Abstract
Glucose-6-phosphate dehydrogenase (G6PD) mutations that result in reduced enzyme activity have been implicated in malarial resistance and constitute one of the best examples of selection in the human genome. In the present study, we characterize the nucleotide diversity across a 5.2-kb region of G6PD in a sample of 160 Africans and 56 non-Africans, to determine how selection has shaped patterns of DNA variation at this gene. Our global sample of enzymatically normal B alleles and A, A−, and Med alleles with reduced enzyme activities reveals many previously uncharacterized silent-site polymorphisms. In comparison with the absence of amino acid divergence between human and chimpanzee G6PD sequences, we find that the number of G6PD amino acid polymorphisms in human populations is significantly high. Unlike many other G6PD-activity alleles with reduced activity, we find that the age of the A variant, which is common in Africa, may not be consistent with the recent emergence of severe malaria and therefore may have originally had a historically different adaptive function. Overall, our observations strongly support previous genotype-phenotype association studies that proposed that balancing selection maintains G6PD deficiencies within human populations. The present study demonstrates that nucleotide sequence analyses can reveal signatures of both historical and recent selection in the genome and may elucidate the impact that infectious disease has had during human evolution.
Introduction
Infectious disease has likely had a large impact on the evolution and differentiation of human populations (e.g., see Cooke and Hill 2001). Therefore, characterization of nucleotide variability in genes that play a role in resistance or susceptibility to infectious disease will be important for understanding how selection shapes patterns of variability and linkage disequilibrium (LD) in the human genome. Although general practice is to identify an association between DNA variants and disease phenotypes in infected individuals, it is also necessary to examine patterns of genetic variation in uninfected individuals, to determine the impact that selection has on the population as a whole. Natural selection can alter levels of nucleotide variability in several ways. For example, balancing selection may maintain allelic diversity within and among populations, whereas directional selection may result in a reduction of nucleotide variability and increased LD at targeted genes relative to that expected under a neutral model. However, it is unclear whether either of these selection regimes is a predominant explanation for patterns of molecular variation in the human genome (Przeworski et al. 2000; Wall and Przeworski 2000; Nachman 2001). Several studies of candidate genes connected to disease phenotypes have identified the impact of natural selection in human populations (Clark et al. 1998; Stephens et al. 1998; Salamon et al. 1999; Fullerton et al. 2000; Nachman and Crowell 2000; Hamblin et al. 2002); however, not all show a “signature” of selection (Harding et al. 1997; Aquadro et al. 2001). Therefore, in continuing to characterize the nucleotide sequence diversity for genes related to disease resistance, we can begin to understand how selection has influenced patterns of variability in the human genome.
One of the most studied infectious diseases is malaria (Greenwood and Mutabingwa 2002), which affects 500 million people each year and is a leading cause of death in the global human population. Although epidemics are prevalent in many areas of the world, Africa is the most strongly affected by this disease (Miller 1994). Archaeological evidence suggests that malaria has had a significant effect on humans only in the past 10,000 years, which is consistent with the advent of agriculture, animal domestication, and increased human population densities in this geographic region (Livingstone 1971). Estimates of genetic diversity in the mosquito vector Anopheles gambiae (Donnelly et al. 2001) and the protozoan parasite Plasmodium falciparum, which causes severe malaria (Anderson et al. 2000; Mu et al. 2002), are in accordance with the predicted recent expansion of human populations and may be due to the coevolution of these organisms. Several human genes show a strong association with malarial resistance (Miller 1994). One major candidate gene encodes glucose-6-phosphate dehydrogenase (G6PD [MIM 305900]), an important “housekeeping” enzyme in the glycolytic pathway for glucose metabolism. G6PD also plays a critical role in maintaining the balance of reduced nicotinamide adenine dinucleotide phosphate (NADPH), a necessary cofactor for cell detoxification. Although several enzymes can recycle this essential cofactor, G6PD is the sole generator of NADPH in the red blood cells and alone may prevent oxidative damage and severe anemia.
G6PD deficiency is the most common known enzymopathy and is estimated to affect 400 million people worldwide. More than 130 different G6PD variants that lead to reduced enzyme activity have been discovered at the DNA level (Luzzatto et al. 2001). G6PD enzyme deficiency is associated with many clinical disorders, such as neonatal jaundice, hemolytic anemia, and several cardiovascular diseases (Beutler 1994). The G6PD locus is located on the telomeric region of the long arm of the X chromosome (at Xq28) and is flanked for 300 kb on each side by Factor VIII and the red/green color-vision genes, which have been widely studied for their association with hemophilia (Toole et al. 1986) and color blindness, respectively (Nathans et al. 1986; Deeb et al. 1992).
Although G6PD deficiency is associated with many clinical disorders, there are three observations that suggest that G6PD deficiencies are selectively maintained. First, G6PD deficiency is strongly associated with the distribution of malarial endemicity, and many variants are found at rare-to-high frequencies in different populations (Vulliamy et al. 1992). A single amino acid replacement is responsible for the classic G6PD A/B polymorphism. The B variant, which possesses normal enzyme activity and dominates in frequency worldwide, is predicted to be the ancestral state by comparison with chimpanzee G6PD (Kay et al. 1992). The A variant, which is due to a derived amino acid replacement in exon 5, possesses 85% enzyme activity and is found in sub-Saharan Africa at frequencies as high as 40% but rarely reaches frequencies >1% outside Africa and the Middle East (Beutler 1994; Ruwende et al. 1995).
Although many other G6PD amino acid replacements have been discovered (Beutler 1994), there are two common variants with severely low enzyme activities. The A− deficiency is the result of an amino acid replacement in exon 4 and is always associated with the amino acid change that gives rise to the A allele. This deficiency has only 12% enzyme activity and is found at frequencies as high as 25% in sub-Saharan Africa, but it is very rare in all other regions except the Mediterranean, where it is found at a frequency of nearly 5% (Beutler 1994). The Med deficiency, which is due to an amino acid replacement in exon 6 on a B allele and possesses only 3% enzyme activity, is typically found at frequencies of 2%–20% in the Mediterranean and as high as 70% in Kurdish Jews (Beutler 1994).
The second observation that argues for an adaptive explanation for G6PD deficiency comes from a study by Ruwende et al. (1995), who have showed that the A− deficiency can reduce the risk of malarial infection by 46%–58% in both heterozygous females and hemizygous males. It is likely that the oxidative stress imposed by G6PD deficiency in the red blood cells also creates a toxic environment for the Plasmodium parasites that cause malaria (Vulliamy et al. 1992; Miller 1994). Although G6PD enzyme deficiency may have detrimental effects, the benefit that it provides in the presence of malaria suggests that G6PD deficiency may be maintained by balancing selection. Ruwende et al. (1995) have found that the A variant, which reduces enzyme activity by 15%, does not significantly deter malarial infection. Therefore, it is possible that only a severe reduction in G6PD activity can act as a successful inhibitor of malarial infection.
The third observation is from a study in which Tishkoff et al. (2001) examined seven RFLPs at the G6PD locus and microsatellite variation in close proximity (within 18 kb downstream), to investigate the extent to which selection for the A− and Med variants has affected LD in this gene region. This study estimates that the A− variant likely arose 3,840–11,760 years before present (BP) and that the Med variant likely arose 1,600–6,640 years BP. Because this is consistent with the estimated time for the spread of severe malaria (Livingstone 1971), it is possible that the A− and Med variants were favored by selection for malarial resistance. The study by Tishkoff et al. (2001) shows that selection has had a large impact on the frequencies of the A− and Med deficiencies; however, we were also interested in what impact selection has had on G6PD nucleotide diversity in a large and random sample of individuals.
In the present study, we present an analysis of DNA sequence variation across a 5.2-kb region of the G6PD locus for both African and non-African groups, to address several questions related to the evolutionary history of G6PD protein polymorphism. To date, all known G6PD amino acid polymorphisms come from studies of individuals who possess G6PD enzyme deficiency. By using a nucleotide sequence approach in randomly selected individuals, we can determine whether there are other amino acid replacement polymorphisms at the nucleotide level, in addition to those that have already been discovered in functional assays. We can also compare the level of silent-site polymorphism in coding and noncoding regions of G6PD with other human genes. Additionally, we were interested in whether patterns of nucleotide diversity at G6PD are consistent with balancing selection for amino acid variation, as predicted by the observed cost and benefit associated with G6PD deficiency (Ruwende et al. 1995).
Finally, we were interested in whether G6PD amino acid polymorphism is consistent with selection for malarial resistance. If this is the case, then we may expect these G6PD variants to be both common and very recent in age, as found for the A− and Med variants (Tishkoff et al. 2001). Although the A variant does not seem to act as a significant barrier to malarial infection (Ruwende et al. 1995), a site-directed mutagenesis experiment found that both the A− and A variants are needed to produce the 12% enzyme activity observed for the A− allele (Town et al. 1992). Therefore, a characterization of the underlying nucleotide variability associated with the A allele may reveal the effects of a yet unknown selective pressure that is responsible for the frequency of this common variant.
