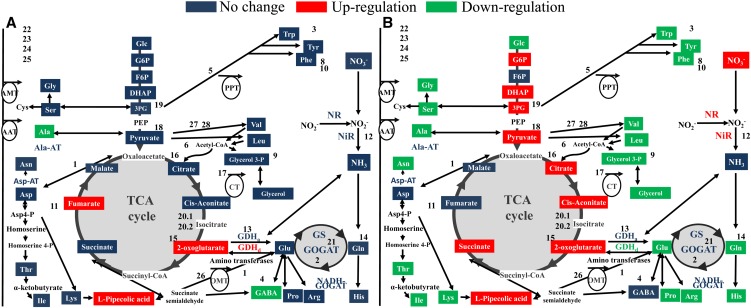
Figure 4.
The responses of C and N metabolism in wild-type and transgenic PSARK::IPT rice plants to water stress. A, Changes in C and N metabolism in transgenic PSARK::IPT rice plants relative to wild-type plants grown under well-watered conditions. B, Changes in C and N metabolism in transgenic PSARK::IPT rice plants relative to wild-type plants grown under water-stress conditions. Metabolites are represented in color boxes and include amino acids (Ala, Arg, Asn, Asp, Glu, Gln, Gly, His, Ile, Leu, Lys, l-pipecolic acid, Met, Phe, Pro, Ser, Thr, Trp, Tyr, and Val), TCA- and glycolysis-related compounds (Glc, Glc-6-P [G6P], Fru-6-P [F6P], dihydroxyacetone phosphate [DHAP], pyruvate, citrate, cis-aconitate, 2-oxoglutarate, succinate, fumarate, malate, glycerol 3-phosphate, and glycerol), and N assimilation compounds (NO3−, NO2−, and NH4+). Assayed enzymes are presented in boldface (NR, NiR, GS, NADH-GOGAT, GDH amination [GDHa] and deamination [GDHd] directions, Ala aminotransferase [Ala-AT], and Asp aminotransferase [Asp-AT]). Numbers represent genes that are differently expressed (listed in Supplemental Fig. S1): 1 (LOC_Os02g14110, aminotransferase), 1.2 (LOC_Os01g08270, aminotransferase), 1.3 (LOC_Os09g28050, aminotransferase), 1.4 (LOC_Os03g18810, aminotransferase), 1.5 (LOC_Os01g55540, aminotransferase), 2 (LOC_Os07g46460, Fd-GOGAT), 3 (LOC_Os02g20360, Tyr aminotransferase), 4 (LOC_Os04g52450, aminotransferase), 5 (LOC_Os08g01410, phosphate/phosphoenolpyruvate translocator), 6 (LOC_Os01g02020, acetyl-CoA acetyltransferase), 7 (LOC_Os07g42600, Ala aminotransferase), 8 (LOC_Os02g07160, glyoxalase family protein), 9 (LOC_Os04g14790, glycerol-3-phosphate dehydrogenase), 10 (LOC_Os02g41680, Phe ammonia-lyase), 11 (LOC_Os04g33300, amino acid kinase), 12 (LOC_Os02g37040, NTP3 [nitrate transporter]), 13 (LOC_Os04g45970, GDH), 14 (LOC_Os01g09000, glutaminyl-tRNA synthetase), 14.2 (LOC_Os07g07260, Gln-dependent NAD), 15 (LOC_Os02g38200, isocitrate dehydrogenase), 15.2 (LOC_Os01g46610, isocitrate dehydrogenase), 16 (LOC_Os02g10070, citrate synthase), 17 (LOC_Os03g05390, citrate transporter), 18 (LOC_Os04g31700, methylisocitrate lyase 2), 19 (LOC_Os10g08550, enolase), 20 (LOC_Os08g09200, aconitate hydratase), 20.2 (LOC_Os03g04410, aconitate hydratase), 21 (LOC_Os04g56400, Gln synthetase), 22.1 (LOC_Os06g16420, amino acid transporter), 22.2 (LOC_Os04g12499, amino acid transporter), 22.3 (LOC_Os06g36210, amino acid transporter), 22.4 (LOC_Os06g43700, amino acid transporter), 22.4 (LOC_Os11g09020, amino acid transporter), 22.5 (LOC_Os12g42850, amino acid permease), 22.6 (LOC_Os03g25869, amino acid permease), 22.7 (LOC_Os08g23440, amino acid permease), 22.8 (LOC_Os10g30090, amino acid permease), 22.9 (LOC_Os11g05690, amino acid permease), 22.10 (LOC_Os04g35540, amino acid transporter), 22.11 (LOC_Os08g03350, amino acid transporter), 22.12 (LOC_Os01g40410, amino acid transporter), 22.13 (LOC_Os01g63854, amino acid transporter), 22.14 (LOC_Os02g44980, amino acid transporter), 22.15 (LOC_Os02g09810, amino acid transporter), 22.16 (LOC_Os04g38680, amino acid transporter), 22.17 (LOC_Os05g50920, amino acid transporter), 22.18 (LOC_Os01g65000, ammonium transporter protein), 23.1 (LOC_Os02g34580, ammonium transporter protein), 23.2 (LOC_Os03g62200, ammonium transporter protein), 23.3 (LOC_Os04g43070, ammonium transporter protein), 24.1 (LOC_Os03g13240, peptide transporter PTR2), 24.2 (LOC_Os06g49250, peptide transporter PTR2), 24.3 (LOC_Os10g02240, peptide transporter PTR2), 24.4 (LOC_Os10g33170, peptide transporter PTR2), 24.5 (LOC_Os03g51050, peptide transporter PTR2), 25.1 (LOC_Os03g13274, NO3 peptide transporter PTR2), 25.2 (LOC_Os10g42900, NO3 peptide transporter PTR3), 26 (LOC_Os11g24450, 2-oxoglutarate/malate translocator), 27 (LOC_Os08g43170, hydroxymethylglutaryl-CoA synthase), and 28 (LOC_Os02g32490, acetyl-CoA synthetase). Red, green, and blue colors indicate higher, lower, and unchanged metabolite content/enzymatic activity, respectively. One-way ANOVA followed by Student’s t test (P ≤ 0.05) was performed using the values (metabolite content, enzyme activity, or gene expression) from six different plants (n = 6) per genotype/treatment.
The total activities of glutamate synthase (GOGAT), glutamine synthetase (GS), and amino acid transferases (Ala and Asp transferases) remained unchanged during water stress in both wild-type and PSARK::IPT plants, suggesting that amino acid synthesis was not affected by water stress (Fig. 4B; Table II). However, differences between wild-type and transgenic PSARK::IPT plants were seen in N assimilation processes under water-stress conditions. The increase in NR and the maintenance of NiR activities in the PSARK::IPT plants during water stress (Table II; Fig. 4B) indicated the positive effects of CK synthesis on N assimilation in transgenic plants. Wild-type plants displayed increased GDH deamination activity and decreased N assimilation (NR and NiR) activities (Table II). The increase in the GDH deamination activity was consistent with the amounts of NH4+ found in wild-type stressed plants (Supplemental Fig. S2). GDH deamination activity fulfills a dual role: providing 2-oxoglutarate to the tricarboxylic acid (TCA) cycle (limited by the reduction of C assimilation in wild-type plants) and providing the required NH4+ for the maintenance of amino acid synthesis (Fig. 4).
