Abstract
Africa presents the most complex genetic picture of any continent, with a time depth for mitochondrial DNA (mtDNA) lineages >100,000 years. The most recent widespread demographic shift within the continent was most probably the Bantu dispersals, which archaeological and linguistic evidence suggest originated in West Africa 3,000–4,000 years ago, spreading both east and south. Here, we have carried out a thorough phylogeographic analysis of mtDNA variation in a total of 2,847 samples from throughout the continent, including 307 new sequences from southeast African Bantu speakers. The results suggest that the southeast Bantu speakers have a composite origin on the maternal line of descent, with ∼44% of lineages deriving from West Africa, ∼21% from either West or Central Africa, ∼30% from East Africa, and ∼5% from southern African Khoisan-speaking groups. The ages of the major founder types of both West and East African origin are consistent with the likely timing of Bantu dispersals, with those from the west somewhat predating those from the east. Despite this composite picture, the southeastern African Bantu groups are indistinguishable from each other with respect to their mtDNA, suggesting that they either had a common origin at the point of entry into southeastern Africa or have undergone very extensive gene flow since.
Introduction
Archaeological and fossil evidence suggests that modern humans originated in Africa during the Middle Stone Age, during the warm phase of oxygen isotope stage 5e (115,000–130,000 years ago) or somewhat earlier (Grün and Stringer 1991). There are signs of modern human behavior from ∼70,000 years ago (Deacon 1989), which become more fully apparent from ∼40,000 years ago, with the onset of the Late Stone Age (Clark 1994). In West Africa, the Middle Stone Age and Late Stone Age are poorly understood, but East, Central, and southern Africa provide evidence of regional traditions dating back to the Acheulian, >200,000 years ago (Phillipson 1993). North Africa has had a distinct history, oriented more towards the Mediterranean, since the late Pleistocene.
The beginning of permanent settlements in Africa can be dated to ∼18,000 years ago, in the favorable environment afforded by the Nile Valley (Phillipson 1993). Agriculture and horticulture arose much later, at several different locations during the Holocene: Egypt (via diffusion from the Near East), the Ethiopian highlands, the Sahel savanna, and West Africa. Each region developed a distinct package of crops—although, with the exception of the guinea fowl (and possibly cattle: Bradley et al. 1996), there are no indigenous sub-Saharan domesticated animals. All of the African domesticates arose north of the equator and were introduced to the south relatively late.
Because of the rather poor state of archaeological understanding, especially within the tropical forest zone, linguistics has played a large role in African prehistory. Greenberg (1963) proposed that continental African languages fall into four major phyla: Niger-Congo (including the Atlantic, Mande, Voltaic, Kwa, Adamawa, and Bantu families), Nilo-Saharan (including east and central Sudanic, Saharan, and Songhai), Afroasiatic (Semitic, Berber, Cushitic, and Chadic), and Khoisan (San and Khoikhoi). It has been suggested that the initial development of the first three families took place somewhere between the Sahara and the equatorial forest (Blench 1993); Niger-Congo and Nilo-Saharan may even share a common ancestor (see Phillipson 1993). The distribution of Khoisan languages may have extended, before the Bantu diaspora, to present-day Ethiopia and Sudan. This is surmised from the presence of small groups of people speaking click-language isolates in Tanzania (Hadza and Sandawe) (Greenberg 1963; Blench 1993) and, more controversially, from the presence of click consonants in some languages in Kenya and Ethiopia (Cavalli-Sforza et al. 1994, and references therein). Also cited in support is archaeological evidence for a putative common Late Stone Age complex (the Wilton) stretching throughout southern and East Africa (a concept which has, however, received pointed criticism; e.g., from Phillipson [1993]), including skeletal remains (Huffman 1982; Newman 1995).
The great majority of subequatorial Africans (>200 million) speak one of >500 closely related Bantu languages. Both the nearest neighbors of Bantu within Niger-Congo and the highest diversity within the Bantu family itself are found in eastern Nigeria and western Cameroon, suggesting that this may have been the “core” area of the Bantu dispersal (Johnston 1913; Greenberg 1972). Using different methodological assumptions, however, Guthrie (1970) suggested that proto-Bantu originated south of the equatorial forest, and some archaeologists have denied altogether that Bantu speakers are recent arrivals in southern Africa (Lwanga-Lunyiigo 1976). Eggert (1993) points to the risk of circularity when linguistic and archaeological evidence are employed together, especially when the latter may be rather scanty.
The consensus view, however, accepts an origin in the Cross River Valley area of western Central Africa (Huffman 1982; Phillipson 1993; Vogel 1994a). Reconstructed proto-Bantu includes words for root crop cultivation and pottery manufacture, which developed in the region during the 3rd millennium b.c., but not stock keeping or iron working. It is therefore assumed that the latter were adopted following the first dispersals during the 3rd millennium. Bantu languages fall into two main sub-groups, west and east (the latter appearing to be of more recent origin), which are thought to have resulted from distinct dispersal routes (Vansina 1995). One group is thought to have moved east, north of the rainforest, although there is no archaeological trail until the Great Lakes region. The second group took a riverine route southward, through the rainforest (possibly facilitated by a more open period during the arid phase 3,000–3,500 years ago; see Maley 1993), with the island of Bioko being one of the earliest areas to be settled.
Metallurgy first appears in the Great Lakes region ∼800 b.c., by which time the presumed Bantu speakers of East Africa had already adopted stock breeding and cereal agriculture from their Nilo-Saharan–speaking neighbors (Vogel 1994b). Archaeological evidence for the “western stream” (believed to be represented by the Kalundu ceramic tradition) is rather sketchy, whereas the “eastern stream” has been studied in some detail (Phillipson 1993). It is thought to appear in the archaeological record as the Early Iron Age Chifumbaze complex to the west of Lake Victoria, ∼2,500 years ago. From this region, a number of related archaeological complexes are thought to track the dispersals of various groups of slash-and-burn agriculturalists eastwards and southwards, reaching the southern savanna ∼1,700 years ago (Phillipson 1993; Whitelaw 1994). In the process, it seems likely that indigenous Khoisan hunters and herders were gradually assimilated or displaced. Characteristic Khoisan clicks are present in several southern African Bantu languages, and Khoisan continued to thrive in the southern regions (which experience a winter-rainfall regime unsuitable for equatorial crops) until the arrival of European colonists in the 15th century. The two streams are thought to have met and intermingled in south Central Africa. By the 7th century a.d., summer-rainfall cultivation using domesticates, originating in equatorial Africa—in combination with stock-breeding (in particular, cattle), metal-working, and new forms of social organization—was spread widely throughout sub-equatorial Africa.
Although progress has undeniably been made, controversy remains on both the issue of proto-Bantu origins and the dispersals themselves; the links between archaeology and linguistics remain largely circumstantial (Phillipson 1993). Furthermore, questions remain concerning the numbers of people involved in the process. For instance, Hiernaux (1968) proposed that the Bantu exodus was initiated by small groups of agriculturists, whereas others favor larger groups (Kuper and van Leynseele 1978). Bakel (1981) suggested that the expansion probably started with a very small group of people with a moderate rate of population growth, whereas Phillipson (1993) has commented: “There can be little doubt that the Chifumbaze complex…was introduced into subequatorial Africa as a result of a substantial and rapid movement of population.”
The contribution of genetics to the debate has only recently begun to take shape. Classical markers suggested that the most important genetic gradient in Africa is north–south, although some indication of the Bantu expansions may be present in the second and fourth principal components (Cavalli-Sforza et al. 1994). However, rather little has emerged from these analyses (MacEachern 2000); it is molecular studies of nonrecombining markers that are likely to have the greatest impact. Both mtDNA and the nonrecombining part of the Y chromosome (NRY) have been used to characterize African populations in recent years.
Studies of mtDNA variation initially focused on low-resolution RFLPs of the entire molecule (Scozzari et al. 1988; Stine et al. 1992) or variation in one or both hypervariable segments (HVS-I and HVS-II) of the fast-evolving control region (Vigilant et al. 1989, 1991; Watson et al. 1996; Bandelt and Forster 1997). More recently, several studies have used high-resolution RFLPs (Chen et al. 1995, 2000), and some have adopted a combined approach (Graven et al. 1995; Soodyall et al. 1996; Watson et al. 1997; Passarino et al. 1998; Alves-Silva et al. 2000; Bandelt et al. 2001; Pereira et al. 2001). Interest has usually focused on the debate concerning modern human origins, although, more recently, the question of the Bantu expansions has received some attention.
Several mtDNA markers have been proposed as signals of Bantu dispersals, although often in the absence of any southern Bantu data. Bandelt et al. (1995) and Chen et al. (1995) suggested haplogroup L1a, part of which (defined by a 9-bp intergenic deletion) was confirmed as an important eastern Bantu marker by Soodyall et al. (1996). Watson et al. (1997) similarly proposed a subset of haplogroup L3b. Subsequently, Alves-Silva et al. (2000) and Bandelt et al. (2001) have proposed (on the basis of analyses of Brazilian mtDNA data) that fragments of haplogroups L2, L3e, and L1e may also be important Bantu mtDNA markers. Bandelt and Forster (1997) highlighted the Khoisan mtDNA pool, which primarily includes members of the ancient haplogroups L1d and L1k, suggesting that extant San groups represent a small and recent splinter from a widespread and ancient Khoisan population (see also Soodyall and Jenkins 1992; Soodyall 1993). (A similar relationship between the Mandenka and the wider West African mtDNA pool was pointed out by Graven et al. 1995.) Pereira et al. (2001) focused specifically on southeastern African Bantu-speaking populations. They found reduced diversity, in comparison with East and West Africans, and confirmed the roles of L1a (both with and without the 9-bp intergenic deletion), L3b, and L3e in the Bantu dispersals. They also highlighted the important role of L2a and estimated a Khoisan assimilation rate in southeast Bantu speakers of ∼5% (L1d). Using L2a, they estimated a founder time of 4,600–16,500 years ago.
Progress has also been made with Y-chromosome analyses. Scozzari et al. (1999) and Underhill et al. (2000, 2001) have proposed that parts of haplogroup E (using the nomenclature of the Y Chromosome Consortium [2002]) have dispersed widely and rather recently through subequatorial Africa and are likely to signal Bantu dispersals. Lineages within this haplogroup form the great majority of NRY lineages in southern African Bantu speakers. Thomas et al. (2000) estimated an expansion time of ∼3,000–5,000 years, on the basis of five microsatellites.
In the present article, we add substantially to the existing data on southeast Bantu speakers by providing HVS-I sequences and complementary RFLP typings of 307 samples from 16 ethnic groups from Mozambique. These allow us to make more precise date estimates, enabling us to test more thoroughly whether the ages of various Bantu-specific subclades are consistent with participation in recent dispersals. Furthermore, we have carried out a comprehensive phylogeographic analysis of all published mtDNA HVS-I sequences, informed by complementary studies that have allocated particular sequence types to well-defined clades in the mtDNA phylogeny (Ingman et al. 2000; Maca-Meyer et al. 2001), on the basis of complete sequences. This allows us to place evidence for more recent demographic changes in sub-Saharan Africa within a Middle and Late Stone Age chronological context, organized by the geographic distribution and dating of various clades (or haplogroups) and subclades within the genealogy. We then attempt to trace the various southeastern Bantu-associated lineages back through the continent, to estimate the extent to which potential source regions in West, Central, and East Africa have contributed to the composition of present-day southeastern Bantu maternal lineages. Finally, we compare our results with recent work on the Y-chromosome genealogy.
Materials and Methods
Sample Collection and DNA Extraction
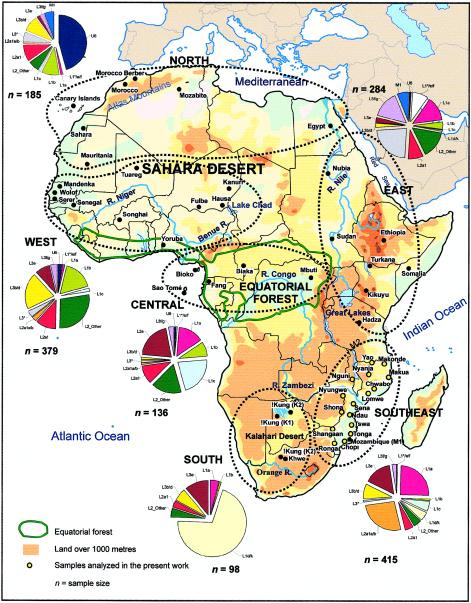
Blood samples were obtained from 307 unrelated individuals belonging to 16 different population groups from southeastern Africa: 10 Yao, 20 Tongas, 22 Shangaan, 27 Chopi, 20 Chwabo, 20 Lomwe, 19 Makonde, 20 Makhwa, 10 Ndau, 11 Nguni, 20 Nyungwe, 20 Nyanja, 21 Ronga, 18 Shona, 21 Sena, and 19 Tswa; all were indigenous Bantu speakers. The geographic origin of each individual's four grandparents and the language of each individual's mother were recorded. The samples were collected mainly in Mozambique, where most of these populations are settled, and, in some cases, in boundary areas (fig. 1; table 1). Appropriate informed consent was obtained from all individuals used in the present study. DNA was extracted as described elsewhere (Salas et al. 1998). Other published data used are listed in table 1. We excluded some sequences from Vigilant et al. (1991) from most analyses, because of sequence ambiguities, and made use of the data of Soodyall (1993) only in a general way, because of the high error rate they seem to exhibit. Where necessary, African populations were grouped geographically into six main continental regions: North, West, East, Central, southeastern, and southern Africa (see table 1; fig. 1). One hundred twenty-three L-types, from a Eurasian HVS-I database of >15,000 individuals, were also considered.
Figure 1.
Map of Africa showing the samples used in the present work. The pie charts represent the haplogroup composition of the main African regions, combining some sub-clades for convenience, and excluding the contribution of haplogroups of non-African origin. Population codes are as defined in table 1.
Table 1.