Subjects and Methods
Population Samples
Sequence variation was surveyed from DNA samples from 216 male individuals from 13 populations (table 1). Our sub-Saharan African sample (labeled as “African”) consists of 160 individuals from 8 different groups. Several of these samples were from similar ethnic groups that individually were small in sample size (often <5); therefore, they were pooled together (see table 1). We also sampled 56 individuals from groups outside sub-Saharan Africa (who are hereafter referred to as “non-African”). Finally, the same G6PD fragment was sequenced from four male chimpanzees (Pan troglodytes troglodytes) for interspecific comparisons. The G6PD locus is found on the X chromosome (at Xq28); therefore, the sampling of males enabled the unambiguous determination of all polymorphic sites (i.e., no heterozygous sites) and complete haplotype phase for all individuals. The present study was approved by the institutional review board at the University of Maryland. All samples were gathered with informed consent.
Table 1.
Population Diversity Estimates
| Sample | na | Sb | πc | Dd | he |
| African: | 160 | 22 | .72 | −.90 | .85 |
| West African: | 105 | 18 | .71 | −.46 | .88 |
| Cameroonf | 16 | 8 | .55 | −.23 | .75 |
| Sierra Leoneg | 44 | 14 | .72 | −.26 | .79 |
| Nigeria | 22 | 10 | .69 | .11 | .79 |
| Bakola Pygmies | 23 | 11 | .71 | −.08 | .88 |
| East African: | 32 | 15 | .71 | −.73 | .92 |
| Hadza | 11 | 9 | .70 | −.28 | .83 |
| Maasai | 10 | 11 | .70 | −.17 | .79 |
| Sandawe | 11 | 12 | .76 | −1.04 | .94 |
| South African Bantu-speakers | 23 | 11 | .76 | .16 | .78 |
| Non-African: | 56 | 8 | .35 | −.46 | .45 |
| Tunisia | 9 | 8 | .65 | −.42 | .54 |
| Cyprus | 9 | 3 | .34 | 1.02 | .69 |
| Lebanon | 12 | 3 | .30 | .78 | .44 |
| South American Andean | 12 | 3 | .35 | 1.47 | .49 |
| Papua New Guinea | 14 | 2 | .07 | −1.46 | .14 |
| Global | 216 | 23 | .73 | −.95 | .82 |
Number of chromosomes.
Number of silent-site SNPs.
Average pairwise sequence differences per site (× 10−3).
Tajima's D statistic (all tests nonsignificant).
Haplotype diversity for B alleles only (see the “Results” section).
Includes the Mandara, Podoko, Uldeme, Bakaka, and Bassa groups.
Includes the Mende, Temne, Creole, Fula, Limba, Loko, and Madingo groups.
PCR and Sequence Determination
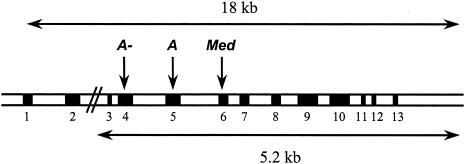
All primers used in the present study are based on GenBank accession number X55448 and are available at S. A. Tishkoff's Web page (Tishkoff Lab at the University of Maryland). PCR was used to amplify the 5.2-kb fragment shown in figure 1. Exon 1 is not translated, and exon 2 is separated from exons 3–13 by the 10 kb of intron 2, which includes many Alu repeats. Although it is possible that additional undiscovered mutations may reside in the 210 bp of exon 2, our analysis focused on the region that spans exons 3–13, where nearly all deficiency mutations recorded to date have been discovered. PCR products were prepared for sequencing by using shrimp alkaline phosphatase and exonuclease I (US Biochemicals). All nucleotide sequence data were obtained using the ABI Big Dye terminator kit and the 3100 automated sequencer (Applied Biosystems), and all variants were confirmed on both strands. Sequence files were aligned using the Sequencher program (version 4.0.5; Gene Codes).
Figure 1.
Diagram of the G6PD locus, spanning ∼18 kb. Exons are shown as blackened boxes, and introns and noncoding regions are shown as unblackened boxes. Intron 2 is nearly 10 kb in length. The 5.2-kb region that was sequenced for this study is shown, as well as the location of the common A−, A, and Med replacement SNPs.
Data Analysis
Samples were initially screened for the A, A−, and Med variants, as described by Tishkoff et al. (2001), and a constructed random sample (CRS) was sequenced on the basis of the estimated population frequencies of the different major alleles (Hudson et al. 1994). For example, the A variant has an average frequency of 20% in Africa. Therefore, our African samples were screened for the A variant, and we randomly sampled 32 A alleles to include in our total of 160 Africans (for a total frequency of 20%). If certain alleles of interest are over- or underrepresented in our sample, then this will significantly influence estimates of nucleotide variability, population structure, age of alleles, and LD at the G6PD locus. By this approach, parameter estimates (i.e., θ and μ) and statistical tests for our sample will reflect the frequencies of major G6PD alleles in natural populations.
This data set will also enable us to make comparisons of LD within African and non-African samples, as well as for deficiency and normal-activity alleles. Stronger LD may be associated with non-African samples because of recent population expansion (Tishkoff et al. 1996, 1998, 2000; Tishkoff and Williams 2002), whereas strong LD may be associated with deficiency alleles if selection has recently increased their frequencies. The Rozas and Rozas (1999) DnaSP program (version 3.53) was used to compute the correlations for all pairwise comparisons between SNPs as R2. Significant associations were identified with χ2 tests by using a Bonferroni correction for multiple comparisons (Sokal and Rohlf 1995). We also used the four-gamete test of Hudson and Kaplan (1985) to estimate the minimum number of recombination events in our sample.
A locus-specific variation estimate based on the number of segregating silent sites (which will include SNPs both at synonymous sites and in introns throughout the study) and corrected by sample size was calculated using Watterson's θ (1975). This estimate was compared with π, which is an estimate based on the average number of pairwise differences at silent sites between all alleles. These two estimates of the parameter 3Neμ (for X-linked genes) were calculated for all populations and allele classes (e.g., B, A, and A−). These two parameter estimates are expected to be equal under a strict model of neutrality, which can be assessed using the test designed by Tajima (1989). Positive D values may be consistent with balancing selection, whereas negative D values suggest directional selection. We were also interested in whether selection on amino acid variants has distorted the genealogical structure for specific allele samples. For example, compared to the overall level of silent polymorphism at G6PD, does our sample of alleles bearing the A− variant contain a significantly lower level of silent variation? Using the coalescent simulator, found in DnaSP, that adopts the approach of Hudson et al. (1994), we constructed 10,000 replicate trees based on the observed θ at G6PD and compared the observed levels of SNP variation for each of the B, A, and A− samples with the simulated data sets.
The ratio of amino acid replacement:silent polymorphisms within humans was compared with the ratio of replacement:silent fixed differences between humans and chimpanzees, using Fisher's exact test of independence (Sokal and Rohlf 1995), as described by McDonald and Kreitman (1991). Because polymorphism and divergence are expected to be correlated, the ratios of these different nucleotide-site comparisons should be equal under neutrality. The Hudson, Kreitman, and Aguade (HKA) test (Hudson et al. 1987) can be applied to test whether the level of silent-site polymorphism at G6PD is consistent with neutrality. If a simple neutral explanation accounts for variation at G6PD, then we may expect that silent-site polymorphism and divergence will be the same across loci. We tested this hypothesis by comparing G6PD with other X-linked loci for which estimates of silent-site polymorphism and divergence were available.
We used the statistic FST, from Hudson et al. (1992), as a relative measure of population differentiation, with all estimates weighted by sample size. We also used the Snn statistic, from Hudson (2000), to measure the amount of genetic differentiation across subpopulations. As Snn approaches 1, a sequence's most closely related sequence is more often found in other sampled populations than in its own population. A permutation test shuffles the populations and reconstructs them on the basis of their original sample sizes. The proportion of samples with Snn larger than or equal to the observed value is the estimated P value. Whereas our estimate of FST is a standard measure of population differentiation, the Snn statistic, using pairwise sequence differences, provides a method to detect genetic differentiation among subpopulations. A Bonferroni test was used to correct for multiple comparisons (Sokal and Rohlf 1995).
Finally, we used two different analyses to examine the evolutionary relationship of G6PD alleles. Although recombination can obscure the true genealogical relationship among alleles, these analyses likely reflect an accurate representation of the genealogical structure of our sample, because we find evidence for only one recombination event (see below). First, using MEGA (version 2.1) (Kumar et al. 2001), we constructed a simple neighbor-joining tree in which chimpanzee sequence was used as an outgroup to visualize both the population and allelic structure at G6PD. This analysis is based only on silent SNPs within and between humans and chimpanzees, thus illustrating the clustering of silent variation and populations independently of replacement SNPs. Second, we used the coalescent analysis outlined by Griffiths and Tavare (1997) and the GENETREE program (available at R. C. Griffiths' Web page [Genetree Software Version 9.0]) to estimate the time to the most recent common ancestor (TMRCA) for our entire sample, as well as for the B, A, and A− alleles. This analysis assumes no homoplasy in our sample; therefore, we removed haplotype N (which accounts for only two individuals in fig. 2) to correct for the single observed recombination event in our sample. This method uses maximum-likelihood (ML) coalescent analyses to estimate the tree topology under the assumptions that mutations are neutral and that allele age, frequency, and intra-allelic variability are correlated (Slatkin and Rannala 1997; Wuif and Donnelly 1999). Although selection has likely altered patterns of DNA variation at G6PD, we can examine the ages of alleles that would be consistent with a model of neutrality, and we can compare this with the results from other studies that have used the same approach (Harding et al. 1997; Harris and Hey 1999, 2001; Jaruzelska et al. 1999; Fullerton et al. 2000).