Regardless of the 2-oxoglutarate input driven by GDH, the wild-type plants subjected to water stress displayed a reduction of glycolysis- and TCA-related metabolites, such as citrate, cis-aconitate, 2-oxoglutarate, succinate, 3-Phosphoglyceric acid, or pyruvate (Table III). The decrease in these metabolites was well correlated with the decreased expression of genes encoding enzymes involved in these pathways, including isocitrate dehydrogenase (LOC_Os02g38200 and LOC_Os01g46610), citrate synthase (LOC_Os02g10070), citrate transporter (LOC_Os03g05390), methylisocitrate lyase 2 (LOC_Os04g31700), enolase (LOC_Os10g08550), and aconitate hydratase (LOC_Os03g04410; Supplemental Fig. S1).
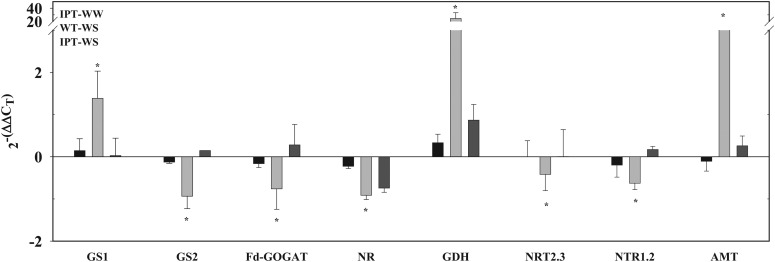
The increased ammonia reassimilation activities in the wild-type plants were paralleled by the increased expression of GS1 (cytosolic Gln synthetase1), GDH, and AMT (NH4+ transporter; Fig. 5). The significant decrease in Gln and Glu contents in the transgenic plants during water stress (Table III) compared with the opposite trend seen in the wild-type plants indicated the capacity of the transgenic plants to maintain protein synthesis during the stress episode. A comparison between wild-type and PSARK::IPT plants revealed a differential expression of genes encoding proteins associated with ribosomal assembly and translation (Supplemental Fig. S2). These results indicated that amino acid accumulation in wild-type plants during stress was a consequence of a decrease in protein synthesis (Supplemental Fig. S2) together with the increase in protein degradation, as supported by the increase in protease activities (Fig. 6B).
Figure 5.
Relative expression of N-related genes in flag leaves of wild-type (WT) and PSARK::IPT transgenic plants grown under well-watered (WW) and water-stress (WS) conditions. Values were calculated and normalized using the transcription elongation factor (LOC_Os03g08060) as an internal control. Expression values were calculated relative to the expression of wild-type plants grown under well-watered conditions (2−ΔΔCt = 0). Values are means ± sd (n = 6). N-related genes are as follows: GS1 (LOC_Os02g50240); GS2 (LOC_Os04g56400); ferredoxin (Fd)-GOGAT (LOC_Os07g46460); NR (LOC_Os08g36480); GDH (LOC_Os04g45970); NTR2.3, high-affinity nitrate transporter2.3 (LOC_Os01g50820); NTR1.2, nitrate transporter1.2 (LOC_Os06g38294); AMT, ammonium transporter (LOC_Os03g62200). Asterisks indicate significant differences between genotypes (*P ≤ 0.05).
Figure 6.
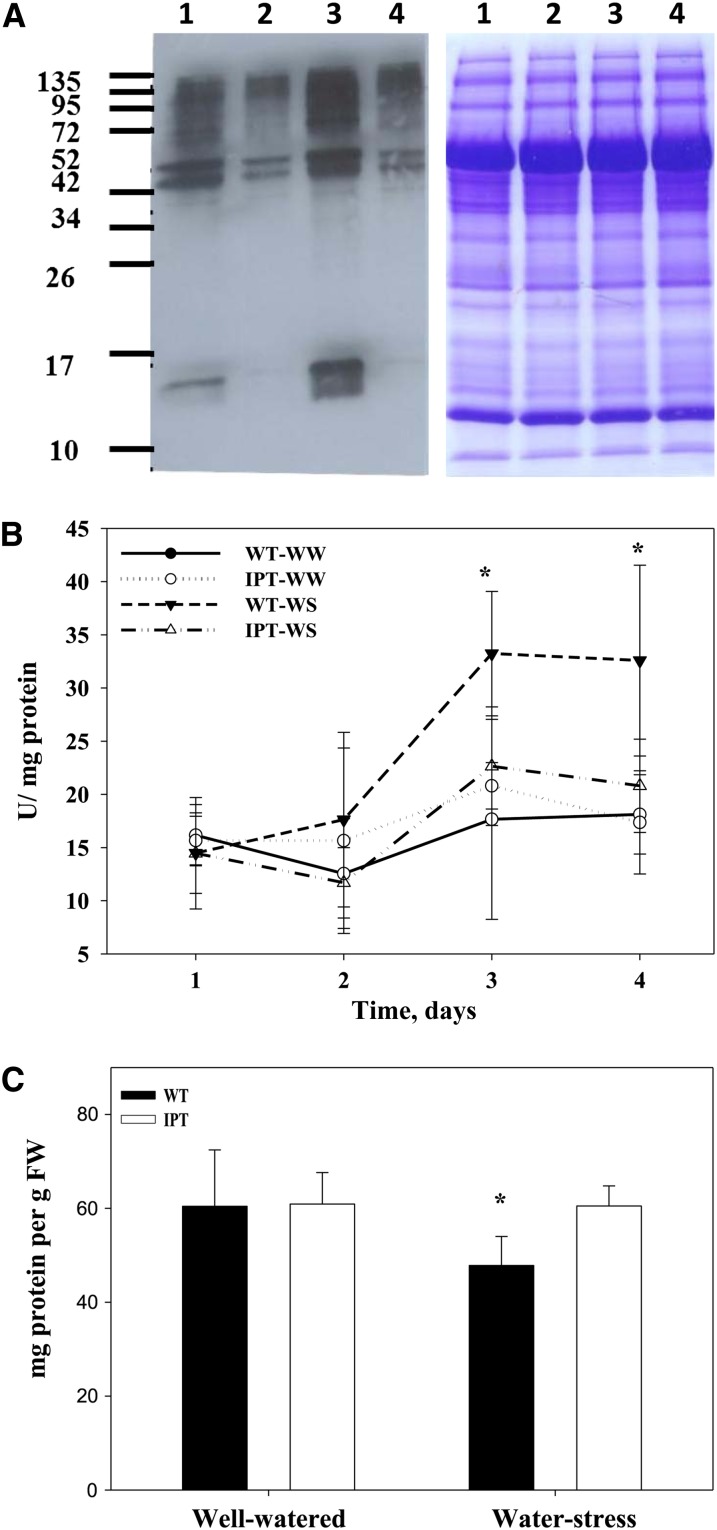
Protein stability and oxidation of wild-type (WT) and transgenic PSARK::IPT rice plants. A, Immunodetection of derivatized protein carbonyl groups using anti-DNP-hydrazone antibody to evaluate oxidative damage of proteins (blot on the left) at well-watered (WW) and water-stress (WS) conditions: wild-type well watered (lane 1), wild-type stressed (lane 2), PSARK::IPT well watered (lane 3), and PSARK::IPT stressed (lane 4). A loading control using SDS-PAGE gels followed by staining with Coomassie Brilliant Blue R-250 is shown at right. B, Protease trypsin-like activity in wild-type well-watered (black circles), wild-type stressed (black triangles), PSARK::IPT well-watered (white circles), and PSARK::IPT stressed (white triangles) rice. C, Protein content of wild-type (black columns) and transgenic PSARK::IPT (white columns) plants grown under well-watered and water-stress conditions. FW, Fresh weight. Values are means ± sd (n = 8). Asterisks indicate significant differences between genotypes (*P ≤ 0.05). [See online article for color version of this figure.]
