Abstract
Background
Anopheles (Kerteszia) cruzii (Diptera: Culicidae) is a primary vector of human and simian malaria parasites in southern and southeastern Brazil. Earlier studies using chromosome inversions, isoenzymes and a number of molecular markers have suggested that An. cruzii is a species complex.
Results
In this study, a multilocus approach using six loci, three circadian clock genes and three encoding ribosomal proteins, was carried out to investigate in more detail the genetic differentiation between the An. cruzii populations from Florianópolis–Santa Catarina (southern Brazil) and Itatiaia–Rio de Janeiro States (southeastern Brazil). The analyses were performed first comparing Florianópolis and Itatiaia, and then comparing the two putative sympatric incipient species from Itatiaia (Itatiaia A and Itatiaia B). The analysis revealed high FST values between Florianópolis and Itatiaia (considering Itatiaia A and B together) and also between the sympatric Itatiaia A and Itatiaia B, irrespective of their function. Also, using the IM program, no strong indication of migration was found between Florianópolis and Itatiaia (considering Itatiaia A and B together) using all loci together, but between Itatiaia A and Itatiaia B, the results show evidence of migration only in the direction of Itatiaia B.
Conclusions
The results of the multilocus analysis indicate that Florianópolis and Itatiaia represent different species of the An. cruzii complex that diverged around 0.6 Mya, and also that the Itatiaia sample is composed of two sympatric incipient species A and B, which diverged around 0.2 Mya. Asymmetric introgression was found between the latter two species despite strong divergence in some loci.
Keywords: Anopheles, Speciation, Complex of cryptic species, Multilocus analysis, Malaria, Mosquitoes
Background
The Anopheles mosquitoes are insects belonging to order Diptera, family Culicidae and subfamily Anophelinae [1]. The genus Anopheles comprises almost 500 formally named species, although many of them are complexes of sibling species [2,3]. This genus is subdivided into six subgenera, among them Kerteszia[1,4]. Anopheles (Kerteszia) cruzii is a Neotropical mosquito highly specialized in using bromeliad plants as breeding reservoirs during the development of immature stages. This bromeliad-breeding mosquito is mainly found in Atlantic forest areas along the Brazilian coast, a habitat rich in plants from the Bromeliaceae family [4-6]. This mosquito is an important vector of human and simian malaria parasites in southern and southeastern Brazil [7,8], essentially in forest environments [9-11].
Different studies have been made for identification of Anopheles species complexes, which are virtually identical in adult morphology, but exhibit differences in their malaria transmission competence, resting habitats, host preference and insecticide resistance [3,12-14]. In this way, many authors have used molecular and genetic data for evolutionary analysis in a number of Anopheles species (An. gambiae, An. fluviatilis, An. funestus, An. barbirostris, An. punctulatus, An. subpictus and others) in an effort to resolve their taxonomic status [2,13,15-21]. Despite all accumulated molecular and genetic data originated from these studies, there are only a few population genetic studies of An. cruzii and most of them suggest that An. cruzii is a species complex. Differences in X chromosome inversions frequencies from populations of southeastern and southern Brazil, suggested the existence of three An. cruzii sibling species [22,23]. On the other hand, isoenzymes indicated two genetically isolated groups, one from Bahia State (northeastern Brazil), and the other from southeastern and southern Brazil (Rio de Janeiro, São Paulo and Santa Catarina States) [24]. These studies led to further analyses using different molecular markers. A fragment of the timeless gene, a locus involved in the control of circadian rhythms, was used as a marker to assess the genetic differentiation among a number of An. cruzii populations. The results indicated that a population of An. cruzii from northeast Brazil constitutes a highly differentiated group when compared with other five populations from the south and southeast regions of the country [25]. In addition, a multilocus approach using six loci, three circadian clock genes and three encoding ribosomal proteins, was implemented to investigate in more detail the genetic differentiation and time of divergence between the populations of the northeast and south Brazil. The analysis revealed very high FST values and fixed differences between these two cryptic species in all six loci, irrespective of their function. This analysis also indicated that they probably have not exchanged migrants since their separation, which was roughly estimated to have occurred around 2.4 million of years ago [26].
Besides, a fragment of the cpr gene, a locus involved in metabolic insecticide resistance and odorant clearance in insects, was used to analyze the divergence between An. cruzii populations from south and southeast Brazil (Florianópolis–SC, Cananéia and Juquitiba–SP, Itatiaia–RJ, Santa Teresa–ES). The cpr gene revealed very high FST values and fixed differences between Itatiaia and the other four populations studied. The data also provided preliminary evidence for the possible occurrence of two sympatric incipient sibling species in Itatiaia, called by the authors Itatiaia A and Itatiaia B [27].
In the current study, a multilocus analysis using six different nuclear gene fragments was performed comparing Florianópolis and Itatiaia populations. Additional multilocus analyses were also carried out considering Itatiaia as two different incipient species (Itatiaia A and Itatiaia B). The aim of the study was to determine if there is gene flow between the putative sibling species and to estimate their divergence time.
Results
One of the assumptions of the Isolation with Migration model used in this study is the absence of recombination within the studied loci. In order to fulfill this requirement, the optimal recombination-filtered block was extracted from each gene alignment. Additional file 1: Table S1 shows the position of the non-recombining (NR) blocks used in this study as well as the putative recombinant sequences that were removed (see Methods). Another assumption of the IM program is that the variation observed in the studied loci is neutral. Therefore, the Tajima [28] and Fu & Li [29] tests of neutrality were used and the results are presented in Additional file 2: Table S2, together with the polymorphism summaries of the six gene fragments. No significant deviations from neutrality were observed in all comparisons. Additional file 2: Table S2 also shows the minimum number of recombination events for each gene (RM) and the length of the whole fragment and for the NR block of each gene (values in parentheses). The alignments of the whole sequences of each gene are presented in Additional file 3: Table S3. All loci include at least one intron of variable size, except the cyc gene fragment, which was composed entirely of an exon. Besides, Additional file 2: Table S2 shows the number of polymorphic sites (S) for each An. cruzii sibling species and two measures of nucleotide diversity: π, based on the average number of pairwise differences and θ, based on the total number of mutations (values for the NR blocks in parentheses). Pairwise estimates of population differentiation between the An. cruzii sibling species are shown in Table 1, which also shows the average number of nucleotide substitutions per site (Dxy), the number of net nucleotide substitutions per site between species (Da) and the distribution of the four mutually exclusive categories of segregating sites observed in each comparison: the number of exclusive polymorphisms for each species (S1 and S2), the number of shared polymorphisms (Ss) and the number of fixed differences (Sf).
Table 1.