Figure 2.
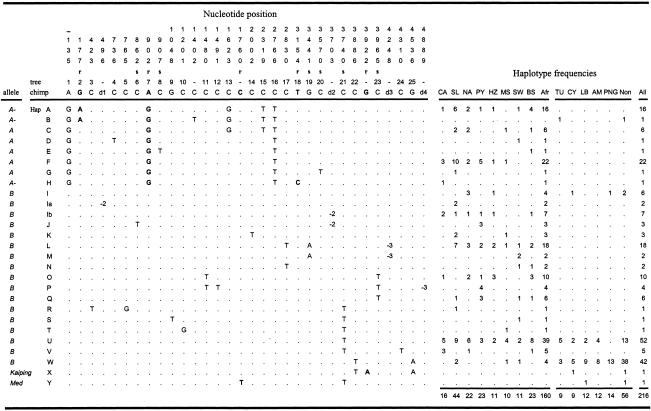
Summary data for 5.2-kb region of G6PD for 216 individuals. All polymorphisms are shown as derived changes as compared with the “chimp” sequence. Nucleotide positions start with the first base pair of exon 3 (the first SNP is in the second intron 135 bp before exon 3). Coding-region SNPs are labeled as “r” and “s,” for replacement and silent sites, respectively; deletions are labeled as “d1”–“d4”; and the row labeled as “tree” refers to the African SNPs that were used in the GENETREE analysis (fig. 5). Although haplotype H was originally typed as an A− allele, it does not have the site 177 SNP that is common to all other A− alleles (see the “Results” section).
Results
G6PD Nucleotide Sequence Variation
A diagram of the 5.2-kb region, on the G6PD gene, that was surveyed in the present study is shown in figure 1. There are 1,428 bp in exons 3–13 (475 codons) and 3,772 bp of noncoding sequence (introns and a portion of the 3′ UTR), for a total of 4,058 effectively silent sites. This number is used to calculate all measures of nucleotide diversity and is equal to the sum of all sites within exons that are silent (i.e., third-base positions) and sites within introns except the two conserved splice sites at the beginning and end of each intron. All SNP data are shown in figure 2 with the haplotype-frequency summary for each of the 13 population samples.
The constructed random sample (CRS) (see the “Subjects and Methods”section) from 160 Africans included 112 B, 32 A, and 16 A− alleles, whereas the sample from 56 non-Africans included 1 Med, 1 A−, and 54 B alleles. A total of 32 variable sites were discovered in the 5.2-kb region. There are four insertion/deletion polymorphisms in introns (comparison with the chimpanzee G6PD implies that all are deletions) and 28 SNPs at 17 intron and 11 coding sites. Of the 11 SNPs in coding regions, 5 replacement SNPs were found at sites 177 (Val→Met change at amino acid residue 68, which results in the A− variant), 902 (Asn→Asp change at amino acid residue 126, which results in the A variant), 1760 (Ser→Phe change at amino acid residue 188, which results in the Med variant), 3154 (Leu→Pro change at amino acid residue 323, which results in a second A− variant), and 3922 (Arg→His change at amino acid residue 463, which results in the Kaiping variant). Earlier functional assays had labeled all alleles that possessed a specific enzyme activity and electrophoretic profile as “A− alleles.” However, DNA analyses revealed that these samples were not all due to the replacement SNP at site 177. For example, a replacement SNP at site 3154 was found in our Cameroon sample; this replacement SNP was originally labeled as “an A− variant,” because it possesses similar enzyme activity to the common A− variant at site 177, and is also found only with the A variant (Beutler 1994). The amino acid replacement that occurs at site 3922 was found in our Cyprus sample; this variant was first labeled as “the Kaiping deficiency” and was initially discovered in China and Laos (Beutler et al. 1992; Chang et al. 1992).
All measures of sequence variability are summarized in tables 1 and 2. Because insertion/deletion polymorphisms likely do not conform to the same expectations as SNPs (i.e., mutation rates may differ), they are omitted from all statistical analyses. On the basis of silent sites for the overall sample of 216 individuals, our two nucleotide-variability measures, θ and π, are 0.094% and 0.073%, respectively, which is consistent with other polymorphism studies (Przeworski et al. 2000; Nachman 2001). Table 1 shows that all Tajima tests are nonsignificant, which suggests that the polymorphism-frequency distribution for our overall sample is consistent with that expected under a simple model of neutrality.
Table 2.
Intra-Allelic Diversity Estimates
| Sample | na | Sb | πc | Dd |
| Global: | ||||
| Be | 165 | 15 | .55 | −.62 |
| A | 32 | 5 | .20 | −.91 |
| A− | 17 | 1 | .03 | −1.15 |
| A/A− | 49 | 6 | .29 | −.30 |
| African: | ||||
| B | 112 | 15 | .51 | −.86 |
| A | 32 | 5 | .20 | −.91 |
| A− | 16 | 0 | .00 | …f |
| A/A− | 48 | 5 | .27 | −.04 |
| Non-Africane | 53 | 3 | .30 | 1.77 |
| Chimpanzee | 4 | 9 | 1.10 | −.79 |
Number of chromosomes.
Number of silent-site SNPs.
Average pairwise sequence differences per site (× 10−3).
Tajima's D statistic (all tests nonsignificant).
Includes only the normal B alleles.
Tajima's test cannot be performed because no silent SNPs are found in this sample.
Table 2 shows that the level of silent-site nucleotide variability associated with the global sample of 165 normal B alleles (π=0.055%) is almost twice that seen for all 49 A/A− alleles (π=0.030%), including 32 A and 17 A− alleles. Our Tajima tests show no significant skew in the polymorphism-frequency distribution for each of the B, A, and A− allelic groups. The global sample of 17 A− alleles segregates only a singleton polymorphism (i.e., a variant that is only found once), which is found at site 1402. Additionally, the A− variant at site 177 is the only difference between all A− and A alleles. In the examination of the variation associated with the different allele classes, the non-African sample was omitted from our DnaSP coalescent simulations, because it contains only a single A− allele. Given the number of silent SNPs in our CRS from 160 Africans (22 silent SNPs), our simulations find that the presence of 15 SNPs in the sample of 112 B alleles is not unusual. However, the association of five silent SNPs with the 32 A alleles (P<.01) and the complete lack of polymorphism in the 16 A− alleles (P < .001) were both rare events in the coalescent simulations. These observations support a recent ancestry for the A− allele class, which is consistent with the microsatellite and LD analyses by Tishkoff et al. (2001). These results also suggest that the A allele may have recently risen to intermediate frequency as well. Finally, not one of the 23 silent SNPs is shared among the A and B clades, and, on average, A and B alleles differ by twice as many silent SNPs as any two B alleles in our sample (4.3 vs. 2.0).
A total of 12 Med alleles sampled from Cyprus and Lebanon were initially sequenced for the same 5.2-kb region to examine the nucleotide variation associated with sequences bearing this specific deficiency variant. For our statistical analyses, only a single individual was included in our non-African CRS to reflect the frequency of the Med variant in this geographic region. Incidentally, all 12 sequences were identical, which reflects a recent ancestry of the Med variant; this recent ancestry is also consistent with Tishkoff et al. (2001).
LD
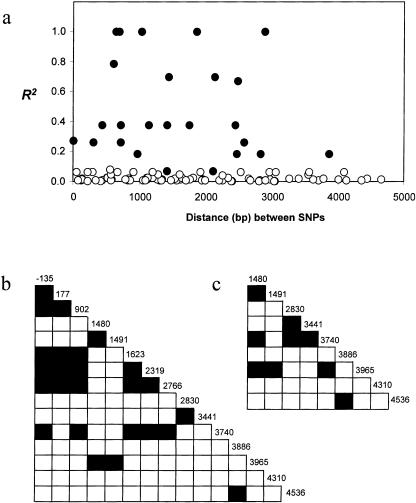
If the frequencies of several SNPs are rare, then LD analyses will lack the power to detect significant associations among sites, and, consequently, this can result in an overestimate of linkage equilibrium among SNPs. In our analysis, we find that our sample size is sufficiently large to detect significant LD with SNPs that have frequencies ⩾3%. Only three SNPs met this criterion in the CRS from 56 non-Africans, and all three pairwise comparisons among them were highly significant (each R2>0.83, P<.0001). In the CRS from 160 Africans, 15 SNPs had a frequency ⩾3%. Figures 3a and 3b show the 25 comparisons, of the possible 105 pairwise comparisons, that are significant (each P<.0001). Because much of this LD might result from differences between the B and A clades, we also tested LD within each allele class. The A alleles have only two SNPs with frequencies >3% (sites 1623 and 2319), and they are in significant LD (P<.0001). There are nine SNPs in the 112 African B alleles that are >3% in frequency, and 9 of the 36 pairwise comparisons among them exhibit significant LD (shown in fig. 3c) (P<.0001). We may expect that strong LD on deficiency alleles is a result of recent selection for malarial resistance; however, we find that significant LD at G6PD is also associated with African B alleles that possess normal enzyme activity. Further evidence for strong LD within our sample is provided by the four-gamete test (Hudson and Kaplan 1985), which reveals only one recombination event (between SNPs 2830 and 3441) in our sample.