As a consequence of protein oxidation, carbonyl groups are added into the side chains of proteins. Protein oxidation is a well-accepted mechanism to target proteins for degradation (Xiong et al., 2007). Wild-type stressed plants displayed an increase in protein oxidation (Fig. 6A) determined by the immunodetection of derivatized carbonyl groups to 2,4-dinitrophenylhydrazone (DNP-hydrazone) in oxidized proteins. Consistent with the increase in protein oxidation seen in wild-type plants during stress, the levels of protease activities showed an increment (Fig. 6B) with a concomitant decrease in flag leaf protein contents (Fig. 6C).
DISCUSSION
We have shown previously that the synthesis of CK, mediated by the expression of IPT under the control of a maturation- and stress-induced promoter (PSARK), led to enhanced drought tolerance of transgenic PSARK::IPT rice (Peleg et al., 2011) and tobacco (Rivero et al., 2007) plants subjected to severe and long-term stress, respectively. Here, we studied the responses of C and N metabolism to drought stress, aiming to characterize physiological responses and metabolic pathways associated with the CK-induced stress tolerance in PSARK::IPT rice plants. Wild-type rice plants under water stress displayed a reduction in stomatal conductance and an inhibition of photosynthetic activity and C assimilation. Lauer and Boyer (1992) demonstrated that, during drought stress, the limitation in C assimilation was due to the reduction of the biochemical capacity of photosynthesis. The authors showed that although a simultaneous reduction of stomatal opening occurred during water deficit, calculated substomatal CO2 concentration (Ci) was maintained or even increased. In the initial stages of water stress, a decrease in stomatal conductance mediated by ABA can limit the internal CO2 concentration used in C assimilation (Zhang and Davies, 1990; Tardieu and Davies, 1993). As the stress progresses, the decrease in photosynthetic activity and the continuation of respiration both contribute to the maintenance (or increase) of internal CO2 in plants subjected to stress (Lawlor and Tezara, 2009). Our results support this concept. The inhibition of photosynthetic activity in wild-type rice plants during stress was accompanied by a decline in stomatal conductance without altering Ci values until the external CO2 concentration reached 1,200 µL L−1 (Fig. 1G), indicating that the impairment of C assimilation was predominantly associated with the biochemical inhibition of photosynthesis. In contrast, and in spite of a reduction in Ci (associated with a reduction in stomatal conductance during water stress), transgenic PSARK::IPT rice maintained photosynthesis at levels similar to those of well-watered plants. Similar results were obtained previously with transgenic tobacco plants expressing PSARK::IPT, where the stress-induced increase in CK resulted in the protection of biochemical processes associated with photosynthesis during water stress (Rivero et al., 2009).
A reciprocal control between C and N assimilation coordinating the production of sugars and amino acids, according to the plant requirements, has been postulated (Kaiser and Brendle-Behnisch, 1991; Foyer et al., 1998, 2004). During water stress, the wild-type plants displayed an increase in vacuolar invertase, cytosolic invertase, and SuSy transcripts and increased invertases and SuSy activities that resulted in higher Glc and Fru contents. The increase in Glc and Fru was not correlated with increases in glycolytic metabolites, indicating that the sugar accumulation together with the increase in free amino acid contents (see below) constitute part of the osmotic adjustment response of wild-type plants to dehydration brought about by water stress (Kameli and Lösel, 1993; Wang et al., 1995; Kim et al., 2000; Merewitz et al., 2011; Nio et al., 2011). On the other hand, the maintenance of photosynthesis in the transgenic PSARK::IPT plants during water stress was well correlated with increased SPS transcription, increased SPS activity, and reduced SuSy and cytosolic invertase activities compared with wild-type plants, leading to increased Suc contents in the flag leaves. This sustained C assimilation in transgenic PSARK::IPT plants during water stress, which was paralleled by an increase in N assimilation, supports the notion of a coordinated regulation of C and N metabolism (Foyer et al., 2004).
Transgenic PSARK::IPT plants displayed higher HK activity and elevated Glc-6-P contents. It has been shown that HK regulates photosynthesis, growth, and senescence. Increased levels of HK expression reduced growth and photosynthesis and induced senescence in tomato (Solanum lycopersicum) and rice plants (Dai et al., 1999; Cho et al., 2009). The complex regulation by HK lies in the existence of interacting factors (hormones, light, or sugar availability) controlling signaling pathways in plants. Interestingly, HK roles in sugar sensing and signaling seem to be independent of the enzyme activity, as demonstrated using catalytically inactive OsHXK alleles of OsHXK5 and OsHXK6 in rice plants (Cho et al., 2009). This notion was further supported by the higher HK activity found in PSARK::IPT plants that correlated well with an increase in Glc-6-P without detrimental effects on photosynthesis, growth, or senescence. On the other hand, higher Glc-6-P contents would enhance the ribulose bisphosphate regeneration capacity during water-stress conditions and maintain C assimilation. Also, high Glc-6-P would sustain enhanced Suc synthesis mediated by SPS activity. Interestingly, a gene encoding the cytosolic form of the enzyme Glc-6-P dehydrogenase (G6PDH; LOC_Os2g38840) was highly up-regulated in the transgenic plants during stress. Since there is a good correlation between G6PDH mRNA levels, protein abundance, and enzyme activities (Hauschild and von Schaewen, 2003), we can assume that levels of G6PDH were also elevated in PSARK::IPT plants. G6PDH supplies the NADPH required for assimilatory processes via the pentose phosphate pathway (Kruger and von Schaewen, 2003). NADPH participates in a number of key reactions, such as ATP production (Liu et al., 2008) and the regeneration of reduced glutathione (Noctor et al., 1998). NADPH is also a limiting factor in NR-mediated N assimilation (Kaiser et al., 2000; Dutilleul et al., 2005). NR showed a dramatic increase under stress conditions in the transgenic plants, indicating that CK might mediate the production of NADPH in the PSARK::IPT plants in response to water stress.
Plants possess complex regulatory machinery that coordinates N assimilation with C metabolism (Nunes-Nesi et al., 2010). This metabolic coordination varies during the day and is controlled by changes in light and/or sink demand (Ainsworth and Bush, 2011). This coordination involves sensing mechanisms for the availability of sugars (Rolland et al., 2002), nitrate and amino acids (Wang et al., 2004), C-N balance (Coruzzi and Zhou, 2001), as well as hormones. CK can affect C and N partitioning (Sakakibara et al., 2006), although the specific mechanisms underlying this regulation remain unknown. One of the most relevant routes proposed for the interaction between CKs and N is the role of CK in the regulation of N uptake and signaling, controlling the N status of the plant (Sakakibara et al., 2006; Kiba et al., 2011). C-N interactions are based on the ability of N to regulate CK biosynthesis in response to N availability (Sakakibara et al., 2006). In PSARK::IPT plants, we found an increased N assimilation capacity that was mediated by the increase in endogenous CK in a C-N-dependent manner.
It has been proposed that CKs play a negative role in the regulation of N uptake-related genes (Kiba et al., 2011). The exogenous application of CK to Arabidopsis (Arabidopsis thaliana) seedlings resulted in the reduced expression of AtNRT genes (encoding nitrate transporters) as well as some ammonium and amino acid transporters (Kiba et al., 2011). A similar CK-induced down-regulation of genes associated with nutrient acquisition was reported in 2-week-old rice seedlings grown hydroponically and treated with exogenously applied CK (Hirose et al., 2007). Since the CK-induced repression of AtNRT genes was observed under both low and high N, it was suggested that the CK effects were independent of the N status of the plant (Kiba et al., 2011). The notion that CKs negatively regulate the expression of genes encoding nitrate transporters should be considered with caution, because in the reports cited above, only some of the NRT genes were down-regulated while others were up-regulated, and most NRTs expressed in the shoots were up-regulated (Kiba et al., 2011). The seedlings used in these studies were very young (3-d-old Arabidopsis seedlings and 14-d-old rice seedlings) and grown under nonphysiological conditions (Arabidopsis seedlings were grown on agar plates containing a growth medium supplemented with Suc; rice seedlings were grown hydroponically). In addition, CK was applied exogenously, and it was probably catabolized via CK oxidase (Paces et al., 1971). Our results showed that, during water stress, the transgenic PSARK::IPT plants displayed enhanced flag leaf nitrate contents, higher NR activity, and sustained ammonium contents. These results were correlated with the higher expression of OsNRT2.3 (LOC_Os01g50820) in the PSARK:IPT transgenic plants, which has been shown previously in rice to be implicated in the long-distance transport of NO3− to shoots (Tang et al., 2012) and the rice ortholog of the Arabidopsis NRT1.2 (LOC_Os06g38294.1), described as a NO3− and ABA transporter able to regulate stomatal opening in Arabidopsis plants (Kanno et al., 2012). The expression of other rice nitrate transporters (LOC_Os08g05910 [ortholog of AtNRT1.1], LOC_Os01g68510 [ortholog of AtNRT1.7], LOC_Os02g47090, LOC_Os02g37040, LOC_Os04g50940, and LOC_Os02g02190) either did not show significant differences between genotypes under both conditions tested or their expression was not detectable in the leaf tissue (i.e. LOC_Os04g50940; data not shown). Our results indicate that the stress-induced CK synthesis, driven by the stress-induced SARK promoter, played a role in maintaining nitrate acquisition in the transgenic plants.
While N assimilation in PSARK::IPT plants was not affected by water stress, wild-type plants displayed a marked inhibition of NR and NiR activities. The inhibition of NR and NiR activities by water stress has been reported previously, and the decrease in activities was dependent on the severity of the stress (Ramanjulu and Sudhakar, 1997). In spite of the inhibition of N assimilation in the wild-type plants, the ammonium content in wild-type and PSARK::IPT plants was similar. Wild-type plants compensated for the inhibition of NiR by increasing the activity of GDH that mediates the deamination of Glu. Increased GDH deamination activity has been associated with catabolic processes during starvation (Krapp et al., 2011) and during the response of rice plants to salt stress (Nguyen et al., 2005). The increase in GDH deamination activity seen in wild-type plants during water stress would increase the amounts of 2-oxoglutarate, contributing to the maintenance of the TCA cycle and providing ammonia needed for amino acid synthesis (Miyashita and Good, 2008).
Sink capacity is the ability of tissues to import and use photosynthates for later growth and biomass production (Ainsworth et al., 2004). The ability of CKs to increase sink capacity has been associated with their role in growth, through the control of cell division (Kuiper, 1993; Werner et al., 2008), photosynthesis (Paul and Foyer, 2001), and N metabolism (Sakakibara et al., 2006). Previously, we showed that the induction of CK synthesis in transgenic PSARK::IPT rice influenced changes in source-sink interactions that resulted in the improvement of yield and grain quality (Peleg et al., 2011). Here, we show that the ability of PSARK::IPT plants to yield during water stress is associated with the maintenance of photosynthetic activity and N assimilation in the flag leaves, supporting the role of CK in improving sink capacity (Bartrina et al., 2011). In rice, flag leaves are the main source for the growing panicles (Mae, 1997), and C and N remobilization from the flag leaves are the main contributors to grain yield (Chen and Wang, 2008). The CK-dependent increase in SPS, NR, and NiR activities in the PSARK::IPT plants during water stress could have contributed to the regulation of C and N supply to the growing sinks. The improvement of the flag leaf sink capacity ensured grain development during post anthesis stages, resulting in increased grain yields in the transgenic plants subjected to water stress during the pre anthesis stage (Peleg et al., 2011).
Under water stress, wild-type plants displayed enhanced protease activities, decreased protein contents, down-regulation of protein synthesis-related genes, and increased free amino acids. The inhibition of protein synthesis in plants growing under water stress is well documented (Hsiao, 1973; Dhindsa and Cleland, 1975), and the stress-induced decline in protein contents is accompanied by the accumulation of free amino acids in plant tissues (Good and Zaplachinski, 1994; Ramanjulu and Sudhakar, 1997). Protein degradation is an important mechanism for N remobilization, and chloroplasts are the major source of cellular proteins (Hörtensteiner and Feller, 2002). During stress, nutrient mobilization driven by the degradation of chloroplast proteins influences the abundance and activity of enzymes that control N and C metabolism (Ishida et al., 2008). Protein degradation in the chloroplast results in an increased amino acid pool (i.e. Glu that serves as GDH substrate) and in the loss of function of plastid enzymes such as GS2 (Kamachi et al., 1992). Thus, under stress, the cytosolic form of GS (GS1) plays and important role in Gln synthesis (Masclaux-Daubresse et al., 2006; Martinelli et al., 2007). These responses were seen in wild-type rice with a reduced expression of GS2 and Fd-GOGAT, both encoding chloroplast enzymes and the up-regulation of GS1 expression, together with the loss of chloroplast function (and the concomitant inhibition of photosynthesis) and the inhibition of primary N assimilation. In the transgenic PSARK::IPT plants, the stress-induced CK synthesis maintained the availability of C and N needed for protein synthesis.