Genetic differentiation between populations
| Locus | Populations | FST | P (FST) | Dxy | Da | Ss | Sf | S1 | S2 |
|---|---|---|---|---|---|---|---|---|---|
|
Timeless |
Florianópolis X Itatiaia |
0.1838 (0.2661) |
0.000 (0.000) |
0.03073 (0.02763) |
0.00564 (0.00735) |
19 (02) |
00 (00) |
44 (19) |
11 (03) |
| |
Florianópolis X Itatiaia A |
0.3154 (0.2876) |
0.000 (0.000) |
0.03058 (0.02813) |
0.01116 (0.00809) |
07 (01) |
02 (00) |
43 (20) |
03 (01) |
| |
Florianópolis X Itatiaia B |
0.1892 (0.3000) |
0.000 (0.000) |
0.02859 (0.02433) |
0.00646 (0.00730) |
09 (01) |
00 (00) |
40 (20) |
09 (00) |
| |
Itatiaia A X Itatiaia B |
0.3418 (0.2081) |
0.0000 (0.0300) |
0.0223 (0.0120) |
0.0076 (0.0025) |
05 (01) |
02 (00) |
05 (04) |
13 (03) |
|
Clock |
Florianópolis X Itatiaia |
0.6707 (0.6340) |
0.000 (0.000) |
0.05159 (0.05930) |
0.03460 (0.03760) |
00 (00) |
01 (01) |
06 (06) |
15 (14) |
| |
Florianópolis X Itatiaia A |
0.7325 (0.7207) |
0.000 (0.000) |
0.05065 (0.05879) |
0.03710 (0.04237) |
00 (00) |
04 (04) |
07 (07) |
06 (05) |
| |
Florianópolis X Itatiaia B |
0.5735 (0.6926) |
0.000 (0.000) |
0.05463 (0.06270) |
0.03133 (0.04343) |
00 (00) |
02 (04) |
06 (07) |
11 (04) |
| |
Itatiaia A X Itatiaia B |
0.1783 (0.0838) |
0.0000 (0.1610) |
0.0282 (0.0308) |
0.0050 (0.0030) |
03 (04) |
00 (00) |
03 (03) |
14 (13) |
|
Cycle |
Florianópolis X Itatiaia |
0.2845 (0.1765) |
0.000 (0.000) |
0.03575 (0.04093) |
0.01017 (0.00723) |
10 (06) |
00 (00) |
12 (07) |
09 (02) |
| |
Florianópolis X Itatiaia A |
0.3510 (0.2236) |
0.000 (0.000) |
0.03600 (0.04284) |
0.01264 (0.00958) |
06 (04) |
00 (00) |
16 (09) |
04 (02) |
| |
Florianópolis X Itatiaia B |
0.2127 (0.0863) |
0.000 (0.029) |
0.03278 (0.03550) |
0.00697 (0.00307) |
08 (04) |
00 (00) |
14 (09) |
04 (00) |
| |
Itatiaia A X Itatiaia B |
0.1523 (0.1524) |
0.0200 (0.1020) |
0.0249 (0.0281) |
0.0038 (0.0043) |
05 (03) |
00 (00) |
05 (02) |
07 (06) |
|
Rp49 |
Florianópolis X Itatiaia |
0.1667 (0.1303) |
0.000 (0.000) |
0.01433 (0.01155) |
0.00239 (0.00151) |
02 (01) |
00 (00) |
09 (09) |
14 (11) |
| |
Florianópolis X Itatiaia A |
0.2985 (0.2228) |
0.000 (0.000) |
0.01147 (0.00860) |
0.00342 (0.00192) |
02 (01) |
00 (00) |
09 (09) |
03 (02) |
| |
Florianópolis X Itatiaia B |
0.3743 (0.1978) |
0.000 (0.000) |
0.01774 (0.01453) |
0.00664 (0.00288) |
00 (00) |
01 (00) |
11 (10) |
08 (07) |
| |
Itatiaia A X Itatiaia B |
0.4784 (0.4865) |
0.0000 (0.0000) |
0.0204 (0.0196) |
0.0098 (0.0096) |
00 (00) |
01 (01) |
05 (05) |
08 (08) |
|
RpS2 |
Florianópolis X Itatiaia |
0.2631 (0.2647) |
0.000 (0.000) |
0.02045 (0.02005) |
0.00538 (0.00531) |
01 (00) |
00 (00) |
17 (15) |
13 (11) |
| |
Florianópolis X Itatiaia A |
0.2961 (0.2539) |
0.000 (0.000) |
0.02066 (0.0199) |
0.00612 (0.0050) |
00 (00) |
00 (00) |
18 (15) |
09 (08) |
| |
Florianópolis X Itatiaia B |
0.4212 (0.4880) |
0.000 (0.000) |
0.01924 (0.0201) |
0.00811 (0.0098) |
01 (00) |
00 (00) |
17 (15) |
03 (03) |
| |
Itatiaia A X Itatiaia B |
0.4494 (0.4730) |
0.0000 (0.000) |
0.0153 (0.0152) |
0.0069 (0.00723) |
01 (01) |
00 (00) |
08 (07) |
03 (02) |
|
RpS29 |
Florianópolis X Itatiaia |
0.7103 (0.7436) |
0.000 (0.000) |
0.05295 (0.07813) |
0.03771 (0.05809) |
01 (00) |
05 (04) |
12 (11) |
24 (16) |
| |
Florianópolis X Itatiaia A |
0.7624 (0.7880) |
0.000 (0.000) |
0.0545 (0.0830) |
0.0415 (0.0654) |
01 (00) |
05 (04) |
13 (12) |
10 (07) |
| |
Florianópolis X Itatiaia B |
0.7363 (0.7536) |
0.000 (0.000) |
0.0536 (0.0758) |
0.0395 (0.0571) |
01 (00) |
04 (03) |
13 (12) |
14 (09) |
| Itatiaia A X Itatiaia B | 0.1819 (0.1675) | 0.0000 (0.0110) | 0.0216 (0.0208) | 0.0039 (0.0035) | 06 (02) | 00 (00) | 05 (03) | 09 (10) |
FST, pair-wise estimates of population differentiation. P-value, significance of FST values (evaluated by 1,000 random permutations). Dxy and Da, average number of nucleotide substitutions per site and the number of net nucleotide substitutions per site between species, respectively [30]. S1, number of polymorphic sites exclusive to Florianópolis. S2, number of polymorphic sites exclusive to Itaparica. Ss and Sf, number of shared polymorphisms and number of fixed differences between the two species, respectively. Numbers in parentheses are related to the non-recombining block (NR) for each locus.
The IM program was used to simultaneously estimate six demographic parameters (θ1, θ2, θA, t, m1, m2) from the An. cruzii sibling species through an "Isolation with Migration" model using multiple loci [31]. As mentioned above, only the NR blocks were used and some recombining sequences were removed before the IM analysis (Additional file 1: Table S1). The analyses (see below) were performed first comparing Florianópolis and Itatiaia, then comparing the two putative sympatric incipient species from Itatiaia [27] and finally comparing those two putative siblings with Florianópolis. For each comparison, four independent runs were performed and among them, the simulations showed good convergence and consistency resulting in complete posterior distributions for all six parameters, except for t and θA in the simulations between Itatiaia A and Itatiaia B (see below).
Finally, gene trees of the sequences from all loci for both whole sequences (see below) and NR blocks (not shown) were estimated using the Neighbor-Joining method (NJ) with very similar results. The most suitable model selected using jModelTest 0.1.1 [32,33] was Kimura 2-parameter [34] for all loci. All trees were performed with 1,000 bootstrap replicates.
Divergence between Florianópolis and Itatiaia
The polymorphism found in both populations was similar in most cases (Additional file 2: Table S2). The larger differences in length between the whole fragment and the NR block were observed for tim and cyc and this was due to the higher number of recombination events identified in these two genes (RM = 13 and 8 respectively). Except for the tim gene, all base substitutions were synonymous or occurred within introns (Additional file 2: Table S2). The few non-synonymous changes found in the tim fragment are described in Rona et al. [25].
Very high FST values were found using the Clk and RpS29 genes between Florianópolis and Itatiaia (0.67 and 0.71, respectively) (Table 1). The other four genes showed FST values ranging from ~ 0.16 to 0.28. In all cases the FST values were significant. Florianópolis and Itatiaia had fixed differences and few or no shared polymorphisms in Clk and RpS29 gene fragments. Among the other genes, there were shared polymorphisms and no fixed differences (Table 1).
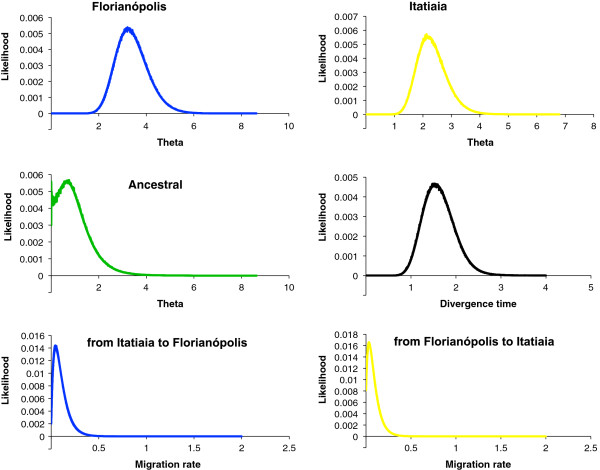
Figure 1 shows the posterior probability distributions for each of the six demographic parameters estimated using IM and Additional file 4: Table S4 summarizes the features from the marginal histograms for each of the parameters in all MCMC runs. The estimates of θ suggest that the effective population size of the ancestral population was smaller than the current Florianópolis and Itatiaia populations indicating that both may have had a history of growth after separation (Figure 1).
Figure 1.
Posterior probability distributions for each of the six demographic parameters estimated using IM for the pairwise comparison Florianópolis vs Itatiaia (A and B). Effective population size for an ancestral and two descendent populations (theta), divergence time between Florianópolis and Itatiaia, and migration rates in both directions. Four IM simulations using different seed numbers were plotted for each parameter estimate (see also Additional file 4: Table S4). All curves are shown including the range of the priors.
Migration parameters have been estimated for all loci together for each population in different IM runs. Our aim was to detect the occurrence of gene flow using the multilocus data. The results have revealed no strong indication of migration in either direction using all loci together.
The divergence time parameter was estimated for all combined loci in four different IM runs (Table 2). This parameter cannot be directly converted to years because the mutation rates in An. cruzii species are unknown. Therefore, an estimate of the divergence time between An. cruzii species was performed using the average of Drosophila synonymous and non-synonymous substitution rates for several nuclear genes (0.0156 and 0.00191 per site per million year respectively) [35]. Using this approach and based on the average of HiSmth values, an estimate of the divergence time between Florianópolis and Itatiaia would be approximately 0.75 Mya (range from 0.51 to 1.1 Mya, based on the average of HPD90Lo and HPD90Hi values).
Table 2.
Divergence time estimates among the An. cruzii populations obtained by IM (based on the average of HiSmth values) and also by the average Da values from the six loci (based on the whole sequence)
| Populations | IM | Da |
|---|---|---|
| Florianópolis x Itatiaia (A and B) |
0.75 Mya |
0.51 Mya |
| Florianópolis x Itatiaia A |
0.70 Mya |
0.59 Mya |
| Florianópolis x Itatiaia B |
0.80 Mya |
0.52 Mya |
| Itatiaia A x Itatiaia B | - | 0.19 Mya |
Since the time parameter did not show good convergence in the IM runs between Itatiaia A and Itatiaia B, this divergence time was estimated using only the Da values.
Another method of estimating the divergence time between these two putative species is to use the same Drosophila synonymous substitution rate mentioned above and the average Da values from the six loci (Table 1). Based on these values, the divergence time between the populations from Florianópolis and Itatiaia was estimated to be approximately 0.51 Mya and 0.62 Mya for the whole sequence and NR blocks, respectively (Table 2).
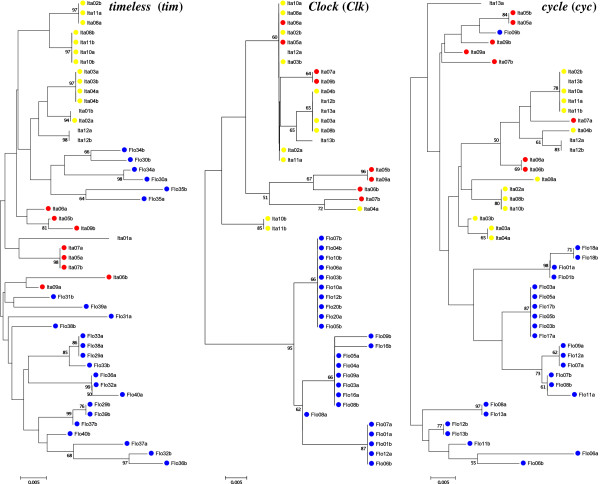
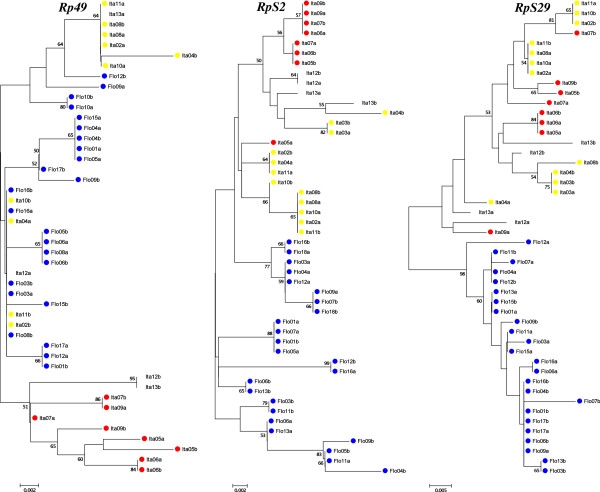
The resulting NJ trees using Clk and RpS29 clearly grouped the sequences from the two sibling species in different clusters with high bootstrap values (Figures 2 and 3).
Figure 2.
Neighbor-joining trees of clock genes nucleotide sequences of Florianópolis and Itatiaia (A and B). Neighbor-joining trees using the three clock genes nucleotide sequences of Florianópolis and Itatiaia (A and B) obtained with Kimura 2-parameter distance for all genes. Numbers on the nodes represent the percentage bootstrap values based on 1,000 replications. Flo: Florianópolis; Ita: Itatiaia; Itatiaia A: yellow circles; Itatiaia B: red circles.
Figure 3.
Neighbor-joining trees of ribosomal protein genes nucleotide sequences of Florianópolis and Itatiaia (A and B). Neighbor-joining trees using the three ribosomal protein genes nucleotide sequences of Florianópolis and Itatiaia (A and B) obtained with Kimura 2-parameter distance for all genes. Numbers on the nodes represent the percentage bootstrap values based on 1,000 replications. Flo: Florianópolis; Ita: Itatiaia; Itatiaia A: yellow circles; Itatiaia B: red circles.
Divergence between Itatiaia A and Itatiaia B
Rona et al. [27] hypothesized that the Itatiaia sample might include two different sets of individuals based on cpr sequences. According to this classification the individuals Ita02, Ita03, Ita04, Ita08, Ita10 and Ita11 belong to Itatiaia A and the mosquitoes Ita05, Ita06, Ita07 and Ita09 belong to Itatiaia B. We therefore used our multilocus data to test the hypothesis that these two Itatiaia groups represent different sympatric incipient species. According to the cpr data, mosquito Ita12 was the only potential "hybrid" while Ita01 and Ita13 were not typed with this gene [27]. Therefore these three mosquitoes were not used in the analysis described below.
Itatiaia A was the least polymorphic, showing the lowest values of θ and π, as well as the smaller number of polymorphic sites (S) in most cases (Additional file 2: Table S2). High and significant FST values were observed between the two sympatric species, ranging from 0.15 to 0.47 (Table 1). In all cases the FST values were significant (except using the NR block of the Clk and cyc genes).
Itatiaia A and Itatiaia B had fixed differences and few or inexistent shared polymorphisms in Rp49 and RpS2 gene fragments, which show the highest values of differentiation (FST = 0.47 and 0.44, respectively). The tim locus also shown a high FST value (FST = 0.34) between these two putative species and fixed differences as well, but shared polymorphisms were also observed in this gene. Among the other loci, there were shared polymorphisms and no fixed differences (Table 1).
Figure 4 shows the posterior probability distributions for each of the six demographic parameters estimated using IM and Additional file 5: Table S5 summarizes the features from the marginal histograms for each of the parameters in all MCMC runs for Itatiaia A and Itatiaia B. Migration parameters have been estimated for all loci together for each population in different IM runs. The results show evidence of migration only in the direction of Itatiaia B.
Figure 4.
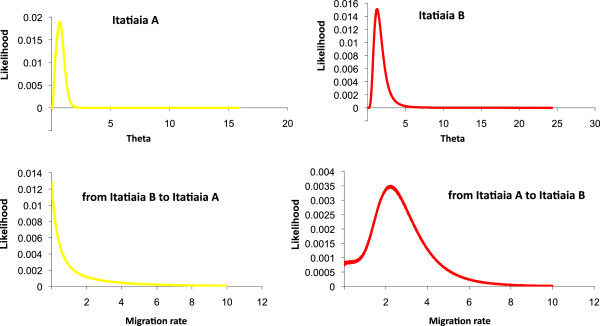
Posterior probability distributions for each of the four demographic parameters estimated using IM for the pairwise comparison Itatiaia A vs Itatiaia B. Effective population size for Itatiaia A and Itatiaia B (theta) and migration rates in both directions. Four IM simulations using different seed numbers were plotted for each parameter estimate (see also Additional file 5: Table S5). All curves are shown including the range of the priors.
Since the time parameter did not show good convergence in the IM runs, the divergence time was estimated between Itatiaia A and Itatiaia B using only the Drosophila synonymous substitution rate and the average Da values from the six loci (Table 1) mentioned above. Based on these values, the divergence time between the populations from Itatiaia A and Itatiaia B was estimated to be approximately 0.19 Mya and 0.16 Mya for the whole sequence and NR blocks, respectively (Table 2).
The NJ trees (Figures 2 and 3) grouped the sequences from the two sibling species in different clusters using tim, RpS2 and Rp49 genes, which show the highest values of differentiation (FST = 0.34, 0.44 and 0.47, respectively). In the RpS2 tree, there is a haplotype (Ita05a) from Itatiaia B among the Itatiaia A sequences. Also, the individuals Ita12 (the only potential "hybrid" typed with cpr gene) and Ita13 (that was not typed with cpr) appeared among Itatiaia A sequences. On the other hand, in the Rp49 tree, these two mosquitoes showed a hybrid aspect, in agreement with cpr gene results, where Ita12 was the only potential "hybrid" [13]. The others, Clk, cyc and RpS29, did not show an evident separation in NJ trees between Itatiaia A and Itatiaia B.
Divergence between Florianópolis vs Itatiaia A and Florianópolis vs Itatiaia B
In addition to the analyses performed between Florianópolis vs Itatiaia (considering Itatiaia A and B together) described above, analyses were also carried out comparing Florianópolis with Itatiaia A and Itatiaia B, separately.
The pairwise estimates of population differentiation between the two groups (Table 1) shown very high FST values using the Clk and RpS29 genes, in agreement with the values found between Florianópolis and Itatiaia (considering Itatiaia A and B together). The further four genes shown FST values ranging from ~ 0.18 to 0.42. In all cases the FST values were significant. Interestingly, the clock genes (tim, cyc and Clk loci) shown the higher FST values between Florianópolis and Itatiaia A, whereas the Rp49 and RpS2 loci shown the higher FST values between Florianópolis x Itatiaia B. The RpS29 showed very high and similar values in both comparisons (FST = 0.73 and 0.76).
Florianópolis and Itatiaia A had fixed differences and few or no shared polymorphisms in Clk, RpS29 and tim gene fragments. Between Florianópolis and Itatiaia B, the loci Clk, RpS29 and Rp49 shown fixed differences and few or inexistent shared polymorphisms (Table 1).
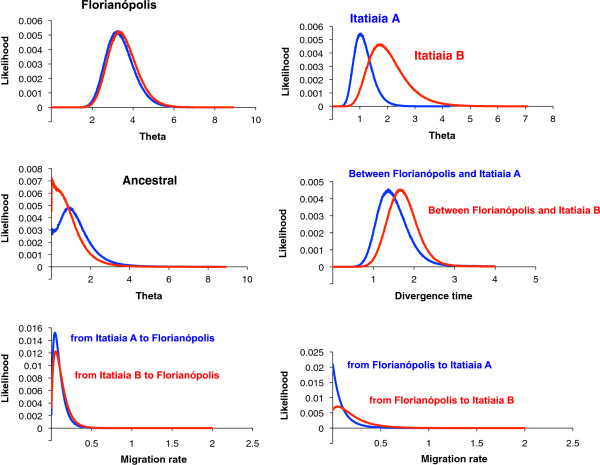
Additional file 6: Table S6 summarizes the features from the marginal histograms for each of the parameters in all MCMC runs between Florianópolis vs Itatiaia A and between Florianópolis vs Itatiaia B, respectively, and Figure 5 shows the posterior probability distributions for each of the six demographic parameters estimated using IM for both groups. The estimates of θ suggest that Itatiaia A and Itatiaia B are smaller than Florianópolis population (Figure 5). Migration parameters have been estimated for all loci together for each population in different IM runs. The results have revealed no indication of migration in either direction using all loci together in both comparisons (Florianópolis vs Itatiaia A and Florianópolis vs Itatiaia B).
Figure 5.
Posterior probability distributions for each of the six demographic parameters estimated using IM for the pairwise comparisons Florianópolis vs Itatiaia A and Florianópolis vs Itatiaia B. Effective population size for an ancestral and two descendent populations (theta), divergence time between Florianópolis and Itatiaia, and migration rates in both directions. Four IM simulations using different seed numbers were plotted for each parameter estimate (see also Additional file 6: Table S6). All curves are shown including the range of the priors.
The divergence time parameter was estimated for all combined loci in four different IM runs. The IM runs indicated similar results between Florianópolis vs Itatiaia A, Florianópolis vs Itatiaia B and Florianópolis vs Itatiaia (considering A and B together), which suggest that the Itatiaia lineage separated from Florianópolis before splitting into A and B (Table 2). Based on the average of HiSmth values, an estimate of the divergence time between Florianópolis and Itatiaia A would be approximately 0.7 Mya and between Florianópolis and Itatiaia B would be approximately 0.8 Mya. Based on Da values, the divergence time between Florianópolis and Itatiaia A and B was estimated to be approximately 0.59 Mya and 0.52 Mya, respectively (Table 2).
Discussion
The multilocus results presented here revealed a high level of differentiation between Florianópolis, Itatiaia A and Itatiaia B indicating that these populations represent indeed different species in the An. cruzii complex. Species as evolutionary lineages are expected to show greater evolutionary independence from one another than are populations within species [36]. Hey & Pinho [36] discuss the use of two measures of evolutionary independence, gene flow and divergence time, for species diagnosis, which help to differentiate intraspecific differences from interspecific differences. The two measures of evolutionary independence were also correlated with FST estimates. For simplicity, the authors suggested a threshold criterion of gene flow < 1, the separation time >1 (based on IM results) and the FST > 0.35 to species diagnosis. Among the six FST values calculated for the pairwise Florianópolis and Itatiaia (including A and B), only two of them are higher than 0.35 (FST = 0.67 and 0.71). Nevertheless, the mean FST among the six loci (0.38) and both measures of evolutionary independence (gene flow and divergence time) are above the threshold criteria. The comparison made between Itatiaia A and Itatiaia B showed similar results, but the migration parameter in the direction of Itatiaia B shown values higher than the suggested threshold, consistent with the idea of incipient speciation.
Previously, we observed no evidence of association between function (circadian x ribosomal genes) in the divergence levels between the sibling species of Florianópolis and Itaparica [26]. In the current work, Clk and RpS29 showed the highest FST values between Florianópolis and Itatiaia (considering Itatiaia A and B together), while Rp49 and RpS2 showed the highest FST values between Itatiaia A and B. Interestingly, these four genes are located on the chromosome 2 of Anopheles gambiae, albeit the three ribosomal protein genes are on the right arm while Clk is on the left arm. Many authors have postulated that chromosomal inversions could promote speciation since their recombination suppressing effects facilitate the maintenance and accumulation of differences between interbreeding populations [37-39]. Ramirez & Dessem [22,23], studying the polytene chromosomes of Anopheles cruzii from south and southeast regions of Brazil, revealed the existence of three putative species, which differed mainly in the banding patterns of the X chromosome. Nevertheless, different autosomal inversions seemed to be associated with each form of X chromosome and it is tempting to speculate that some of the markers we used and showed high levels of differentiation between the sibling species are located within or nearby the inversions studied by Ramirez & Dessem [22,23]. Future in situ hybridization experiments using these markers as probes might confirm if that is the case.
The observation of a putative hybrid [27] and the varying degrees of differentiation observed in the six loci between Itatiaia A and Itatiaia B, suggest that these two siblings are in a process of incipient speciation. Similar results were found in the Anopheles gambiae complex [40-42] that maintains a genome in a mosaic form, with regions of low and high divergence. These variations among the different regions of the genome in the divergence between the An. cruzii siblings have two main explanations: i) the maintenance of ancestral polymorphisms as these cryptic species have separated recently and ii) differential introgression in different genomic regions between the species reflecting locations where gene flow occurs freely or is restricted by selection.
The two conflicting but no mutually exclusive hypotheses of retention of ancestral polymorphism and introgression between closely related species are often difficult to distinguished. Introgression can sometimes be excluded based on the geographic separation of two species and in this case, the shared polymorphisms can be classified as ancestral [43]. This is perhaps the situation found between the allopatric Florianópolis and Itatiaia (A and B) with no strong indication of migration, in either direction, as suggested by the IM results. On the other hand, the introgression hypothesis is of course more likely if the species are sympatric. This is probably the case of Itatiaia A and Itatiaia B, where the introgression hypothesis seem confirmed by the IM results that revealed nonzero values of migration parameter between the latter species, only in the direction of Itatiaia B.
Asymmetric introgression has been documented in a variety of taxa [44-48], including mosquitoes. For example, Donnelly et al. [43], comparing haplotype frequencies in allopatric and sympatric populations of An. arabiensis and An. gambiae, found unidirectional introgression, from the former into the latter species, only in sympatric populations. Gomes et al. [49] found differential introgression in the Culex pipiens complex, in spite of the high levels of genetic differentiation between its forms.
Marsden et al. [42] found similar results in the An. gambiae complex, between M and S molecular forms, and they argue, as one of the possible explanations, that the asymmetric introgression may be a consequence of the differences in relative abundance of the two taxa (S form–the genetic recipient–was more common than M form–the genetic donor–in all sites assessed), which could result in higher levels of backcrossing between the hybrids and the more abundant species. Our results are also consistent with this explanation as the estimates of population sizes and migration parameters from the IM program suggest that Itatiaia A (the genetic donor) has a smaller estimated population size than Itatiaia B (the genetic recipient).
Marsden et al. [42] also proposed that the asymmetric introgression can contribute to the maintenance of differentiation between M and S, since the unidirectional movement of nuclear genes from the M into the S form would prevent homogenization of their gene pools because of the conservation of unique polymorphism within the S form and a lack of admixture in M. Indeed, the observed genetic diversity was considerably lower in Itatiaia A than in Itatiaia B.
The estimated divergence time (based on the Da values) between Itatiaia A and Itatiaia B is ~ 0.2 Mya and between Florianópolis and Itatiaia (including A and B) is ~ 0.6 Mya. This indicates an earlier speciation process splitting the populations of Florianópolis and Itatiaia, followed by a more recent separation between the two sympatric species in the latter locality.
The time of divergence of these three sibling species as well as that between the Bahia and the south and southeast populations (~ 2.4 Mya) [26], points to the importance of Pleistocene environmental changes (glaciations and inter-glaciations periods) [50] as factors in the diversification of An. cruzii species complex. Since temperature and water are essential for the development of Anopheles spp. immature stages [4], the hypothesis that Pleistocene environmental changes driving Anopheles diversification seems plausible. Furthermore, in the case of An. cruzii (Kerteszia subgenus), its larval development is associated with water trapped in Bromeliads plants, which are restricted to the rainforest [4,7]. So, a number of studies show that several malaria vectors, including An. cruzii, have revealed patterns of genetic divergence associated with the Pleistocene environmental changes [26,51-55].
The process of successive rainforest contractions and expansions was a very important consequence of these environmental modifications possibly favoring the differentiation and speciation of forest obligate species [56-58]. Accordingly, a number of studies of closely related species demonstrated the importance of the Pleistocene climatic changes in shaping the Brazilian Atlantic forest biodiversity [59,60], since patterns of endemicity in many forest obligate species are concordant in geographic distribution [59,61-63]. For example, Mata et al. [64], studying the molecular phylogeny and biogeography of the eastern Tapaculo birds in Brazilian Atlantic forest, corroborated the importance of the Bahia refuge as an avian center of endemism and also concluded that the southeast (Serra da Mantiqueira, where Itatiaia is located) lineage is genetically different from the southern populations.
Turchetto-Zolet et al. [65] studying the molecular variation patterns in Schizolobium parahyba (Fabaceae), a widespread tree in the Brazilian Atlantic forest, found high levels of genetic diversity in populations from the southeast region and low levels in southern populations. They argue that the low genetic diversity in the south may be the result of a founder effect followed by a range expansion after glacial periods. Lorenz-Lemke et al. [66] and Palma-Silva et al. [67] also reported expansion toward the south in Atlantic forest plants. However, in the case of An. cruzii, Florianópolis (south sibling species) showed a higher level of genetic diversity than the Itatiaia (southeast) sympatric sibling species. Therefore, it is likely that there was a persistence of An. cruzii populations in the south during the contraction of the forest, as was proposed for toads [60], instead of a southern colonization of the Atlantic forest from northern regions.
Conclusions
In summary, the results obtained with the studies of the An. cruzii complex suggest so far three main events of cladogenesis originating the different sibling species. The first one originated the Bahia (Itaparica) species and occurred toward the end of the Pliocene (~2.4 Mya) [25,26], the second that occurred around 0.6 Mya probably separated the Itatiaia lineage from other south and southeastern populations [25,27], and finally the most recent event (around 0.2 Mya) originated the new sympatric incipient species found in this locality. Analysis of a number of other populations of An. cruzii from Rio de Janeiro State, where Itatiaia is located, might allow us to determine the distribution of these two new sibling species and to investigated whether they might have possibly originated by a process of sympatric or parapatric speciation.
Methods
Molecular analysis
The mosquitoes used in this study were females captured in Florianópolis, Santa Catarina State (SC) (27°31′S / 48°30′W) and Itatiaia, Rio de Janeiro State (RJ) (22°27′S / 44°36′W). They were identified on the basis of their morphology according to Consoli and Lourenço-de-Oliveira [4]. For the molecular analysis, 12 individuals from Florianópolis and 10 to 12 from Itatiaia were used for each of the six gene fragments analyzed. Three of the fragments used are orthologues of Drosophila melanogaster genes involved in the control of circadian rhythms: timeless (tim), Clock (Clk) and cycle (cyc); and three code for ribosomal proteins: Rp49 (Ribosomal protein 49, known also as RpL32–Ribosomal protein L32), RpS2 (Ribosomal protein S2) and RpS29 (Ribosomal protein S29).
The sequences of the six genes from Florianópolis and the sequences of the tim gene from Itatiaia were those previously published by our group [25,26] (Accession numbers: GU016330–GU016569 and FJ408732–FJ408865, respectively). The sequences of the other five genes from Itatiaia were obtained by PCR, cloning and sequencing as described below.
The primers used in this study were those previously published by our group [25,26]. The An. cruzii genomic DNA extracted according to Jowett [68] was used in PCR reactions carried out in an Eppendorf Mastercycler® thermocycler using the proofreading Pfu DNA polymerase (Biotools).
PCR products were purified and cloned using either Zero Blunt TOPO PCR cloning kit (Invitrogen) or pMOS Blue vector blunt-ended cloning kit (GE Healthcare). Sequencing of positive clones was carried out in an ABI Prism 3730 DNA sequencer at the Oswaldo Cruz Institute using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems). The identity of the cloned fragments was determined by BlastX analysis using GenBank database [69].
