Abstract
The methylation of lysine residues in the N-terminal tails of histones is a highly conserved mechanism that regulates critical functions of chromatin, such as the control of gene expression. Using a biochemical approach, we recently identified new methylation marks on the histone H4 tail in budding yeast at lysines 5, 8 and 12, catalyzed by the previously-uncharacterized enzyme Set5. Genetic studies revealed that Set5 functions in cellular processes that also rely on the global chromatin modifying complexes COMPASS and NuA4, which methylate H3 lysine 4 and acetylate H4 lysines 5, 8 and 12, respectively. The identification of new methylation events on the H4 tail raises many intriguing questions regarding their function and their interaction with known histone modifications. Here, we analyze the insights gained about the new enzyme Set5 and the implications for new functionality added to the H4 tail.
Keywords: lysine methylation, histone, chromatin, Set5, COMPASS, NuA4, acetylation, yeast
Lysine Methylation Signaling at Chromatin
The covalent post-translational modification (PTM) of histones, including acetylation, methylation, phosphorylation and ubiquitylation, is a critical mechanism to direct fundamental DNA templated processes such as transcription and DNA repair.1 The dynamic marking of histones is controlled by a set of enzymes charged with either adding or removing these PTMs. Specifically, methylation of lysine (K) residues is performed by lysine methyltransferases (KMTs), which can add a mono-, di- or tri-methyl mark to the lysine side chain.2 The addition of a methyl mark establishes a platform for the binding and activity of chromatin effector proteins. These methyl-lysine binding proteins are stabilized at chromatin in a manner specified by the extent and context of the methylation, and they transduce a biological signal to affect a response at chromatin.3 Histone methyl marks therefore act as signals that orchestrate proper programming of the genome, and aberrant methylation signaling is implicated in the initiation and progression of many human diseases.4,5
Lysine methyltransferases exist in two evolutionarily-conserved structural classes: the SET (named for the Drosophilia Su(var) 3–9, Enhancer of Zeste and Trithorax proteins) domain family and the seven-β strand family.6,7 To date, KMTs that methylate histones are largely defined by the catalytic SET domain, whereas only one histone KMT, Dot1, possesses the seven-β strand domain.1 The human proteome contains greater than 50 KMTs of the SET domain family, while budding yeast has 12 members.8 The catalytic activity and substrate specificity of the majority of the human enzymes remains unknown, and there are four remaining yeast enzymes for which no substrates have been described. Furthermore, proteomic analysis of both yeast and mammalian cells highlight the existence of uncharacterized methylated histone species for which there is no identified KMT.9,10 The extensive study of the canonical histone methylation sites H3K4, H3K36 and H3K79, and their cognate KMTs in yeast—Set1, Set2 and Dot1–has provided essential insight in to the function of these marks in humans.11 Therefore, evolutionarily conserved mechanisms of chromatin function can be revealed through the identification and investigation of new methylation events in yeast.
We have recently discovered the existence of methyl marks on the H4 tail at lysines 5, 8 and 12, catalyzed by the enzyme Set5 in budding yeast. In vitro analysis revealed that Set5 is a monomethyltransferase and in vivo studies showed that monomethylation of these H4 residues is SET5-dependent in cells.12 This work identifies the first known substrate for the enzyme Set5 and demonstrates that H4 is subject to lysine methylation on the functionally-important residues K5, K8 and K12. Here, we will highlight the insights gained regarding the newly-characterized enzyme Set5 and discuss the possible functions for and implications of methylation of the H4 tail lysines 5, 8 and 12.
Set5: A New Yeast Histone Methyltransferase
Set5 was a previously uncharacterized member of the SET-domain family from budding yeast. Unlike the majority of yeast SET domain KMTs, Set5, along with Set6, contains a split SET domain and has two consecutive zinc fingers, one canonical and one unique.13,14 These structural elements and sequence comparisons indicate that Set5 is orthologous to the mammalian SMYD family of lysine methyltransferases, which likewise contain split SET domains and a zinc finger domain known as MYND.6 Recent work from our lab has shown that SMYD3 is an H4K5 methyltransferase both in vitro and in human cells.15 This is the first demonstration that methylation of H4K5 exists in human cells and indicates functional conservation of Set5′s activity. Importantly, SMYD3 has been implicated in tumorigenesis: knockdown of SMYD3 inhibits proliferation and anchorage-independent growth of cancer cell lines15,16 and overexpression of SMYD3 has been observed in liver, breast and rectal carcinomas.17,18 The study of Set5 and H4 methylation in yeast may therefore uncover conserved mechanisms that contribute to SMYD3-dependent oncogenesis in human cells.