In conclusion, stress-induced CK synthesis in the PSARK::IPT transgenic plants enhanced the sink capacity of the flag leaf through a CK-dependent coordinated regulation of C and N metabolism that maintained photosynthetic activity and nitrate assimilation during the water deficit episode.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of wild-type rice (Oryza sativa japonica ‘Kitaake’) and transgenic plants expressing PSARK::IPT were germinated on moist germination paper for 4 d at 28°C in the dark. Seedlings were then transplanted into 8-L pots filled with a mix of 80% sand and 20% peat. Two plants were planted per pot and placed in water trays. Greenhouse conditions were kept at 12-h/12-h day/night under an illumination of 1,200 μmol photons m−2 s−1 at 30°C/20°C. Plants were fertilized with a solution 50% N:phosphorus:potasium (20:10:20) and 50% ammonium sulfate (total of 0.5 g of N) every 10 d until panicle initiation. Water stress treatments were applied at pre anthesis (end of booting stage toward panicle emerging) by withholding water. Plants were rewatered when visual stress symptoms (i.e. leaf rolling) appeared in the transgenic plants.
Physiological Analysis
For photosynthesis and chlorophyll fluorescence measurements, rates of CO2 assimilation were determined in flag leaves of wild-type and PSARK::IPT plants after 3 d of stress using a LI-COR 6400-40 with an integrated fluorescence chamber head (Li-COR). The leaf cuvette was set at photosynthetic photon flux density of 1,200 µmol m−2 s−1, relative humidity of 70%, and 30°C. For measurements of net CO2 assimilation rate (A) versus Ci (A/Ci curves), the CO2 concentration in the leaf cuvette was set at 11 levels (50, 100, 200, 300, 370, 400, 600, 800, 1,000, 1,200, and 1,500 µmol m−2 s−1). A/Ci curves were fitted in SigmaPlot version 11 (Systat Software) using a nonlinear exponential rise to maximum model based on the equations calculated by Farquhar et al. (1980) and subsequently modified by Sharkey (1985) Jmax, Vcmax, and TPU were calculated using the Photosyn Assistant software (Dundee Scientific) based on the model developed by Farquhar et al. (1980). Chlorophyll fluorescence measurements of Fv/Fm (Butler, 1978) were used to assess the maximum quantum efficiency of PSII in dark-adapted plants for 20 min, following manufacturer settings (Li-COR). Stomatal conductance was measured using a LI-COR 6400-40. Measurements of flag leaf RWC were taken from wild-type and PSARK::IPT plants grown under well-watered and water-stress conditions. Flag leaves were sampled before dawn and placed in a preweighed air-tight vial. Vials were weighted to obtain leaf fresh weight (FW), after which the samples were immediately hydrated to full turgidity (6 h). The leaf samples were weighted again to obtain turgid weight (TW). Samples were then oven dried at 80°C for 24 h, and dry weight (DW) was determined. RWC was calculated according to the equation:
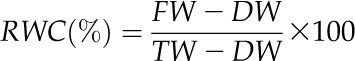 |
Soluble Sugar Quantification
Leaves were collected in the early morning (7–8 am) and immediately frozen in liquid N. The frozen samples were freeze dried, and 0.1 g was used for the soluble sugar extraction as described previously (Peleg et al., 2011). The samples were dissolved in distilled, deionized water, filtered (0.45 µm), and subsequently analyzed by high-performance anion-exchange chromatography on a HPLC apparatus using a Carbohydrate Analysis column (Aminex HPX-87P 300X7.8 mm). Starch quantification was determined as described by Smith and Zeeman (2006). The release of Glc was determined at 340 nm using a Glc assay (GAHK20; Sigma-Aldrich) with a spectrophotometer (DU-640; Beckman Coulter). Starch content was calculated as 90% of Glc content.
NO3− and NH4+ Contents
Leaf freeze-dried tissue (0.01 g) already ground was used for NH4+ and NO3− quantification. NH4+ was determined according to Forster (1995). Absorbance was measured at 660 nm in a spectrophotometer (DU-640; Beckman Coulter). NO3− was determined according to Doane and Horwath (2003) by reducing nitrate to nitrite using vanadium (III) chloride followed by the addition of sulfanilamide and N-(1-naphthyl)-ethylenediaminedihydrochloride. Subsequent colorimetric nitrite analysis was done at 540 nm in a spectrophotometer (DU-640; Beckman Coulter). Nitrite analysis was done without the use of vanadium (III) chloride and then was subtracted from the nitrite content determined for NO3− analysis. Standard curves were obtained using KNO3 as a standard.
Amino Acid Quantification
Leaf frozen tissue was ground and freeze dried. Free amino acids were extracted as described previously by Hacham et al. (2002). Tissue (0.1 g) was ground in the presence of 400 µL of water:chloroform:methanol (3:5:12, v/v) and centrifuged for 2 min at 12,000g. This step was repeated twice, the supernatants were combined, and a mixture of 200 µL of chloroform and 300 µL of water was added. After 2 min of centrifugation at 14,000g, the supernatant was collected and dried. Determination of free amino acids was carried out on a Hitachi L-8900 amino acid analyzer.
Metabolite Profiling
Metabolites were analyzed in flag leaves of wild-type and PSARK::IPT plants grown under well-watered and water-stress conditions. Samples (n = 6) were submitted to Metabolon. The sample preparation and analysis process was carried out as described previously (Lawton et al., 2008; Evans et al., 2009; Oliver et al., 2011). Compounds were identified by comparison with library entries of purified standards or recurrent unknown entities using Metabolon’s proprietary software. The identification of known chemical entities was based on comparison with metabolomics library entries of purified standards (DeHaven et al., 2010).
Enzyme Assays
Enzyme activities were determined in flag leaves of wild-type and transgenic PSARK::IPT plants subjected to 3 d of water stress and collected in the early morning (7–8 am). Flag leaf homogenates were obtained from leaf tissue collected 2 to 10 cm from the apical meristem. Enzyme activities are expressed as moles of metabolite generated/consumed per milligram of protein per unit of time.
SPS activity measurements were based on Klann et al. (1993). Sugars were quantified using Anthrone (Van Handel, 1968), and absorbance was measured using a microplate reader (Synergy MxMonochromator-Based Multi-Mode Microplate Reader; BIOTek) at 620 nm.
SuSy activity was determined in the direction of Suc synthesis as well as Suc degradation. Suc synthesis mediated by Suc synthase was determined according to Klann et al. (1993). Suc cleavage was assayed as described by Zrenner et al. (1995). Absorbance was measured using a microplate reader (Synergy MxMonochromator-Based Multi-Mode Microplate Reader; BIOTek) at 540 nm using Fru as a standard.
Cell wall invertase, cytosolic invertase, and vacuolar invertase were assayed according to Roitsch et al. (1995). Controls were boiled before incubation with the reaction buffer. Reaction was stopped using 100 µL of 1 m NaOH, and reducing sugars were determined after reaction time (Miller, 1959).
HK, trehalase, total amylolytic, and α-amylase activities were measured according to Wiese et al. (1999), Müller et al. (1995), and Lee et al. (2009), respectively.