African Samples Used in the Present Study[Note]
| Ethnic Group (Code) | Geographical Origin | n | Reference | Language | LanguageFamily(Branch)a |
| America: | |||||
| “African American” (Af) | USA | 110 | Parsons et al./Armed Forces Institute of Pathology (see hvrBase Web site) and Vigilant et al. (1991) | English | IE |
| “White Brazilian” (B1) | Brazil | 245 | Alves-Silva et al. (2001) | Portuguese | IE |
| African–Brazilian (B2) | Brazil, S and NE | 42 | Bortolini et al. (1997) | Portuguese | IE |
| Chocó (Co) | Colombia | 49 | Authors' unpublished data | Spanish | IE |
| Mexican (Mx) | Mexico | 87 | Green et al. (2000) | Spanish | IE |
| Garífuna (Ga) | Belize, Panama | 44 | Authors' unpublished data | Spanish | IE |
| Caribbean (Cr) | Caribbean | 28 | Monsalve et al. (1997) | Spanish (?) | IE |
| Dominican (St) | Santo Domingo, Dominican Republic | 127 | Torroni et al. (2001) and Torroni et al. (unpublished data) | Spanish | IE |
| North Africa: | |||||
| West Saharan (Sh) | Western Sahara | 25 | Rando et al. (1998) | ND | ND |
| Mauritanian (Ma) | Mauritania | 30 | Rando et al. (1998) | ND | ND |
| Moroccan (Mc) | Morocco; Souss Valley | 50 | Brakez et al. (2001) | ND | ND |
| Berber (Mo) | Morocco, N | 60 | Rando et al. (1998) | Berber | AA |
| Moroccan, not Berber (Mr) | Morocco, N | 32 | Rando et al. (1998) | Non-Berber/Arabic (?) | AA |
| Egyptian (Eg) | Egypt | 68 | Krings et al. (1999) | Arabic/Coptic | AA |
| Mozabite (Mb) | Algeria | 85 | Côrte-Real et al. (1996) | Berber | AA |
| Canarian (Ca) | Canary Islands | 300 | Pinto et al. (1996) and Rando et al. (1998) | Spanish | IE |
| West Africa: | |||||
| Hausa (Ha) | Nigeria, Niger | 20 | Watson et al. (1997) | Hausa (Chadic) | AA |
| Kanuri (Ka) | Nigeria, Niger | 14 | Watson et al. (1997) | Kanuri (Saharan) | NS |
| Fulbe (Fu) | Nigeria, Niger, Benin, Cameroon, Burkina Faso | 60 | Watson et al. (1997) | Fufulbe (West Atlantic) | NK |
| Songhai (So) | Nigeria, Niger, Mali | 10 | Watson et al. (1997) | Songhai | NS |
| Tuareg (Tu) | Nigeria, Niger, Mali | 23 | Watson et al. (1997) | Tamasheq (Berber) | AA |
| Yoruba (Yo) | Nigeria | 34 | Watson et al. (1997) and Vigilant et al. (1991) | Yoruba | NK |
| Senegalese (Sn) | Senegal | 50 | Rando et al. (1998) | Various | NK |
| Serer (Sr) | Senegal | 23 | Rando et al. (1998) | Serer (?) | NK |
| Wolof (Wo) | Senegal | 48 | Rando et al. (1998) | French/Arabic (?) | NK |
| Mandenka (Mn) | Senegal | 119 | Graven et al. (1995) | Mande | NK |
| Central Africa: | |||||
| Bubi (Bo) | Equatorial Guinea | 45 | Mateu et al. (1997) | Bantu | NK |
| São Tomé (Sa) | São Tomé and Príncipe | 50 | Mateu et al. (1997) | Portuguese/Creole | IE |
| Fang (Fg) | Equatorial Guinea | 11 | Pinto et al. (1996) | Bantu | NK |
| Mbuti (Mt) | Democratic Republic of Congo | 20 | Vigilant et al. (1991) | Nilo-Saharan | NS |
| Biaka (Bi) | Central African Republic | 17 | Vigilant et al. (1991) | Bantu | NK |
| East Africa: | |||||
| Hadza (Hz) | Tanzania | 12 | Vigilant et al. (1991) | Hadza (“Khoisan”) | KH |
| Turkana (Tk) | Kenya | 37 | Watson et al. (1997) | Turkana (East Sudanic) | NS |
| Somalian (Sm) | Kenya, Somalia, Ethiopia | 27 | Watson et al. (1997) | Somali (Cushitic) | AA |
| Kikuyu (Ki) | Kenya | 24 | Watson et al. (1997) | Kikuyu (Bantu) | NK |
| Nubian (Nu) | Sudan, Egypt | 80 | Krings et al. (1999) | Nubian | NS |
| Nuba, Shilio, Duba, Nuer (Su) | Sudan, S | 76 | Krings et al. (1999) | Nilotic | NS/NK |
| Ethiopian (Et) | Ethiopia | 74 | Thomas et al. (2002) | Amharic | AA |
| Southeastern Africa: | |||||
| Bantu-speaking people (M1) | Mozambique, | 109 | Pereira et al. (2001) | Bantu | NK |
| Yao (a) | Mozambique, N; Tanzania, S | 10 | Present study | Yao (Bantu) | NK |
| Tonga (b) | Mozambique, SE | 20 | Present study | Gitonga (Bantu) | NK |
| Shangaan (c) | Mozambique, SW; Zimbabwe, S; South Africa, NE; Swaziland | 22 | Present study | Shangaan (Bantu) | NK |
| Chopi (d) | Mozambique, SE | 27 | Present study | Chopi (Bantu) | NK |
| Chwabo (e) | Mozambique, NE | 20 | Present study | Chwabo (Bantu) | NK |
| Lomwe (f) | Mozambique, NE | 20 | Present study | Lomwe (Bantu) | NK |
| Makonde (g) | Mozambique, N; Tanzania, S | 19 | Present study | Makonde (Bantu) | NK |
| Makhuwa (h) | Mozambique, NE | 20 | Present study | Makhuwa (Bantu) | NK |
| Ndau (i) | Mozambique, Central E | 19 | Present study | Ndau (Bantu) | NK |
| Nguni (j) | Mozambique, W; Malawi, E | 11 | Present study | Zulu (Bantu) | NK |
| Nyungwe (k) | Mozambique, W; Zimbabwe, E | 20 | Present study | Nyungwe (Bantu) | NK |
| Nyanja (l) | Mozambique, N; Malawi, E | 20 | Present study | Nyanja (Bantu) | NK |
| Ronga (m) | Mozambique, S; Swaziland | 21 | Present study | Ronga (Bantu) | NK |
| Shona (n) | Mozambique, Central; Zimbabwe, E | 18 | Present study | Shona (Bantu) | NK |
| Sena (o) | Mozambique, Central | 21 | Present study | Sena (Bantu) | NK |
| Tswa (p) | Mozambique, SE | 19 | Present study | Tswa (Bantu) | NK |
| Southern Africa: | |||||
| !Kung (K1) | Botswana | 24 | Vigilant et al. (1991) | Zhu/twasi (Khoisan) | KH (north) |
| !Kung (K2) | South Africa | 43 | Chen et al. (2000) | Khoisan | KH (south?) |
| Khwe (Kw) | South Africa | 31 | Chen et al. (2000) | Khoisan | KH (south?) |
Note.— Additional codes for Euroasiatic samples used in figures 4–9: Ch = China; Bd = Bedouin; Ye = Yemen; Iq = Iraq; Sy = Syria; Jw = west Jordan; Je = east Jordan; Pl = Palestine; Ty = Turkey; Ku = Kurds; Bl = Bulgaria; Ka = Kabardia; Ab = Albania; Ts = Tuscany; Rm = Rome; Tr = Sicily; Sd = Sardinia; Gal = Galicia (Spain); Bs = Basque Country (Spain); Pt = Portugal; Sp = Spain; Sw = Sweden; Po = Poland; Ge = Germany; Ny = Norway; Ic = Iceland; Fr = France; Sco = Scotland (UK); Fn = Finland. Code M2 will be used to refer to the whole Mozambique sample new to the present work (all ethnic groups taken together).
NS= Nilo-Saharan; AA = Afro–Asiatic; IE = Indo–European; NK = Niger–Kordofanian; KH = Khoisan; ND = not determined or not reported by the author; ? = the language is assumed but not clearly specified in the corresponding reference.
Amplification and Sequencing of HVS-I
HVS-I amplification was carried out in a Perkin Elmer 480-A Thermocycler. The temperature profile for 32 cycles of amplification was 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. Primers and PCR strategy were as described by Wilson et al. (1995). PCR product purification and sequencing were performed as in Salas et al. (1998). For those sequences containing a homopolymeric cytosine stretch from positions 16184 to 16193 (usually associated with length heteroplasmy), additional amplification and sequencing were performed using primers L16209 (5′-CCC CAT GCT TAC AAG CAA GT-3′) and H16164 (5′-TTT GAT GTG GAT TGG GTT T-3′).
Restriction Enzyme Analysis
All samples were analyzed for six selected RFLP sites, which help distinguish major African mtDNA haplogroups: 2349 MboI (L3e), 3592 HpaI (L1 and L2), 8616 MboI (L3d), 10084 TaqI (L3b), 10397 AluI (M), and 10871 MnlI (N). Sequences belonging to haplogroup L2 were additionally typed for 3693 MboI (L2d), 4157 AluI (L2b), 13803 HaeIII (L2a), and 13957 HaeIII (L2c) (Torroni et al. 2001). The resulting fragments were resolved through electrophoresis in standard polyacrylamide gels followed by silver staining.
Phylogeographic and Population-Genetic Analyses
For all data analyzed, a string of 276 bp belonging to HVS-I, from positions 16090 to 16365 (Anderson et al. 1981), was used. However, RFLP and HVS-II data (and some HVS-I information outside this region) available for some subset of samples were used in situations in which HVS-I information alone was insufficient to allocate a sequence to its haplogroup. Where we refer to some position out of this standard region, it is indicated by the polymorphism within brackets. Length variation will not be considered in the present article. Transitions in HVS-I are recorded, in the tables and figures, by their position in the Cambridge Reference Sequence (CRS) (Anderson et al. 1981) minus 16,000; transversions are accompanied by a suffix specifying the variant base.
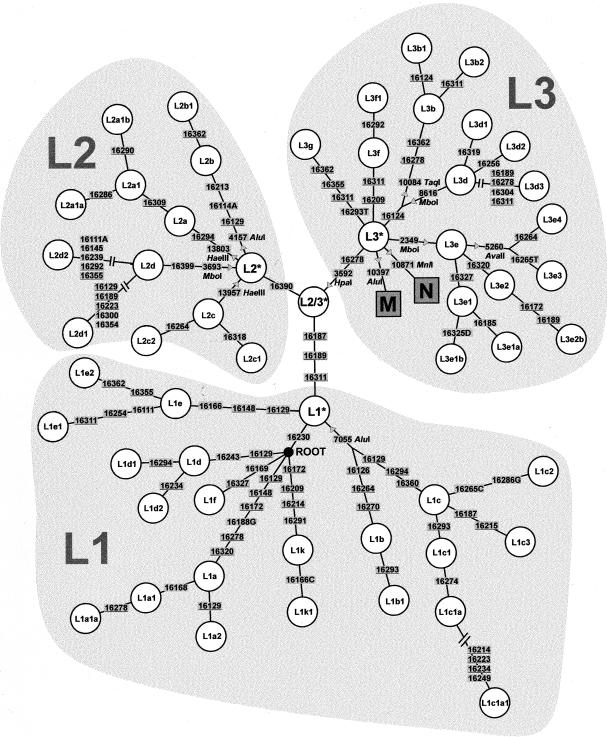
The haplogroup classification in the present article (shown in the skeleton tree of fig. 2) is based on the phylogenetic analyses performed by Chen et al. (1995), Watson et al. (1997), Rando et al. (1998, 1999), Quintana-Murci et al. (1999), Alves-Silva et al. (2000), Bandelt et al. (2001), Pereira et al. (2001), and Torroni et al. (2001). mtDNA clades that originated in sub-Saharan Africa include L1a through L1k, L2, and L3A (defined as all members of L3 not included in haplogroups M or N: Rando et al. 1998). These are here informally referred to as “L-haplogroups” or “L-types,” although they do not together form a clade. Haplogroup U6 is predominantly North African (Macaulay et al. 1999; Rando et al. 1998), whereas M1 may have originated in East Africa (Quintana-Murci et al. 1999).
Figure 2.
mtDNA skeleton showing a schematic phylogeny of African haplogroups used in the present paper to classify HVS-I sequences. The skeleton includes HVS-I and some coding-region RFLPs (with an arrow pointing in the direction of site gain).
We compiled a database of published L-type HVS-I sequences (both African and non-African) and used them to construct phylogenetic networks (Bandelt et al. 1995, 1999), by hand and by use of the program Network 3.1 (Bandelt et al. 1999). Haplogroup U6 and M1 sequences were also compiled. We developed a strategy for approximating the phylogeny by combining smaller subnetworks, in the following manner:
-
1.
A preliminary allocation of the database sequences to haplogroups was carried out according to the existing mtDNA skeleton (Richards and Macaulay 2000; Pereira et al. 2001).
-
2.
A more detailed skeleton network was estimated for each haplogroup, using only those sequence types present more than once in the database (see Richards et al. 1996).
-
3.
This was confirmed by repeating step (2) after eliminating substitutions at hypermutable positions, to obtain a phylogeny based exclusively on stable positions.
-
4.
All HVS-I sequences in the database were screened and grouped according to the motifs identified from these analyses.
-
5.
Networks were constructed for these related grouped sequences using all the variable positions in the haplotypes and the RM algorithm.
The networks were further resolved, in many cases, using additional information—for instance, on the mutability of different positions. It should be noted that subclades identified solely on the basis of fast-evolving sites in HVS-I, such as 16293 in certain contexts, may need revision in the light of future complete-sequence data.
The time to the most recent common ancestor (TMRCA) of clades in the phylogeny was estimated as described by Forster et al. (1996) and Saillard et al. (2000). Founder times were estimated for each of the major southeastern African founder types, identified as sequence types matching types in the potential source regions (of East and West Africa). For a detailed discussion of the assumptions involved, see Richards et al. (2000).
Sequence diversity was estimated as [n/(n-1)](1-Σki=1p2i), where pi is the frequency of each of the k different sequences in the sample. Haplogroup diversity was similarly calculated. The average number of nucleotide pairwise differences, the number of segregating sites, and Tajima’s D statistic (Tajima 1989) were evaluated. Principal component (PC) analysis was performed on the basis of the haplogroup composition (relative frequencies) in the various population samples (considering L-haplogroups plus U6 and M1). Initially, all populations were included. Subsequently, outliers in North, Central, and southern Africa, as well as the very small sample of Biaka, were excluded. The apportionment of genetic variation between and within populations was estimated by AMOVA (Excoffier et al. 1992), by means of the Arlequin package.
Results
Genetic Diversity in Southeastern African Populations
We determined HVS-I mtDNA sequences from each of the 307 southeastern African individuals (table 2). Of the 115 different HVS-I mtDNA sequence types, 67 (58%; 95% credible region [CR] .491–.669) were found only once, whereas several occur at high frequencies: three types (from haplogroups L1a and L2) account for 32% (95% CR .270–.373) of the total. These three types are common in almost all of the 16 populations analyzed.
Table 2.
HVS-I Sequences and Coding-Region RFLPs in 307 Individuals Belonging to 16 Different Bantu-Speaking Populations from Southeastern Africa[Note]
|
RFLP Typing |
Populations |
||||||||||||||||||||||||||||
| Haplotype ID | HVS-I Haplotypes | 2349j | 3592h | 8616j | 10084l | 10397a | 10871z | 3693j | 4157a | 13803e | 13957e | a | b | c | d | e | f | g | h | i | j | k | l | m | n | o | p | Sample Size | Haplogroup |
| H001 | 111 223 266 290 319 362 | − | − | + | − | − | + | # | # | # | # | 1 | 1 | A | |||||||||||||||
| H002 | 129 148 172 187 188G 189 223 230 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1a | |||||||||||||||
| H003 | 129 148 172 187 189 223 230 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1a | |||||||||||||||
| H004 | 129 148 168 172 187 188G 189 223 230 305 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1a1 | |||||||||||||||
| H005 | 129 148 168 172 187 188G 189 223 230 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | 2 | L1a1 | ||||||||||||||
| H006 | 093 129 148 168 172 187 188G 189 223 230 278 293 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | 2 | L1a1a | ||||||||||||||
| H007 | 129 148 168 172 187 188A 189 223 230 278 293 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | 1 | 1 | 4 | L1a1a | ||||||||||||
| H008 | 129 148 168 172 187 188G 189 209 223 230 278 293 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1a1a | |||||||||||||||
| H009 | 129 148 168 172 187 188G 189 223 230 239 278 293 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1a1a | |||||||||||||||
| H010 | 129 148 168 172 187 188G 189 223 230 278 293 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | 1 | 2 | 1 | 1 | 3 | 2 | 1 | 1 | 14 | L1a1a | ||||||
| H011 | 129 148 168 172 187 188G 189 223 230 278 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | 1 | 1 | 1 | 5 | L1a1a | |||||||||||
| H012 | 093 148 172 187 188G 189 214 223 230 289 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | 2 | L1a2 | ||||||||||||||
| H013 | 093 148 172 187 188G 189 214 223 230 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1a2 | |||||||||||||||
| H014 | 093 148 172 187 188G 189 223 230 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | 2 | 4 | L1a2 | |||||||||||||
| H015 | 148 172 187 188G 189 223 230 256 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1a2 | |||||||||||||||
| H016 | 148 172 187 188G 189 223 230 311 320 | − | + | + | − | − | − | # | # | # | # | 3 | 2 | 3 | 3 | 1 | 7 | 3 | 6 | 4 | 1 | 2 | 3 | 1 | 1 | 6 | 46 | L1a2 | |
| H017 | 148 172 188A 189 223 230 311 320 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1a2 | |||||||||||||||
| H018 | 126 187 189 223 264 270 278 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | 2 | L1b | ||||||||||||||
| H019 | 126 187 189 209 223 264 270 278 293 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1b1 | |||||||||||||||
| H020 | 126 187 189 223 264 270 278 293 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1b1 | |||||||||||||||
| H021 | 129 187 189 212 223 278 294 311 360 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1c | |||||||||||||||
| H022 | 117 129 172 173 188A 189 223 256 278 291 293 294 311 360 368 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1c1 | |||||||||||||||
| H023 | 129 163 187 189 209 223 278 293 294 311 360 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1c1 | |||||||||||||||
| H024 | 129 172 173 188A 189 223 256 278 293 294 311 360 368 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1c1 | |||||||||||||||
| H025 | 129 187 189 223 274 278 292G 293T 296delC 311 360 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1c1a | |||||||||||||||
| H026 | 129 187 189 223 274 278 293 294 311 360 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | 2 | L1c1a | ||||||||||||||
| H027 | 093 129 187 189 223 265C 278 286G 294 311 320 360 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1c2 | |||||||||||||||
| H028 | 129 145 187 189 213 223 234 265C 278 286G 294 311 360 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | 1 | 3 | L1c2 | |||||||||||||
| H029 | 129 169 187 189 223 265C 278 286G 294 311 360 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1c2 | |||||||||||||||
| H030 | 129 187 189 260 265C 286A 294 311 360 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1c2 | |||||||||||||||
| H031 | 093 114 129 183C 189 215 223 278 294 311 355 360 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1c3 | |||||||||||||||
| H032 | 129 182C 183C 189 215 223 278 294 311 360 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1c3 | |||||||||||||||
| H033 | 129 183C 189 215 223 278 294 311 360 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | 2 | L1c3 | ||||||||||||||
| H034 | 069 129 169 189 212 223 230 243 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1d | |||||||||||||||
| H035 | 129 145 187 189 223 230 243 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1d | |||||||||||||||
| H036 | 129 169 189 223 230 243 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1d | |||||||||||||||
| H037 | 129 172 187 189 212 223 230 243 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1d | |||||||||||||||
| H038 | 129 187 189 223 230 243 284C 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1d | |||||||||||||||
| H039 | 129 187 189 223 230 243 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1d | |||||||||||||||
| H040 | 187 189 223 230 243 274 278 290 300 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1d | |||||||||||||||
| H041 | 187 189 223 230 243 311 343 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1d | |||||||||||||||
| H042 | 129 179 187 189 223 230 243 290 311 362 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1d | |||||||||||||||
| H043 | 129 187 189 223 230 239 243 294 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | 2 | L1d1 | ||||||||||||||
| H044 | 129 189 223 230 239 243 294 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1d1 | |||||||||||||||
| H045 | 187 189 223 230 234 243 294G 311 | − | + | + | − | − | − | # | # | # | # | 1 | 1 | L1d2 | |||||||||||||||
| H046 | 189 192 223 278 294 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | 1 | 3 | L2a | |||||||||||||
| H047 | 223 234 249 278 294 295 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | 2 | L2a | ||||||||||||||
| H048 | 129 189 223 278 293 294 309 311 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1 | |||||||||||||||
| H049 | 131 183C 189 223 225 234 278 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1 | |||||||||||||||
| H050 | 182C 183C 189 223 278 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1 | |||||||||||||||
| H051 | 182C 183C 189 192 223 278 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1 | |||||||||||||||
| H052 | 189 192 223 278 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 2 | 3 | L2a1 | ||||||||||||||
| H053 | 189 223 278 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | 2 | L2a1 | ||||||||||||||
| H054 | 223 278 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 8 | L2a1 | |||||||||
| H055 | 085 189 192 223 270 278 286 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1a | |||||||||||||||
| H056 | 092 223 278 286 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1a | |||||||||||||||
| H057 | 223 278 286 291 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1a | |||||||||||||||
| H058 | 223 278 286 294 309 390 | − | + | + | − | − | − | + | − | + | + | 5 | 2 | 2 | 2 | 4 | 2 | 2 | 4 | 1 | 24 | L2a1a | |||||||
| H059 | 223 278 286 294 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1a | |||||||||||||||
| H060 | 092 182C 183C 189 223 278 290 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1b | |||||||||||||||
| H061 | 150 182C 183C 223 278 290 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1b | |||||||||||||||
| H062 | 168 182C 183C 189 223 278 290 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1b | |||||||||||||||
| H063 | 182C 183C 189 192 223 278 290 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | 2 | 1 | 5 | L2a1b | ||||||||||||
| H064 | 182C 183C 189 223 264 278 290 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1b | |||||||||||||||
| H065 | 182C 183C 189 223 278 290 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1b | |||||||||||||||
| H066 | 182C 183C 189 223 278 290 294 309 320 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1b | |||||||||||||||
| H067 | 182C 183C 189 223 278 290 294 309 390 | − | + | + | − | − | − | + | − | + | + | 4 | 4 | 5 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 28 | L2a1b | |||
| H068 | 182C 183C 189 223 290 294 309 390 | − | + | + | − | − | − | + | − | + | + | 1 | 1 | L2a1b | |||||||||||||||
| H069 | 114A 129 213 223 274 278 390 | − | + | + | − | − | − | + | + | − | + | 1 | 1 | 2 | L2b | ||||||||||||||
| H070 | 114A 129 213 223 278 354 390 | − | + | + | − | − | − | + | + | − | + | 1 | 1 | 2 | L2b | ||||||||||||||
| H071 | 223 264 278 311 390 | − | + | + | − | − | − | + | − | − | − | 1 | 1 | 2 | L2c | ||||||||||||||
| H072 | 129 189 223 278 300 354 390 399 | − | + | + | − | − | − | − | − | − | + | 1 | 1 | L2d1 | |||||||||||||||
| H073 | 129 189 278 300 311 354 390 399 | − | + | + | − | − | − | − | − | − | + | 1 | 1 | 2 | L2d1 | ||||||||||||||
| H074 | 223 | − | − | + | − | − | − | # | # | # | # | 2 | 1 | 1 | 4 | L3* | |||||||||||||
| H075 | 093 124 223 278 309 362 | − | − | + | + | − | − | # | # | # | # | 1 | 1 | L3b | |||||||||||||||
| H076 | 093 124 223 278 362 | − | − | + | + | − | − | # | # | # | # | 1 | 1 | 2 | L3b | ||||||||||||||
| H077 | 124 148 223 278 362 | − | − | + | + | − | − | # | # | # | # | 1 | 1 | L3b | |||||||||||||||
| H078 | 124 223 261 362 | − | − | + | + | − | − | # | # | # | # | 1 | 1 | L3b | |||||||||||||||
| H079 | 124 223 278 362 | − | − | + | + | − | − | # | # | # | # | 1 | 1 | L3b | |||||||||||||||
| H080 | 124 223 278 305 311 362 | − | − | + | + | − | − | # | # | # | # | 1 | 1 | L3b2 | |||||||||||||||
| H081 | 124 223 278 311 362 | − | − | + | + | − | − | # | # | # | # | 1 | 1 | L3b2 | |||||||||||||||
| H082 | 093 124 223 | − | − | − | − | − | − | # | # | # | # | 1 | 1 | L3d | |||||||||||||||
| H083 | 124 182C 183C 189 223 | − | − | − | − | − | − | # | # | # | # | 1 | 1 | 2 | L3d | ||||||||||||||
| H084 | 124 223 | − | − | − | − | − | − | # | # | # | # | 1 | 1 | 1 | 3 | L3d | |||||||||||||
| H085 | 124 223 278 | − | − | − | − | − | − | # | # | # | # | 1 | 1 | L3d | |||||||||||||||
| H086 | 124 223 292 | − | − | − | − | − | − | # | # | # | # | 1 | 1 | L3d | |||||||||||||||
| H087 | 124 223 284C 319 | − | − | − | − | − | − | # | # | # | # | 1 | 1 | L3d1 | |||||||||||||||
| H088 | 124 223 319 | − | − | − | − | − | − | # | # | # | # | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 10 | L3d1 | |||||||||
| H089 | 124 223 319 362 | − | − | − | − | − | − | # | # | # | # | 1 | 1 | L3d1 | |||||||||||||||
| H090 | 124 183C 189 223 243 278 304 311 | − | − | − | − | − | − | # | # | # | # | 1 | 1 | L3d3 | |||||||||||||||
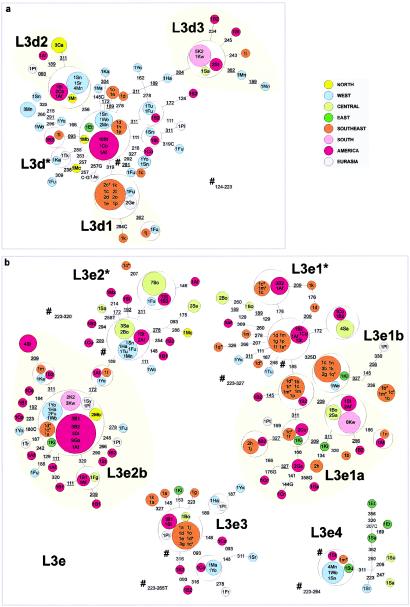
| H091 | 176 209 223 327 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e1 | |||||||||||||||
| H092 | 176 223 327 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e1 | |||||||||||||||
| H093 | 209 223 327 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e1 | |||||||||||||||
| H094 | 223 278 327 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e1 | |||||||||||||||
| H095 | 223 327 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | 1 | 1 | 1 | 5 | L3e1 | |||||||||||
| H096 | 134 185 209 223 311 327 | + | − | + | − | − | − | # | # | # | # | 2 | 2 | L3e1a | |||||||||||||||
| H097 | 185 186 209 223 327 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e1a | |||||||||||||||
| H098 | 185 209 223 311 320 327 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e1a | |||||||||||||||
| H099 | 185 223 311 | + | − | + | − | − | − | # | # | # | # | 2 | 1 | 3 | L3e1a | ||||||||||||||
| H100 | 185 223 311 327 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e1a | |||||||||||||||
| H101 | 185 223 327 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | 1 | 1 | 4 | L3e1a | ||||||||||||
| H102 | 223 239 325delT | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e1b | |||||||||||||||
| H103 | 223 325delT 327 | + | − | + | − | − | − | # | # | # | # | 3 | 1 | 2 | 1 | 1 | 8 | L3e1b | |||||||||||
| H104 | 183C 189 223 320 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e2 | |||||||||||||||
| H105 | 172 183C 189 223 320 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e2b | |||||||||||||||
| H106 | 172 183C 189 223 311 320 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e2b | |||||||||||||||
| H107 | 145 223 265T | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e3 | |||||||||||||||
| H108 | 223 265T | + | − | + | − | − | − | # | # | # | # | 1 | 1 | 1 | 3 | 1 | 1 | 8 | L3e3 | ||||||||||
| H109 | 223 265T 327 | + | − | + | − | − | − | # | # | # | # | 1 | 1 | 2 | L3e3 | ||||||||||||||
| H110 | 265T | + | − | + | − | − | − | # | # | # | # | 1 | 1 | L3e3 | |||||||||||||||
| H111 | 129 166 209 223 311 | − | − | + | − | − | − | # | # | # | # | 1 | 1 | L3f | |||||||||||||||
| H112 | 129 209 223 311 | − | − | + | − | − | − | # | # | # | # | 1 | 1 | L3f | |||||||||||||||
| H113 | 209 223 291 311 | − | − | + | − | − | − | # | # | # | # | 2 | 2 | L3f | |||||||||||||||
| H114 | 209 223 311 | − | − | + | − | − | − | # | # | # | # | 2 | 1 | 3 | L3f | ||||||||||||||
| H115 | 209 218 223 256 292 311 | − | − | + | − | − | − | # | # | # | # | 1 | 1 | L3f1 | |||||||||||||||
| Total | 10 | 20 | 22 | 27 | 20 | 20 | 19 | 20 | 19 | 11 | 20 | 20 | 21 | 18 | 21 | 19 | 307 | ||||||||||||
Note.— Variant positions from the CRS are shown, from 16024 to 16408 (minus 16000). The columns after each HVS-I sequence type represent the status for the enzymes used in the present work (see below). All individuals were typed for the first six restriction polymorphisms indicated in the table, and only those sequences belonging to haplotype L2 were typed for four additional positions. A plus sign (+) indicates the presence of the restriction site, a minus sign (−) indicates its absence, and a number sign (#) indicates that its presence or absence was not determined. Haplogroups were assigned according to HVS-I motifs and RFLP status, as discussed in the text. Enzyme codes: a = AluI; e = HaeIII; h = HpaI; j = MboI; l = TaqI; and z = MnlI. Code for the populations: see table 1.
To summarize the variability accumulated in the southeastern African sequences, sequence diversity was computed for all the populations analyzed and was compared with that of other African populations (table 3). Most populations analyzed here showed similar values for this index, ranging from 1.000 in Nguni to 0.890 in Sena. Almost all these ethnic groups displayed similar diversity; the average sequence-diversity value for the southeastern African sample is 0.957. These numbers are comparable with those of other African regions (see for instance, western and eastern populations in table 3). The average number of pairwise nucleotide differences (M=8.03 for the whole sample; table 3) was also similar to those of other sub-Saharan populations. Tajima’s D values are slightly negative but are nonsignificant.
Table 3.