In yeast, the only KMT known to have more than one substrate is Set1, which targets both histone H3 and the kinetochore component Dam1.19,20 It remains to be determined if Set5 has additional substrates, however in vitro experiments showed that Set5 is capable of methylating both histone H2A and the histone variant H2A.Z,12 although we have been unable to detect these methyl marks in cells (data not shown). The acetyltransferase Esa1, which is responsible for H4 K5, K8 and K12 acetylation, also targets histone H2A and H2A.Z both in vitro and in vivo,21,22 suggesting that, at the very least, there are likely to be parallels between the structural mechanisms directing substrate specificity for these two enzymes. Set5 interacts with chromatin in cells, indicating that it may be methylating H4 at chromatin. However, a significant fraction of Set5 is not chromatin-associated and is cytoplasmic.12 This subcellular distribution suggests that Set5′s access to chromatin may be regulated via nucleo-cytoplasmic shuttling, that it potentially targets newly-synthesized H4 in the cytoplasm, or that it has additional cytoplasmic substrates. Further biochemical and structural studies are needed to reveal the mechanisms directing Set5′s substrate selection and specificity, as well as to understand the regulatory control of its enzymatic activity.
Set5 Adds New Functionality to the H4 Tail
Histone modifications do not function in isolation, but rather they act in concert with other modifications, histone variants and chromatin-binding proteins to regulate chromatin structure and dynamics. In our recent work, we also sought to understand the function of H4 methylation by Set5 and place it in context with known histone modifications. Genetic interaction studies revealed that Set5 functions in parallel and/or compensating pathways to two global chromatin-modifying complexes: the COMPASS complex, which contains the H3 K4 methyltransferase Set1,23,24 and the NuA4 complex, which contains the H4 K5, K8 and K12 acetyltransferase Esa1.22,25 These results suggest that Set5′s methylation of H4 is likely to impact on cellular processes that also require H3K4 methylation and H4 K5, K8 and K12 acetylation.
Functional cooperation between Set5 and Set1
The H3K4 tri-methyltransferase Set1 is the catalytic subunit of the COMPASS complex, a highly conserved chromatin-regulatory complex that functions in the control of gene expression in both active and silent regions of the genome.23,24 H3K4 tri-methylation marks transcription start sites and is a hallmark for active gene expression,26,27 but it can also act as a signaling platform stabilizing repressive activities at chromatin28 and function in maintaining heterochromatin boundaries,29,30 indicating that it has multiple unique roles throughout the genome. We observed that combined deletion of SET5 and SET1 led to decreased fitness in response to cellular stress,12 suggesting the possibility that Set5 functions with Set1 to regulate gene expression. Although genome-wide microarray analysis did not indicate a global role for Set5 in transcription alone, it remains an open question whether Set5 may function with Set1 in specific cellular or genomic contexts to regulate transcription. Studies to investigate the role of Set5 in stress-dependent gene expression and at unique genomic regions, such as heterochromatin-euchromatin boundaries, may more precisely define the functional interaction between Set5 and Set1.