For NR and NiR, leaf frozen tissue (0.1 g) was ground in the presence of 1 mL of buffer containing 50 mm KH2PO4-KOH buffer, pH 7.5, 2 mm EDTA, 2 mm dithiothreitol, and 1% polyvinylpolypyrrolidone. Extracts were centrifuged at 20,000g for 20 min at 4°C. NR activity was measured according to Kaiser and Lewis (1984) with some modifications. The reaction was initiated by the addition of 700 µL of reaction buffer (50 mm KH2PO4-KOH buffer, pH 7.5, 10 mm KNO3, and 0.1 mm NADH) to 100 µL of total soluble proteins. Samples were incubated at 28°C for 15 min, and controls were boiled before incubation with the reaction buffer. The reactions were stopped, and NO2− was determined following the method established by Hageman (1971). NiR activity measurements were performed according to Lillo (1984).
NADH-GOGAT and GS were determined according to Matoh and Takahashi (1982) and O’Neal and Joy (1973), respectively.
Both amination and deamination activities of GDH were assessed according to protocols described previously by Loyola-Vargas and de Jimenez (1984). Asp and aminotransferase activities were assayed according to Lillo (1984).
Protein Quantification
The Bradford assay (Bradford, 1976) was used for protein quantification using bovine serum albumin as a standard.
Oxidative Modification of Proteins
Protein oxidation was evaluated using the OxyBlot Kit (Millipore). Immunodetection of derivatized carbonyl groups to DNP-hydrazone was performed following kit protocol.
Proteinase Activity Assay
Trypsin-like activity was determined using the QuantiCleave Protease Assay Kit (Pierce Chemical). Leaf homogenates were prepared using 50 mm HEPES-KOH, pH 7.5, 2 mm EDTA, 5 mm MgCl2, 0.05% Triton X-100, 5 mm β-mercaptoethanol, and 1% polyvinylpolypyrrolidone (w/v). Fifty microliters of the extract was used for the assays following the manufacturer’s instructions. Absorbance was measured at 450 nm. Blanks were prepared without the addition of substrate (succinyl casein solution).
Quantitative PCR Analysis
RNA was extracted from flag leaves of wild-type and transgenic PSARK::IPT rice plants under well-watered and water-stress conditions. First-strand complementary DNA synthesis, primer design, and quantitative PCR were performed as described before (Peleg et al., 2011). The different sets of primers used for the amplification of the different target genes are listed in Supplemental Table S1. Analysis of the relative gene expression was performed according to the Comparative Cycle Threshold Method as described by Livak and Schmittge (2001).
Processing Microarray Data
Normalization and statistical analyses were performed using the JMP Genomics (version 5.1) statistical package (SAS Institute) as described before (Peleg et al., 2011). The CELL data files of the 12 Affymetrix GeneChip results of this study were deposited in the public microarray database National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) with accession number GSE 23211.
Statistical Analysis
The JMP (version 8.0) statistical package (SAS Institute) was used for statistical analyses. ANOVA was employed to test the effect of genotype on trait. The LSMeans t test (Least Square Means) was used to compare means by genotype/treatment at a probability level of 5%. Levels of significance are represented by asterisks as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; n.s. indicates not significant. The experiments were based on a randomized complete block design using six replicates.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Heat map of the list of genes involved in C and N metabolism
Supplemental Figure S2. Heat map of the list of genes encoding enzymes involved in protein synthesis.
Supplemental Table S1. List of Primers Used in qPCR (Real Time PCR).
Supplementary Material
Glossary
- CK
cytokinin
- C
carbon
- N
nitrogen
- ABA
abscisic acid
- RWC
relative water content
- Vcmax
maximum rate of carboxylation
- Jmax
maximum rate of electron transport
- TPU
triose phosphate utilization
- Fv/Fm
ratio of variable to maximal chlorophyll fluorescence
- SPS
sucrose phosphate synthase
- SuSy
sucrose synthase
- GDH
glutamate dehydrogenase
- NR
nitrate reductase
- NiR
nitrite reductase
- GOGAT
glutamate synthase
- GS
glutamine synthetase
- TCA
tricarboxylic acid
- DNP-hydrazone
2,4-dinitrophenylhydrazone
- Ci
calculated substomatal CO2 concentration
- G6PDH
Glc-6-P dehydrogenase
- A
assimilation rate
- HK
hexokinase
References
- Ainsworth EA, Bush DR. (2011) Carbohydrate export from the leaf: a highly regulated process and target to enhance photosynthesis and productivity. Plant Physiol 155: 64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth EA, Rogers A, Nelson R, Long SP. (2004) Testing the “source-sink” hypothesis of down-regulation of photosynthesis in elevated [CO2] in the field with single gene substitutions in Glycine max. Agric Meteorol 122: 85–94 [Google Scholar]
- Bartrina I, Otto E, Strnad M, Werner T, Schmülling T. (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Butler WL. (1978) Energy distribution in the photochemical apparatus of photosynthesis. Annu Rev Plant Physiol 29: 345–378 [Google Scholar]
- Chen H, Jiang JG. (2010) Osmotic adjustment and plant adaptation to environmental changes related to drought and salinity. Environ Rev 18: 309–319 [Google Scholar]
- Chen HJ, Wang SJ. (2008) Molecular regulation of sink-source transition in rice leaf sheaths during the heading period. Acta Physiol Plant 30: 639–649 [Google Scholar]
- Cho JI, Ryoo N, Eom JS, Lee DW, Kim HB, Jeong SW, Lee YH, Kwon YK, Cho MH, Bhoo SH, et al. (2009) Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol 149: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi GM, Zhou L. (2001) Carbon and nitrogen sensing and signaling in plants: emerging ‘matrix effects.’ Curr Opin Plant Biol 4: 247–253 [DOI] [PubMed] [Google Scholar]
- Dai N, Schaffer A, Petreikov M, Shahak Y, Giller Y, Ratner K, Levine A, Granot D. (1999) Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell 11: 1253–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WJ, Zhang J. (1991) Root signals and the regulation of growth and development of plants in drying soil. Annu Rev Plant Physiol Plant Mol Biol 42: 55–76 [Google Scholar]
- DeHaven C, Evans A, Dai H, Lawton K. (2010) Organization of GC/MS and LC/MS metabolomics data into chemical libraries. Cheminformatics 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa RS, Cleland RE. (1975) Water stress and protein synthesis. I. Differential inhibition of protein synthesis. Plant Physiol 55: 778–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doane TA, Horwath WR. (2003) Spectrophotometric determination of nitrate with a single reagent. Anal Lett 36: 2713–2722 [Google Scholar]
- Dutilleul C, Lelarge C, Prioul JL, De Paepe R, Foyer CH, Noctor G. (2005) Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol 139: 64–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. (2009) Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 81: 6656–6667 [DOI] [PubMed] [Google Scholar]
- Farquhar GD, Caemmerer S, Berry JA. (1980) A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149: 78–90 [DOI] [PubMed] [Google Scholar]
- Forster JC (1995) Soil sampling, handling, storage and analysis. In A Kassem, N Paolo, eds, Methods in Applied Soil Microbiology and Biochemistry. Academic Press, London, pp 49–121 [Google Scholar]