Figure 3.
Measurements and plots of LD and association. a, Relationship between the LD measure as R2 and the distance (in bp) between 15 SNPs found at frequencies >3% in the African sample from 160 individuals. Blackened circles indicate the 25 associations, of the 105 possible, that are significant at the 0.01% level by a χ2 test and Bonferroni correction. b, Plot of the association among the 15 African SNPs for 160 individuals. Blackened boxes indicate significant LD at the 0.01% level, and numbers denote the nucleotide positions of SNPs in figure 2. c, Plot of LD for the 112 African B alleles. Blackened boxes indicate significance at the 0.01% level for 9 of the possible 36 associations, for the nine SNPs found at frequencies >3% in the African sample.
Population Differentiation
Our estimates of silent-site nucleotide variability for the CRS from 160 Africans are 0.095% and 0.072% for θ and π, respectively (table 1). We find that our African sample exhibits twice as much genetic diversity as our non-African sample (π=0.035%), and, of the 28 SNPs found in our global sample, only 1 silent and 2 replacement singletons are unique to the non-African sample. Although there is a threefold difference in sample size between our African and non-African samples, this is accounted for in all population parameter estimates (e.g., θ, π, and h) and therefore cannot explain the differences in nucleotide or haplotype diversity between the two samples.
Because our CRS is based on our a priori knowledge of the average frequencies of the A and A− variants in African and non-African populations, it is statistically invalid to subsequently test for differences in frequency for the A and A− variants across our population samples. We may expect that selection acting on the replacement SNPs determines their geographic variation and that silent SNPs in our sample will also show the same pattern of geographic variation because of the strong LD across the G6PD gene; however, we expect that the haplotype structure composed of silent SNPs within each of the B, A, and A− classes will reflect neutral processes, such as gene flow and drift (Berry and Kreitman 1993; Verrelli and Eanes 2001; Hamblin et al. 2002). We were interested in examining both the A and B allele classes for population structure. However, the A allele class segregates only two SNPs (sites 1623 and 2319) and is not found outside our African sample; therefore, we used only the global sample of 165 B alleles.
Table 1 shows that the African B haplotype diversity (0.85) is almost twice that of the non-African sample (0.45). The FST estimates in table 3 show that, although 13%–16% of the total diversity is found between the African and non-African samples, the differences among African populations are very small. The Hudson (2000) Snn statistical analysis finds significant population structure between African and non-African samples independently of the A and A− variants. Although there is little difference in genetic diversity among African groups, we do find significant heterogeneity within the West African sample. A further inspection of the West African samples shows that the Pygmy sample is genetically different (P<.01), which is consistent with other studies of this ethnic group (Cavalli-Sforza et al. 1996) and may reflect reduced gene flow between Pygmies and other Africans.
Table 3.
Summary Statistics of Population Structure
| Sample Contrasta | FST | Snnb | P |
| All (13) | .159 | .190 | .0001 |
| Among Africans (8) | .021 | .228 | .0001 |
| Among African regions (3) | .000 | .472 | .1710 |
| Among all regions (4): | .128 | .508 | .0001 |
| West vs. East Africans | .000 | .626 | .1320 |
| West vs. South Africans | .000 | .685 | .3180 |
| West vs. non-Africans | .151 | .793 | .0001 |
| East vs. South Africans | .000 | .461 | .8040 |
| East vs. non-Africans | .131 | .757 | .0001 |
| South vs. non-Africans | .160 | .804 | .0001 |
| Among East Africans (3) | .022 | .418 | .0840 |
| Among West Africans (4) | .030 | .415 | .0001 |
| Among non-Africans (5) | .107 | .212 | .1930 |
| Africans vs. non-Africans (2) | .127 | .805 | .0001 |
Number of population samples in contrast is given in parentheses. “African regions” denotes the West, East, and South African groups; “all regions” denotes the three African groups and the non-African sample. Only B alleles were included (see the “Results” section).
Hudson's (2000) Snn statistic. Significance was assessed by permutation tests (P<.001 [in boldface italics]; see the “Subjects and Methods” section).
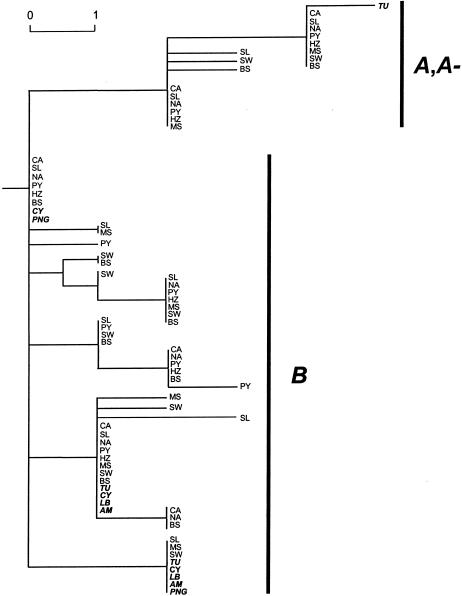
Although all replacement SNPs were omitted from our neighbor-joining analysis, figure 4 shows two distinct clades for the A/A− and B alleles. This clustering of silent variation independently of the A and A− replacement SNPs demonstrates both the strong LD between silent SNPs and the lack of shared variation between the A and B clades. Figure 4 also indicates that A, A−, and B haplotypes are spread across the population samples; therefore, it is unlikely that the lack of shared variation between the two clades is a result of reduced gene flow. Finally, figure 4 demonstrates the lack of haplotype diversity for the non-African sample and shows that this sample is largely nested within African lineages.
Figure 4.
Neighbor-joining tree, based on only silent substitutions among 216 individuals and rooted with chimpanzee G6PD sequence. Each haplotype is represented only once for each time it is found in a different population. Scale indicates one substitution. Non-African haplotypes are shown in boldface italics. The A and A− haplotypes differ by only a single replacement SNP; therefore, except for the single Tunisian A− allele that possesses a silent SNP, there is no distinct A− haplotype clade. CA = Cameroon; SL = Sierra Leone; NA = Nigeria; PY = Bakola Pygmies; HZ = Hadza; MS = Maasai; SW = Sandawe; BS = South African Bantu-speakers; TU = Tunisia; CY = Cyprus; LB = Lebanon; AM = South American Andean; PNG = Papua New Guinea.
Interspecific Comparisons
The first observation from our analysis of chimpanzee G6PD sequences is that the B variant is the ancestral state. In our chimpanzee sample, we discovered nine silent SNPs (two of which were within exons) and no replacement SNPs, in addition to an 8-bp insertion polymorphism and a 21-bp deletion polymorphism (both of which were in introns). Our estimate of G6PD nucleotide variability from four individuals (π=0.110%) is much greater than that found in humans but is consistent with other polymorphism studies in chimpanzees (Deinard and Kidd 2000; Kaessmann et al. 2001; Ebersberger et al. 2002). Our analysis of interspecific divergence between chimpanzees and humans finds 44 and 0 fixed differences at silent and replacement sites, respectively. Table 4 contrasts this interspecific divergence with the magnitude of intraspecific polymorphism found in our human samples by using a McDonald-Kreitman test of neutrality (McDonald and Kreitman 1991). Our global sample shows a significant result, which is also seen when the African and non-African samples are independently tested. Because the A− allele contributes two replacement SNPs and is found only once in our non-African sample, the inclusion of this single allele alone may result in a significant test. However, if we remove the A− allele from our non-African sample, then our McDonald-Kreitman test is still significant with three silent and two replacement SNPs from the remaining 55 individuals (P<.01).
Table 4.
McDonald-Kreitman Tests of Neutrality
| Samplea | Silent | Replacement | Fisher's Exact Test |
| Africans (n=160): | |||
| Polymorphic | 22 | 3 | P<.05 |
| Fixed | 44 | 0 | |
| Non-Africans (n=56): | |||
| Polymorphic | 8 | 4 | P<.001 |
| Fixed | 44 | 0 | |
| Global (n=216): | |||
| Polymorphic | 23 | 5 | P<.01 |
| Fixed | 44 | 0 |
Numbers of polymorphisms and fixed differences found at both silent and replacement (i.e., those that change the amino acid) sites are contrasted with a 2×2 Fisher's exact test of independence (see the “Subjects and Methods” section).
In general, our estimates of silent-site variation within humans (π = 0.073%) and silent-site divergence from chimpanzees (1.1%) are both consistent with other genes (see Przeworski et al. 2000; Nachman 2001). Using HKA tests, we contrasted G6PD with other X-linked loci in table 5. It is difficult to compare the G6PD polymorphism and divergence for our non-African sample with other loci, since other studies often include very different non-African populations. In addition, because we find significant genetic differentiation between our African and non-African samples, we cannot combine all individuals from these two geographic regions to statistically test for differences in silent variation between G6PD and other loci (e.g., see Begun and Aquadro 1993). Table 5 shows that our HKA tests for X-linked loci typed in African samples are nonsignificant, thus indicating that neither an unusual level of silent-site polymorphism within humans nor silent-site divergence with chimpanzees can explain our significant McDonald-Kreitman tests. It is apparent that the number of human G6PD replacement SNPs is significantly greater than we would expect from a neutral model of evolution.
Table 5.
HKA Tests of G6PD versus Other X-Linked Loci for African Samples
| Locus | na | Sitesb | Sc | Dd | HKA χ2 | Reference |
| PDAH1 | 16 | 3,788 | 24 | 44 | 1.250 | Harris and Hey (1999) |
| ZFX | 113 | 1,089 | 7 | 13 | .048 | Jaruzelska et al. (1999) |
| Xq13.3 | 23 | 10,163 | 24 | 95 | .254 | Kaessmann et al. (2001) |
| Dmd (int44) | 10 | 3,000 | 15 | 27 | 1.700 | Nachman and Crowell (2000) |
| FIX | 16 | 3,731 | 6 | 37 | .750 | Harris and Hey (2001) |
| FY (regVIII) | 16 | 1,440 | 6 | 54 | 2.064 | Hamblin et al. (2002) |
Number of chromosomes sampled.
Number of silent sites at sampled locus.
Number of silent-site SNPs.
Divergence measured as number of silent-site differences between chimpanzees and humans.
Estimating the Ages of G6PD Variants
We were primarily interested in estimating the TMRCA for the B, A, and A− allele classes; therefore, we include only the African sample in our GENETREE analysis of the age of G6PD SNPs for two reasons. First, it is apparent from the present study, as well as others (e.g., Tishkoff and Williams 2002), that non-Africans represent a subset of the genetic diversity found within African populations, which suggests that the common ancestor for our non-African sample is rooted within Africa. Second, our non-African sample can contribute very little to our estimate of the age of the A variant, given that this variant was found only once in our non-African sample. For this analysis, we used the observed number of silent differences between humans and chimpanzees as our locus-specific estimate of the neutral mutation rate. If we take the average of the two within-species nucleotide-variation estimates from π and subtract this from the amount of divergence between species, then our estimate of the net sequence divergence is 43 fixed differences. Under the assumptions of a human-chimpanzee divergence time of ∼5 million years (Myr) BP (Horai et al. 1992) and of ∼20 years per generation, the estimated neutral mutation rate per generation (μ) along each species' lineage is 9×10-5. We used the GENETREE ML estimate of the parameter θ and our μ estimate from above to calculate the effective population size (Ne) from the relationship θ=3Neμ (X linked) as 19,800 for our African sample. Figure 5 shows the results for all 25 African SNPs, and table 6 summarizes age estimates of specific interest.
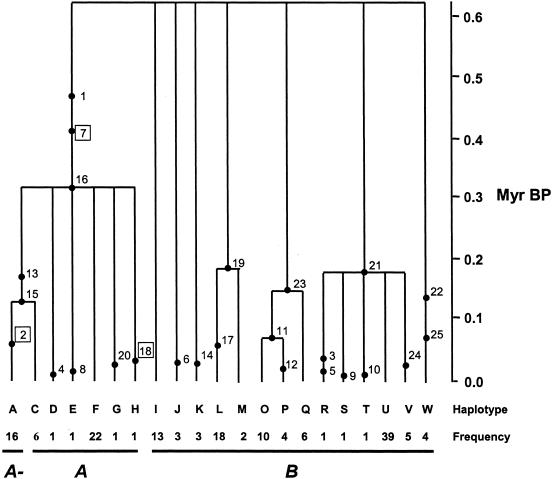
Figure 5.
GENETREE results for 158 African individuals (haplotype N is omitted; see the “Subjects and Methods” section). The number and letter codes for the SNPs and haplotypes, respectively, are found in figure 2. All replacement SNPs are shown in boxes. SNPs 2 and 7 designate the common A− and A variants, respectively. Although haplotype H was originally labeled as “an A− allele” on the basis of functional analyses, it does not have the common SNP 2 (see the “Results” section). The order of SNPs in time is arbitrary when they are found on the same branch (e.g., SNPs 1, 7, and 16), and ages for all variants are mean estimates.
Table 6.
Summary of GENETREE Estimates
| Sample | na | θMLb | Nec | TMRCAd | 95% CI |
| African | 158 | 5.35 | 19,800 | 620 | 480–760 |
| B lineage | 110 | 3.40 | 13,000 | 426 | 172–680 |
| A lineage | 32 | 1.73 | 6,400 | 316 | 72–560 |
| Non-Africane | 55 | 1.00 | 3,700 | 241 | 52–430 |
Number of chromosomes sampled after predicted recombinants were omitted (see the “Subjects and Methods” section).
ML estimate of nucleotide variability per locus from the GENETREE analysis.
Effective population size derived from θML and μ=9×10-5/locus/generation (see the “Results” section).
Mean age estimate (in kyr) derived from Ne, with a generation time of 20 years (see the “Results” section).
The single A− allele is omitted (see the “Results” section).
Our coalescent analysis enables us to estimate the ages of SNPs (i.e., the A and A− variants), as well as the coalescent time back to a common ancestor for specific haplotype groups (i.e., the TMRCA for each of the B and A clades). We have estimated the TMRCA of our African sample at 620 thousand years (kyr) BP (95% CI 480–760 kyr), which is similar to estimates from other gene genealogies for African samples (Harding et al. 1997; Fullerton et al. 2000). Our estimate of the TMRCA for our 112 African B alleles (426±254 kyr BP) is not significantly different from the TMRCA for the entire sample. We also find that the coalescent time for the A clade (316±244 kyr BP, which is the point at which our sampled A alleles coalescence to a common ancestor) overlaps with that of the B clade. Because the TMRCAs for each of the A and B clades are not significantly different, it is likely that the divergence within these two clades occurred during the same relative time period. This time period also includes our estimate for the origin of the A variant (420±120 kyr BP). Using the same methodology, we find, for our non-African B sample, an ancestry that is more recent (241±189 kyr BP) than that for our African B sample. This is likely explained by a recent founding and a smaller effective population size of non-African populations, which is consistent with other studies using the same approach (Harding et al. 1997; Harris and Hey 1999, 2001). Although our estimates for the ages of the A− (45±20 kyr BP) and Med (10±25 kyr BP) variants are slightly higher than that found by Tishkoff et al. (2001), they represent a very recent origin for these deficiencies relative to that found for all other G6PD alleles.
Discussion
In screening large samples from human populations, initial studies had as their primary goal the identification of new G6PD deficiencies based on enzymatic properties. Although such screening was necessary for understanding the functional implications of mutations in this gene region, especially in relationship to malarial endemicity, we were interested in characterizing the amino acid variation in a random sample of individuals. Therefore, in an effort to better understand how selection has shaped G6PD diversity globally, we have sampled 56 non-African and 160 African individuals, together representing the largest study, to date, of nuclear DNA diversity for sub-Saharan Africans. By using this sequence-based approach, we have identified amino acid changes at the DNA level in a large sample, and we have compared these changes with findings from functional assays. The present study finds five replacement SNPs, which have all been documented in previous studies and are associated with reduced enzyme activity compared with normal B alleles. The absence of any amino acid variation other than that responsible for the common deficiencies implies that there is very little hidden G6PD amino acid replacement polymorphism at high frequency.
The Signature of Balancing Selection
Compared with other loci, at G6PD, we see a typical level of silent-site fixation (1.1%) between chimpanzees and humans, which implies a normal level of fixation of neutral mutations at this locus. If much of the human G6PD protein variation is simply neutral, then we may expect that, like the silent-site variation, some of this neutral amino acid variation increases in frequency and contributes to fixation between humans and chimpanzees. However, the absence of amino acid fixation suggests that there has been strong historical purifying selection against G6PD amino acid polymorphism and that this has been operating in both species. Given the lack of amino acid fixation, the significant excess of G6PD amino acid polymorphism that we find segregating within human populations is all the more surprising.