Dissecting the interplay between H4 methylation and acetylation at K5, K8 and K12
In addition to participating in crosstalk with modification of the H3 tail, the methylation of H4 by Set5 will likely impact modifications of the H4 tail. In budding yeast, the H4 tail is acetylated at lysines 5, 8, 12 and 16 by histone acetyltransferases (HATs), and removal of these marks is performed by histone deactylases (HDACs).31 Unlike methylation, acetylation of histone tails neutralizes the positive charge of the lysine residue. This provides two means by which acetylation can affect transactions at chromatin: (1) acting as a unique binding site for chromatin effector proteins, and (2) changing the charge state such that nucleosome-DNA or nucleosome-nucleosome interactions, and potentially higher-order chromatin structure, are altered.31 Studies in yeast have demonstrated that acetylation marks at lysines 5, 8 and 12 are correlated with one another, whereas acetylation at K16 is largely distinct.32,33 Specifically, H4K16ac is known to be a key regulator of silent chromatin,34-36 but also functions in other pathways, such as the response to DNA damage.37 Acetylation of H4 at K5, K8 and K12 has been implicated in transcription, chromatin assembly and DNA damage repair.38-40 In investigating each of these processes, site-specific mutation of the individual lysine residues revealed that lysines 5, 8 and 12 often possess similar functionality, can substitute for one another and are not likely to recruit opposing activities to chromatin.38,40,41 Despite the presence of this functional redundancy, unique roles for these modifiable lysines have been demonstrated, such as the recent discovery that H4 K12 has a specific function in the establishment of telomeric chromatin.42
Methylation and acetylation of the same lysine residue are mutually exclusive. Numerous lysines within the core histones are known to be sites of both methylation and acetylation, including H3K4 in yeast and humans, and H3K9, H3K27, H3K36 and, recently, H3K56 in humans.1,43 Analysis of the genomic pattern of the methyl and acetyl marks for each of these residues reveals that they do not co-exist in the same regions and generally have opposing functions.43,44 One exception to this is the overlapping patterns of acetylation and methylation at H3K4 in yeast and the observation that H3K4me directly regulates the localization of H3K4ac in this context.45
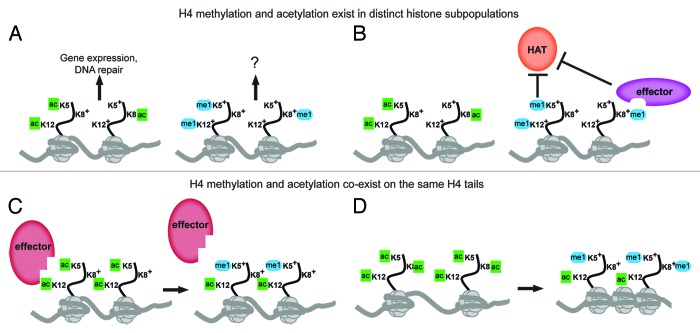
The H4 lysines 5, 8 and 12 possess a number of unique properties that distinguish them from these other sites of histone modification. Primarily, they are a cluster of three lysines which can all be modified by the same HAT, Esa1, or the same KMT, Set5, and they can functionally substitute for one another. This raises a number of questions regarding the coordinated regulation of acetylation and methylation of these residues. Do methylation and acetylation exist in distinct subpopulations of H4 or do they co-exist on the same H4 tail? Are their activities inhibitory or complementary to one another? The distribution of methylated H4 within the genome remains to be determined, however, there are multiple possible scenarios that could describe its relationship to acetylated H4 (Fig. 1). If methylation exists primarily on unacetylated H4 tails (i.e., methylation and acetylation are in different subpopulations of histones), the H4 methyl marks may function independently from acetylation at chromatin (Fig. 1A). However, it is also plausible that methylation may act as a counterpoint to acetylation through either active (the recruitment of effector proteins) or passive (the mere presence at the same lysines) inhibition of subsequent acetylation reactions (Fig. 1B). Alternatively, if methylation and acetylation co-exist on different lysines of the same H4 tail, methylation has the potential to impact both acetyl-lysine binding proteins (e.g., bromodomain proteins) (Fig. 1C) and the chromatin compaction and higher order structures mediated by the charge neutralization of acetylation (Fig. 1D).
Figure 1. The relationship between acetylation and methylation of H4 tail lysines 5, 8 and 12. (A-B) Acetyl and methyl marks on H4 may exist in distinct histone subpopulations. Acetylation of K5, K8 and K12 has been implicated in the control of gene expression and DNA damage repair, whereas the function of methylation remains unknown. Methylation may act independently of acetylation in the genome (A) or inhibit acetylation on K5, K8 and K12 either directly through blocking HAT activity or through the recruitment of effector proteins that may inhibit HAT activity (B) or potentially promote HDAC activity. (C-D) Acetylation and methylation of K5, K8 and K12 may co-exist on different lysines of the same H4 tail. Combinations of methyl and acetyl marks on the same H4 tail may affect the binding of chromatin effectors that recognize acetyl-lysine moieties, potentially interfering with their binding to chromatin (C) or modulating the acetyl-lysine state to promote or stabilize binding. The combination of methylation and acetylation at K5, K8 and K12 may also influence chromatin compaction or folding (D). Acetylation of histone tails is thought to lead to a decondensed chromatin state through the neutralization of the positive charge of the lysine, but the addition of methylation may promote or allow for compaction of the chromatin by maintaining the charge at H4 tail lysines, which could subsequently influence higher order folding.