Diversity Indices for HVS-I in African (or African-Influenced) Populations[Note]
| Population | Code | n | K(K/n)a | S(S/l)b | H (SE)c | Md | De |
| North Africa: | |||||||
| West Saharans | Sh | 25 | 20 (80.0) | 28 (10.1) | .973 (.022) | 5.09 | −1.17 |
| Mauritanian | Ma | 30 | 22 (73.3) | 28 (10.1) | .970 (.018) | 5.83 | −.63 |
| Moroccan (Souss region) | Mc | 50 | 33 (66.0) | 34 (12.3) | .959 (.018) | 4.30 | −1.52 |
| Berber (Morocco) | Mo | 60 | 38 (63.3) | 47 (17.0) | .963 (.015) | 4.44 | −1.97 |
| Moroccan not Berber | Mr | 32 | 29 (90.6) | 44 (15.9) | .988 (.014) | 5.84 | −1.70 |
| Egypt | Eg | 68 | 57 (83.8) | 62 (22.5) | .989 (.006) | 6.61 | −1.72 |
| Berber (Algeria) | Mb | 85 | 29 (34.1) | 35 (12.7) | .942 (.010) | 4.73 | −1.02 |
| Canarians | Ca | 300 | 127 (42.3) | 89 (32.2) | .965 (.006) | 4.89 | −2.02f |
| West Africa: | |||||||
| Hausa | Ha | 20 | 19 (95.0) | 30 (10.9) | .995 (.018) | 5.77 | −1.25 |
| Kanuri | Ka | 14 | 13 (92.9) | 32 (11.6) | .989 (.031) | 6.90 | −1.35 |
| Fulbe | Fu | 60 | 38 (54.3) | 43 (15.6) | .972 (.010) | 6.82 | −.98 |
| Songhai | So | 10 | 9 (90.0) | 28 (10.1) | .978 (.054) | 8.49 | −.68 |
| Tuareg | Tu | 23 | 21 (91.3) | 39 (14.1) | .992 (.015) | 6.75 | −1.40 |
| Yoruba | Yo | 34 | 31 (91.2) | 43 (15.6) | .995 (.009) | 7.25 | −1.18 |
| Senegalese | Sn | 50 | 42 (84.0) | 41 (14.9) | .989 (.008) | 6.24 | −1.08 |
| Serer | Sr | 23 | 21 (91.3) | 40 (14.5) | .992 (.015) | 8.09 | −.98 |
| Wolof | Wo | 48 | 39 (81.3) | 42 (15.2) | .991 (.006) | 7.50 | −.84 |
| Mandenka | Mn | 119 | 47 (39.5) | 48 (17.4) | .965 (.007) | 5.62 | −1.24 |
| Central Africa: | |||||||
| Bubi | Bo | 45 | 16 (35.5) | 30 (10.9) | .910 (.020) | 7.09 | .11 |
| São Tomé | Sa | 50 | 32 (64.0) | 46 (16.7) | .973 (.011) | 7.86 | −.92 |
| Fang | Fg | 11 | 10 (90.9) | 30 (10.9) | .982 (.046) | 8.36 | −.85 |
| Mbuti Pygmy | Mt | 20 | 9 (45.0) | 16 (5.8) | .858 (.054) | 5.12 | .50 |
| Biaka Pygmy | Bi | 17 | 8 (47.1) | 20 (7.2) | .890 (.043) | 7.81 | 1.27 |
| East Africa: | |||||||
| Turkana | Tk | 37 | 33 (89.2) | 54 (19.6) | .991 (.010) | 9.52 | −1.05 |
| Somali | Sm | 27 | 24 (88.9) | 41 (14.9) | .991 (.013) | 6.90 | −1.32 |
| Kikuyu | Ki | 24 | 22 (91.7) | 45 (16.3) | .993 (.014) | 8.17 | −1.30 |
| Nubia | Nu | 80 | 47 (58.8) | 61 (22.1) | .970 (.009) | 7.42 | −1.41 |
| Sudan | Su | 76 | 63 (82.9) | 73 (26.4) | .993 (.004) | 8.33 | −1.58 |
| Ethiopia | Et | 74 | 62 (83.8) | 73 (26.4) | .994 (.003) | 8.43 | −1.66 |
| Southeastern Africa: | |||||||
| Yao | a | 10 | 8 (80.0) | 20 (7.2) | .933 (.077) | 7.16 | .06 |
| Tonga | b | 20 | 14 (70.0) | 29 (10.5) | .947 (.034) | 7.45 | −.35 |
| Shangaan | c | 22 | 17 (77.3) | 35 (12.7) | .961 (.029) | 8.52 | −.53 |
| Chopi | d | 27 | 18 (66.6) | 32 (11.6) | .954 (.025) | 7.15 | −.79 |
| Chwabo | e | 20 | 15 (75.0) | 30 (10.9) | .942 (.043) | 7.76 | −.32 |
| Lomwe | f | 20 | 12 (60.0) | 28 (10.1) | .879 (.065) | 8.06 | −.06 |
| Makonde | g | 19 | 12 (63.2) | 25 (9.1) | .942 (.032) | 6.90 | −.42 |
| Makhwa | h | 20 | 12 (60.0) | 32 (11.6) | .905 (.053) | 9.06 | −.21 |
| Ndau | i | 19 | 15 (78.9) | 30 (10.9) | .959 (.036) | 8.82 | −.15 |
| Nguni | j | 11 | 11 (100) | 21 (7.6) | 1.000 (.039) | 7.91 | .24 |
| Nyungwe | k | 20 | 16 (80.0) | 34 (12.3) | .974 (.025) | 9.03 | −.34 |
| Nyanja | l | 20 | 12 (60.0) | 25 (9.1) | .937 (.033) | 7.53 | .11 |
| Ronga | m | 21 | 18 (85.7) | 36 (13.0) | .986 (.019) | 8.57 | −.56 |
| Shona | n | 18 | 16 (88.9) | 37 (13.4) | .987 (.023) | 8.86 | −.81 |
| Sena | o | 21 | 11 (52.4) | 22 (8.0) | .890 (.049) | 7.00 | .55 |
| Tswa | p | 19 | 16 (84.2) | 26 (9.4) | .977 (.027) | 7.01 | −.23 |
| SE Africa Bantu | M2 | 307 | 115 (37.5) | 72 (26.1) | .957 (.006) | 8.03 | −1.09 |
| SE Africa Bantu | M1 | 109 | 49 (44.9) | 57 (20.7) | .960 (.008) | 7.76 | −1.05 |
| Southern Africa: | |||||||
| !Kung | K1 | 24 | 9 (37.5) | 16 (5.8) | .830 (.053) | 2.97 | −1.10 |
| !Kung | K2 | 43 | 12 (27.9) | 31 (11.2) | .812 (.045) | 7.30 | −.04 |
| Khwe | Kw | 31 | 10 (32.6) | 34 (12.3) | .884 (.029) | 8.75 | .10 |
Note.— For sources, see table 1.
K = number of different sequences found and percentage of sample size in brackets.
S = number of segregating sites and percentage of all sites in brackets. l = length of the sequence (in bp).
H = sequence diversity.
M = average number of pairwise differences.
D = Tajima’s D statistic.
.01<P⩽.05. All other values of D were not significantly different from zero.
PC Analysis
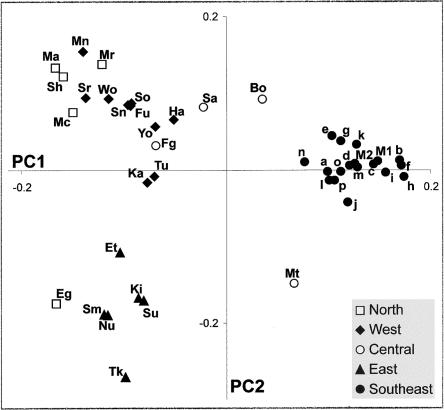
The most striking feature of the PC plot (fig. 3) is the tight clustering of all the southeastern African populations through the first PC (PC1), which accounts for 15% of the variance; even those populations with small sample sizes form part of this cluster. This indicates the high similarity of these ethnic groups, as suggested by the diversity indices and their distribution in the phylogenetic networks. Their closest neighbors in PC1 are the Mbuti, and the Bioko and São Tomé islanders, all classified as Central Africans in the present analysis. (By contrast, the Equatorial Guinean Fang fall within the West African cluster in both PC1 and PC2.) East, West, and even North Africans cluster together towards the opposite pole. The main haplogroups responsible for PC1 are the western L1b, the southeastern L2a1b, the southeastern/eastern L1a, and the eastern L3*.
Figure 3.
Plot showing the first two principal components of haplogroup frequency profiles for the African samples (population codes as in table 1).
PC2 (11% of the genetic variance), by contrast, clusters southeastern Africans with West Africans and clusters the Mbuti with East Africans. Again, North Africans tend to cluster with West Africans, suggesting that the sub-Saharan component of North Africans originates primarily from West rather than East Africa (as expected, on geographical grounds). Unlike other North Africans, Egyptians are closer to East than to West Africans. (Note that, if Eurasian haplogroups were included, North and West Africans would be much more clearly distinguished, since, in the former, the major contribution is from European and Near Eastern mtDNAs [Rando et al. 1999].) PC2 has a large contribution from the eastern lineage groups L3g and L3*; however, L2a, L1b1a, and L3e2* also make a similar contribution.
Apportionment of Genetic Variance
The AMOVA analysis performed on the 16 Bantu-speaking populations analyzed in the present work showed that almost all the genetic variation (98.8%) was found to be within populations, with the remaining 1.2% between populations (but not significantly different from 0; P=.103). These results again reflect the very high level of genetic homogeneity among these populations.
AMOVA analysis was also applied to the whole African data set, using several designs:
-
1.
Taking all the African populations separately, 79.2% of the variability occurs within populations, whereas 20.8% of the variability occurs between populations.
-
2.
Grouping the populations by main geographic areas, 10.6% between groups, 12.5% between populations within groups, and 76.9% for variance within groups.
-
3.
Considering the main groups of African languages (Afroasiatic, Niger-Congo, Nilo-Saharan, and Khoisan), similar values were obtained for the variation within groups (76.8%), but 18.9% was found to occur between populations within groups, with the remaining 4.3% corresponding with differences between groups. (This last was not significantly different from 0; P=.068.)
-
4.
When populations were grouped into Bantu versus non-Bantu, a similar apportionment of genetic variation was found: 74.9% within populations, 17.2% among populations within groups, and 7.9% among groups.
Therefore, it seems that, in Africa, geography plays an important role in defining differences between the main groups, whereas language plays a lesser role.
Phylogeography of African mtDNA Variation
We have constructed phylogenetic networks for each of the major sub-Saharan African (“L-type”) haplogroups, to investigate the mtDNA data for phylogeographical patterns (figs. 4–9). All sequences were first classified on the basis of HVS-I motifs and any available HVS-II or RFLP data (table 4). With the exception of one sequence (H001) belonging to the east Asian/Native American haplogroup A (which exactly matches one individual from the Brazilian sample of Alves-Silva et al. 2000), all southeastern African sequence types could be classified as L-types.
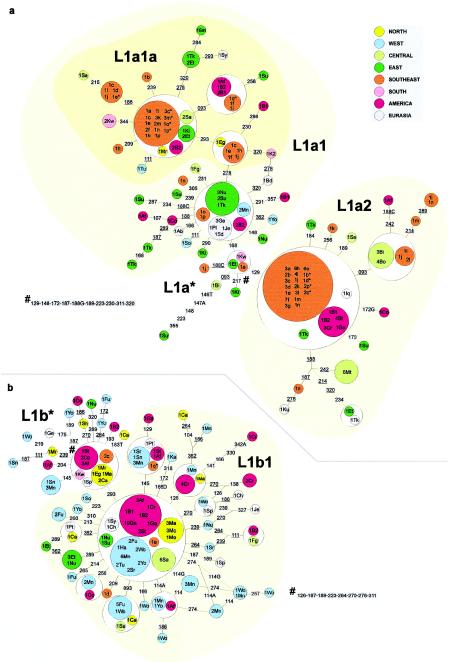
Figure 4.
Networks of (a) L1a and (b) L1b lineages. Circle sizes are proportional to the haplotype frequency in the sample.
Figure 9.
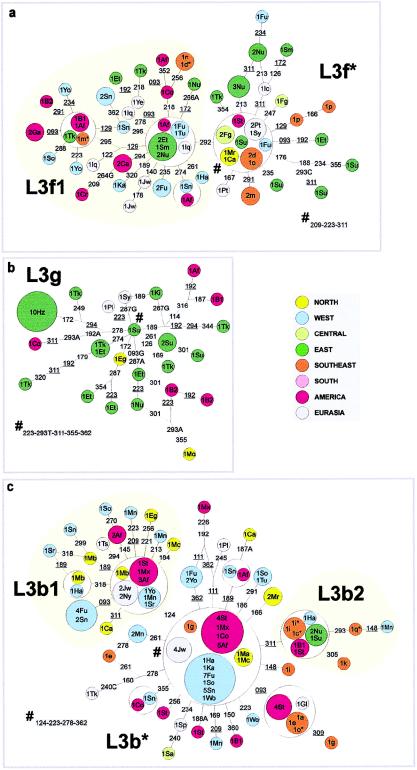
Networks of (a) L3d and (b) L3e lineages
Table 4.
Haplogroup Composition of Southeastern African Samples[Note]
|
Ethnic Group |
||||||||||||||||||
| Haplogroup | a | b | c | d | e | f | g | h | i | j | k | l | m | n | o | p | qa | Total (Frequency)b |
| A | 1 | 1 (.0024) | ||||||||||||||||
| L1a* | 1 | 1 | 2 (.0048) | |||||||||||||||
| L1a1* | 1 | 1 | 1 | 3 (.0072) | ||||||||||||||
| L1a1a | 1 | 1 | 3 + 3 | 1 + 1 | 2 + 1 | 4 | 3 | 2 | 2 | 3 | 1 | 2 + 3 | 1 | + 1 | 1 + 1 | 27 + 10 (.0889) | ||
| L1a2 | 3 | 2 + 1 | 3 + 2 | 3 + 1 | 1 | 7 | 3 | 6 | 6 | 2 | 3 | 5 | 2 | 3 | 6 | + 2 | 55 + 6 (.1466) | |
| L1b* | 1 | 1 | 2 (.0048) | |||||||||||||||
| L1b1 | 1 | 1 | + 1 | 2 + 1 (.0072) | ||||||||||||||
| L1c* | 1 | 1 (.0024) | ||||||||||||||||
| L1c1* | + 1 | + 1 | 1 | 1 | 1 | 3 + 2 (.0120) | ||||||||||||
| L1c1a | 1 | 1 | 1 | 3 (.0072) | ||||||||||||||
| L1c2 | + 1 | 2 | 1 | 1 | 1 | 1 | + 1 | 6 + 2 (.0192) | ||||||||||
| L1c3 | 1 | 1 | 1 | 1 | + 1 | 4 + 1 (.0120) | ||||||||||||
| L1d* | 1 | 1 + 1 | 1 | 1 + 1 | 2 + 2 | 1 | 1 + 1 | + 1 | 8 + 6 (.0335) | |||||||||
| L1d1 | 1 | 1 + 1 | 2 | 4 + 1 (.0120) | ||||||||||||||
| L1d2 | 1 | + 1 | 1 + 1 (.0048) | |||||||||||||||
| L1e2 | + 1 | + 1 | + 2 (.0048) | |||||||||||||||
| L2a* | 1 | + 1 | 1 | 1 | 3 | 1 | 7 + 1 (.0192) | |||||||||||
| L2a1* | 1 + 2 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 3 | 1 + 1 | 2 + 1 | + 1 | 17 + 5 (.0529) | |||||
| L2a1a | + 1 | + 6 | 1 | 5 | 2 | 1 | 2 + 1 | 2 | 4 | 3 + 4 | 2 | 4 | 2 | + 1 | 28 + 13 (.0986) | |||
| L2a1b | 7 + 2 | 4 + 10 | 7 + 5 | 2 + 1 | 4 | 2 + 2 | 1 | 1 | 1 | 2 + 4 | 2 | 2 + 1 | 3 + 2 | + 1 | 38 + 28 (.1587) | |||
| L2b* | 1 | 1 | 1 | 1 + 1 | + 1 | 4 + 2 (.0144) | ||||||||||||
| L2c2 | + 1 | + 1 (.0024) | ||||||||||||||||
| L2c* | 1 | 1 | 2 (.0048) | |||||||||||||||
| L2d1 | 1 | 1 | 1 | 3 (.0072) | ||||||||||||||
| L3* | 2 | 1 | 1 | 4 (.0096) | ||||||||||||||
| L3b* | 1 | 2 | 2 | 1 | + 1 | 6 + 1 (.0168) | ||||||||||||
| L3b2 | + 1 | 1 + 1 | 1 | + 1 | 2 + 3 (.0120) | |||||||||||||
| L3d* | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 8 (.0192) | ||||||||||
| L3d1 | 1 + 2 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 12 + 2 (.0336) | |||||||||
| L3d3 | 1 | 1 (.0024) | ||||||||||||||||
| L3e1* | 1 + 1 | 2 + 1 | + 1 | 1 | 1 | 1 | 2 + 1 | 1 | + 1 | 9 + 5 (.0336) | ||||||||
| L3e1a | 2 | 1 + 1 | + 1 | 1 | 4 | + 1 | 1 | 1 + 1 | 1 | 1 | 12 + 4 (.0384) | |||||||
| L3e1b | 4 + 1 | 1 + 1 | 2 | 1 | + 1 | 1 | + 1 | 9 + 4 (.0312) | ||||||||||
| L3e2* | 1 | 1 (.0024) | ||||||||||||||||
| L3e2a | + 1 | + 1 (.0024) | ||||||||||||||||
| L3e2b | 1 | + 1 | + 1 | 1 | 2 + 2 (.0096) | |||||||||||||
| L3e3 | 3 | + 1 | 1 + 1 | 1 | 4 | 1 | 1 | 1 | 12 + 2 (.0312) | |||||||||
| L3e4 | + 1 | + 1 (.0024) | ||||||||||||||||
| L3f* | 2 | 2 | 1 | 2 | 7 (.0168) | |||||||||||||
| L3f1 | + 1 | + 1 | 1 | 1 + 2 (.0072) | ||||||||||||||
| Total | 10 | 20 + 8 | 22 + 35 | 27 + 12 | 20 + 3 | 20 | 19 | 20 + 2 | 19 + 4 | 12 | 20 | 20 | 21 + 21 | 18 | 21 + 4 | 19 + 8 | + 12 | 307 + 109 = 416 |
Note.— Number after “+” indicates those individuals taken from Pereira et al. (2001).
q includes those individuals from Pereira et al. (2001) who lacked a clear assignment to some ethnic group; it includes Manhembane, Suwase, Fulana, and other uncharacterized individuals.
Last column indicates total numbers for each row and in brackets the corresponding frequencies for each haplogroup, calculated over the total number of individuals (n=416).
Our study is hampered by the lack of sufficient characters for maximal resolution of the genealogy and will benefit when complete mtDNA sequences become widely available. However, an advantage is the large number of African HVS-I data now available, which have not been brought together in a single analysis before (although substantial analyses have been made of North African data and of a few individual haplogroups [Bandelt et al. 2001; Torroni et al. 2001]). Furthermore, these can now be interpreted in the light of an improved knowledge of the topology of the African mtDNA tree that has come from combined HVS/RFLP studies and from the published complete (or nearly complete) sequences (Ingman et al. 2000; Maca-Meyer et al. 2001; Torroni et al. 2001; Herrnstadt et al. 2002).
Outgroup comparisons with chimpanzees for complete sequences (Ingman et al. 2000; Maca-Meyer et al. 2001) and with Neanderthals for HVS-I and HVS-II (Krings et al. 1997, 2000; Ovchinnikov et al. 2000) indicate that the root of the mtDNA tree lies within the cluster of sub-Saharan mtDNAs called “L1” by Chen et al. (1995). This paraphyletic group (“paragroup” in the terminology of Brehm et al. 2002) has been subdivided into a number of clades: L1a, L1b’c, L1d, L1e, L1f, and L1k (fig. 2). The remainder of the phylogeny consists of haplogroups L2 and L3. L3 includes all Eurasian variation, as well as much variation that is exclusively African. L3 includes the sub-Saharan paragroup L3*, two sub-Saharan haplogroups, L3b’d and L3e, and two Eurasian haplogroups, M and N (Quintana-Murci et al. 1999; Richards and Macaulay 2000). Here, we adopt the convention of Rando et al. (1998), referring to L3A to distinguish the African L3 lineages from haplogroups M and N.
The distribution of the major indigenous African haplogroups across the different regions of the continent is displayed in figure 1, and divergence times are shown in table 5. Divergence times may be underestimates, since it is unlikely that all recurrent mutations can be reconstructed at these time depths.
Table 5.
TMRCA, Sequence Diversity, and Average Number of Pairwise Differences of mtDNA L-Haplogroups
| Haplogroup | TMRCA (SE) | Ha (SE) | Mb |
| L1a | 40,350 (16,250) | .881 (.021) | 2.49 |
| L1a1 | 33,350 (16,600) | .699 (.079) | 1.50 |
| L1a2 | 8,300 (3,650) | .542 (.059) | 1.11 |
| L1a1a | 27,350 (17,950) | .724 (.058) | 1.23 |
| L1b | 30,550 (16,250) | .853 (.024) | 1.88 |
| L1c | 59,650 (11,800) | .968 (.010) | 5.53 |
| L1c1 | 53,350 (12,750) | .933 (.034) | 3.80 |
| L1c2 | 44,100 (10,650) | .986 (.015) | 4.37 |
| L1c3 | 19,250 (5,750) | .636 (.115) | 1.68 |
| L1d | 49,600 (13,450) | .910 (.019) | 4.11 |
| L1e | 82,950 (24,900) | .936 (.037) | 5.03 |
| L2 | 70,100 (15,300) | .974 (.002) | 4.78 |
| L2a | 55,150 (19,350) | .954 (.004) | 3.29 |
| L2a1 | 33,700 (13,400) | .928 (.006) | 2.54 |
| L2b | 31,600 (11,200) | .945 (.015) | 2.63 |
| L2c | 27,500 (7,250) | .901 (.022) | 2.64 |
| L2d | 121,900 (34,200) | .910 (.044) | 6.82 |
| L3 | 61,300 (11,650) | .983 (.002) | 5.16 |
| L3b | 21,600 (6,850) | .955 (.011) | 2.93 |
| L3d | 30,250 (8,450) | .921 (.015) | 2.77 |
| L3e | 49,250 (11,750) | .952 (.007) | 3.85 |
| L3e1 | 32,150 (11,450) | .914 (.017) | 2.58 |
| L3e1a | 26,750 (12,000) | .848 (.049) | 2.30 |
| L3e2 | 37,400 (18,350) | .827 (.033) | 2.15 |
| L3e2b | 9,150 (3,100) | .627 (.071) | 1.09 |
| L3e3 | 14,150 (4,500) | .779 (.077) | 1.30 |
| L3e4 | 24,200 (10,400) | .657 (.138) | 2.38 |
| L3f | 36,300 (12,800) | .948 (.016) | 2.83 |
| L3f1 | 28,650 (8,650) | .941 (.025) | 2.52 |
| L3g | 45,100 (12,500) | .996 (.043) | 4.63 |
H = sequence diversity.
M = average number of pairwise differences.
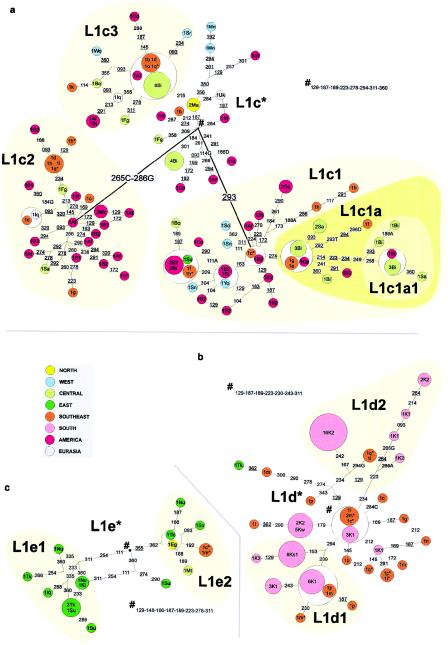
Paragroup L1
The paragroup L1 includes the MRCA of human mtDNA, which is at least 150,000–170,000 years old (Horai et al. 1995; Ingman et al. 2000). Haplogroup L1a (fig. 4a) is common (∼20%–25%) in East, Central, and southeastern Africa, and is almost absent in North, West, and southern Africa. The main subclade, L1a1, is ∼33,350 (SE 16,600) years old and is quite starlike, with a predominantly East/southeastern African distribution and a root type that is common in East Africa. There has been considerable drift on several derived types in southeastern Africa. The second principal subclade, L1a2, is ∼8,300 (SE 3,650) years old and is predominantly Central African, occurring in both Biaka and Mbuti, and, again, several types (in particular, the root type) appear at elevated frequency in southeastern Africa.
An East African origin of L1a seems likely, given that Central African types tend to be more derived in the tree. The expansion into the tropical forest zone of Central Africa (primarily involving L1a2) may have been quite early, sometime within the past 10,000 years, since it comprises a high proportion of both Mbuti and Biaka mtDNAs. The tropical forest had probably already reached something like its present-day extent in Central Africa by ∼10,000 years ago (Adams and Faure 1997). However, there may have been a retreat during the arid phase ∼3,000 years ago, which may have facilitated either the expansion of L1a2 into Central Africa or the Bantu expansions or both (Maley 1993; Adams and Faure 1997).
L1a seems likely to have been brought to southeastern Africa by the eastern stream of the Bantu expansion, having been picked up in East Africa. This is supported by its presence in the Bantu-speaking East African Kikuyu, and, in particular, by a match between a Kikuyu lineage and one of the commonest southeastern African types (within L1a1a). A second possibility would be that the L1a lineages in southeastern Africa were brought directly from a region close to the source of the Bantu languages in western Central Africa or from some intermediate position on the western stream route through Central Africa. The analysis of Soodyall et al. (1996) may help to distinguish these possibilities. They showed an association between an intergenic COII/tRNALys 9-bp deletion and a subset of L1a types lacking the transitions from the CRS at both 16129 and 16168—that is, within L1a2. This deletion is common in southeastern African Bantu speakers, as well as some East and Central African groups. It was absent not only in all Khoisan groups but also in virtually all southwestern African Bantu speakers (with the exception of three Ambo individuals from Namibia, for whom a southeastern Bantu origin was proposed; see also Soodyall and Jenkins 1993). They propose a Central rather than an East African origin for the deletion; we concur that, although L1a seems most likely to have originated in East Africa, L1a2 may have emerged in Central Africa.
The presence of L1a in São Tomé and Bioko may also have a more recent explanation, since many slaves were moved during the last millennium to these Atlantic islands from Mozambican sources (Newman et al. 1995). A predominantly East African origin for L1a types also explains its relative scarcity in America, in comparison with other African types. Most American representatives of L1a, in fact, match types from southeastern Africa, and probably derive directly from that region.
The two major founder candidate sequence types in L1a—in L1a1a and L1a2, respectively—date to 1,900 (SE 750) and 800 (SE 550) years. The average age for the two founder types is 1,100 (SE 400) years. This is consistent with the formation of east Bantu communities in the Lake Victoria region around the last century b.c. and the first few centuries a.d. (Phillipson 1993). L1a may therefore have been introduced into the Bantu community by assimilation of East African non-Bantu speakers, rather than being dispersed from western Central Africa. This suggests that approximately a quarter of the lineages in southeastern Bantu have an East (or eastern central) African origin.
L1b (fig. 4b) has a completely different geographical distribution within Africa. It is concentrated in West Africa, with some overflow into Central and North Africa (particularly geographically adjacent areas, connected by the West African coastal pathway) but little in East, southeastern, or southern Africa. It is also common in African Americans (∼27% of all L1b-types in the database), in agreement with the known importance of the West African coast to the Atlantic slave trade. A simple interpretation would therefore attribute a West African origin to L1b, with significant diffusion into North and Central Africa. However, because the coalescence time of L1b is estimated at only ∼30,000 years—whereas its sister clade, L1c, is estimated at ∼60,000 years old—a recent bottleneck and re-expansion in West Africa may have shaped the evolution of L1b. Given the likely origin of its sister clade L1c in Central Africa, a Central African origin seems plausible for L1b as well.
Haplogroup L1c (fig. 5a) is less starlike than L1a and L1b, with three major well-defined subclades and high internal diversity. The geographic distribution of L1c is especially interesting. More than one-third of L1c haplotypes in our database belong to African Americans, and few of them show matches with continental Africans. The great majority of the remainder of L1c comes from Central Africans, with a few in the west and the southeast. There are virtually none in the east or south; of the “Pygmy” groups sampled, only the western group (the Biaka) have L1c. Representatives in West Africa are restricted to two derived subclades, suggesting an expansion westwards relatively late in the evolution of the haplogroup. It is notable, however, that the southeastern representatives tend to be most closely related to Central African types and include types in clusters not present in West Africa.
Figure 5.
Networks of (a) L1c, (b) L1d, and (c) L1e lineages
This suggests that the origin of L1c can be placed somewhere in Central Africa towards the Atlantic west coast, in the uncharacterized areas of Angola and the Congo delta, to the south of the putative Bantu homeland, on the route of the “western stream” of the Bantu expansion. A West African origin for the African American L1c types is unlikely, because American types do not match with West African ones, this region being the best represented in the database.
Haplogroup L1d (fig. 5b) is nonstarlike and characterizes Khoisan groups (Bandelt and Forster 1997), where it represents about half of the total haplogroup composition for the southern African samples (!Kung and Khwe). L1d is additionally found at ∼5% in the southeastern African samples (see also Pereira et al. 2001), and there is a single East African L1d type from Lake Turkana.
This distribution strongly implies an origin for L1d amongst the ancestors of the Khoisan, long before the arrival of Bantu speakers in the region. The Turkana sample may represent a relict of a former wider distribution of the Khoisan; however, since this type is now seen to be derived from a southeastern African type, recent gene flow from the southeast is more likely. This surmise is supported by the evidence that the Khoisan-speaking Hadza of Tanzania (admittedly a small sample from a small population) resemble East Africans in bearing haplogroups L3g and L2; they lack any L1d or L1k that might suggest that they are a relict of a widespread “genetically” Khoisan population. This is also the case for classical markers (Cavalli-Sforza et al. 1994).
Assuming that L1d in the southeastern Bantu speakers represents assimilation of Khoisan lineages, there is no evidence for a major founder effect during the process. It might have taken place at any time since the arrival of the Bantu speakers, or as a result of recurrent gene flow into the Bantu population. Since it is only present at ∼5%, the level of gene flow (or acculturation) seems to have been rather slight in this area, although it is much higher in parts of South Africa (according to the data of Soodyall 1993; see our “Discussion” section). There is a complete lack of the second major Khoisan haplogroup, L1k, in the southeastern Bantu-speaking sample.
The minor but ancient haplogroup L1e is restricted almost solely to East Africa, with minor gene flow of one subclade into Central Africa (Mbuti) and southeastern Bantu speakers. Even in East Africa, L1e only accounts for ∼6% of the data. L1f types (network not shown) are present at even lower frequencies in the database and, again, only in East Africa.
L1k sequences (network not shown) have been found exclusively in southern African Khoisan-speaking populations (at ∼20%) and, like those from L1d, are probably indigenous. Given the high levels of drift evidenced in both L1d and L1k, we can speculate that they may represent the survivors of many more L1 lineages that have become extinct—perhaps quite recently, as their territories were encroached upon by metal-using agriculturalists, both African and European.
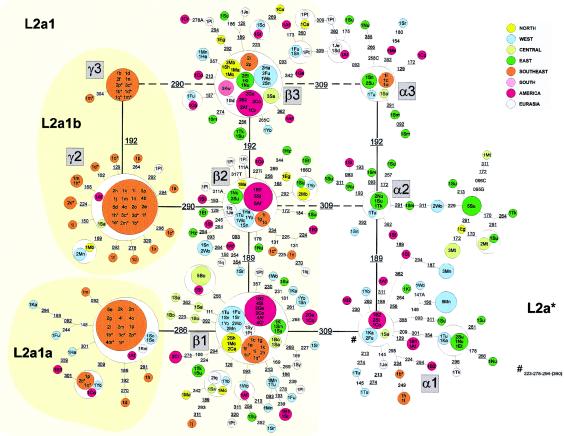
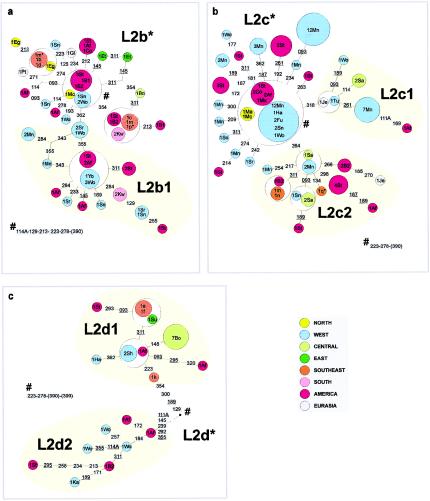
Haplogroup L2
Haplogroup L2 (figs. 6 and 7) is commonly subdivided into four main subclades, L2a through L2d (Chen et al. 2000; Pereira et al. 2001; Torroni et al. 2001). L2c cannot be distinguished from L2* without HVS-II information (325 in HVS-II) or coding-region mutations, although some of its subclades have distinctive HVS-I motifs. Among the southeastern Africans typed for this study (table 1), we found no L2* mtDNAs (in agreement with Torroni et al. 2001). The great majority belong to L2a (fig. 6), the most frequent and widespread mtDNA cluster in Africa (nearly a quarter of all indigenous types), as well as in African Americans.
Figure 6.
Network of L2a lineages
Figure 7.
Networks of (a) L2b, (b) L2c, and (c) L2d lineages
We have attempted partly to disentangle the structure of L2a, retaining as irreducible on present evidence three major squares close to the root of the cluster. These reticulations link eight main clusters by single-step mutations. We assume that the main reticulations of the network are due to the existence of rapid transitions at positions 16189 and 16192 (Howell et al. 2000), which approach saturation due to the high time depth of African lineages. We also assume that position 16309 is more stable than the two known fast sites and therefore is not responsible for the main reticulations. On these grounds, clusters α1-α2-α3, as well as β1-β2-β3, might be collapsed into two main clusters, one of them with the basal motif of L2a and the other harboring the transition at 16309 (L2a1). Several instances in which 16309 must nevertheless evolve in parallel can then be read off the network.
There are two L2a clusters well represented in southeastern Africans, L2a1a and L2a1b, both defined by transitions at quite stable HVS-I positions. Both of these appear to have an origin in West Africa (as indicated by the distribution of matching or neighboring types), and to have undergone dramatic expansion either in southeastern Africa or in a population ancestral to present-day southeastern Africans. L2a1b almost certainly includes the 16192T-derived subcluster, which is exclusively present in the southeast. The very recent starbursts in subclades L2a1a and L2a2 suggest a signature for the Bantu expansions, as also suggested by Pereira et al. (2001). The L2a1a founder candidate dates to 2,700 (SE 1,200) years ago. For L2a1b there is a rather older age estimate of 8,850 years, but this has an enormous standard error (SE 4,600 years) as a result of the early 16192 branch (Pereira et al. 2001). If we assume a starlike tree by suppressing the 16192 variant (effectively assuming that this is a third founder type), the age is 5,250 (SE 1,600) years. An average age estimate, under the assumption of two founders in L2a, is 6,600 (SE 3,000) years or, under the assumption of three founders, 3,750 (SE 900) years. Thus, it appears that the founder ages for L2a are significantly older than for L1a, consistent with the phylogeographical picture, with an earlier West African origin for the L2a lineages of southeastern Africa and a more recent East African origin for the L1a lineages. Indeed, the age of the L2a founders in southeastern Africa is consistent with an origin in the earliest Bantu dispersal from the Cameroon plateau, 3,500 years ago (Phillipson 1993).
It is difficult to trace the origin of L2a with any confidence. The deepest part of L2a, represented by clusters α1-α3, is most common in East Africa. However, the diversity and TMRCA are similar in East (61,250 [SE 13,500] years) and West (54,100 [SE 17,087] years) Africa. The diversity accumulated separately in East and West Africa, estimated from the main shared founder types (and disregarding the possibility of subsequent gene flow), is again similar in the two regions, at ∼14,000 years (14,100 years [SE 5,100], and 13,800 years [SE 4,700], respectively), suggesting a separation shortly after the Last Glacial Maximum. An easterly origin for L2a also faces the following difficulties: that the other subclades of L2 (L2b, L2c, and L2d) have a clear western distribution, and that L2d diverges earlier in the mtDNA phylogeny than L2a (Torroni et al. 2001). A possible solution would be an origin for L2a somewhere between east and west, followed by dispersals in both directions along the Sahel corridor.
Haplogroups L2b, L2c, and L2d appear to be largely confined to West and western Central Africa (and African Americans), with only minor occurrences of a few derived types in the southeast. L2b also shows isolated occurrences in the east and as far north as Iberia. Therefore, an origin for all three in West and western Central Africa seems likely. Complete sequence data indicate that L2d is the oldest of the four subclades of L2, diverging before L2a, and that L2b and L2c are sister clades that diverged more recently (Torroni et al. 2001). The estimated divergence times, ranging from ∼120,000 years, for L2d, through 55,000 years, for L2a, and ∼30,000 years, for L2b and L2c, with an estimated overall age for L2 of ∼70,000 years, are consistent with this pattern. In the light of this, it is scarcely surprising that tracing its place of origin is problematic. At such an age, it seems perhaps unlikely that L2d should have diverged in West Africa, but, given the period of potential drift and extinction, the data are certainly consistent with a Central African origin. A single type in the subclade L2d1, not seen in the southeastern Africans but present at high frequency in the Bubi of Bioko, may represent a trace of this.
L2 contributes 36% (95% CR .316–.408) to the southeastern Bantu population. If we sum this with the other major southeastern haplogroups of clear West African origin, L3b and L3d, the combined contribution of a putative West African source is ∼44% (95% CR .398–.493).
Paragroup L3A
We here define two previously unlabeled subclades of L3A, L3f, and L3g. The lineages remaining within L3* represent ∼20% of all L3A types in Africa. Although they are distributed throughout the continent, they reach the highest frequencies in East Africa, where they account for about half of all types from this region. This frequency profile suggests an origin for L3 in East Africa (Watson et al. 1997). This is supported by the evidence that the out-of-Africa migration, which took place from a source in East Africa 60,000–80,000 years ago, gave rise only to L3 lineages outside Africa.
Both L3f (fig. 8a) and L3g (fig. 8b) are rare and also appear to have an East African origin. L3f* and L3g are virtually restricted to East Africa (with some dispersal into Central Africa, southeastern Africa, and the Near East). The subclade L3f1 appears to have spread at an early date into West Africa and is correspondingly also better represented in African Americans. Of particular interest is the fact that, as noted above, the small Hadza sample from Tanzania are largely L3g, with a small fraction of L2: they entirely lack the Khoisan-diagnostic L1d and L1k lineages.
Figure 8.
Networks of (a) L3f, (b) L3g, and (c) L3b lineages
By contrast, the commoner haplogroup L3b (fig. 8c) is predominantly West African, with a substantial representation again in African Americans. It has spilled over into North Africa and on into the Near East. There is very little dispersal into either East Africa or even Central Africa, but several derived types are present in southeastern Africa.
Its sister clade, haplogroup L3d (fig. 9a), is also mainly West African and African American. A number of types are found in southeastern Africa, including one type (in L3d1), matching a Fulbe lineage, at considerably elevated frequency. A second type (in L3d3) is not seen in our southeastern African sample but occurs at high frequency in the south, in both Khwe and !Kung, and matches a type apparently found at high frequency in the Herero (Vigilant et al. 1991; not included in the network here because of sequence ambiguities). This likely arose in the Bantu population and spread later into the Khoisan speakers, since a single one-step derivative is present in the southeast. This weighs against a pre-Bantu expansion into the south, which would only be supported if some clades outside L1d and L1k, with a northern origin, were present in Khoisan speakers but not in Bantu speakers. The arrival in the southeast indeed appears to have been very recent, since most southeastern (and non-indigenous southern) types have West African matches or derivatives.
L3e (fig. 9b) is the most widespread, frequent, and ancient of the African L3 clades, comprising approximately one-third of all L3 types in sub-Saharan Africa. This haplogroup has recently been dissected in some detail by Bandelt et al. (2001), who suggest an origin for the haplogroup in the Central Africa/Sudan region ∼45,000 years ago. As they recognized, L3e1 in particular is common amongst southeastern African Bantu speakers, along with some L3e2 and L3e3 lineages. L3e also represents approximately one-third of all African mtDNA lineages in Brazil. Alves-Silva et al. (2000) therefore hypothesized that it might be a common component of the (as yet unsampled) Angolan mtDNA pool, from where it may have been carried to Brazil during the slave trade.
L3e1 is distributed throughout sub-Saharan Africa, but it is especially common in southeastern Africa. This clade appears to have a west Central African origin and is rare among West Africans, although it is well represented among African Americans. Several southeastern African types are shared with East African Bantu-speaking Kikuyu from Kenya. This suggests that L3e1 may have spread into Kenya via the eastern stream from a Cameroon source population (best represented in this data set by Bioko and São Tomé) or from some Central African source. It subsequently dispersed into the southeast (although, with so little data, back migration into Kenya cannot be ruled out). The African American types may be the result of direct transportation from Mozambique, given the lack of West African representatives. One L3e1a type is also present at elevated frequency in the Khwe, but, since it matches two Herero and also has a direct derivative in the southeast, this again appears to have been the result of gene flow from Bantu speakers, even though the type has not been sampled in that group.
L3e2 is more frequent in Central and West Africa. It is not possible to distinguish L3e2a without HVS-II information (a transition at np 198), and, as this information is not available in most sequences in the database, we have incorporated L3e2a into L3e2* in figure 9b. L3e2* appears not to have been transferred to the southeast, with one exception. L3e2* is found mainly in Central Africa, and the derived subclade L3e2b is found primarily in West Africa, with a clear founder type within L3e2*. This indicates a range expansion from Central into West Africa (∼9,000 years ago). Other instances of such expansions (for example, in haplogroup L2) may be undetectable, at present, because of poor phylogenetic resolution. Few L3e2b types are found in southeastern Africa, but a great many are present in African Americans.
Finally, there are two small sister clades, L3e3 and L3e4. L3e3 is primarily West African, but with its root type present at elevated frequency in the southeast and with some southeastern African derivatives. There is also a Kikuyu derivative, again raising a possible connection with the eastern stream. L3e4 is present in East, Central, and West Africa, with one individual in the southeast, but is too rare to draw conclusions from.
Assuming that L3e and L1e are both of entirely Central African provenance, we can estimate an approximate Central African contribution to the southeastern Bantu speakers of ∼21% (95% CR .171–.248). However, it is not clear (in the absence of further data from the region) whether these lineages should be attributed to assimilation in the forest zone or whether the Cameroon source region could have given rise to both the “West African” and “Central African” lineages found in the southeastern African Bantu speakers.
Discussion
mtDNA in Southeastern African Populations
We analyzed a total of 16 different Bantu-speaking populations from the vicinity of Mozambique. The results indicate that these populations are highly homogenous with respect to their mtDNA, as reflected in all of the diversity indices, AMOVA, and the PC analysis. The phylogeographic analyses also reflect this degree of homogeneity, since almost all the ethnic groups are equally distributed within the southeastern African mtDNA pool. Several additional analyses (neighbor-joining trees, UPGMA [unweighted pair-group method using arithmetic averages], split decomposition, etc.) showed the same pattern (data not shown). This homogeneity may either be the result of a common origin, of high levels of gene flow between the groups, or of both.
Interestingly, despite the evidence for pronounced founder effects in the networks, substantial diversity has been maintained in these groups; indeed, they include most of the haplogroups present throughout sub-Saharan Africa. This implies that substantial numbers must have been involved in the dispersal, even by the end point of the process. This perhaps lends support to the archaeological view that the Bantu expansion was not a single population movement, despite its rapidity, but rather a complex process involving many short-range dispersals. The formation of the different ethnic groups in the southeast would likely postdate their arrival (or several arrivals) in the region.
mtDNA and the Middle/Late Stone Age
Approaching the study of sub-Saharan African mtDNA variation is a daunting task, as the time depth of lineages within the continent is considerably greater than elsewhere. Although all Eurasian mtDNA lineages coalesce on a single founder type at the root of haplogroup L3, ∼80,000 years ago (Watson et al. 1997), the coalescence time of African mtDNAs extends at least twice as far into the past (Ingman et al. 2000). The task is made particularly difficult because most African mtDNA data sets comprise solely HVS-I sequences, which experience high levels of recurrent mutation at high time depths. This tends to foreshorten coalescence times and render tree reconstruction problematic. However, the situation has been greatly improved in recent years by the publication of several complete mtDNA sequence data sets, which, along with high-resolution RFLP analyses, can be combined with HVS-I data to render a more accurate tree. We have taken advantage of the increased knowledge that has resulted, to estimate plausible phylogenetic networks for all of the major sub-Saharan African haplogroups and to undertake a phylogeographic analysis of all of the available HVS-I data. At the same time, we have added substantially to the database of southeastern African Bantu populations, to shed further light on one of the most important demographic upheavals in African history, the Bantu expansion.
Despite the high time depth of the African mtDNA tree as a whole, African haplogroups rarely exceed an age of 100,000 years when dated on the basis of accumulated HVS-I variation, and many more are of the order of ∼50,000 years or so. This implies that mtDNA may provide little information about demographic events that took place >100,000 years ago and probably rather little information for the following 50,000 years or so. Some of the deepest clades in the phylogeny may date to a much more recent period, suggesting that population fragmentation and re-expansion may have had a major impact on the ancestry of modern sub-Saharan Africans. Nevertheless, most of the major clades have restricted distributions within Africa, the signals for which have not been erased despite substantial subsequent gene flow between regions, and the order of mutations is often known with some confidence, so that a relative time frame can be applied.
The poverty of information that can be retrieved, as the TMRCA is approached, also makes it rather difficult to establish a geographical center of gravity for the origin of modern humans (which, on fossil evidence, is thought to have been completed ∼130,000 years ago, perhaps in Ethiopia; see Grün and Stringer 1991). Five major branches radiate from the MRCA, at the level of resolution considered here, of which two are East or east Central African and two southern African (found primarily in Khoisan speakers); the fifth leads to the rest of the tree. Although the Khoisan retain only two major haplogroups from their pre–Bantu-contact period, this pattern does not preclude an origin for modern humans in southern Africa, followed by a subsequent penetration (before ∼40,000 years ago) into East and Central Africa, where modern humans then flourished to a greater extent than their fellows in the south. However, an early southern offshoot from an East African source population is at least as likely.
The distributions and ages of L1a, L1c/L3e, and L1d testify to the habitation of East, Central, and southern Africa, respectively, by modern humans, by ∼40,000 years ago. Similarly, L1b, L3b, and L3d imply that West Africa has been inhabited since at least 20,000–30,000 years ago. The main puzzle is the almost ubiquitous haplogroup L2a, which we suggest may have become prevalent somewhere in north Central Africa, spreading both east and west along the Sahel belt ∼20,000 years ago at the peak of the LGM (or somewhat earlier). We recognize, however, that the origins of these haplogroups may be more ancient than we can trace (L2, for example, may be well >70,000 years old) and that, in such cases, evidence of the earlier distribution of these clusters may have been erased by subsequent demographic processes.
An important influence on the subsequent genetic landscape of the continent is likely to have been the LGM. Paleovegetational studies have indicated that, between 30,000 and 11,000 years ago, much of the continent was extremely arid (Adams and Faure 1997). The Sahara advanced hundreds of kilometers further south, and the equatorial rainforests were reduced to a small fraction of their present size, leaving open woodland and savanna in much of the Congo basin. This may have formed a refuge area from which modern humans later dispersed: some with haplogroup L2a east and west, with L1b west; perhaps even some with L1a east and L1d southward. The origins of these expansions may lie earlier, at the beginnings of the Later Stone Age, ∼40,000 years ago. Archaeological evidence has demonstrated substantial human activity in the equatorial forest area—for example, in Cameroon and Equatorial Guinea, 35,000 years ago (Martí et al. 2001).
It is worth noting that the mtDNA data do not support the clustering of sub-Saharan Africans into (pre-Holocene) geographical races, as assumed by many authors (Hiernaux 1975; Newman 1995), if only because the so-called “Pygmies” clearly do not form a coherent group. The westerly Biaka sample includes only L1a and L1c, and the more easterly Mbuti include only L1a (shared with the Biaka), L1e and L2. Therefore, the Biaka tend to resemble other Central African populations, whereas the Mbuti more closely resemble those from East Africa, although both groups are much reduced in diversity in comparison with neighboring populations. It is also notable that the Tanzania Khoisan-speaking Hadza resemble other East Africans rather than southern African Khoisan speakers. Both results appear to be consistent with the results from classical markers (Cavalli-Sforza et al. 1994).
mtDNA and Bantu Dispersals
The mtDNA pool of present-day southeastern Africa seems to comprise at least two distinct components: ancestral types carried by the people living in the region before the arrival of Bantu speakers and types brought with the Bantu speakers themselves. Khoisan lineages comprise only ∼5% of the total lineages in the Bantu speakers of Mozambique. The 5% may represent either assimilation on arrival or subsequent gene flow; it is difficult to distinguish the two, but the evidence suggests that no pronounced founder effect has occurred in the L1d lineages, possibly making the second possibility more likely. The data of Soodyall (1993) indicate that the Dama similarly have only ∼5% Khoisan lineages but that the southern African Xhosa and Zulu may have ∼25% and ∼50% Khoisan lineages, respectively (also all L1d). This much higher level of assimilation is consistent with the presence of Khoisan click consonants in both languages. In southern Africa, Khoisan speakers themselves appear to have experienced high levels of assimilation of Bantu lineages: ∼23% in the Vasikela !Kung (Chen et al. 2000), ∼24% in the Sekele !Kung (Soodyall 1993), and ∼61% in the Khwe, consistent with their similar physical appearance to southern African Bantu speakers (Chen et al. 2000).
Of the lineages brought in by the Bantu speakers themselves, about half appear to have an origin in West Africa ∼4,000 years ago, about one-fourth in East Africa ∼2,000 years ago, and about one-fifth have a Central or west Central African origin. The East African component is virtually restricted to eastern Bantu speakers, implying that they were the result of assimilation by the eastern stream while en route, which, moreover, involved one or several severe founder effects. This is likely to have taken place in the Great Lakes region, as prelude to the expansion of Urewe ware and the widespread Chifumbaze complex witnessed in the archaeological record.
The Central African lineages may be the result of assimilation in the rainforests, or they may (given the intermediate position of Cameroon between west and Central Africa) have formed part of the founder group, along with the West African lineages. If so, nearly three-quarters of southeastern Bantu lineages can be traced back to the putative “homeland” region in West/Central Africa postulated on linguistic and archaeological grounds. However, the founder effects on Central African types appear to be much less severe than on those from the east and west, which may imply a more gradual process of assimilation within the rainforests. Indeed, part of L1a (the subclade L1a2) may have a proximal origin in the rainforests rather than in East Africa, given its prevalence in both Mbuti and Biaka. More data from Central Africa are needed to resolve this issue.
It is also likely that a number of lineages have arrived as a result of more recent gene flow. Most haplogroups exhibit a number of minor types in southeastern Africa, indicating that founder effects were either insufficiently severe to eliminate all of the variation save for a few major founders or that subsequent gene flow has taken place. Particularly curious are those from western haplogroups such as L1b, which seem unlikely to have taken part in the eastern Bantu expansion, since no major founders survive. It is possible that these have arisen from exchange with western Bantu groups (which perhaps arrived carrying L1b and other western clades). It is also likely that recent gene flow has taken place from East Africa as a result of the Islamic coastal trading system.
No published HVS-I sequences are available from southwest Bantu speakers. However, we undoubtedly have some representation of this part of Africa in the African American lineages, since, during the Atlantic slave trade of the 15th–19th centuries, many people were forcibly moved to America from this part of Africa. It is thought that Angola was the second most important country that contributed to the Atlantic trade (>2.5 million people; see Thomas 1997). This reasoning has been used by Alves-Silva et al. (2000) to predict a large proportion of L3e and L1c lineages in Angola, as well as some L2 lineages, but no L1d or L1k lineages, on the basis of their prevalence in the present-day Brazilian population (although L1d/k might be present at low levels, as in the southeast). We can add the lack of L1a (or at least L1a2) on the basis of the argument of Soodyall et al. (1996), and lack of other east-specific clusters such as L1f. Our networks confirm the large numbers of L1c and L3e types present in African Americans that lack matches in the existing mtDNA database for Africa. Since the main source regions for the Atlantic slave trade were West Africa, southeastern Africa, and Angola, the likelihood that these represent a fair sample of the Angolan Bantu population remains. The lack of major founder effects in both L3e and L1c is again striking if we consider that they may be of western Bantu origin. We can speculate that the western Bantu expansion, which dispersed through the tropical forest zone, may have been a more gradual process, and involved more assimilation of indigenous lineages in the forest zone, than the eastern stream. Furthermore, L1c in particular may have been introduced to the southeastern Bantu groups via the western stream (or the western component of the eastern stream), rather than having been carried directly eastward from the source region in the earliest dispersals.
Comparisons with the Y Chromosome
The suggestion of Hammer et al. (2001) that the majority of African NRY lineages (haplogroup E) had an Upper Paleolithic Asian origin has now been called into question (if not decisively ruled out), because it was based on poorly resolved data (Underhill et al. 2001). However, Cruciani et al. (2002) have also identified the signature of a probable ancient migration from Asia into north Cameroon, in the form of a derived form of haplogroup R. This clade is not found in present-day western Eurasia or anywhere else in Africa, with the likely exception of Egypt (at 13%; see Scozzari et al. 1999), but it occurs in north Cameroon at a frequency of ∼40%. A very recent immigration event from North Africa, perhaps mediated by Fulbe or other pastoralists, may explain this pattern; the Fulbe in the mtDNA database (from Nigeria) show the presence of two West African U6 types, a U5 type (found otherwise only in Senegal), and an apparently indigenous West African subclade of haplogroup H. Although the H and U lineages combined make up only ∼18% of the Fulbe sample, they resemble the NRY haplogroup R lineages in indicating an ancient Eurasian origin, despite the absence of sequence matches in the modern European sample. The NRY haplogroup R and the mtDNA haplogroup H and U6 lineages may have originated in North African Berbers, some of whose NRY lineages may have arrived in North Africa from the Near East sometime between ∼20,000 and ∼50,000 years ago, as hypothesized for mtDNA haplogroup U6 (Rando et al. 1998; Macaulay et al. 1999). A recent arrival in north Cameroon from North Africa would explain both the ancient Asian origin hypothesized by Cruciani et al. (2002) and the geographically restricted distribution in that area that they document.
The NRY displays a lower diversity in African populations than mtDNA (Hammer et al. 2001). There are two major deep branches, haplogroups A and B. Haplogroup A is frequent in East and southern Africa, with subclades concentrated in the Khoisan and East Africa, and haplogroup B throughout sub-Saharan Africa. The most frequent African clade, however, is haplogroup E, which seems likely to have spread from East Africa within the last 50,000 years to West Africa. Subclades then dispersed to North Africa and Central/southern Africa (Scozzari et al. 1999; Underhill et al. 2000; Bosch et al. 2001; Hammer et al. 2001; Underhill et al. 2001).
One particular widespread derived subclade, E3a, has been implicated in the Bantu expansion; it has rather little diversity and forms the majority of lineages in Central and southern African Bantu-speaking samples (Scozzari et al. 1999; Underhill et al. 2001). This subclade occurs at a frequency of ∼63% in the southern African Bantu speakers of Underhill et al. (2000), with one predominant haplotype and its one-step derivative (Underhill et al. 2001). These two haplotypes (hts 24 and 22 in the work of Underhill et al. [2001] and Cruciani et al. [2002]) occur at ∼84% in south Cameroon (Cruciani et al. 2002). Both haplotypes are widespread in West and Central Africa, but this evidence is clearly consistent with a Cameroon origin for southern African Bantu speakers. A minor E3a type also present in southern African Bantu speakers (ht 27) occurs at ∼9% in Cameroon and is almost absent in other African groups sampled, so the case for a south Cameroon origin is even stronger in this case.
The third major haplogroup E type present in these southern African Bantu speakers, within the subclade E2 (15% in Underhill et al. 2000), is present in Burkina Faso and north Cameroon (hts 40–41, Cruciani et al. 2002), as well as Khoisan speakers and also Central African “Pygmies” (Underhill et al. 2000). Again, a West African origin, with dispersal through Central Africa and some introgression into Khoisan groups seems very likely. E3 as a whole amounts to 81% of the southern African Bantu speakers of Underhill et al. (2000, 2001).
Haplogroup B lineages are distributed throughout Africa, and one haplotype (ht 12) occurs in the southern African Bantu speakers at 13% (Underhill et al. 2000). This type is also present in south Cameroon at low frequencies, and therefore may also have spread south from there with Bantu dispersals. This would raise the putative West African component of the NRY lineages in this sample to 94%, concentrated on three main haplotypes. Again, however, the type is rather widespread and could possibly have been brought south from elsewhere. Haplogroup A, which most likely represents Khoisan introgression, occurs at just 6% in the southern African Bantu speakers, a strikingly similar level to our mtDNA data from southeastern Africa.
Overall, therefore, there does seem to have been higher drift on the male than on the female line during the Bantu dispersals, resulting in a reduced variety of incoming haplogroup and haplotypes. This may in part be accounted for by higher levels of assimilation on the female side, possibly from both Central and (in the case of the eastern Bantu) East Africa, or it may reflect a larger female effective population size in the dispersing groups. Different mating patterns and cultural practices between males and females may have played a part in these processes.