A significant excess of amino acid polymorphism is typically explained by either balancing selection or slightly deleterious selection. If amino acid polymorphisms are completely deleterious, then they are removed by selection almost immediately. However, if purifying selection against deleterious amino acid variants within species is sufficiently weak because amino acid variation is “slightly” deleterious, then these amino acid variants may remain polymorphic at low frequencies, but eventually selection will keep them from high frequencies and fixation (Ohta 1992). Although this theory is in accordance with the possession, by several genes, of rare amino acid polymorphisms (Nachman et al. 1996; Nielsen and Weinreich 1999; Harding et al. 2000; Fay et al. 2001; Smirnova et al. 2001), other genes do not show this pattern (Clark et al. 1998; Salamon et al. 1999; Fullerton et al. 2000; Koda et al. 2001). Amino acid polymorphisms at G6PD are not all rare in frequency, and, in fact, many are found to be as high as 40%–70% in frequency in specific populations. For example, in addition to the common A− and Med variants in African and Mediterranean regions, respectively, the Union variant, from Melanesia (Ganczakowski et al. 1995), and the Orissa variant, from India (Kaeda et al. 1995), are common to their respective geographic regions. Although this does not imply that all amino acid polymorphism is adaptive, both the high-frequency and positive-fitness attributes of enzyme deficiencies (Ruwende et al. 1995) convincingly reject a slightly deleterious selection model at G6PD.
Although the association between enzyme deficiencies and geographic regions of malarial prevalence largely suggested balancing selection, it was unclear how this might impact variation both within and between species at the nucleotide level. Our comparison of human and chimpanzee G6PD nucleotide sequences implies that there has been no protein change for this enzyme possibly for the past 5 Myr. Therefore, the abundant G6PD amino acid polymorphism is likely the result of a relatively recent change in selective pressures in human populations. Detrimental amino acid polymorphisms that would be selectively removed in the effectively larger population of Africa may be neutral and can accumulate in the relatively smaller effective population of non-Africans. This argument has been used to explain the presence, in non-African populations, of rare melanocortin-1 receptor amino acid polymorphisms that are absent in African populations (Harding et al. 2000). In contrast, our McDonald-Kreitman analysis finds a significant excess of G6PD amino acid polymorphism in both African and non-African populations. If this amino acid polymorphism in non-African populations simply reflected a recent increase in neutral variation, then we might also expect a relative increase in silent-site variation; however, we actually find an eightfold reduction of silent-site variation in non-Africans as compared with Africans. In addition, our contrasts of G6PD with other X-linked loci by using HKA tests indicate that silent polymorphism in African populations is not unusually high (which was also recently found by Saunders et al. [in press]). This suggests that G6PD amino acid polymorphism in both African and non-African populations is maintained by selection.
Global Patterns of Population Differentiation
Our analysis of silent variation at G6PD in figure 4, showing that global populations likely had an ancestral root within Africa, and our coalescent analysis, showing a large difference in Ne between Africans and non-Africans, are both consistent with other studies (Harding et al. 1997; Harris and Hey 1999, 2001; Jaruzelska et al. 1999; Fullerton et al. 2000). This difference in genetic diversity between these two geographic regions is often explained by the colonization of non-African populations by an effectively smaller number of individuals emerging out of Africa (Tishkoff and Williams 2002). Although our estimates of Ne for both African and non-African populations may differ compared with other studies, different genes are expected to show contrasting patterns of genetic diversity because Ne likely varies across genomic regions as a result of selection, drift, different recombination rates, and the stochastic variance that is associated with population sampling (Hey 1997; Hey and Harris 1999).
Although there are several documented cases in which population-specific G6PD variants have been found in other geographic regions (see Beutler 1994), including our finding of the Kaiping variant (of Southeast Asia) in Cyprus, this observation likely reflects limited gene flow across non-African populations. We might expect that gene flow plays a large role in the distribution of adaptive variation to different populations and that restricted gene flow would hinder the evolution of human traits, such as resistance to disease. However, our analysis suggests that selection does not necessarily favor specific G6PD amino acid variants per se but that enzyme deficiency in general is adaptive. This is seen from the convergence of populations toward a similar enzyme-deficiency phenotype that is the result of unique mutations at the G6PD locus. Therefore, in spite of limited gene flow, recent balancing selection can effectively maintain both normal and deficiency G6PD phenotypes in natural populations, because of repeated amino acid mutation. This is an example of convergent evolution due to a common selective pressure across populations and is analogous to the pattern of variation observed for β-globin hemopathologies, such as sickle cell anemia and thalassemia.
A general observation from other genetic diversity analyses of mtDNA and nuclear genes, as well as microsatellites, implies that African populations may have been historically subdivided (Tishkoff and Williams 2002), yet we find very little genetic differentiation among our African samples. However, the extent to which subdivision will be detected will depend on the relative time of population divergence, gene flow, the underlying level of genetic variability, and, most important, the differential strength of selection across the genome. Therefore, studies with highly variable markers (i.e., microsatellite haplotypes) may be more informative for the detection of population structure in Africa (Tishkoff et al. 1996, 1998, 2000; Jorde et al. 1997; Tishkoff and Williams 2002). Nonetheless, the observation that A− alleles sampled from geographically diverse African groups possess the same SNP and microsatellite haplotype backgrounds indicates that both gene flow and selection have maintained this enzyme deficiency across Africa (Tishkoff et al. 2001).
The Ages of G6PD Amino Acid Variants
We were interested in how our coalescent analyses of nucleotide variation would compare with the approach of Tishkoff et al. (2001) in using LD and microsatellite variation. The analyses of Tishkoff et al. (2001) estimated the ages of the A− and Med variants to be 3,840–11,760 and 1,600–6,640 years BP, respectively. The absence of fixed silent SNPs between A and A− alleles and the significantly low level of silent variation associated with the sample of A− alleles (as found by the DnaSP coalescent simulations) support a recent origin and rapid increase in frequency for the A− variant, which has an estimated age of 45±20 kyr BP from our GENETREE analysis. The lack of nucleotide sequence variation associated with our sample of 12 Med alleles and the estimated age of the Med variant as 10±25 kyr BP from our GENETREE analysis are also consistent with a recent ancestry for this deficiency. In contrast to our study of DNA sequence variation, Tishkoff et al. (2001) used faster-evolving microsatellite markers to more accurately resolve the ages of the A− and Med variants. Tishkoff et al. (2001) also considered selection models to fit the pattern of variation associated with the A− and Med alleles, whereas our standard coalescent model assumes strict neutrality. Therefore, although deficiency alleles that are high in frequency may appear to be older in origin, their frequencies may simply be a result of recent selection. The relative lack of nucleotide variation associated with the A− and Med alleles indicates that this may be the case. Our significant McDonald-Kreitman tests also suggest that the general pattern of G6PD amino acid polymorphism is consistent with relatively recent balancing selection.
Although the A variant does not appear to be recently derived, the coalescent time for the entire A clade is consistent with the estimated time of divergence of African and non-African populations within the past 100,000 years (Tishkoff and Williams 2002). It is possible that the A variant is old in origin but that the divergence of the A clade may have been recent and may have occurred sometime after the colonization of non-African groups. Several derived SNPs are associated with the A clade; however, our Tajima tests find no skew toward rare SNPs that may be expected if selection has recently increased the frequency of the A variant. Simulations show that Tajima's test does possess the statistical power to detect distortions in the polymorphism-frequency spectrum (Braverman et al. 1995); however, this test may be weak if selection has been relatively recent (Simonsen et al. 1995). Therefore, if selection on the A and A− alleles has been recent, then our Tajima tests are likely nonsignificant because there has been little time for SNPs to accumulate on these haplotype backgrounds. In contrast, our DnaSP coalescent simulations find that both the A and A− clades segregate significantly lower levels of SNP variation given the overall level of variation at G6PD. If the A variant was historically lower in frequency and has recently become more common because of selection, then this may have provided little opportunity for recombination between A and B alleles and may explain the association of several derived SNPs (i.e., sites −135, 1623, 2319, and 2766) with A alleles.
Although several G6PD variants may be under selection for resistance to malaria, our analysis of the A variant suggests that this SNP predates the estimated age at which severe malaria has likely had a major impact in humans. It is possible that there was a period of time when malaria was less severe; therefore, selection for G6PD deficiencies with low, but not severely reduced, enzyme activity maintained the A variant at low frequencies. With the expansion of human populations and the rapid growth of agriculture, selection for more-extreme G6PD deficiencies (i.e., the A and Med variants) may have become necessary as severe malaria became highly prevalent. As with the CCR5-Δ32 polymorphism, which is not recent in origin but is strongly associated with resistance to HIV infection (Martin et al. 1998), it is also possible that the A variant had been historically maintained by an unknown selective pressure in African populations. This possibility may require additional studies of the A variant, to identify an association with malarial resistance or with another function altogether. Finally, Saunders et al. (in press) find significant LD between the A− allele and genes as far as 550 kb away. Therefore, future analyses of haplotype variation at other genes in close proximity may elucidate the effect that selection on the A and A− variants has had on LD in this region.
Depending on the surrounding environment, G6PD variants can be beneficial or detrimental. We may expect that selection for malarial resistance initially drives up the frequencies of new deficiencies but also that balancing selection for normal enzyme activity eventually maintains these deficiency alleles at intermediate frequencies. Therefore, it is unlikely that these amino acid polymorphisms will become fixed. Instead, some form of spatially or temporally varying selective pressure may maintain A, A−, and Med deficiency alleles, in addition to normal B alleles, across geographic regions. Because selection for the A− and Med variants has been recent, analyses of microsatellite haplotypes were informative for the demonstration of the recent ancestry of these alleles (Tishkoff et al. 2001). In contrast, because SNP mutation rates are lower, they are more informative for the investigation of historical events that occur deep in the genealogical history of genes—such as the fixation of silent and replacement SNPs between humans and chimpanzees.