- Foyer C, Ferrario-Méry S, Huber S (2004) Regulation of carbon fluxes in the cytosol: coordination of sucrose synthesis, nitrate reduction and organic acid and amino acid biosynthesis. In R Leegood, T Sharkey, S Caemmerer, eds, Photosynthesis: Physiology and Metabolism, Vol 9. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 177–203 [Google Scholar]
- Foyer CH, Valadier MH, Migge A, Becker TW. (1998) Drought-induced effects on nitrate reductase activity and mRNA and on the coordination of nitrogen and carbon metabolism in maize leaves. Plant Physiol 117: 283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AG, Zaplachinski ST. (1994) The effects of drought stress on free amino acid accumulation and protein synthesis in Brassica napus. Physiol Plant 90: 9–14 [Google Scholar]
- Guivarc’h A, Rembur J, Goetz M, Roitsch T, Noin M, Schmülling T, Chriqui D. (2002) Local expression of the ipt gene in transgenic tobacco (Nicotiana tabacum L. cv. SR1) axillary buds establishes a role for cytokinins in tuberization and sink formation. J Exp Bot 53: 621–629 [DOI] [PubMed] [Google Scholar]
- Hacham Y, Avraham T, Amir R. (2002) The N-terminal region of Arabidopsis cystathionine γ-synthase plays an important regulatory role in methionine metabolism. Plant Physiol 128: 454–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman RH. (1971) Nitrate reductase. Methods Enzymol 23: 497–503 [Google Scholar]
- Hauschild R, von Schaewen A. (2003) Differential regulation of glucose-6-phosphate dehydrogenase isoenzyme activities in potato. Plant Physiol 133: 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H. (2007) Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol 48: 523–539 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Feller U. (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53: 927–937 [DOI] [PubMed] [Google Scholar]
- Hsiao TC. (1973) Plant responses to water stress. Annu Rev Plant Physiol Plant Mol Biol 24: 519–570 [Google Scholar]
- Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T. (2008) Mobilization of Rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J, Lewis O. (1984) Nitrate reductase and glutamine synthetase activity in leaves and roots of nitrate-fed Helianthus annuus L. Plant Soil 77: 127–130 [Google Scholar]
- Kaiser WM, Brendle-Behnisch E. (1991) Rapid modulation of spinach leaf nitrate reductase activity by photosynthesis. I. Modulation in vivo by CO2 availability. Plant Physiol 96: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WM, Kandlbinder A, Stoimenova M, Glaab J. (2000) Discrepancy between nitrate reduction rates in intact leaves and nitrate reductase activity in leaf extracts: what limits nitrate reduction in situ? Planta 210: 801–807 [DOI] [PubMed] [Google Scholar]
- Kamachi K, Yamaya T, Hayakawa T, Mae T, Ojima K. (1992) Vascular bundle-specific localization of cytosolic glutamine synthetase in rice leaves. Plant Physiol 99: 1481–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameli A, Lösel DM. (1993) Carbohydrates and water status in wheat plants under water stress. New Phytol 125: 609–614 [DOI] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M. (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA 109: 9653–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Kudo T, Kojima M, Sakakibara H. (2011) Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J Exp Bot 62: 1399–1409 [DOI] [PubMed] [Google Scholar]
- Kim JY, Mahé A, Brangeon J, Prioul JL. (2000) A maize vacuolar invertase, IVR2, is induced by water stress: organ/tissue specificity and diurnal modulation of expression. Plant Physiol 124: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann EM, Chetelat RT, Bennett AB. (1993) Expression of acid invertase gene controls sugar composition in tomato (Lycopersicon) fruit. Plant Physiol 103: 863–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Berthomé R, Orsel M, Mercey-Boutet S, Yu A, Castaings L, Elftieh S, Major H, Renou JP, Daniel-Vedele F. (2011) Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol 157: 1255–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause GH, Somersalo S, Osmond CB, Briantais JM, Schreiber U. (1989) Fluorescence as a tool in photosynthesis research: application in studies of photoinhibition, cold acclimation and freezing stress and discussion. Philos Trans R Soc Lond B Biol Sci 323: 281–293 [Google Scholar]
- Kruger NJ, von Schaewen A. (2003) The oxidative pentose phosphate pathway: structure and organisation. Curr Opin Plant Biol 6: 236–246 [DOI] [PubMed] [Google Scholar]
- Kuiper D. (1993) Sink strength: established and regulated by plant growth regulators. Plant Cell Environ 16: 1025–1026 [Google Scholar]
- Kuppu S, Mishra N, Hu R, Sun L, Zhu X, Shen G, Blumwald E, Payton P, Zhang H. (2013) Water-deficit inducible expression of a cytokinin biosynthetic gene IPT improves drought tolerance in cotton. PLoS ONE 8: e64190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer MJ, Boyer JS. (1992) Internal CO2 measured directly in leaves: abscisic acid and low leaf water potential cause opposing effects. Plant Physiol 98: 1310–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DW, Tezara W. (2009) Causes of decreased photosynthetic rate and metabolic capacity in water-deficient leaf cells: a critical evaluation of mechanisms and integration of processes. Ann Bot (Lond) 103: 561–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton KA, Berger A, Mitchell M, Milgram KE, Evans AM, Guo L, Hanson RW, Kalhan SC, Ryals JA, Milburn MV. (2008) Analysis of the adult human plasma metabolome. Pharmacogenomics 9: 383–397 [DOI] [PubMed] [Google Scholar]
- Lee KW, Chen PW, Lu CA, Chen S, Ho TH, Yu SM. (2009) Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci Signal 2: ra61. [DOI] [PubMed] [Google Scholar]
- Lillo C. (1984) Diurnal variations of nitrite reductase, glutamine synthetase, glutamate synthase, alanine aminotransferase and aspartate aminotransferase in barley leaves. Physiol Plant 61: 214–218 [Google Scholar]
- Liu YJ, Norberg FE, Szilágyi A, De Paepe R, Åkerlund HE, Rasmusson AG. (2008) The mitochondrial external NADPH dehydrogenase modulates the leaf NADPH/NADP+ ratio in transgenic Nicotiana sylvestris. Plant Cell Physiol 49: 251–263 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–∆∆CT Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Loyola-Vargas VM, de Jimenez ES. (1984) Differential role of glutamate dehydrogenase in nitrogen metabolism of maize tissues. Plant Physiol 76: 536–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mae T. (1997) Physiological nitrogen efficiency in rice: nitrogen utilization, photosynthesis, and yield potential. Plant Soil 196: 201–210 [Google Scholar]
- Martinelli T, Whittaker A, Bochicchio A, Vazzana C, Suzuki A, Masclaux-Daubresse C. (2007) Amino acid pattern and glutamate metabolism during dehydration stress in the ‘resurrection’ plant Sporobolus stapfianus: a comparison between desiccation-sensitive and desiccation-tolerant leaves. J Exp Bot 58: 3037–3046 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot (Lond) 105: 1141–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, Lelandais M, Grandjean O, Kronenberger J, Valadier MH, Feraud M, Jouglet T, Suzuki A. (2006) Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol 140: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoh T, Takahashi E. (1982) Changes in the activities of ferredoxin- and NADH-glutamate synthase during seedling development of peas. Planta 154: 289–294 [DOI] [PubMed] [Google Scholar]
- Merewitz EB, Gianfagna T, Huang B. (2011) Protein accumulation in leaves and roots associated with improved drought tolerance in creeping bentgrass expressing an ipt gene for cytokinin synthesis. J Exp Bot 62: 5311–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GL. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31: 426–428 [Google Scholar]
- Miyashita Y, Good AG. (2008) Glutamate deamination by glutamate dehydrogenase plays a central role in amino acid catabolism in plants. Plant Signal Behav 3: 842–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, Deleu C, Larher F, Bouchereau A. (2006) The lysine-ketoglutarate reductase-saccharopine dehydrogenase is involved in the osmo-induced synthesis of pipecolic acid in rapeseed leaf tissues. Plant Physiol Biochem 44: 474–482 [DOI] [PubMed] [Google Scholar]
- Müller J, Boller T, Wiemken A. (1995) Effects of validamycin A, a potent trehalase inhibitor, and phytohormones on trehalose metabolism in roots and root nodules of soybean and cowpea. Planta 197: 362–368 [Google Scholar]
- Nguyen HT, Shim IS, Kobayashi K, Usui K. (2005) Regulation of ammonium accumulation during salt stress in rice (Oryza sativa L.) seedlings. Plant Prod Sci 8: 397–404 [Google Scholar]
- Nio SA, Cawthray GR, Wade LJ, Colmer TD. (2011) Pattern of solutes accumulated during leaf osmotic adjustment as related to duration of water deficit for wheat at the reproductive stage. Plant Physiol Biochem 49: 1126–1137 [DOI] [PubMed] [Google Scholar]
- Noctor G, Arisi AC, Jouanin L, Kunert KJ, Rennenberg H, Foyer CH. (1998) Glutathione: biosynthesis, metabolism and relationship to stress tolerance explored in transformed plants. J Exp Bot 49: 623–647 [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Oliver MJ, Guo L, Alexander DC, Ryals JA, Wone BW, Cushman JC. (2011) A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus. Plant Cell 23: 1231–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neal D, Joy KW. (1973) Glutamine synthetase of pea leaves. I. Purification, stabilization, and pH optima. Arch Biochem Biophys 159: 113–122 [DOI] [PubMed] [Google Scholar]
- Paces V, Werstiuk E, Hall RH. (1971) Conversion of N-(Δ-isopentenyl)adenosine to adenosine by enzyme activity in tobacco tissue. Plant Physiol 48: 775–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MJ, Foyer CH. (2001) Sink regulation of photosynthesis. J Exp Bot 52: 1383–1400 [DOI] [PubMed] [Google Scholar]
- Peleg Z, Blumwald E. (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 14: 290–295 [DOI] [PubMed] [Google Scholar]
- Peleg Z, Reguera M, Tumimbang E, Walia H, Blumwald E. (2011) Cytokinin-mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water-stress. Plant Biotechnol J 9: 747–758 [DOI] [PubMed] [Google Scholar]
- Qin H, Gu Q, Zhang J, Sun L, Kuppu S, Zhang Y, Burow M, Payton P, Blumwald E, Zhang H. (2011) Regulated expression of an isopentenyltransferase gene (IPT) in peanut significantly improves drought tolerance and increases yield under field conditions. Plant Cell Physiol 52: 1904–1914 [DOI] [PubMed] [Google Scholar]
- Ramanjulu S, Sudhakar C. (1997) Drought tolerance is partly related to amino acid accumulation and ammonia assimilation: a comparative study in two mulberry genotypes differing in drought sensitivity. J Plant Physiol 150: 345–350 [Google Scholar]
- Rivero RM, Gimeno J, Van Deynze A, Walia H, Blumwald E. (2010) Enhanced cytokinin synthesis in tobacco plants expressing PSARK:IPT prevents the degradation of photosynthetic protein complexes during drought. Plant Cell Physiol 51: 1929–1941 [DOI] [PubMed] [Google Scholar]
- Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E. (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104: 19631–19636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero RM, Shulaev V, Blumwald E. (2009) Cytokinin-dependent photorespiration and the protection of photosynthesis during water deficit. Plant Physiol 150: 1530–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T. (1999) Source-sink regulation by sugar and stress. Curr Opin Plant Biol 2: 198–206 [DOI] [PubMed] [Google Scholar]
- Roitsch T, Bittner M, Godt DE. (1995) Induction of apoplastic invertase of Chenopodium rubrum by d-glucose and a glucose analog and tissue-specific expression suggest a role in sink-source regulation. Plant Physiol 108: 285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J. (2002) Sugar sensing and signaling in plants. Plant Cell (Suppl) 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronzhina ES, Mokronosov AT. (1994) Source-sink relations and the role of cytokinins in the regulation of transport and partitioning of organic-substances in plants. Russ J Plant Physiol 41: 396–406 [Google Scholar]
- Sakakibara H. (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57: 431–449 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N. (2006) Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci 11: 440–448 [DOI] [PubMed] [Google Scholar]
- Sharkey TD. (1985) Photosynthesis in intact leaves of C3 plants: physics, physiology and rate limitations. Bot Rev 51: 53–105 [Google Scholar]
- Smith AM, Zeeman SC. (2006) Quantification of starch in plant tissues. Nat Protoc 1: 1342–1345 [DOI] [PubMed] [Google Scholar]
- Tang Z, Fan X, Li Q, Feng H, Miller AJ, Shen Q, Xu G. (2012) Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol 160: 2052–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F, Davies WJ. (1993) Integration of hydraulic and chemical signalling in the control of stomatal conductance and water status of droughted plants. Plant Cell Environ 16: 341–349 [Google Scholar]
- Van Handel E. (1968) Direct microdetermination of sucrose. Anal Biochem 22: 280–283 [DOI] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM. (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Quebedeaux B, Stutte G. (1995) Osmotic adjustment: effect of water stress on carbohydrates in leaves, stems and roots of apple. Funct Plant Biol 22: 747–754 [Google Scholar]
- Werner T, Holst K, Pörs Y, Guivarc’h A, Mustroph A, Chriqui D, Grimm B, Schmülling T. (2008) Cytokinin deficiency causes distinct changes of sink and source parameters in tobacco shoots and roots. J Exp Bot 59: 2659–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A, Gröner F, Sonnewald U, Deppner H, Lerchl J, Hebbeker U, Flügge UI, Weber A. (1999) Spinach hexokinase I is located in the outer envelope membrane of plastids. FEBS Lett 461: 13–18 [DOI] [PubMed] [Google Scholar]
- Wilkinson S, Davies WJ. (2010) Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant Cell Environ 33: 510–525 [DOI] [PubMed] [Google Scholar]
- Wilkinson S, Kudoyarova GR, Veselov DS, Arkhipova TN, Davies WJ. (2012) Plant hormone interactions: innovative targets for crop breeding and management. J Exp Bot 63: 3499–3509 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. (2007) Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 143: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Zhang J, Davies WJ. (1990) Changes in the concentration of ABA in xylem sap as a function of changing soil water status can account for changes in leaf conductance and growth. Plant Cell Environ 13: 277–285 [Google Scholar]
- Zrenner R, Salanoubat M, Willmitzer L, Sonnewald U. (1995) Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J 7: 97–107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.