Acknowledgments
We express our appreciation to the original blood donors analyzed in the present work. We also thank E. Valverde and J. Cobreiro for their help with the collection of the samples, and we thank the Universidad Eduardo Mondlane and the Hospital Militar of Maputo (Mozambique) for their help. We thank Hans-Jürgen Bandelt, David Phillipson, and James Newman for critically reading the manuscript; Antonio Torroni for access to unpublished information on the sample from Santo Domingo; Luisa Pereira for supplying additional information on her Mozambique sample; and Fulvio Cruciani and Rosaria Scozzari for advice on the Y-chromosome comparisons. A.S. has a research contract with the University of Santiago de Compostela. V.M. was supported by a Research Career Development Fellowship from the Wellcome Trust. This work was supported by grants from the Ministerio de Educación y Ciencia (DGCYT-P4. BIO2000-0145-P4-02) and the Xunta de Galicia (PGIDT-01-PXI-20806-PR).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Arlequin 2.0: A Software for Population Genetic Data Analysis; http://anthropologie.unige.ch/arlequin/
- Fluxus Engineering Web site, http://www.fluxus-engineering.com (for Network 3.1 software)
- hvrBase, http://www.zi.biologie.uni-muenchen.de/science/mtdna/hvrbase/
References
- Adams JM, Faure H (eds) (1997) Review and atlas of palaeovegetation: preliminary land ecosystem maps of the world since the Last Glacial Maximum. Oak Ridge National Laboratory, TN (published online only: http://www.esd.ornl.gov/ern/qen/adams1.html)
- Alves-Silva J, Santos M, Guimarães P, Ferreira AC, Bandelt H-J, Pena SD, Prado VF (2000) The ancestry of Brazilian mtDNA lineages. Am J Hum Genet 67:444–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organisation of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Bakel MA (1981) The “Bantu” expansion: demographic models. Curr Anth 22:688–691 [Google Scholar]
- Bandelt H-J, Alves-Silva J, Guimarães P, Santos M, Brehm M, Pereira L, Coppa A, Larruga JM, Rengo C, Scozzari R, Torroni A, Prata MJ, Amorim A, Prado VF, Pena SDJ (2001) Phylogeography of the human mitochondrial L3e: a snapshot of African prehistory and Atlantic slave trade. Ann Hum Genet 65:549–563 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P (1997) The myth of bumpy hunter-gatherer mismatch distributions. Am J Hum Genet 61:980–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Sykes BC, Richards MD (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blench (1993) Recent developments in African language classification. In: Shaw T, Sinclair P, Andah B, Okpoko A (eds) The archaeology of Africa: foods, metals and towns. Routledge, London [Google Scholar]
- Bortolini MC, Zago MA, Salzano FM, Silva WA, Bonatto SL, da Silva MC, Weimer TA (1997) Evolutionary and anthropological implications of mitochondrial DNA variation in African Brazilian populations. Hum Biol 69:141–159 [PubMed] [Google Scholar]
- Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J (2001) High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet 68:1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley DG, MacHugh DE, Cunningham P, Loftus RT (1996) Mitochondrial diversity and the origins of African and European cattle. Proc Natl Acad Sci USA 14:5131–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakez Z, Bosch E, Izaabel H, Akhayat O, Comas D, Bertranpetit J, Calafell F (2001) Human mitochondrial DNA sequence variation in the Moroccan population of the Souss Area. Ann Hum Biol 28:295–307 [DOI] [PubMed] [Google Scholar]
- Brehm A, Pereira L, Bandelt H-J, Prata MJ, Amorim A (2002) Mitochondrial portrait of the Cabo Verde archipelago: the Senegambian outpost of the Atlantic slave trade. Ann Hum Genet 66:49–60 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) History and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Chen YS, Olckers A, Schurr TG, Kogelnik AM, Huoponen K, Wallace DC (2000) mtDNA variation in the South African Kung and Khwe—and their genetic relationships to other African populations. Am J Hum Genet 66:1362–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, Torroni A, Excoffier L, Santachiara-Benerecetti AS, Wallace DC (1995) Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. Am J Hum Genet 57:133–149 [PMC free article] [PubMed] [Google Scholar]
- Clark JD (1994) Africa from the appearance of Homo sapiens sapiens to the beginnings of food production. In: De Laet SJ (ed) The history of humanity. Vol. I. Routledge, London, pp 191–206 [Google Scholar]
- Côrte-Real HB, Macaulay VA, Richards MB, Hariti G, Issad MS, Cambon-Thomsen A, Papiha S, Bertranpetit Y, Sykes BC (1996) Genetic diversity in the Iberian Peninsula determined from mitochondrial sequence analysis. Ann Hum Genet 60:331–350 [DOI] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, Modiano D, Holmes S, Destro-Bisol G, Coia V, Wallace DC, Oefner PJ, Torroni A, Cavalli-Sforza LL, Scozzari R, Underhill PA (2002) A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 70:1197–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon HJ (1989) Late Pleistocene palaeolecology and archaeology in the Southern Cape, South Africa. In: Mellars P, Stringer C (eds) The human revolution: behavioural and biological perspectives in the origins of modern humans. Edinburgh University Press, Edinburgh, pp 547–564 [Google Scholar]
- Eggert MKH (1993) Central Africa and the archaeology of the equatorial rainforest: reflections on some major topics. In: Shaw T, Sinclair P, Andah B, Okpoko A (eds) The archaeology of Africa: foods, metals and towns. Routledge, London, pp 289–329 [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt H-J (1996) Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed] [Google Scholar]
- Graven L, Passarino G, Semino O, Boursot P, Santachiara-Benerecetti S, Langaney A, Excoffier L (1995) Evolutionary correlation between control region sequence and restriction polymorphisms in the mitochondrial genome of a large Senegalese Mandenka sample. Mol Biol Evol 12:334–345 [DOI] [PubMed] [Google Scholar]
- Green LD, Derr JN, Knight A (2000) mtDNA affinities of the peoples of North-Central Mexico. Am J Hum Genet 66:989–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JH (1963) The languages of Africa. Indiana University Press, Bloomington, IN [Google Scholar]
- Greenberg JH (1972) Linguistic evidence concerning Bantu origins. J Afr Hist 13:189–216 [Google Scholar]
- Grün R, Stringer CB (1991) Electron spin resonance dating and the evolution of modern humans. Archaeometry 33:153–199 [Google Scholar]
- Guthrie M (1970) Contributions from comparative Bantu studies to the prehistory of Africa. In: Dalby D (ed) Language and history in Africa. Frank Cass, London, pp 20–33 [Google Scholar]
- Hammer MF, Karafet TM, Redd AJ, Jarjanazi H, Santachiara-Benerecetti S, Soodyall H, Zegura SL (2001) Hierarchical patterns of global human Y-chromosome diversity. Mol Biol Evol 18:1189–1203 [DOI] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N (2002) Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 70:1152–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiernaux J (1968) Bantu expansion: the evidence from physical anthropology confronted with linguistic and archaeological evidence. J Afr Hist 9:505–515 [Google Scholar]
- Hiernaux J (1975) The people of Africa. Scribner’s, New York, NY [Google Scholar]
- Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N (1995) Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs. Proc Natl Acad Sci USA 7:532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell N, Smejkal CB (2000) Persistent heteroplasmy of a mutation in the human mtDNA control region: hypermutation as an apparent consequence of simple-repeat expansion/contraction. Am J Hum Genet 66:1589–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman TN (1982) Archaeology and the ethnohistory of the African Iron Age. Annu Rev Anthropol 11:133–150 [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408:708–713 [DOI] [PubMed] [Google Scholar]
- Johnston HH (1913) A survey of the ethnography of Africa and the former racial and tribal migrations in that continent. J R Anthropol Inst 43:345–421 [Google Scholar]
- Krings M, Capelli C, Tschentscher F, Geisert H, Meyer S, von Haeseler A, Grossschmidt K, Possnert G, Paunovic M, Pääbo S (2000) A view of Neandertal genetic diversity. Nat Genet 26:144–146 [DOI] [PubMed] [Google Scholar]
- Krings M, Salem AE, Bauer K, Geisert H, Malek AK, Chaix L, Simon C, Welsby D, Di Rienzo A, Utermann G, Sajantila A, Pääbo S, Stoneking M (1999) mtDNA analysis of Nile River Valley populations: a genetic corridor or a barrier to migration? Am J Hum Genet 64:1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S (1997) Neandertal DNA sequences and the origin of modern humans. Cell 11:19–30 [DOI] [PubMed] [Google Scholar]
- Kuper A, van Leynselle P (1978) Social anthropology and the Bantu expansions. Africa 48:335–352 [Google Scholar]
- Lwanga-Lunyiigo S (1976) The Bantu problem reconsidered. Curr Anthropol 17:282–286 [Google Scholar]
- Maca-Meyer N, González AM, Larruga JM, Flores C, Cabrera VM (2001) Major genomic mitochondrial lineages delineate early human expansions. BMC Genet 2:13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonné-Tamir B, Sykes B, Torroni A (1999) The emerging tree of west Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacEachern S (2000) Genes, tribes, and African history. Curr Anthropol 41:357–384 [PubMed] [Google Scholar]
- Maley J (1993) Climatic and vegetational history of the equatorial regions. In: Shaw T, Sinclair P, Andah B, Okpoko A (eds) The archaeology of Africa: foods, metals and towns. Routledge, London, pp 43–52 [Google Scholar]
- Martí R, Mercader-Florín J (2001) Edad de piedra en los bosques Centroafricanos. Investigación y Ciencia 303:30–31 [Google Scholar]
- Mateu E, Comas D, Calafell F, Pérez-Lezaun A, Abade A, Bertranpetit J (1997) A tale of two islands: population history and mitochondrial DNA sequence variation of Bioko and São Tomé, Gulf of Guinea. Ann Hum Genet 61:507–518 [DOI] [PubMed] [Google Scholar]
- Monsalve MV, Hagelberg E (1997) Mitochondrial DNA polymorphisms in Carib people of Belize. Proc R Soc Lond B Biol Sci USA 264:1217–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JL (1995) The peopling of Africa. Yale University Press, New Haven, CT [Google Scholar]
- Ovchinnikov IV, Gotherstrom A, Romanova GP, Kharitonov VM, Liden K, Goodwin W (2000) Molecular analysis of Neanderthal DNA from the northern Caucasus. Nature 30:490–493 [DOI] [PubMed] [Google Scholar]
- Thomas MG, Weale ME, Jones AL, Richards M, Smith A, Redhead N, Torroni A, Scozzari R, Gratrix F, Tarekegn A, Wilson JF, Capelli C, Bradman N, Goldstein DB (2002) Founding mothers of Jewish communities: geographically separated Jewish groups were independently founded by very few female ancestors. Am J Hum Genet 70:1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarino G, Semino O, Quintana-Murci L, Excoffier L, Hammer M, Santachiara-Benerecetti AS (1998) Different genetic components in the Ethiopian population, identified by mtDNA and Y-chromosome polymorphisms. Am J Hum Genet 62:420–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Macaulay V, Torroni A, Scozzari R, Prata MJ, Amorin A (2001) Prehistoric and historic traces in the mtDNA of Mozambique: insights into the Bantu expansions and the slave trade. Ann Hum Genet 65:439–458 [DOI] [PubMed] [Google Scholar]
- Phillipson DW (1993) African archaeology. Cambridge University Press, Cambridge [Google Scholar]
- Pinto F, González AM, Hernández M, Larruga JM, Cabrera VM (1996) Genetic relationship between the Canary Islanders and their African and Spanish ancestors inferred from mitochondrial DNA sequences. Ann Hum Genet 60:321–330 [DOI] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt H-J, Passarino G, McElreavey K, Santachiara-Benerecetti AS (1999) Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23:437–441 [DOI] [PubMed] [Google Scholar]
- Rando JC, Cabrera VM, Larruga JM, Hernández M, González AM, Pinto F, Bandelt H-J (1999) Phylogeographic patterns of mtDNA reflecting the colonization of the Canary Islands. Ann Hum Genet 63:413–428 [DOI] [PubMed] [Google Scholar]
- Rando JC, Pinto F, González AM, Hernández M, Larruga JM, Cabrera VM, Bandelt H-J (1998) Mitochondrial DNA analysis of northwest African populations reveals genetic exchanges with European, Near-Eastern, and sub-Saharan populations. Ann Hum Genet 62:531–550 [DOI] [PubMed] [Google Scholar]
- Richards M, Côrte-Real H, Forster P, Macaulay V, Wilkinson-Herbots H, Demaine A, Papiha S, Hedges R, Bandelt H-J, Sykes B (1996) Paleolithic and neolithic lineages in the European mitochondrial gene pool. Am J Hum Genet 59:185–203 [PMC free article] [PubMed] [Google Scholar]
- Richards M, Macaulay V (2000) Genetic data and the colonization of Europe: genealogies and founders. In: Renfrew C, Boyle K (eds) Archaeogenetics: DNA and the population prehistory of Europe. McDonald Institute for Archaeological Research, Cambridge, pp 139–151 [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, et al (2000) Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby S (2000) mtDNA Variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 67:718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Comas D, Lareu MV, Bertranpetit J, Carracedo A (1998) mtDNA analysis of the Galician population: a genetic edge of European variation. Eur J Hum Genet 6:365–375 [DOI] [PubMed] [Google Scholar]
- Scozzari R, Cruciani F, Santolamazza P, Malaspina P, Torroni A, Sellitto D, Arredi B, Destro-Bisol G, de Stefano G, Rickards O, Martinez-Labarga C, Modiano D, Biondi G, Moral P, Olckers A, Wallace DC, Novelletto A (1999) Combined use of biallelic and microsatellite Y-chromosome polymorphisms to infer affinities among African populations. Am J Hum Genet 65:829–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scozzari R, Torroni A, Semino O, Sirugo G, Brega A, Santachiara-Benerecetti AS (1988) Genetic studies on the Senegal population. I. Mitochondrial DNA polymorphisms. Am J Hum Genet 43:534–544 [PMC free article] [PubMed] [Google Scholar]
- Soodyall H (1993) Mitochondrial DNA polymorphisms in southern African populations. PhD thesis, University of Witwatersrand, Johannesburg [Google Scholar]
- Soodyall H, Jenkins T (1992) Mitochondrial DNA polymorphisms in Khoisan populations from Southern Africa. Ann Hum Genet 56:315–324 [DOI] [PubMed] [Google Scholar]
- Soodyall H, Jenkins T (1993) Mitochondrial DNA polymorphisms in Negroid populations from Namibia: new light on the origins of the Dama, Herero and Ambo. Ann Hum Biol 20:477–485 [DOI] [PubMed] [Google Scholar]
- Soodyall H, Vigilant L, Hill AV, Stoneking M, Jenkins T (1996) mtDNA control-region sequence variation suggests multiple independent origins of an “Asian-specific” 9-bp deletion in sub-Saharan Africans. Am J Hum Genet 58:595–608 [PMC free article] [PubMed] [Google Scholar]
- Stine OC, Dover GJ, Zhu D, Smith KD (1992) The evolution of two west African populations. J Mol Evol 34:336–344 [DOI] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H (1997) The slave trade—the history of the Atlantic slave trade: 1440–1870. Macmillan, London [Google Scholar]
- Thomas MG, Parfitt T, Weiss DA, Skorecki K, Wilson JF, le Roux M, Bradman N, Goldstein DB (2000) Y chromosomes traveling south: the Cohen modal haplotype and the origins of the Lemba—the “Black Jews of Southern Africa.” Am J Hum Genet 66:674–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, Calderon FL, Simionati B, Valle G, Richards M, Macaulay V, Scozzari R (2001) Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet 69:1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 [DOI] [PubMed] [Google Scholar]
- Vansina J (1995) New linguistic evidence and the “Bantu expansion.” J Afr Hist 36:173–195 [Google Scholar]
- Vigilant L, Pennington R, Harpending H, Kocher TD, Wilson AC (1989) Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci USA 86:9350–9354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC (1991) African populations and the evolution of mitochondrial DNA. Science 253:1503–1507 [DOI] [PubMed] [Google Scholar]
- Vogel JO (1994a) Bantu expansion. In: Vogel JO (ed) Encyclopedia of precolonial Africa. AltaMira Press, Walnut Creek, pp 435–438 [Google Scholar]
- Vogel JO (1994b) Eastern and south-central African iron age. In: Vogel JO (ed) Encyclopedia of precolonial Africa. AltaMira Press, Walnut Creek, pp 439–444 [Google Scholar]
- Watson E, Bauer K, Aman R, Weiss G, von Haeseler A, Pääbo S (1996) mtDNA sequence diversity in Africa. Am J Hum Genet 59:437–444 [PMC free article] [PubMed] [Google Scholar]
- Watson E, Forster P, Richards M, Bandelt H-J (1997) Mitochondrial footprints of human expansions in Africa. Am J Hum Genet 61:691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw G (1994) Southern African Iron Age. In: Vogel JO (ed) Encyclopedia of precolonial Africa. AltaMira Press, Walnut Creek, pp 444–455 [Google Scholar]
- Wilson MR, DiZinno JA, Polanskey D, Replogle J, Budowle B (1995) Validation of mitochondrial DNA sequencing for forensic casework analysis. Int J Legal Med 108:68–74 [DOI] [PubMed] [Google Scholar]
- Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]