At least eight clones were sequenced for each mosquito. Sequences were edited and in most cases consensus sequences representing the two alleles were generated. In a number of individuals only one haplotype was observed among the eight sequences and in these cases mosquitoes were classified as homozygotes. The probability of incorrectly classifying a heterozygote as a homozygote individual with this procedure is less than 1%. The sequences from homozygote mosquitoes were duplicated prior to analysis. However, when carried out without duplicating homozygote sequences, the analysis rendered similar results. Sequences were submitted to GenBank (Accession numbers: JX129234–JX129351).
DNA sequence analysis
The sequences were aligned using ClustalX software [70] and population genetics analysis was carried out using DNASP4.0 [71] and PROSEQ v 2.91 [72] softwares.
The jModelTest version 0.1.1 [32,33] was used to find the most suitable model for each gene evolution. Models selected by the Bayesian Information Criterion (BIC) were favored and used in the phylogenetic analysis, carried out using MEGA 4.0 [73].
The IM program is an implementation of the Isolation with Migration model and is based on the MCMC (Markov Chain Monte Carlo) simulations of genealogies [31,74]. It simultaneously estimates six demographic parameters from multilocus data: effective population size for an ancestral and two descendent populations (θA, θ1, and θ2, respectively), divergence time (t) and migration parameters in both directions (m1 and m2). Initial IM runs were performed in order to establish appropriate upper limits for the priors of each demographic parameter. These preliminary simulations generated marginal distributions that facilitated the choice of parameter values used in the final IM analyses. The convergence was assessed through multiple long runs (four independent MCMC runs with different seed numbers were carried out with at least 30,000,000 recorded steps after a burn-in of 100,000 steps) and by monitoring the ESS values, the update acceptance rates and the trend lines.
The Infinite Sites model [75] was chosen as the mutation model in the IM simulations because the two species are closely related and all genes are nuclear.
The optimal recombination-filtered block was extracted from each gene alignment using the IMGC program, which also removes haplotypes that represent likely recombinant sequences [76,77].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LDPR participated in data generation and analysis, and drafted the manuscript. She also helped capture mosquitoes in Florianópolis. CJCP carried out the capture and morphological identification of mosquitoes. AAP is the principal investigator, participated in its design and coordination, and helped to write the manuscript. All authors read and approved the final manuscript.
Supplementary Material
NR blocks and sequences excluded from the IM analysis. Edition of sequences prior to IM analysis using the IMGC program and based on alignments presented in Additional file 3: Table S3. NR blocks, fragment positions of the non-recombining blocks used in the analyses; Removed sequences, the putative recombinant sequences removed before the IM analysis.
Polymorphism summaries of An. cruzii sibling species. RM, the minimum number of recombination events; n, number of DNA sequences of each sibling species; S, number of polymorphic (segregating) sites; θ, nucleotide diversity based on the total number of mutations (Eta); π, nucleotide diversity based on the average number of pair-wise differences; DT, Tajima’s D [28]; DFL, Fu & Li’s D [29] and FFL, Fu & Li’s F [29], based on Eta (total number of mutations). No significant deviations from neutrality were observed after Bonferroni correction. Numbers in parentheses are related to the non-recombining block (NR) for each locus.
Alignments of the DNA sequences of the timeless, Clock, cycle, Rp49, RpS2 and RpS29 gene fragments from Florianópolis and Itatiaia. The translated amino acid sequences are shown above the alignments and the introns are highlighted in grey. Dots represent identity and dashed represent gaps. Flo: individuals from Florianópolis; Ita: individuals from Itatiaia.
Summarized features of the marginal histograms for each parameter for the pairwise comparison Florianópolis vs Itatiaia.
Summarized features of the marginal histograms for each parameter for the pairwise comparison Itatiaia A vs Itatiaia B.
Summarized features of the marginal histograms for each parameter to the pairwise comparison Florianópolis vs Itatiaia A (A) and Florianópolis vs Itatiaia B (B).
Contributor Information
Luísa DP Rona, Email: luisadrp@yahoo.com.br.
Carlos J Carvalho-Pinto, Email: carlospinto@ccb.ufsc.br.
Alexandre A Peixoto, Email: luisadrp@yahoo.com.br.
Acknowledgements
The authors are indebted to Robson Costa da Silva for his technical assistance and to PDTIS-FIOCRUZ for use of its DNA sequencing facility. This work was supported by grants from the Howard Hughes Medical Institute, FIOCRUZ, FAPERJ and CNPq.
References
- Harbach RE. The culicidae (Diptera): a review of taxonomy, classification and phylogeny. Zootaxa. 2007;1668:591–638. [Google Scholar]
- Krzywinski J, Besansky NJ. Molecular systematics of Anopheles: from sub- genera to subpopulations. Annu Rev Entomol. 2003;48:111–139. doi: 10.1146/annurev.ento.48.091801.112647. [DOI] [PubMed] [Google Scholar]
- Loaiza JR, Bermingham E, Sanjur OI, Scott ME, Bickersmith SA, Conn JE. Review of genetic diversity in malaria vectors (Culicidae: anophelinae) Infect Genet Evol. 2012;12:1–12. doi: 10.1016/j.meegid.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Consoli RAGB, Lourenço-de-Oliveira R. Principais mosquitos de importância sanitária no Brasil. Rio de Janeiro: Ed. Fiocruz; 1994. [Google Scholar]
- Marques GRAM, Forattini OP. Aedes albopictus em bromélias de solo em Ilhabela, litoral do estado de São Paulo. Rev Saude Publica. 2005;39:548–552. doi: 10.1590/s0034-89102005000400005. [DOI] [PubMed] [Google Scholar]
- Marrelli MT, Malafronte RS, Sallum MA, Natal D. Kerteszia subgenus of Anopheles associated with the Brazilian Atlantic rainforest: current knowledge and future challenges. Malar J. 2007;6:127. doi: 10.1186/1475-2875-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachou RG. Anofelinos do Brasil: comportamento das espécies vetoras de malária. Rev Bras Malariol Doencas Trop. 1958;10:145–181. [Google Scholar]
- Deane LM, Ferreira-Neto JA, Deane SP, Silveira IP. Anopheles (Kerteszia) cruzii, a natural vector of the monkey malaria parasites, Plasmodium simium and Plamodium brasilianum. Trans R Soc Trop Med Hyg. 1970;64:647. doi: 10.1016/0035-9203(70)90088-x. [DOI] [PubMed] [Google Scholar]
- Forattini OP, Kakitani I, Marques GRAM, Brito M. Formas imaturas de anofelíneos em recipientes artificiais. Rev Saude Publica. 1998;32:189–191. doi: 10.1590/S0034-89101998000200015. [DOI] [PubMed] [Google Scholar]
- Rezende HR, Cerutti C Jr, Santos CB. Aspectos atuais da distribuição geográfica de Anopheles (Kerteszia) cruzii DYAR & KNAB, 1908 no Estado do Espírito Santo, Brasil. Entomol Vectores. 2005;12:123–126. doi: 10.1590/S0328-03812005000100011. [DOI] [Google Scholar]
- Laporta GZ, Ramos DG, Ribeiro MC, Sallum MA. Habitat suitability of Anopheles vector species and association with human malaria in the Atlantic forest in south-eastern Brazil. Mem Inst Oswaldo Cruz. 2011;1:239–245. doi: 10.1590/s0074-02762011000900029. [DOI] [PubMed] [Google Scholar]
- Marrelli MT, Honorio NA, Flores-Mendoza C, Lourenco-de-Oliveira R, Marinotti O, Kloetzel JK. Comparative susceptibility of two members of the Anopheles oswaldoi complex, An. oswaldoi and An. konderi to infection by Plasmodium vivax. Trans R Soc Trop Med Hyg. 1999;93:381–384. doi: 10.1016/S0035-9203(99)90123-2. [DOI] [PubMed] [Google Scholar]
- Della Torre A, Costantini C, Besansky NJ, Caccone A, Petrarca V, Powell JR, Coluzzi M. Speciation within Anopheles gambiae–the glass is half full. Science. 2002;298:115–117. doi: 10.1126/science.1078170. [DOI] [PubMed] [Google Scholar]
- Riehle MM, Guelbeogo WM, Gneme A, Eiglmeier K, Holm I, Bischoff E, Garnier T, Snyder GM, Li X, Markianos K, Sagnon N, Vernick KD. A cryptic subgroup of Anopheles gambiae is highly susceptible to human malaria parasites. Science. 2011;331:596–598. doi: 10.1126/science.1196759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris DE. Genetic markers for study of the anopheline vectors of human malaria. Int J Parasitol. 2002;32:1607–1615. doi: 10.1016/S0020-7519(02)00189-3. [DOI] [PubMed] [Google Scholar]
- Djadid ND, Gholizadeh S, Aghajari M, Zehi AH, Raeisi A, Zakeri S. Genetic analysis of rDNA-ITS2 and RAPD loci in field populations of the malaria vector, Anopheles stephensi (Diptera: Culicidae): implications for the control program in Iran. Acta Trop. 2006;97:65–74. doi: 10.1016/j.actatropica.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Bass C, Williamson MS, Field LM. Development of a multiplex real-time PCR assay for identification of members of the Anopheles gambiae species complex. Acta Trop. 2008;107:50–53. doi: 10.1016/j.actatropica.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Paredes-Esquivel C, Donnelly MJ, Harbach RE, Townson H. A molecular phylogeny of mosquitoes in the Anopheles barbirostris Subgroup reveals cryptic species: implications for identification of disease vectors. Mol Phylogenet Evol. 2009;50:141–151. doi: 10.1016/j.ympev.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Chandra G, Bhattacharjee I, Chatterjee S. A review on Anopheles subpictus Grassi--a biological vector. Acta Trop. 2010;115:142–154. doi: 10.1016/j.actatropica.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Seah IM, Ambrose L, Cooper RD, Beebe NW. Multilocus population genetic analysis of the Southwest Pacific malaria vector Anopheles punctulatus. Int J Parasitol. 2013;43:825–835. doi: 10.1016/j.ijpara.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Coetzee M, Koekemoer LL. Molecular systematics and insecticide resistance in the major African malaria vector Anopheles funestus. Annu Rev Entomol. 2013;58:393–412. doi: 10.1146/annurev-ento-120811-153628. [DOI] [PubMed] [Google Scholar]
- Ramirez CC, Dessen EM. Chromosomal evidence for sibling species of the malaria vector Anopheles cruzii. Genome. 2000;43:143–151. doi: 10.1139/g99-103. [DOI] [PubMed] [Google Scholar]
- Ramirez CC, Dessen EM. Chromosome differentiated populations of Anopheles cruzii: evidence for a third sibling species. Genetica. 2000;108:73–80. doi: 10.1023/A:1004020904877. [DOI] [PubMed] [Google Scholar]
- Carvalho-Pinto CJ, Lourenço-de-Oliveira R. Isoenzymatic analysis of four Anopheles (Kerteszia) cruzii (Diptera: Culicidae) populations of Brazil. Mem Inst Oswaldo Cruz. 2004;99:471–475. doi: 10.1590/s0074-02762004000500002. [DOI] [PubMed] [Google Scholar]
- Rona LD, Carvalho-Pinto CJ, Gentile C, Grisard EC, Peixoto AA. Assessing the molecular divergence between Anopheles (Kerteszia) cruzii populations from Brazil using the timeless gene: further evidence of a species complex. Malar J. 2009;8:60. doi: 10.1186/1475-2875-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rona LD, Carvalho-Pinto CJ, Mazzoni CJ, Peixoto AA. Estimation of divergence time between two sibling species of the Anopheles (Kerteszia) cruzii complex using a multilocus approach. BMC Evol Biol. 2010;10:91. doi: 10.1186/1471-2148-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rona LD, Carvalho-Pinto CJ, Peixoto AA. Molecular evidence for the occurrence of a new sibling species within the Anopheles (Kerteszia) cruzii complex in south-east Brazil. Malar J. 2010;9:33. doi: 10.1186/1475-2875-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Li WH. Molecular evolution. Sunderland: Sinauer Associates; 1997. [Google Scholar]
- Hey J, Pinho C. Population genetics and objectivity in species diagnosis. Evolution. 2012;66:1413–1429. doi: 10.1111/j.1558-5646.2011.01542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala FJ, Coluzzi M. Chromosome speciation: humans, drosophila, and mosquitoes. Proc Natl Acad Sci U S A. 2005;102:6535–6542. doi: 10.1073/pnas.0501847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg L. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Nosil P. Chromosomal inversions and species differences: when are genes affecting adaptive divergence and reproductive isolation expected to reside within inversions? Evolution. 2009;63:3061–3075. doi: 10.1111/j.1558-5646.2009.00786.x. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang-Sattler R, Blandin S, Ning Y, Blass C, Dolo G, Touré YT, Delle Torre A, Lanzaro GC, Steinmetz LM, Kafatos FC, Zheng L. Mosaic genome architecture of the Anopheles gambiae species complex. PLoS One. 2007;2:e1249. doi: 10.1371/journal.pone.0001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden CD, Lee Y, Nieman CC, Sanford MR, Dinis J, Martins C, Rodrigues A, Cornel AJ, Lanzaro GC. Asymmetric introgression between the M and S forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridization. Mol Ecol. 2011;20:4983–4994. doi: 10.1111/j.1365-294X.2011.05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly MJ, Pinto J, Girod R, Besansky NJ, Lehmann T. Revisiting the role of introgression vs shared ancestral polymorphisms as key processes shaping genetic diversity in the recently separated sibling species of the Anopheles gambiae complex. Heredity. 2004;92:61–68. doi: 10.1038/sj.hdy.6800377. [DOI] [PubMed] [Google Scholar]
- Nevado B, Fazalova V, Backeljau T, Hanssens M, Verheyen E. Repeated unidirectional introgression of nuclear and mitochondrial DNA between four congeneric Tanganyikan cichlids. Mol Biol Evol. 2011;28:2253–2267. doi: 10.1093/molbev/msr043. [DOI] [PubMed] [Google Scholar]
- Khimoun A, Burrus M, Andalo C, Liu ZL, Vicédo-Cazettes C, Thébaud C, Pujol B. Locally asymmetric introgressions between subspecies suggest circular range expansion at the Antirrhinum majus global scale. J Evol Biol. 2011;24:1433–1441. doi: 10.1111/j.1420-9101.2011.02276.x. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Brys R, Honnay O, Roldan-Ruiz I. Asymmetric gene introgression in two closely related Orchis species: evidence from morphometric and genetic analyses. BMC Evol Biol. 2012;12:178. doi: 10.1186/1471-2148-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbout J, Yeh FC, Bousquet J. Large-scale asymmetric introgression of cytoplasmic DNA reveals Holocene range displacement in a North American boreal pine complex. Ecol Evol. 2012;2:1853–1866. doi: 10.1002/ece3.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beysard M, Perrin N, Jaarola M, Heckel G, Vogel P. Asymmetric and differential gene introgression at a contact zone between two highly divergent lineages of field voles (Microtus agrestis) J Evol Biol. 2012;25:400–408. doi: 10.1111/j.1420-9101.2011.02432.x. [DOI] [PubMed] [Google Scholar]
- Gomes B, Sousa CA, Novo MT, Freitas FB, Alves R, Côrte-Real AR, Salgueiro P, Donnelly MJ, Almeida AP, Pinto J. Asymmetric introgression between sympatric molestus and pipiens forms of Culex pipiens (Diptera: Culicidae) in the Comporta region. Portugal. BMC Evol Biol. 2009;9:262. doi: 10.1186/1471-2148-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantolla AU Earth’s climate history: servicio central de publicaciones del gobierno vasco 2003. 24082460
- Mirabello L, Conn JE. Population analysis using the nuclear white gene detects Pliocene/Pleistocene lineage divergence within Anopheles nuneztovari in South America. Med Vet Entomol. 2008;22:109–119. doi: 10.1111/j.1365-2915.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- O’Loughlin SM, Okabayashi T, Honda M, Kitazoe Y, Kishino H, Somboon P, Sochantha T, Nambanya S, Saikia PK, Dev V, Walton C. Complex population history of two Anopheles dirus mosquito species in South Asia suggests the influence of Pleistocene climate change rather than human- mediated effects. J Evol Biol. 2008;21:1555–1569. doi: 10.1111/j.1420-9101.2008.01606.x. [DOI] [PubMed] [Google Scholar]
- Loaiza JR, Scott ME, Bermingham E, Rovira J, Conn JE. Evidence for Pleistocene population divergence and expansion of Anopheles albimanus in southern Central America. Am J Trop Med Hyg. 2010;82:156–164. doi: 10.4269/ajtmh.2010.09-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loaiza JR, Scott ME, Bermingham E, Sanjur OI, Wilkerson R, Rovira J, Gutiérrez LA, Correa MM, Grijalva MJ, Birnberg L, Bickersmith S, Conn JE. Late Pleistocene environmental changes lead to unstable demography and population divergence of Anopheles albimanus in the northern Neotropics. Mol Phylogenet Evol. 2010;57:1341–1346. doi: 10.1016/j.ympev.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan K, Linton YM, Somboon P, Saikia P, Dev V, Socheat D, Walton C. Inter-specific gene flow dynamics during the Pleistocene-dated speciation of forest-dependent mosquitoes in Southeast Asia. Mol Ecol. 2010;19:2269–2285. doi: 10.1111/j.1365-294X.2010.04635.x. [DOI] [PubMed] [Google Scholar]
- Haffer J. Speciation in Amazonian forest birds. Science. 1969;165:131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- Vasconcelos PM, Becker TA, Renne PR, Brimhall GH. Age and duration of weathering by 40K-40Ar and 40Ar/39Ar analysis of potassium-manganese oxides. Science. 1992;258:451–455. doi: 10.1126/science.258.5081.451. [DOI] [PubMed] [Google Scholar]
- Ravelo AC, Andreasen DH, Lyle M, Olivarez Lyle A, Wara MW. Regional climate shifts caused by gradual global cooling in the Pliocene epoch. Nature. 2004;429:263–267. doi: 10.1038/nature02567. [DOI] [PubMed] [Google Scholar]
- Carnaval AC, Hickerson MJ, Haddad CF, Rodrigues MT, Moritz C. Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science. 2009;323:785–789. doi: 10.1126/science.1166955. [DOI] [PubMed] [Google Scholar]
- Thomé MT, Zamudio KR, Giovanelli JG, Haddad CF, Baldissera FA Jr, Alexandrino J. Phylogeography of endemic toads and post-Pliocene persistence of the Brazilian Atlantic forest. Mol Phylogenet Evol. 2010;55:1018–1031. doi: 10.1016/j.ympev.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Marroig G, Cropp S, Cheverud JM. Systematics and evolution of the Jacchus group of marmosets (Platyrrhini) Am J Phys Anthropol. 2004;123:11–22. doi: 10.1002/ajpa.10146. [DOI] [PubMed] [Google Scholar]
- Grazziotin FG, Monzel M, Echeverrigaray S, Bonatto SL. Phylogeography of the Bothrops jararaca complex (Serpentes: Viperidae): past fragmentation and island colonization in the Brazilian Atlantic forest. Mol Ecol. 2006;15:3969–3982. doi: 10.1111/j.1365-294X.2006.03057.x. [DOI] [PubMed] [Google Scholar]
- Conn JE, Mirabello L. The biogeography and population genetics of neotropical vector species. Heredity. 2007;99:245–256. doi: 10.1038/sj.hdy.6801002. [DOI] [PubMed] [Google Scholar]
- Mata H, Fontana CS, Maurício GN, Bornschein MR, de Vasconcelos MF, Bonatto SL. Molecular phylogeny and biogeography of the eastern Tapaculos (Aves: Rhinocryptidae: Scytalopus, Eleoscytalopus): cryptic diversification in Brazilian Atlantic Forest. Mol Phylogenet Evol. 2009;53:450–462. doi: 10.1016/j.ympev.2009.07.017. [DOI] [PubMed] [Google Scholar]
- Turchetto-Zolet AC, Cruz F, Vendramin GG, Simon MF, Salgueiro F, Margis-Pinheiro M, Margis R. Large-scale phylogeography of the disjunct Neotropical tree species Schizolobium parahyba (Fabaceae-Caesalpinioideae) Mol Phylogenet Evol. 2012;65:174–182. doi: 10.1016/j.ympev.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Lorenz-Lemke AP, Muschner VC, Bonatto SL, Cervi AC, Salzano FM, Freitas LB. Phylogeographic inferences concerning evolution of Brazilian Passiflora actinia and P. elegans (Passifloraceae) based on ITS (nrDNA) variation. Ann Bot. 2005;95:799–806. doi: 10.1093/aob/mci079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Silva C, Lexer C, Paggi GM, Barbará T, Bered F, Bodanese-Zanettini MH. Range-wide patterns of nuclear and chloroplast DNA diversity in Vriesea gigantea (Bromeliaceae), a neotropical forest species. Heredity (Edinb) 2009;103:503–512. doi: 10.1038/hdy.2009.116. [DOI] [PubMed] [Google Scholar]
- Jowett T. In: Drosophila, a practical approach. Roberts DB, editor. Oxford: IRL press; 1998. Preparation of nucleic acids; pp. 347–371. [Google Scholar]
- GenBank database. http://www.ncbi.nlm.nih.gov/BLAST/
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Filatov DA, Charlesworth D. DNA polimorphism, haplotype structure and balancing selection in the Leavenworthia PgiC locus. Genetics. 1999;153:1423–1434. doi: 10.1093/genetics/153.3.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Hey lab distributed software: evolutionary genetics; IM. 2009. http://lifesci.rutgers.edu/~heylab/HeylabSoftware.htm#IM.
- Kimura M. The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics. 1969;61:893–903. doi: 10.1093/genetics/61.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerner AE, Cox MP, Hammer MF. Recombination-filtered genomic datasets by information maximization. Bioinformatics. 2007;23:1851–1853. doi: 10.1093/bioinformatics/btm253. [DOI] [PubMed] [Google Scholar]
- Hammer lab, IMGC Online. http://hammerlab.biosci.arizona.edu/imgc_online.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NR blocks and sequences excluded from the IM analysis. Edition of sequences prior to IM analysis using the IMGC program and based on alignments presented in Additional file 3: Table S3. NR blocks, fragment positions of the non-recombining blocks used in the analyses; Removed sequences, the putative recombinant sequences removed before the IM analysis.
Polymorphism summaries of An. cruzii sibling species. RM, the minimum number of recombination events; n, number of DNA sequences of each sibling species; S, number of polymorphic (segregating) sites; θ, nucleotide diversity based on the total number of mutations (Eta); π, nucleotide diversity based on the average number of pair-wise differences; DT, Tajima’s D [28]; DFL, Fu & Li’s D [29] and FFL, Fu & Li’s F [29], based on Eta (total number of mutations). No significant deviations from neutrality were observed after Bonferroni correction. Numbers in parentheses are related to the non-recombining block (NR) for each locus.
Alignments of the DNA sequences of the timeless, Clock, cycle, Rp49, RpS2 and RpS29 gene fragments from Florianópolis and Itatiaia. The translated amino acid sequences are shown above the alignments and the introns are highlighted in grey. Dots represent identity and dashed represent gaps. Flo: individuals from Florianópolis; Ita: individuals from Itatiaia.
Summarized features of the marginal histograms for each parameter for the pairwise comparison Florianópolis vs Itatiaia.
Summarized features of the marginal histograms for each parameter for the pairwise comparison Itatiaia A vs Itatiaia B.
Summarized features of the marginal histograms for each parameter to the pairwise comparison Florianópolis vs Itatiaia A (A) and Florianópolis vs Itatiaia B (B).