Because of the historical association between Plasmodium and mammals, many Plasmodium species have adapted to specific primate hosts. However, unlike humans that are infected by P. falciparum, many nonhuman primate species are infected by other Plasmodium species that are associated with less-severe forms of malaria (Ollomo et al. 1997). In comparison with human G6PD, our chimpanzee sample has a higher level of silent variation yet segregates no amino acid variation. Although this sample is small, it may be the case that G6PD amino acid variation is not adaptive in chimpanzees, because malaria may be rare and less detrimental in this species. Future analyses of G6PD nucleotide sequence variation in nonhuman primates may reveal how selection impacts closely related species that are exposed to varying degrees of Plasmodium infection.
In accordance with other studies, we have revealed that selection for disease resistance can be associated with a complex pattern of nucleotide variability (Clark et al. 1998; Fullerton et al. 2000; Kidd et al. 2000; Nachman and Crowell 2000; Hamblin et al. 2002; Tishkoff and Williams 2002). To determine how selection has impacted different genomic regions, one may need to employ a combination of genetic markers with different mutation rates (i.e., microsatellites and SNPs). In addition, it is imperative to examine patterns of coevolution among Plasmodium, Anopheles, and human genomes in the development of new vaccines and disease-prevention tactics (Hoffman et al. 2002; Miller et al. 2002). Finally, it has been proposed that common diseases—such as hypertension, diabetes, and obesity—are results of historical selection for mutations beneficial in an ancestral environment but detrimental in modern environments (i.e., “thrifty” genotypes). Therefore, characterization of selection in the human genome will be important for the identification of loci that are associated with common diseases. Analysis of genetic variation at G6PD, a locus thought to be under strong selection, indicates that certain tests of selection may lack the power to reject neutrality. However, our comparisons of intraspecific variation with interspecific divergence, as well as analyses of microsatellite variation and LD, reflect the signature of recent balancing selection at G6PD.
Acknowledgments
We thank M. Stoneking, T. Jenkins, A. Lane, N. Salem, E. Chouery, A. Megarbane, V. Delague, E. Tarazona, A. Awomoyi, and A. Gessain, for providing samples and assistance; R. Hudson and L. Matzkin, for their analytical advice; M. Saunders, M. Hammer, and M. Nachman, for helpful comments and for sharing unpublished results; and two anonymous reviewers, for helpful comments on the manuscript. This project was funded by a Burroughs Wellcome Fund Career Award, a David and Lucille Packard Career Award, and National Science Foundation grant BCS-9905396 (all to S.A.T.). B.C.V. was partially supported by National Science Foundation Integrative Graduate Education and Research Traineeships training grant BCS-9987590.
Electronic Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for accession number X55448)
- Genetree Software Version 9.0, http://www.stats.ox.ac.uk/~griff/software.html (for the GENETREE program)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for G6PD [MIM 305900])
- Tishkoff Lab at the University of Maryland, http://www.life.umd.edu/biology/tishkofflab/ (for G6PD primers)
References
- Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP (2000) Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol 17:1467–1482 [DOI] [PubMed] [Google Scholar]
- Aquadro CF, Bauer DuMont V, Reed FA (2001) Genome-wide variation in the human and fruitfly: a comparison. Curr Opin Genet Dev 11:627–634 [DOI] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF (1993) African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature 365:548–550 [DOI] [PubMed] [Google Scholar]
- Berry AJ, Kreitman M (1993) Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics 134:869–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E (1994) G6PD deficiency. Blood 84:3613–3636 [PubMed] [Google Scholar]
- Beutler E, Westwood B, Kuhl W, Hsia YE (1992) Glucose-6-phosphate dehydrogenase variants in Hawaii. Hum Hered 42:327–329 [DOI] [PubMed] [Google Scholar]
- Braverman JM, Hudson RR, Kaplan NL, Langley CH, Stephan W (1995) The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics 140:783–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1996) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Chang J-G, Chiou S-S, Perng L-I, Chen T-C, Liu T-C, Lee L-S, Chen P-H, Tang T-K (1992) Molecular characterization of glucose-6-phosphate dehydrogenase (G6PD) deficiency by natural and amplification created restriction sites: five mutations account for most G6PD deficiency cases in Taiwan. Blood 80:1079–1082 [PubMed] [Google Scholar]
- Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengård J, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF (1998) Haplotype structure and population genetic inferences from nucleotide sequence variation in human lipoprotein lipase. Am J Hum Genet 63:595–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke GS, Hill AVS (2001) Genetics of susceptibility to human infectious disease. Nat Rev Genet 2:967–977 [DOI] [PubMed] [Google Scholar]
- Deeb SS, Lindsey DT, Hibiya Y, Sanocki E, Winderickx J, Teller DY, Motulsky AG (1992) Genotype-phenotype relationships in human red/green color-vision defects: molecular and psychophysical studies. Am J Hum Genet 51:687–700 [PMC free article] [PubMed] [Google Scholar]
- Deinard AS, Kidd KK (2000) Identifying conservation units within captive chimpanzee populations. Am J Phys Anthropol 111:25–44 [DOI] [PubMed] [Google Scholar]
- Donnelly MJ, Licht MC, Lehmann T (2001) Evidence for recent population expansion in the evolutionary history of the malaria vectors Anopheles arabiensis and Anopheles gambiae. Mol Biol Evol 18:1353–1364 [DOI] [PubMed] [Google Scholar]
- Ebersberger I, Metzler D, Schwarz C, Pääbo S (2002) Genomewide comparison of DNA sequences between humans and chimpanzees. Am J Hum Genet 70:1490–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wyckoff GJ, Wu C-I (2001) Positive and negative selection on the human genome. Genetics 158:1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton SM, Clark AG, Weiss KM, Nickerson DA, Taylor SL, Stengård JH, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF (2000) Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am J Hum Genet 67:881–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganczakowski M, Town M, Bowden DK, Vulliamy TJ, Kaneko A, Clegg JB, Weatherall DJ, Luzzatto L (1995) Multiple glucose-6-phosphate dehydrogenase–deficient variants correlate with malaria endemicity in the Vanuatu Archipelago (southwestern Pacific). Am J Hum Genet 56:294–301 [PMC free article] [PubMed] [Google Scholar]
- Greenwood B, Mutabingwa T (2002) Malaria in 2002. Nature 415:670–672 [DOI] [PubMed] [Google Scholar]
- Griffiths RC, Tavare S (1997) Computational methods for the coalescent. In: Donnelly P, Tavare S (eds) Progress in population genetics and human evolution. Springer-Verlag, New York, pp 165–182 [Google Scholar]
- Hamblin MT, Thompson EE, Di Rienzo A (2002) Complex signatures of natural selection at the Duffy blood group locus. Am J Hum Genet 70:369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding RM, Fullerton SM, Griffiths RC, Bond J, Cox MJ, Schneider JA, Moulin DS, Clegg JB (1997) Archaic African and Asian lineages in the genetic ancestry of modern humans. Am J Hum Genet 60:772–789 [PMC free article] [PubMed] [Google Scholar]
- Harding RM, Healy E, Ray AJ, Ellis NS, Flanagan N, Todd C, Dixon C, Sajantila A, Jackson IJ, Birch-Machin MA, Rees JL (2000) Evidence for variable selective pressures at MC1R. Am J Hum Genet 66:1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EE, Hey J (1999) X chromosome evidence for ancient human histories. Proc Natl Acad Sci USA 96:3320–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2001) Human populations show reduced DNA sequence variation at the Factor IX locus. Curr Biol 11:774–778 [DOI] [PubMed] [Google Scholar]
- Hey J (1997) Mitochondrial and nuclear genes present conflicting portraits of human origins. Mol Biol Evol 14:166–172 [DOI] [PubMed] [Google Scholar]
- Hey J, Harris EE (1999) Population bottlenecks and patterns of human polymorphism. Mol Biol Evol 16:1423–1426 [DOI] [PubMed] [Google Scholar]
- Hoffman SL, Subramanian GM, Collins FH, Venter JC (2002) Plasmodium, human and Anopheles genomics and malaria. Nature 415:702–709 [DOI] [PubMed] [Google Scholar]
- Horai S, Satta Y, Hayasaka K, Kondo R, Inoue T, Ishida T, Hayashi S, Takahata N (1992) Man's place in Hominoidea revealed by mitochondrial DNA genealogy. J Mol Evol 35:32–43 [DOI] [PubMed] [Google Scholar]