An additional component to the relationship between H4 methylation and acetylation is their relative abundance in chromatin. H4 is highly acetylated in vivo, with over 80% of H4 molecules possessing at least one acetylated lysine. H4K16ac is the most abundant species, but approximately 30% of acetylated H4 contains K5ac, 25% contains K8ac and over 50% contains K12ac.46 In contrast, we expect H4 methyl marks to be in low abundance, as they are difficult to detect in vivo and mass spectrometry analysis revealed their presence in only a very small fraction of purified histones (data not shown). This suggests that, unlike the global role for H4 acetylation, H4 methylation is more likely to be playing a role in modulating chromatin functions in a site- or context-specific manner. Furthermore, any interactions between methylation and acetylation of K5, K8 and K12 are not likely to be occurring on a genome-wide scale, but rather, methylation may be influencing acetyl-lysine functions in specific and unique environments throughout the genome.
To investigate the relationship between H4 methylation and acetylation, we generated yeast strains lacking both SET5 and YNG2, a component of the NuA4 complex required for maintaining global H4 acetylation levels.47 These cells have decreased cell growth compared with cells lacking only one of these genes.12 Although we cannot disregard the possibility that this phenotype is dependent on other substrates for either Set5 or the NuA4 complex, this suggests functional cooperation between acetylation and methylation of H4 K5, K8 and K12 to promote cellular fitness. These data therefore argue that it is unlikely that H4 methylation and acetylation oppose one another, but rather that they function in similar pathways, which could occur independently of one other, or on the same tail. Studies of the genomic distribution of H4 methyl marks relative to acetyl marks and detailed molecular analysis of mutants lacking both methylation and acetylation will be required to elucidate the complex interplay between these modifications, and to understand the function of H4 K5, K8 and K12 methylation by Set5.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/20695
References
- 1.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green EM, Gozani O. Everybody's welcome: the big tent approach to epigenetic drug discovery. Drug Discov Today Ther Strateg. 2011;••• doi: 10.1016/j.ddstr.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurdistani SK. Histone modifications in cancer biology and prognosis. Prog Drug Res. 2011;67:91–106. doi: 10.1007/978-3-7643-8989-5_5. [DOI] [PubMed] [Google Scholar]
- 6.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katz JE, Dlakić M, Clarke S. Automated identification of putative methyltransferases from genomic open reading frames. Mol Cell Proteomics. 2003;2:525–40. doi: 10.1074/mcp.M300037-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Petrossian TC, Clarke SG. Uncovering the human methyltransferasome. Mol Cell Proteomics. 2011;10:M110–, 000976. doi: 10.1074/mcp.M110.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–28. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat Res. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol. 2006;7:657–66. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 12.Green EM, Mas G, Young NL, Garcia BA, Gozani O. Methylation of H4 lysines 5, 8 and 12 by yeast Set5 calibrates chromatin stress responses. Nat Struct Mol Biol. 2012;19:361–3. doi: 10.1038/nsmb.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Böhm S, Frishman D, Mewes HW. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 1997;25:2464–9. doi: 10.1093/nar/25.12.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porras-Yakushi TR, Whitelegge JP, Miranda TB, Clarke S. A novel SET domain methyltransferase modifies ribosomal protein Rpl23ab in yeast. J Biol Chem. 2005;280:34590–8. doi: 10.1074/jbc.M507672200. [DOI] [PubMed] [Google Scholar]
- 15.Van Aller GS, Reynoird N, Barbash O, Huddleston M, Liu S, Zmoos A-F, et al. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics. 2012;7:340–3. doi: 10.4161/epi.19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang SZ, Luo XG, Shen J, Zou JN, