Abstract
A set of 87 multicase families with systemic lupus erythemathosus (SLE) from European (Iceland, Sweden, England, Norway, Italy, and Greece) and recently admixed (Mexico, Colombia, and the United States) populations were genotyped and analyzed for 62 microsatellite markers on chromosome 1. By parametric two-point linkage analysis, six regions (1p36, 1p21, 1q23, 1q25, 1q31, and 1q43) were identified that have LOD scores of Z⩾1.50, with different contributions, depending on the population of origin of the families (European or admixed American). All of the regions have been described previously and have therefore been confirmed in this analysis. The locus at 1q31 showed a significant three-point LOD score of Z=3.79 and was contributed by families from all populations, with several markers and under the same parametric model. Analysis of a known mutation in the CD45 gene did not support the role that this mutation plays in disease. We conclude that the locus at 1q31 contains a major susceptibility gene, important to SLE in general populations.
Introduction
Systemic lupus erythemathosus (SLE [MIM 152700]) is one of several common autoimmune diseases, such as psoriasis (MIM 177900), rheumatoid arthritis (MIM 180300), Graves disease (MIM 275700), diabetes type 1 (MIM 222100), and multiple sclerosis (MIM 126200). Approximately 4%–5% of the population is affected with some type of autoimmune disease, thus making it a major health problem. Females are generally affected more often than male individuals are (Hochberg 1987; Vyse and Todd 1996).
Little is known about the predisposing factors and mechanisms leading to SLE, but some of these may involve loss of immunologic tolerance to intracellular antigens, primarily nuclear antigens. Autoantibodies to such components represent a main feature of SLE.
Family-based studies show a 10%–12% increased risk for relatives of patients with SLE, and asymptomatic relatives often have immunologic abnormalities (Hochberg 1987; Lawrence et al. 1987). The familial aggregation (λ value) for SLE is estimated to be between 20 and 58 (Hochberg 1985, 1987; Lawrence et al. 1987; Gudmundsson and Steinsson 1990; Jonsson et al. 1990). Concordance rates between MZ twins vary from 25% to 69%, which is >10 times higher than that between DZ twins (1%–2%) (Deapen et al. 1992; Jarvinen and Aho 1994). Together, these data indicate that there is a relatively high genetic component behind SLE and point toward an oligogenic background, with several susceptibility genes acting on disease expression along with environmental factors.
During recent years, six complete (Gaffney et al. 1998, 2000; Moser et al. 1998; Shai et al. 1999; Gray-McGuire et al. 2000; Lindqvist et al. 2000) and five partial (Tsao et al. 1997, 1999, 2001; Moser et al. 1999; Graham et al. 2001) scans of the human genome have been performed on multicase families with SLE. A total of 48 potential susceptibility loci have been identified. However, only six regions linked to SLE met the Lander and Kruglyak (1995) threshold of genomewide significance (maximum LOD score [Z] ⩾3.3): 1q23-24 (Moser et al. 1998), 1q41-43 (SLEB1 [MIM 601744]) (Moser et al. 1998; Tsao et al. 1999), 2q37 (SLEB2 [MIM 605218]) (Lindqvist et al. 2000), 4p16-15.2 (SLEB3 [MIM 605480]) (Moser et al. 1998), 6p21-11 (Gaffney et al. 1998), and 16q13 (Gaffney et al. 1998).
Chromosome 1 is replete with genes of relevance to immune responses, and this may be one of the reasons why linkage to SLE has been detected at so many regions on this chromosome. At least seven potential loci have been linked to SLE on chromosome 1 (identified as having a LOD score of 1.5). These are at 1p36, 1p21, 1p13, 1q21, 1q23-24, 1q25, and 1q31. Furthermore, a broad region at 1q41-43 probably consists of two or more loci. Only 1q23-24 and 1q41-43 have been linked with a significant LOD score (i.e., >3.30), and only the 1q41-43 region has been confirmed, in a different study, as having at least a suggestive LOD score (i.e., >2.20).
Not surprisingly, when different studies are compared, different results are obtained. One explanation for this divergence is that different analytical methods, genetic maps, and markers have been used. Another explanation for the divergence in results between studies is that SLE is a multifactorial and heterogeneous disease with presumably different contributing susceptibility factors. Striking differences in incidence, prevalence, and the clinical pattern between different ethnic populations found in several epidemiological studies support this view (Greenwood 1968; Samanta et al. 1992; Hopkinson et al. 1993, 1994, 2000; Adebajo and Davis 1994; Johnson et al. 1995; McCarty et al. 1995; Karlson et al. 1997; Malaviya et al. 1997; Molina et al. 1997; Wang et al. 1997; Bae et al. 1998; Molokhia et al. 2001). Although the studies have used different methods of assessment that make it difficult to compare them, a rough picture emerges in which at least African American, African Caribbean, and Chinese populations have higher incidences of SLE (two- to fourfold higher) than patients from different European populations. No epidemiological data are available for the Mexican or Mexican American populations. In general, the data indicate that environmental factors, as well as genetic heterogeneity between and within populations, influences the susceptibility to SLE. The varying results reflect, at least in part, that different studies have had access to families from different ethnicities and geographical regions. To analyze chromosome 1, we used 62 microsatellite markers in a new set of 87 multicase families from European and recently admixed American populations.
Families, Material, and Methods
Families
Eighty-seven multicase families from 11 countries in Europe and the Americas (mainly Mexico and Colombia) were recruited through international multicenter collaborations. Multicase families were included when at least two individuals in the families were considered as having SLE by fulfilling four or more of the 1982 American College of Rheumatology criteria (Tan et al. 1982). Individuals having either fewer than four clinical manifestations or other manifestations compatible with unspecific autoimmune disease and all healthy individuals aged <30 years were assigned the phenotype “unknown” in the analyses. The families participating in the study provided informed consent, and permission from the local ethical committees was obtained. The families included both nuclear families and extended pedigrees. Seven of the 10 Icelandic families and 11 of the 14 Swedish families constitute an initial set used in our previous genome scan (Lindqvist et al. 2000) and are therefore overlapping.
Genotyping
Genotyping was performed using fluorescent-labeled primers for microsatellite markers. The primers were dye-conjugated with FAM, HEX, or TET (Thermo Hybaid, Interactiva Division) or NED (Applied Biosystems). The PCR amplification was performed using a Peltier thermal cycler (PTC-225; MJ Research). Each reaction was performed in a volume of 10 μl (containing 17 ng genomic DNA, 1–2.5 mM MgCl2, 1 × PCR buffer [Applied Biosystems], 200 μM each dNTP [Amersham Pharmacia Biotech], 1.5 pmol each primer, and 0.17 U AmpliTaq Gold DNA Polymerase [Applied Biosystems]). PCR amplification consisted of an initial step at 95°C for 10 min; 9 cycles of denaturation at 95°C for 30 s, annealing at 50°C, 55°C, or 60°C for 45 s, and extension at 72°C for 45 s; 19 cycles of denaturation at 89°C for 15 s, annealing at 50°C, 55°C, or 60°C for 45 s, and extension at 72°C for 45 s; and a final step at 72°C for 7 min.
Products from up to eight different PCRs were diluted 1:10 (1:5 for products labeled with HEX and NED) and were pooled together. Two microliters of the pooled reactions were then mixed with 2 μl TAMRA size standard GS-350 or GS-500 and were diluted 1:2 with loading buffer (50 mg/ml blue dextran; 25 mM EDTA) (Applied Biosystems). The pooled reactions were denaturated at 95°C for 1 min and were loaded on an ABI 377 DNA analyzer (Applied Biosystems) with a 4% polyacrylamide gel and a 36-cm well-to-read plate. The data were collected with the GeneScan analysis software, version 3.1 (Applied Biosystems).
Alternatively, the PCR amplification was performed using ABI 877 integrated thermal cyclers (Applied Biosystems) under the same conditions as described above, and the products were pooled automatically. After pooling, 1 μl of the pooled reactions was mixed with 10 μl ROX size standard 400 HD or 500 HD, was diluted 1:56 with HI-DI formamide (Applied Biosystems), was denaturated at 95°C for 1 min, and was loaded on an ABI 3700 DNA analyzer (Applied Biosystems). The data were collected using the GeneScan analysis software, version 3.5.1 (Applied Biosystems).
Allele calling was performed using Genotyper, version 2.1 (Applied Biosystems). Inconsistent inheritance, bin, and allele calling was performed using the GAS software package, version 2.0 (Alan Young, Oxford University). The allele frequencies were conservatively calculated from the family material—first for all of the families jointly and second for each of the two geographical subgroups, with all individuals typed in the families.
Fifty-nine of the 62 markers used in this study were positioned according to the Marshfield sex-averaged genetic map (Broman et al. 1998). Three markers (D1S170, D1S171, and D1S3469) were not found in the genetic map but were located, using BLAST, in the Ensembl human genome sequence (version 5.29.1; May 12, 2002, freeze) (Hubbard et al. 2002), and were positioned in the genetic map according to their distance to neighboring markers in the genome sequence. Four of the markers (D1S1609, D1S1679, D1S2675, and D1S2844) had a different order in the genetic map compared to the genome sequence, and, for consistency, the genetic map was used.
Statistical Analysis
To use all available information from the pedigrees, we selected a parametric-LOD-score method. Two-point and multipoint LOD scores were calculated by using the ANALYZE software package, version 2.1 (Terwilliger 1995), and the FASTLINK software package, version 4.0P (Cottingham et al. 1993; Schäffer et al. 1994).
All linkage analyses were calculated under the assumption of locus heterogeneity by using the admixture test (Smith 1963), which was performed by the HOMOG routine, version 3.35, in the ANALYZE software (Ott 1991). This functions as a simple two-locus test in which only one or the other (or neither) of two unlinked susceptibility loci (with the same genetic risk and inheritance model) have an effect in each family. The proportion of families with linkage for a given marker is expressed as α.
We used three screening inheritance models, as described in our original genome-scan study of Scandinavian populations (Lindqvist et al. 2000). These were designated to fit the assumption of a “common” or a “rare” single dominant disease allele (with frequencies of 0.02 and 0.002, respectively) or a “common” single recessive disease allele (with a frequency of 0.03 in the population). To take into consideration the different prevalences for male and female patients with SLE, we established two liability classes in which females had eight times higher penetrance than males did. The male and female penetrance values, respectively, in the dominant (50% and 6.25%) and the recessive (70% and 8.75%) models were set so that they, together with the disease-allele frequency, fitted the observed prevalence of SLE found in northern European populations (Gudmundsson and Steinsson 1990). We also used the screening inheritance models applied in the genome scan by Moser et al. (1998), also using a parametric-LOD-score method. Their screening models are based on the assumption of an“autoimmune” disease allele with a frequency of p=0.10 (Bias et al. 1986). The models had the following penetrances: two models had a “high” penetrance in both sexes (90% in the dominant model and 100% in the recessive model), two models had a “low” penetrance in both sexes (50% in both the dominant model and the recessive model), and two models had a high penetrance in females and a low penetrance in males (95% and 49%, respectively, in both the dominant model and the recessive model).
Genotyping of the CD45 Exon 4 Mutation
The primers used for the detection of the CD45 (MIM 151460) exon 4 G77C mutation were as follows: forward primer, 5′-ATTTATTTTGTCCTTCTCCCA-3′, and reverse primer: 5′-GTTAACAACTTTTGTGTGCC-3′ (Thermo Hybaid). The PCR amplification was performed with a Peltier thermal cycler (PTC-225; MJ Research). Each reaction was performed in a volume of 10 μl (containing 34 ng genomic DNA, 1.5 mM MgCl2, 1 × PCR buffer [Applied Biosystems], 200 μM each dNTP [Amersham Pharmacia Biotech], 0.17 U AmpliTaq Gold DNA Polymerase [Applied Biosystems], and 1.5 pmol each primer). The PCR amplification consisted of an initial step at 95°C for 10 min; 11 cycles of denaturation at 95°C for 30 s, annealing at 54°C for 45 s, and extension at 72°C for 45 s; 22 cycles of denaturation at 89°C for 15 s, annealing at 54°C for 45 s, and extension at 72°C for 45 s; and a final step at 72°C for 7 min. The 10 μl of amplified PCR product was digested with 1 U of MspI restriction enzyme (New England Biolabs) and 1 × NE-Buffer2 (New England Biolabs), in a total volume of 15 μl. The reaction was incubated at 37°C for 3 h. The enzyme was denaturated at 64°C for 20 min. The digested DNA was blotted on a 3% SeaKem LE agarose gel (BioWhittaker Molecular Applications) and was stained with ethidium bromide.
Results
The geographical distribution and the characteristics of the families are shown in table 1. The mean female:male ratio among all affected individuals was 7.3:1. Forty-four families were from Europe, and 37 families were from the Americas, mainly from Mexico (n=30); a separate group of 6 families were Chinese from Singapore. We decided to analyze the families from the Americas together and to consider them as a group of recent admixture (Gorodezky et al. 2001; Cerda-Flores et al. 2002), in order to have a group comparable to the European families. The six Chinese families were considered only when all families were analyzed jointly. In total, 576 individuals were genotyped, of whom 191 were affected.
Table 1.
Total Number of Individuals and Number of Patients with SLE, Distributed among Countries and Geographical Areas Used in the Analysis[Note]
| Country | Families | Typed | Total AffectedIndividuals | AffectedMales | AffectedFemales | Female:MaleRatio |
| Eu: | ||||||
| Swedena | 14 | 73 | 34 | 5 | 29 | 5.8:1 |
| Icelanda | 10 | 110 | 25 | 3 | 22 | 7.3:1 |
| Italya | 6 | 40 | 12 | 3 | 9 | 3.0:1 |
| Norwaya | 6 | 29 | 13 | 0 | 13 | … |
| Englanda | 6 | 19 | 12 | 0 | 12 | … |
| Greecea | 2 | 10 | 4 | 1 | 3 | 3.0:1 |
| Overall | 44 | 281 | 100 | 12 | 88 | 7.3:1 |
| Am: | ||||||
| Mexicob | 30 | 205 | 65 | 7 | 58 | 8.3:1 |
| Colombiab,c | 4 | 40 | 7 | 0 | 7 | … |
| United Statesa,c | 2 | 8 | 5 | 0 | 5 | … |
| Jamaicac | 1 | 4 | 2 | 0 | 2 | … |
| Overall | 37 | 257 | 79 | 7 | 72 | 10.9:1 |
| Singapored | 6 | 38 | 12 | 4 | 8 | 2.0:1 |
| Overall | 87 | 576 | 191 | 23 | 168 | 7.3:1 |
Note.— Eu = European families; Am = American admixed families; All = all families.
Whites.
Mestizos (mainly Native American and Spanish admixture).
African Caribbeans/Americans.
Chinese.
In a first step, an initial screening for microsatellites along the whole of chromosome 1 was performed using 26 markers from the Cooperative Human Linkage Center/Weber screening set, version 6. The markers had ⩾80% of individuals typed, an average polymorphism information content (PIC) value of 0.73, and a mean intermarker distance of 10.81 cM.
Altogether, 17 markers gave LOD scores ⩾1.50. Together, these were distributed around seven chromosomal regions identified by either one or several markers, separated from each other by <15 cM. The results are shown in table 2.
Table 2.
Summary of Chromosomal Regions Linked to SLE, as Detected for Each Geographical Subset or for All Families Jointly[Note]
|
LOD Score (Mode, α Value) |
|||||
| Regiona | Marker | Distanceb(cM) | Eu | Am | All |
| p36.33 | D1S243 | 0 | .42 (R, .24) | 3.06 (R, .46) | 2.10 (R, .24) |
| p21.1 | D1S2626 | 136.34 | .01 (D, .98) | 1.51 (D, .42) | .51 (D, .14) |
| p21.1 | D1S1631 | 136.88 | neg | 2.17 (D, 1.00) | .27 (D, 1.00) |
| q23.1 | D1S1595 | 161.05 | .27 (R, .14) | 2.24 (R, 1.00) | 1.96 (R, .42) |
| q24.2 | D1S2844 | 175.03 | 1.65 (R, .23) | .70 (D, 1.00) | 1.38 (R, .15) |
| q25.1 | D1S1589 | 192.05 | 2.23 (D, 1.00) | .03 (R, .33) | .88 (D, 1.00) |
| q25.2 | D1S212 | 193.76 | 1.71 (D, 1.00) | .13 (R, .11) | .67 (D, 1.00) |
| q31.3 | D1S413 | 212.44 | 1.65 (R, .24) | 1.86 (R, .73) | 2.73 (R, .36) |
| q31.3 | D1S1660 | 212.44 | 1.88 (R, .25) | 2.08 (R, .37) | 3.33 (R, .26) |
| q32.1 | D1S2738 | 215.17 | 1.41 (R, .21) | 1.63 (R, .50) | 2.17 (R, .25) |
| q32.3 | D1S425 | 231.11 | 1.76 (R, .34) | 1.31 (D, .36) | 1.86 (D, .37) |
| q43 | D1S547 | 267.51 | .48 (R, .11) | .64 (R, .21) | 1.54 (R, .12) |
Note.— The values give the two-point LOD scores, assuming heterogeneity, and “neg” denotes LOD score ⩽0.0. Values in boldface italic give the highest maximum LOD score, Z, obtained for this marker. Inside the parentheses, “R” denotes recessive model, “D” denotes dominant model, and the value given is the α value at that marker. Eu = European families; Am = American admixed families; All = all families.
Cytogenetic region in Ensembl physical map.
Sex-averaged distance from the top in the Marshfield genetic map.
A region on the short arm of chromosome 1, at 1p36, was detected with one marker contributed only by American families (Z=3.06, under the recessive model). Also, a region at 1p21 showed contribution only from American families for two markers, together spanning 0.5 cM with a dominant model (Z=1.51 and Z=2.17). In the long arm of chromosome 1, the region 1q23-q24 was detected with both American (Z=2.24) and European (Z=1.65) families. Both use a recessive inheritance model, but with two different markers, together spanning an interval of 14 cM. Only families from Europe contributed to the linkage at 1q25. The linkage was positive with two markers, spanning a 2-cM interval (Z=2.23 and Z=1.71, under the dominant model). At 1q31, three markers gave the highest LOD scores when all of the families were analyzed together (Z = 2.73, Z = 3.33, and Z = 2.17), all with the same recessive model of inheritance. Linkage at 1q32.3 (Z=1.86) was identified with a single marker for all families using a dominant model contributed by both groups (Z=1.22 [European families] and Z=1.31 [American families]), but the European families showed linkage using recessive models as well (Z = 1.76). Finally, we also detected linkage with a single marker located at 1q43 (Z=1.54, under the recessive model). This marker showed linkage only when all families were analyzed jointly. Unfortunately, amplification for adjacent markers was not obtained, giving a gap of 20 cM to the nearest marker.
In a second step, 36 markers were included to fine map the 1p36 and 1q31 regions and to reduce gaps from the initial screening, giving a total set of 62 markers. With all markers combined, the average PIC value was 0.74, and the mean intermarker distance was reduced to 2.89 cM. At 1p36, a second marker, 1 cM from the original marker, confirmed the exclusive contribution by the American families (Z=1.61), but with a lower LOD score and a dominant model (table 3).
Table 3.
Fine Mapping of 1p36 by Two-Point Analysis, as Detected for Each Geographical Subset or for All Families Jointly[Note]
|
LOD Score (Mode, α Value) |
|||||
| Regiona | Marker | Distanceb(cM) | Eu | Am | All |
| p36.33 | D1S243 | 0 | .42 (R, .24) | 3.06 (R, .46) | 2.10 (R, .24) |
| p36.32 | D1S171 | 1.0c | neg | 1.61 (D, .95) | .61 (D, .16) |
| p36.32 | D1S468 | 4.22 | .06 (R, .66) | .67 (D, .56) | .53 (R, .08) |
| p36.32 | D1S2660 | 10.78 | neg | .51 (D, 1.00) | .11 (D, .07) |
| p36.31 | D1S2795 | 11.87 | .67 (R, .24) | neg | .01 (R, .02) |
| p36.31 | D1S214 | 14.04 | neg | .01 (D, 1.00) | .01 (D, 1.00) |
| p36.23 | D1S1612 | 16.22 | .04 (R, .04) | 1.37 (D, 1.00) | .82 (D, 1.00) |
| p36.22 | D1S434 | 29.93 | neg | .41 (D, 1.00) | .04 (D, 1.00) |
| p36.13 | D1S170 | 40.0c | .23 (D, 1.00) | 1.35 (D, 1.00) | 1.20 (D, 1.00) |
| p36.13 | D1S552 | 45.33 | .01 (D, 1.00) | .47 (D, 1.00) | .17 (D, 1.00) |
Note.— The values give the two-point LOD scores, assuming heterogeneity, and “neg” denotes LOD score ⩽0.0. Values in boldface italic give the highest maximum LOD score, Z, obtained for this marker. Inside the parentheses, “R” denotes recessive model, “D” denotes dominant model, and the value given is the α value at that marker. Eu = European families; Am = American admixed families; All = all families.
Cytogenetic region in the Ensembl physical map.
Sex-averaged distance from the top in the Marshfield genetic map.
Genetic distance estimated from Ensembl genome sequence.
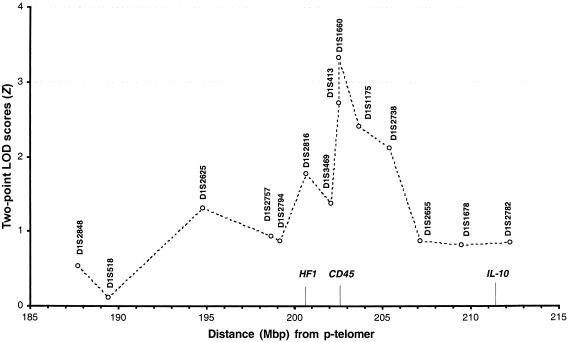
In contrast, several of the additional markers at 1q31 gave LOD scores >1.50 in all families, assuming the same recessive model as with the original markers (fig. 1). A three-point analysis was performed for all of the families, with D1S1660 and D1S1175. A maximum-multipoint-LOD-score Z value of 3.79 was obtained under the assumption of heterogeneity and with the same recessive model that gave the highest LOD scores in the two-point analyses.
Figure 1.
Fine mapping of the SLE-susceptibility locus at 1q31 in a set of 44 European (from Iceland, Sweden, England, Norway, Italy, and Greece) and 37 recently admixed American (from Mexico, Colombia, and the United States) multicase families with SLE. The markers in this figure are analyzed with a two-point LOD, assuming for heterogeneity and with the same recessive screening model. Dots are joined by lines for the sake of clarity. The markers are located according to the physical location (in Mbp) in Ensembl human sequence. The location of interesting candidate genes is indicated along the X-axis.
In a final step, since, at this point, we considered 1q31 to be a confirmed susceptibility locus for SLE, we analyzed CD45, one of the strongest candidate genes located within the region. CD45 is a protein-tyrosine-phosphatase receptor that has recently been shown to have a possible role in autoimmunity (Majeti et al. 2000). A mutation in a splicing-silencer element on exon 4, at position 77 (C/G), has recently been found that is associated with multiple sclerosis in three independent sets of German patients (Jacobsen et al. 2000). This mutation affects the splicing of exon 4, leading to the aberrant expression of CD45 isoforms on activated and memory T cells. To determine whether the C77G mutation is associated with SLE in the present set of families, we genotyped 70 patients with SLE from the families that contributed to the LOD score. The mutation was found in the heterozygous form in two of three individuals with SLE from a single Swedish family, thus excluding this mutation as a contributor to the LOD score that we observed. It cannot be excluded that other mutations presently unknown in CD45 could contribute to the linkage or that a gene nearby is involved in SLE susceptibility at this major locus.
Discussion
In the present study, we provide significant evidence for linkage to a locus at 1q31. Furthermore, our results show that there is a large extent of genetic heterogeneity at most of the loci where we observed linkage. For each locus, linkage was contributed by a small proportion of families. The exceptions were D1S1631 (on 1p21), both markers on 1q23 in American families, and both markers on 1q25 in European families. In these, all families contributed to the linkage observed.
The European families in the present study contributed minimally to the linkage at previously defined regions on chromosome 1 in U.S. populations (1p36, 1p13, 1p21, 1q23-24, and 1q41), partly explaining why our own previous genome scan on Nordic families did not detect these (Lindqvist et al. 2000). In contrast, the admixed American families unambiguously confirmed (with a threshold LOD score of Z=1.5) nearly all of the previously identified regions, with the exceptions of 1p13 and 1q41. A summary of the loci identified in the present study and in previous studies is shown in table 4.
Table 4.
LOD Scores (Z) for Markers at SLE-Susceptibility Regions in Chromosome 1, Identified in the Present Study and in Other Mapping Studies[Note]
|
LOD Score (Mode) |
|||||||
| Regiona | Marker | Distanceb(cM) | UCLAc | OKd | USCe | MNf | UPg |
| 1p36 | D1S243 | 0 | 3.06 Am (R) | ||||
| D1S468 | 4.22 | 1.58 Mex | 1.06 All | ||||
| 1p21-22 | D1S2868 | 126.16 | 1.41 All | ||||
| D1S1631 | 136.88 | 2.17 Am (D) | |||||
| 1p13 | D1S252 | 150.27 | 1.40 EA (R)h | 1.53 All | |||
| 1q23.1 | D1S1595 | 161.05 | 2.24 Am (R) | ||||
| 1q23.3 | D1S484 | 169.84 | p = .0008 | 1.51 All | |||
| FcγRIIA | 170.29 | 3.37 AA (R) | |||||
| 1q24.2 | D1S2844 | 175.03 | 1.65 Eu (R) | ||||
| 1q25 | D1S1589 | 192.05 | 2.23 Eu (D) | ||||
| Lamc1 | ∼199 | 2.04 All (D) | |||||
| 1q31 | D1S1660 | 212.44 | 3.33 All (R) | ||||
| 1q32.3 | D1S425 | 231.11 | 1.86 All (D) | ||||
| 1q41 | D1S229 | 237.73 | p = .0005 | 1.46 EA (D) | 1.33 All | ||
| D1S2616 | 239.09 | 1.23 All | |||||
| D1S549 | 239.66 | 3.30 All | |||||
| 1q42 | D1S3462 | 247.23 | 3.50 AA (D) | ||||
| D1S235 | 254.64 | 1.92 All | |||||
| 1q43 | D1S2785 | 266.27 | 2.41 All | ||||
| D1S547 | 267.51 | 1.54 All (R) | |||||
Note.— Inside the parentheses, “R” denotes recessive model, and “D” denotes dominant model. Am = admixed Americans; Mx = Mexican Americans; EA = European Americans; All = all families; AA = African Americans; Eu = Europeans.
Cytogenetic region in Ensembl physical map.
Sex-averaged distance (in cM) from the top in the Marshfield genetic map.
Shai et al. 1999. Zlr has been converted to LOD score: LOD=Z2lr/2ln10.
Present study.
Estimated from figure 2 in Moser et al. 1998.
We previously identified 1q31 in the subset of Swedish multicase families, using D1S1660 (Lindqvist et al. 2000). In the present study, this marker showed the highest LOD score with all families analyzed jointly in the two-point analysis (Z=3.33, under the recessive model). When a three-point analysis was performed using D1S1660 and the neighboring marker D1S1175, the LOD score increased to Z=3.79 for D1S1660. Two other markers in this region also gave LOD scores of Z>2.00, when the same recessive inheritance model was used.
Two interesting candidate genes are located near the highest LOD-score peak—complement factor H (HF1 [MIM 134370]) and protein-tyrosine-phosphatase receptor C (CD45, or PTPRC) (fig. 1). In fact, both of the markers (D1S1660 and D1S413) that give the highest LOD scores are placed within the CD45 gene. CD45 is involved in the regulation of the antigen-induced signaling of naive B and T cells (Trowbridge and Thomas 1994). CD45-knockout mice are severely immunodeficient because of impaired positive selection in the thymus, leading to the presence of very few T cells in the periphery. Also, the antigen-receptor–mediated signal transduction is significantly reduced in both T and B cells (Kishihara et al. 1993; Byth et al. 1996). In humans, two children with severe combined immunodeficiency have been found who have deletions in the CD45 gene (Cale et al. 1997; Kung et al. 2000; Tchilian et al. 2001b). Both children expressed very low numbers of CD45, resembling the CD45-knockout mice. Thus, the presence of CD45 is necessary for the normal functioning of the immune system. In contrast, a “knock-in” mouse with a single-point mutation in the CD45 gene caused lymphoproliferation, autoimmune nephritis, and autoantibody production (Majeti et al. 2000). Majeti et al. (2000) speculated that the mutation prevents CD45 from reducing its own positive regulatory activity, which would lower the threshold necessary for an antigen-receptor–mediated signal to start activation. This indicates that regulation of CD45 is critical and can lead to autoimmune disease if it is disturbed.