- Hudson RR (2000) A new statistic for detecting genetic differentiation. Genetics 155:2011–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Bailey K, Skarecky D, Kwiatowski J, Ayala FJ (1994) Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics 136:1329–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL (1985) Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP (1992) Estimation of levels of gene flow from DNA sequence data. Genetics 132:583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaruzelska J, Zietkiewicz E, Batzer M, Cole DEC, Moisan J-P, Scozzari R, Tavare S, Labuda D (1999) Spatial and temporal distribution of the neutral polymorphisms in the last ZFX intron: analysis of the haplotype structure and genealogy. Genetics 152:1091–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde LB, Rogers AR, Bamshad M, Watkins WS, Krakowiak P, Sung S, Kere J, Harpending HC (1997) Microsatellite diversity and the demographic history of modern humans. Proc Natl Acad Sci USA 94:3100–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeda JS, Chhotray GP, Ranjit MR, Bautista JM, Reddy PH, Stevens D, Naidu JM, Britt RP, Vulliamy TJ, Luzzatto L, Mason PJ (1995) A new glucose-6-phosphate dehydrogenase variant, G6PD Orissa (44 Ala→Gly), is the major polymorphic variant in tribal populations in India. Am J Hum Genet 57:1335–1341 [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Wiebe V, Weiss G, Pääbo S (2001) Great ape DNA sequences reveal a reduced diversity and an expansion in humans. Nat Genet 27:155–156 [DOI] [PubMed] [Google Scholar]
- Kay AC, Kuhl W, Prchal JT, Beutler E (1992) The origin of glucose-6-phosphate dehydrogenase (G6PD) polymorphisms in Afro-Americans. Am J Hum Genet 50:394–398 [PMC free article] [PubMed] [Google Scholar]
- Kidd JR, Pakstis AJ, Zhao H, Lu R-B, Okonofua FE, Odunsi A, Grigorenko E, Batsheva B-T, Friedlaender J, Schulz LO, Parnas J, Kidd KK (2000) Haplotypes and linkage disequilibrium at the phenylalanine hydroxylase locus, PAH, in a global representation of populations. Am J Hum Genet 66:1882–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koda Y, Tachida H, Pang H, Liu Y, Soejima M, Ghaderi AA, Takenaka O, Kimura H (2001) Contrasting patterns of polymorphisms at the ABO-secretor gene (FUT2) and plasma α(1,3)fucosyltransferase gene (FUT6) in human populations. Genetics 158:747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software, Arizona State University, Tempe, AZ [DOI] [PubMed] [Google Scholar]
- Livingstone FB (1971) Malaria and human polymorphisms. Annu Rev Genet 5:33–64 [DOI] [PubMed] [Google Scholar]
- Luzzatto L, Mehta A, Vulliamy TJ (2001) In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease. McGraw-Hill, New York, pp 4517–4553 [Google Scholar]
- Martin MP, Dean M, Smith MW, Winkler C, Gerrard B, Michael NL, Lee B, Doms RW, Margolick J, Buchbinder S, Goedert JJ, O'Brien TR, Hilgartner MW, Vlahov D, O'Brien SJ, Carrington M (1998) Genetic acceleration of AIDS progression by a promoter variant at CCR5. Science 282:1907–1911 [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654 [DOI] [PubMed] [Google Scholar]
- Miller LH (1994) Impact of malaria on genetic polymorphism and genetic diseases in Africans and African Americans. Proc Natl Acad Sci USA 91:2415–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LH, Baruch DI, Marsh K, Doumbo OK (2002) The pathogenic basis of malaria. Nature 415:673–679 [DOI] [PubMed] [Google Scholar]
- Mu J, Duan J, Makova KD, Joy DA, Huynh CQ, Branch OH, Li WH, Su XZ (2002) Chromosome-wide SNPs reveal an ancient origin for Plasmodium falciparum. Nature 18:323–326 [DOI] [PubMed] [Google Scholar]
- Nachman MW (2001) Single nucleotide polymorphisms and recombination rate in humans. Trends Genet 17:481–485 [DOI] [PubMed] [Google Scholar]
- Nachman MW, Brown WM, Stoneking M, Aquadro CF (1996) Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics 142:953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL (2000) Contrasting evolutionary histories of two introns of the Duchenne muscular dystrophy gene, Dmd, in humans. Genetics 155:1855–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness DS (1986) Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science 232:193–202 [DOI] [PubMed] [Google Scholar]
- Nielsen R, Weinreich DM (1999) The age of nonsynonymous and synonymous mutations in animal mtDNA and implications for the mildly deleterious theory. Genetics 153:497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T (1992) The nearly neutral theory of molecular evolution. Ann Rev Ecol Syst 23:263–286 [Google Scholar]
- Ollomo B, Karch S, Bureau P, Elissa N, Georges AJ, Millet P (1997) Lack of malaria parasite transmission between apes and humans in Gabon. Am J Trop Med Hyg 56:440–445 [DOI] [PubMed] [Google Scholar]
- Przeworski M, Hudson RR, Di Rienzo A (2000) Adjusting the focus on human variation. Trends Genet 16:296–302 [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175 [DOI] [PubMed] [Google Scholar]
- Ruwende C, Khoo SC, Snow RW, Yates SNR, Kwiatkowski D, Gupta S, Warn P, Allsopp CEM, Gilbert SC, Peschu N, Newbold CI, Greenwood BM, Marsh K, Hill AVS (1995) Natural selection of hemi- and heterozygotes for G6PD deficiency in Africa by resistance to severe malaria. Nature 376:246–249 [DOI] [PubMed] [Google Scholar]
- Salamon H, Klitz W, Easteal S, Gao X, Erlich HA, Fernandez-Vina M, Trachtenburg EA, McWeeney SK, Nelson MP, Thomson G (1999) Evolution of HLA class II molecules: allelic and amino acid site variability across populations. Genetics 152:393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders MA, Hammer MF, Nachman MW. Nucleotide variability at G6PD and the signature of malarial selection in humans. Genetics (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen KL, Churchill GA, Aquadro CF (1995) Properties of statistical tests of neutrality for DNA polymorphism data. Genetics 141:413–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Rannala B (1997) Estimating the age of alleles by use of intraallelic variability. Am J Hum Genet 60:447–458 [PMC free article] [PubMed] [Google Scholar]
- Smirnova I, Hamblin MT, McBride C, Beutler B, Di Rienzo A (2001) Excess of rare amino acid polymorphisms in the Toll-like receptor 4 in humans. Genetics 158:1657–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ (1995) Biometry. W. H. Freeman, San Francisco [Google Scholar]
- Stephens JC, Reich DE, Goldstein DB, Shin HD, Smith MW, Carrington M, Winkler C, et al (1998) Dating the origin of the CCR5-Δ32 AIDS-resistance allele by the coalescence of haplotypes. Am J Hum Genet 62:1507–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Dietzsch E, Speed W, Pakstis AJ, Kidd JR, Cheung K, Bonne-Tamir B, Santachiara-Benerecetti AS, Moral P, Krings M (1996) Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science 271:1380–1387 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Goldman A, Calafell F, Speed WC, Deinard AS, Bonne-Tamir B, Kidd JR, Pakstis AJ, Jenkins T, Kidd KK (1998) A global haplotype analysis of the myotonic dystrophy locus: implications for the evolution of modern humans and for the origin of myotonic dystrophy mutations. Am J Hum Genet 62:1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Pakstis AJ, Stoneking M, Kidd JR, Destro-Bisol G, Sanjantila A, Lu RB, Deinard AS, Sirugo G, Jenkins T, Kidd KK, Clark AG (2000) Short tandem-repeat polymorphism/Alu haplotype variation at the PLAT locus: implications for modern human origins. Am J Hum Genet 67:901–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, Destro-Bisol G, Drousiotou A, Dangerfield B, Lefranc G, Loiselet J, Piro A, Stoneking M, Tagarelli A, Tagarelli G, Touma EH, Williams SM, Clark AG (2001) Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science 293:455–462 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM (2002) Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet 3:611–621 [DOI] [PubMed] [Google Scholar]
- Toole JJ, Pittman DD, Orr EC, Murtha P, Wasley LC, Kaufman RJ (1986) A large region (approximately equal to 95 kDa) of human factor VIII is dispensable for in vitro procoagulant activity. Proc Natl Acad Sci USA 83:5939–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town M, Bautista JM, Mason PJ, Luzzatto L (1992) Both mutations in G6PD A− are necessary to produce the G6PD deficient phenotype. Hum Mol Genet 1:171–174 [DOI] [PubMed] [Google Scholar]
- Verrelli BC, Eanes WF (2001) Clinal variation for amino acid polymorphisms at the Pgm locus in Drosophila melanogaster. Genetics 158:1649–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliamy TJ, Mason P, Luzzatto L (1992) The molecular basis of glucose-6-phosphate dehydrogenase deficiency. Trends Genet 8:138–142 [DOI] [PubMed] [Google Scholar]
- Wall JD, Przeworski M (2000) When did the human population size start increasing? Genetics 155:1865–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA (1975) On the number of segregating sites in genetic models without recombination. Theor Popul Biol 7:256–276 [DOI] [PubMed] [Google Scholar]
- Wuif C, Donnelly P (1999) Conditional genealogies and the age of a neutral mutant. Theor Popul Biol 56:183–201 [DOI] [PubMed] [Google Scholar]