Different types of leukocytes express different numbers of multiple CD45 isoforms. The isoforms are obtained through differential splicing of exons 4–6 (Streuli et al. 1987). Normally, B cells and naive T cells express the high-molecular-weight CD45RA isoform, and activated and memory T cells express the low-molecular-weight CD45RB and CD45RO isoforms. A C77G mutation in a splicing element on exon 4 has been found to prevent normal splicing of this exon (Schwinzer and Wonigeit 1990; Thude et al. 1995; Lynch and Weiss 2001; Tchilian et al. 2001a). This results in the aberrant expression of the high-molecular-weight isoform in activated and memory T cells. Recently, this mutation was identified in patients with the autoimmune disease multiple sclerosis in three independent sets of Germans, but later studies have been unable to confirm this association (Jacobsen et al. 2000; Barcellos et al. 2001). Association was also not found in other autoimmune disorders, such as diabetes type 1 and Graves disease (Tchilian et al. 2002; Wood et al. 2002). We were also unable to find an association between the C77G mutation and the patients with SLE in the families that we studied. Abnormalities in the CD45 phosphatase activity have been described in SLE (Takeuchi et al. 1997), and other mutations may play a role in this disease. Therefore, the CD45 gene cannot be excluded as a candidate gene, and further research is required.
The second-highest LOD score (Z=3.06, under the recessive model) identified in the present study was at 1p36. Mexican families contributed most to this genetic linkage, whereas no linkage was found in the European families. This result is in agreement with previous findings made by the University of Southern California (USC) study (Shai et al. 1999), whose region was attributed to Mexican American families with a LOD score of NPL=2.7 (Z=1.58) (table 4). This result and the present study indicate that a susceptibility locus for SLE is located at 1p36 and has its greatest effect in families of Mexican origin. The TNFR2 gene (MIM 191191), located within 1p36, has been considered to be a candidate gene for this region, and studies of this gene have found a polymorphism (196M/R) associated with patients with SLE in Japan (Komata et al. 1999; Morita et al. 2001). Our multipoint analyses point toward the most telomeric side of this locus, but the lack of more microsatellite markers make it difficult for us to define the region better at this point.
Three other regions—1p21-22, 1q23, and 1q25—were unambigously identified in the present study, with LOD scores ⩾2.2, thereby confirming earlier studies (Moser et al. 1998; Shai et al. 1999; Gray-McGuire et al. 2000; Tsao et al. 2001) (table 4).
When analyzed in the American families, two markers indicated a linkage at 1p21, with the strongest signal of Z=2.17, under a dominant model. As for 1p36, this region was previously identified by the study of Mexican American and European American families (NPL=2.56 [Z=1.41]) (Shai et al. 1999). There is no evident candidate gene for this locus, but several other genome scans for other autoimmune diseases have shown linkage for multiple sclerosis (Sawcer et al. 1996) and Crohn disease (MIM 266600) (Hugot et al. 1996).
In the present study, two markers 14 cM from each other confirmed the 1q23 region. When our results are compared with earlier studies, linkage to this interval is found with LOD scores >1.5 in most studies, with two exceptions being the University of Minnesota (MN) study (Gaffney et al. 1998, 2000) and our genome scan (the University of Uppsala [UP] study) (Lindqvist et al. 2000). Most (>80%) of the families in the former study were originally from the Minnesota area, the population of which has, to a great extent, immigrated from northern Europe, and the lack of linkage in the MN study was speculated to be an effect of ethnic heterogeneity (Gaffney et al. 1998). This hypothesis is in agreement with our own previous results, which were based only on Swedish and Icelandic families and in which we did not detect linkage to 1q23. In addition, none of the northern European families included in the present study contributed to linkage to 1q23, and the European contribution to this locus that we detected was from Italians and Greeks (data not shown).
Linkage to 1q23 was first identified in the Oklahoma Medical Research Foundation (OK) study (Moser et al. 1998). The gene FcγRIIA (MIM 146790) was the marker giving the highest linkage (Z=3.37, under the recessive model) when the families were stratified for African American ancestry. Although it had a LOD score of only NPL=2.56 (Z=1.51) when all of the Mexican and European American families were analyzed together, the 1q23 susceptibility region was considered to have been confirmed by the USC study (Shai et al. 1999). The most important candidate genes in this region are the low-affinity Fcγ receptors. Previous studies (Botto et al. 1996; Salmon et al. 1996; Wu et al. 1997; Manger et al. 1998; Song et al. 1998) have identified association with FcγRIIA and FcγRIIIA (MIM 146740) in various populations, although some controversy still exists as to whether any of the low-affinity Fcγ receptors are genes for SLE susceptibility or whether the associated alleles are in linkage disequilibrium with closely linked genes.
Two markers gave linkage at 1q25 in European families, under the assumption of a dominant model, with LOD scores of Z = 2.23 and Z = 1.71, respectively. This confirms the linkage at 1q25 found in the OK study (Moser et al. 1998), by using a polymorphism in the Lamc1 gene (MIM 150290) and a dominant model on all families (Z = 2.04). Together, the markers span a region of 7 cM. This region is 22 cM telomeric from the low-affinity Fcγ receptors but is only 13 cM above 1q31 and is therefore not clearly separated from the latter region. However, we consider this to be a different region, because the highest LOD scores at 1q31 were identified with a recessive model of inheritance, whereas linkage to 1q25 was obtained with a dominant model.
In the present study, we found linkage to 1q32.3 with only one marker (D1S425). The highest LOD score was obtained for all families analyzed jointly (Z=1.86, under the dominant model). Although its contribution was from both family groups, the European families also gave a relatively high LOD score in a recessive model (Z=1.76). This could suggest the presence of genetic heterogeneity for this locus. D1S425 is located 7 cM telomeric from D1S229, which was the first marker used when linkage was found in the 1q41 region by the University of California–Los Angeles (UCLA) study (Tsao et al. 1997). The UCLA group also found association with a microsatellite located in the PARP gene (MIM 173870) (Tsao et al. 1999). Despite several studies (Gaffney et al. 1998, 2000; Moser et al. 1998, 1999; Graham et al. 2001) that also show linkage to 1q41, our own unpublished results (as well as those of Delrieu et al. [1999] and Criswell et al. [2000]) could not detect association by using the PARP gene microsatellite (data not shown). That several independent studies fail to replicate the association between PARP and SLE, together with the linkage data from the present study and most other studies, indicates that PARP is not a major contributor to this locus.
Our findings at the 1q43 region support earlier findings, mainly in Mexican families, made in the USC study (NPL=3.33 [Z=2.41]) (Shai et al. 1999). It is difficult to interpret the findings from the fine mapping in the MN study (Gaffney et al. 2000; Graham et al. 2001). This group found linkage at D1S235 (Z = 1.92), a marker that is located ∼13 cM centromeric from our finding but only 7 cM from the locus, at 1q42, that had been found in African American families in the OK study (Moser et al. 1998; Gray-McGuire et al. 2000). Clearly, thorough fine mapping is required within 1q41-43 to determine the status of this interval.
In summary, we have studied a new set of families from defined populations, and we have confirmed regions that were previously identified for SLE, on chromosome 1. We have also shown that the 1q31 region is a major SLE-susceptibility locus that may contain a major gene of importance for the development of this complex disease in general populations.
Acknowledgments
We would like to acknowledge the technical support given by Inger Jonasson, Jenny Jonsson, and Anne-Sofie Strand, from the Uppsala Genotyping Center, as well as technical help from Susanne Lindberg, Irja Johansson, and Paula Jalonen. This work was supported by grants from the Swedish Science Council (12763), BIOMED II (BMH4-98-3489), the Swedish Medical Society, the Gustav V 80-Year Jubilee Foundation, the Börje Dahlin Foundation, and the Swedish Foundation for Strategic Research. Lupus UK is also acknowledged.
The Collaborative Group on the Genetics of SLE: Antonio Iglesias, Eduardo Egea, and Gloria de Egea (Colombia); Ignacio Garcia de la Torre (Mexico); Ralph Williams Jr. (United States); Kok-Yok Fong (Singapore); Mauro Galeazzi, Sergio Migliarese, Domenico Sebastiani, and Ornella de Pitá (Italy); and K. Boki, Maria Kastorida, and H. Moutsopoulos (Greece).
The BIOMED II Collaboration on the Genetics of SLE and Sjögrens Syndrome: Helga Kristjánsdóttir, Kristján Steinsson, and Gerdur Gröndal (Iceland); Roland Jonsson and Anne-Isine Bolstad (Norway); Elisabet Svennungsson, Iva Gunnarsson, Ingrid Lundberg, Gunnar Sturfelt, and Lennart Truedsson (Sweden); and Caroline Gordon (United Kingdom).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- Ensembl Genome Browser, http://www.ensembl.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SLE [MIM 152700], psoriasis [MIM 177900], rheumatoid arthritis [MIM 180300], Graves disease [MIM 275700], diabetes type 1 [MIM 222100], multiple sclerosis [MIM 126200], SLEB1 [MIM 601744], SLEB2 [MIM 605218], SLEB3 [MIM 605480], CD45 [MIM 151460], HF1 [MIM 134370], TNFR2 [MIM 191191], Crohn disease [MIM 266600], FcγRIIA [MIM 146790], FcγRIIIA [MIM 146740], Lamc1 [MIM 150290], and PARP [MIM 173870])
References
- Adebajo A, Davis P (1994) Rheumatic diseases in African blacks. Semin Arthritis Rheum 24:139–153 [DOI] [PubMed] [Google Scholar]
- Bae SC, Fraser P, Liang MH (1998) The epidemiology of systemic lupus erythematosus in populations of African ancestry: a critical review of the “prevalence gradient hypothesis.” Arthritis Rheum 41:2091–2099 [DOI] [PubMed] [Google Scholar]
- Barcellos LF, Caillier S, Dragone L, Elder M, Vittinghoff E, Bucher P, Lincoln RR, Pericak-Vance M, Haines JL, Weiss A, Hauser SL, Oksenberg JR (2001) PTPRC (CD45) is not associated with the development of multiple sclerosis in U.S. patients. Nat Genet 29:23–24 [DOI] [PubMed] [Google Scholar]
- Bias WB, Reveille JD, Beaty TH, Meyers DA, Arnett FC (1986) Evidence that autoimmunity in man is a Mendelian dominant trait. Am J Hum Genet 39:584–602 [PMC free article] [PubMed] [Google Scholar]
- Botto M, Theodoridis E, Thompson EM, Beynon HL, Briggs D, Isenberg DA, Walport MJ, Davies KA (1996) Fc gamma RIIa polymorphism in systemic lupus erythematosus (SLE): no association with disease. Clin Exp Immunol 104:264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byth KF, Conroy LA, Howlett S, Smith AJ, May J, Alexander DR, Holmes N (1996) CD45-null transgenic mice reveal a positive regulatory role for CD45 in early thymocyte development, in the selection of CD4+CD8+ thymocytes, and B cell maturation. J Exp Med 183:1707–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cale CM, Klein NJ, Novelli V, Veys P, Jones AM, Morgan G (1997) Severe combined immunodeficiency with abnormalities in expression of the common leucocyte antigen, CD45. Arch Dis Child 76:163–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda-Flores RM, Villalobos-Torres MC, Barrera-Saldana HA, Cortes-Prieto LM, Barajas LO, Rivas F, Carracedo A, Zhong Y, Barton SA, Chakraborty R (2002) Genetic admixture in three Mexican Mestizo populations based on D1S80 and HLA-DQA1 loci. Am J Hum Biol 14:257–263 [DOI] [PubMed] [Google Scholar]
- Cottingham RW, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Criswell LA, Moser KL, Gaffney PM, Inda S, Ortmann WA, Lin D, Chen JJ, Li H, Gray-McGuire C, Neas BR, Rich SS, Harley JB, Behrens TW, Seldin MF (2000) PARP alleles and SLE: failure to confirm association with disease susceptibility. J Clin Invest 105:1501–1502 [PubMed] [Google Scholar]
- Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, Walker A, Mack TM (1992) A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum 35:311–318 [DOI] [PubMed] [Google Scholar]
- Delrieu O, Michel M, Frances C, Meyer O, Michel C, Wittke F, Crassard I, Bach JF, Tournier-Lasserve E, Piette JC, GRAID Research Group (1999) Poly(ADP-ribose) polymerase alleles in French Caucasians are associated neither with lupus nor with primary antiphospholipid syndrome. Arthritis Rheum 42:2194–2197 [DOI] [PubMed] [Google Scholar]
- Gaffney PM, Kearns GM, Shark KB, Ortmann WA, Selby SA, Malmgren ML, Rohlf KE, Ockenden TC, Messner RP, King RA, Rich SS, Behrens TW (1998) A genome-wide search for susceptibility genes in human systemic lupus erythematosus sib-pair families. Proc Natl Acad Sci USA 95:14875–14879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney PM, Ortmann WA, Selby SA, Shark KB, Ockenden TC, Rohlf KE, Walgrave NL, Boyum WP, Malmgren ML, Miller ME, Kearns GM, Messner RP, King RA, Rich SS, Behrens TW (2000) Genome screening in human systemic lupus erythematosus: results from a second Minnesota cohort and combined analyses of 187 sib-pair families. Am J Hum Genet 66:547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodezky C, Alaez C, Vazquez-Garcia MN, de la Rosa G, Infante E, Balladares S, Toribio R, Perez-Luque E, Munoz L (2001) The genetic structure of Mexican Mestizos of different locations: tracking back their origins through MHC genes, blood group systems, and microsatellites. Hum Immunol 62:979–991 [DOI] [PubMed] [Google Scholar]
- Graham RR, Langefeld CD, Gaffney PM, Ortmann WA, Selby SA, Baechler EC, Shark KB, Ockenden TC, Rohlf KE, Moser KL, Brown WM, Gabriel SE, Messner RP, King RA, Horak P, Elder JT, Stuart PE, Rich SS, Behrens TW (2001) Genetic linkage and transmission disequilibrium of marker haplotypes at chromosome 1q41 in human systemic lupus erythematosus. Arthritis Res 3:299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-McGuire C, Moser KL, Gaffney PM, Kelly J, Yu H, Olson JM, Jedrey CM, Jacobs KB, Kimberly RP, Neas BR, Rich SS, Behrens TW, Harley JB (2000) Genome scan of human systemic lupus erythematosus by regression modeling: evidence of linkage and epistasis at 4p16-15.2. Am J Hum Genet 67:1460–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood BM (1968) Autoimmune disease and parasitic infections in Nigerians. Lancet 2:380–382 [DOI] [PubMed] [Google Scholar]
- Gudmundsson S, Steinsson K (1990) Systemic lupus erythematosus in Iceland 1975 through 1984: a nationwide epidemiological study in an unselected population. J Rheumatol 17:1162–1167 [PubMed] [Google Scholar]
- Hochberg MC (1985) The incidence of systemic lupus erythematosus in Baltimore, Maryland, 1970–1977. Arthritis Rheum 28:80–86 [DOI] [PubMed] [Google Scholar]
- ——— (1987) The application of genetic epidemiology to systemic lupus erythematosus. J Rheumatol 14:867–869 [PubMed] [Google Scholar]
- Hopkinson ND, Doherty M, Powell RJ (1993) The prevalence and incidence of systemic lupus erythematosus in Nottingham, UK, 1989–1990. Br J Rheumatol 32:110–115 [DOI] [PubMed] [Google Scholar]
- ——— (1994) Clinical features and race-specific incidence/prevalence rates of systemic lupus erythematosus in a geographically complete cohort of patients. Ann Rheum Dis 53:675–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkinson ND, Jenkinson C, Muir KR, Doherty M, Powell RJ (2000) Racial group, socioeconomic status, and the development of persistent proteinuria in systemic lupus erythematosus. Ann Rheum Dis 59:116–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, et al (2002) The Ensembl genome database project. Nucleic Acids Res 30:38–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, Dupas JL, Van Gossum A, Orholm M, Bonaiti-Pellie C, Weissenbach J, Mathew CG, Lennard-Jones JE, Cortot A, Colombel JF, Thomas G (1996) Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature 379:821–823 [DOI] [PubMed] [Google Scholar]
- Jacobsen M, Schweer D, Ziegler A, Gaber R, Schock S, Schwinzer R, Wonigeit K, Lindert RB, Kantarci O, Schaefer-Klein J, Schipper HI, Oertel WH, Heidenreich F, Weinshenker BG, Sommer N, Hemmer B (2000) A point mutation in PTPRC is associated with the development of multiple sclerosis. Nat Genet 26:495–499 [DOI] [PubMed] [Google Scholar]
- Jarvinen P, Aho K (1994) Twin studies in rheumatic diseases. Semin Arthritis Rheum 24:19–28 [DOI] [PubMed] [Google Scholar]
- Johnson AE, Gordon C, Palmer RG, Bacon PA (1995) The prevalence and incidence of systemic lupus erythematosus in Birmingham, England: relationship to ethnicity and country of birth. Arthritis Rheum 38:551–558 [DOI] [PubMed] [Google Scholar]
- Jonsson H, Nived O, Sturfelt G, Silman A (1990) Estimating the incidence of systemic lupus erythematosus in a defined population using multiple sources of retrieval. Br J Rheumatol 29:185–188 [DOI] [PubMed] [Google Scholar]
- Karlson EW, Daltroy LH, Lew RA, Wright EA, Partridge AJ, Fossel AH, Roberts WN, Stern SH, Straaton KV, Wacholtz MC, Kavanaugh AF, Grosflam JM, Liang MH (1997) The relationship of socioeconomic status, race, and modifiable risk factors to outcomes in patients with systemic lupus erythematosus. Arthritis Rheum 40:47–56 [DOI] [PubMed] [Google Scholar]
- Kishihara K, Penninger J, Wallace VA, Kundig TM, Kawai K, Wakeham A, Timms E, et al (1993) Normal B lymphocyte development but impaired T cell maturation in CD45− exon6 protein tyrosine phosphatase-deficient mice. Cell 74:143–156 [DOI] [PubMed] [Google Scholar]
- Komata T, Tsuchiya N, Matsushita M, Hagiwara K, Tokunaga K (1999) Association of tumor necrosis factor receptor 2 (TNFR2) polymorphism with susceptibility to systemic lupus erythematosus. Tissue Antigens 53:527–533 [DOI] [PubMed] [Google Scholar]
- Kung C, Pingel JT, Heikinheimo M, Klemola T, Varkila K, Yoo LI, Vuopala K, Poyhonen M, Uhari M, Rogers M, Speck SH, Chatila T, Thomas ML (2000) Mutations in the tyrosine phosphatase CD45 gene in a child with severe combined immunodeficiency disease. Nat Med 6:343–345 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lawrence JS, Martins CL, Drake GL (1987) A family survey of lupus erythematosus. 1. Heritability. J Rheumatol 14:913–921 [PubMed] [Google Scholar]
- Lindqvist AK, Steinsson K, Johanneson B, Kristjánsdóttir H, Àrnasson A, Gröndal G, Jonasson I, Magnusson V, Sturfelt G, Truedsson L, Svenungsson E, Lundberg I, Terwilliger JD, Gyllensten UB, Alarcón-Riquelme ME (2000) A susceptibility locus for human systemic lupus erythematosus (hSLE1) on chromosome 2q. J Autoimmun 14:169–178 [DOI] [PubMed] [Google Scholar]
- Lynch KW, Weiss A (2001) A CD45 polymorphism associated with multiple sclerosis disrupts an exonic splicing silencer. J Biol Chem 276:24341–24347 [DOI] [PubMed] [Google Scholar]
- Majeti R, Xu Z, Parslow TG, Olson JL, Daikh DI, Killeen N, Weiss A (2000) An inactivating point mutation in the inhibitory wedge of CD45 causes lymphoproliferation and autoimmunity. Cell 103:1059–1070 [DOI] [PubMed] [Google Scholar]
- Malaviya AN, Chandrasekaran AN, Kumar A, Shamar PN (1997) Systemic lupus erythematosus in India. Lupus 6:690–700 [DOI] [PubMed] [Google Scholar]
- Manger K, Repp R, Spriewald BM, Rascu A, Geiger A, Wassmuth R, Westerdaal NA, Wentz B, Manger B, Kalden JR, van de Winkel JG (1998) Fcgamma receptor IIa polymorphism in Caucasian patients with systemic lupus erythematosus: association with clinical symptoms. Arthritis Rheum 41:1181–1189 [DOI] [PubMed] [Google Scholar]
- McCarty DJ, Manzi S, Medsger TA Jr, Ramsey-Goldman R, LaPorte RE, Kwoh CK (1995) Incidence of systemic lupus erythematosus: race and gender differences. Arthritis Rheum 38:1260–1270 [DOI] [PubMed] [Google Scholar]
- Molina JF, Molina J, Garcia C, Gharavi AE, Wilson WA, Espinoza LR (1997) Ethnic differences in the clinical expression of systemic lupus erythematosus: a comparative study between African-Americans and Latin Americans. Lupus 6:63–67 [DOI] [PubMed] [Google Scholar]
- Molokhia M, McKeigue PM, Cuadrado M, Hughes G (2001) Systemic lupus erythematosus in migrants from West Africa compared with Afro-Caribbean people in the UK. Lancet 357:1414–1415 [DOI] [PubMed] [Google Scholar]
- Morita C, Horiuchi T, Tsukamoto H, Hatta N, Kikuchi Y, Arinobu Y, Otsuka T, Sawabe T, Harashima S, Nagasawa K, Niho Y (2001) Association of tumor necrosis factor receptor type II polymorphism 196R with systemic lupus erythematosus in the Japanese: molecular and functional analysis. Arthritis Rheum 44:2819–2827 [DOI] [PubMed] [Google Scholar]
- Moser KL, Gray-McGuire C, Kelly J, Asundi N, Yu H, Bruner GR, Mange M, Hogue R, Neas BR, Harley JB (1999) Confirmation of genetic linkage between human systemic lupus erythematosus and chromosome 1q41. Arthritis Rheum 42:1902–1907 [DOI] [PubMed] [Google Scholar]
- Moser KL, Neas BR, Salmon JE, Yu H, Gray-McGuire C, Asundi N, Bruner GR, Fox J, Kelly J, Henshall S, Bacino D, Dietz M, Hogue R, Koelsch G, Nightingale L, Shaver T, Abdou NI, Albert DA, Carson C, Petri M, Treadwell EL, James JA, Harley JB (1998) Genome scan of human systemic lupus erythematosus: evidence for linkage on chromosome 1q in African-American pedigrees. Proc Natl Acad Sci USA 95:14869–14874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1991) Analysis of human genetic linkage, revised ed. Johns Hopkins University Press, Baltimore, MD [Google Scholar]
- Salmon JE, Millard S, Schachter LA, Arnett FC, Ginzler EM, Gourley MF, Ramsey-Goldman R, Peterson MG, Kimberly RP (1996) Fc gamma RIIA alleles are heritable risk factors for lupus nephritis in African Americans. J Clin Invest 97:1348–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta A, Roy S, Feehally J, Symmons DP (1992) The prevalence of diagnosed systemic lupus erythematosus in whites and Indian Asian immigrants in Leicester City, UK. Br J Rheumatol 10:679–682 [DOI] [PubMed] [Google Scholar]
- Sawcer S, Jones HB, Feakes R, Gray J, Smaldon N, Chataway J, Robertson N, Clayton D, Goodfellow PN, Compston A (1996) A genome screen in multiple sclerosis reveals susceptibility loci on chromosome 6p21 and 17q22. Nat Genet 13:464–468 [DOI] [PubMed] [Google Scholar]
- Schäffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Heredity 44:225–237 [DOI] [PubMed] [Google Scholar]
- Schwinzer R, Wonigeit K (1990) Genetically determined lack of CD45R− T cells in healthy individuals: evidence for a regulatory polymorphism of CD45R antigen expression. J Exp Med 171:1803–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai R, Quismorio FP Jr, Li L, Kwon OJ, Morrison J, Wallace DJ, Neuwelt CM, Brautbar C, Gauderman WJ, Jacob CO (1999) Genome-wide screen for systemic lupus erythematosus susceptibility genes in multiplex families. Hum Mol Genet 8:639–644 [DOI] [PubMed] [Google Scholar]
- Smith CAB (1963) Testing for heterogeneity of recombination fraction values in human genetics. Ann Hum Genet 27:175–182 [DOI] [PubMed] [Google Scholar]
- Song YW, Han CW, Kang SW, Baek HJ, Lee EB, Shin CH, Hahn BH, Tsao BP (1998) Abnormal distribution of Fc gamma receptor type IIa polymorphisms in Korean patients with systemic lupus erythematosus. Arthritis Rheum 41:421–426 [DOI] [PubMed] [Google Scholar]
- Streuli M, Hall LR, Saga Y, Schlossman SF, Saito H (1987) Differential usage of three exons generates at least five different mRNAs encoding human leukocyte common antigens. J Exp Med 166:1548–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Pang M, Amano K, Koide J, Abe T (1997) Reduced protein tyrosine phosphatase (PTPase) activity of CD45 on peripheral blood lymphocytes in patients with systemic lupus erythematosus (SLE). Clin Exp Immunol 109:20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ (1982) The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 25:1271–1277 [DOI] [PubMed] [Google Scholar]
- Tchilian EZ, Dawes R, Ramaley PA, Whitworth JA, Yuldasheva N, Wells SR, Watera C, French N, Gilks CF, Kunachiwa W, Ruzibakiev R, Leetrakool N, Carrington CV, Ramdath DD, Gotch F, Stephens HA, Hill AV, Beverley PC (2002) A CD45 polymorphism associated with abnormal splicing is absent in African populations. Immunogenetics 53:980–983 [DOI] [PubMed] [Google Scholar]
- Tchilian EZ, Wallace DL, Imami N, Liao HX, Burton C, Gotch F, Martinson J, Haynes BF, Beverley PC (2001a) The exon A (C77G) mutation is a common cause of abnormal CD45 splicing in humans. J Immunol 166:6144–6148 [DOI] [PubMed] [Google Scholar]
- Tchilian EZ, Wallace DL, Wells RS, Flower DR, Morgan G, Beverley PC (2001b) A deletion in the gene encoding the CD45 antigen in a patient with SCID. J Immunol 166:1308–1313 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD (1995) A powerful likelihood method for the analysis of linkage disequilibrium between trait loci and one or more polymorphic marker loci. Am J Hum Genet 56:777–787 [PMC free article] [PubMed] [Google Scholar]
- Thude H, Hundrieser J, Wonigeit K, Schwinzer R (1995) A point mutation in the human CD45 gene associated with defective splicing of exon A. Eur J Immunol 25:2101–2106 [DOI] [PubMed] [Google Scholar]
- Trowbridge IS, Thomas ML (1994) CD45: an emerging role as a protein tyrosine phosphatase required for lymphocyte activation and development. Annu Rev Immunol 12:85–116 [DOI] [PubMed] [Google Scholar]
- Tsao BP, Cantor RM, Grossman JM, Shen N, Teophilov NT, Wallace DJ, Arnett FC, Hartung K, Goldstein R, Kalunian KC, Hahn BH, Rotter JI (1999) PARP alleles within the linked chromosomal region are associated with systemic lupus erythematosus. J Clin Invest 103:1135–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao BP, Cantor RM, Kalunian KC, Chen CJ, Badsha H, Singh R, Wallace DJ, Kitridou RC, Chen SL, Shen N, Song YW, Isenberg DA, Yu CL, Hahn BH, Rotter JI (1997) Evidence for linkage of a candidate chromosome 1 region to human systemic lupus erythematosus. J Clin Invest 99:725–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao BP, Grossman JM, Kim SK, Wallace DJ, Chen C-J, Lau CS, Ginzler EM, Goldstein R, Shen N, Kalunian KC, Arnett FC, Hahn BH, Cantor RM (2001) Linkage and interaction of loci on 1q23 and IBD1 may contribute to susceptibility to systemic lupus erythematosus (SLE). Am J Hum Genet Suppl 69:2024 [DOI] [PubMed] [Google Scholar]
- Vyse TJ, Todd JA (1996) Genetic analysis of autoimmune disease. Cell 85:311–318 [DOI] [PubMed] [Google Scholar]
- Wang F, Wang CL, Tan CT, Manivasagar M (1997) Systemic lupus erythematosus in Malaysia: a study of 539 patients and comparison of prevalence and disease expression in different racial and gender groups. Lupus 6:248–253 [DOI] [PubMed] [Google Scholar]
- Wood JP, Bieda K, Segni M, Herwig J, Krause M, Usadel KH, Badenhoop K (2002) CD45 exon 4 point mutation does not confer susceptibility to type 1 diabetes mellitus or Graves' disease. Eur J Immunogenet 29:73–74 [DOI] [PubMed] [Google Scholar]
- Wu J, Edberg JC, Redecha PB, Bansal V, Guyre PM, Coleman K, Salmon JE, Kimberly RP (1997) A novel polymorphism of FcγRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J Clin Invest 100:1059–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]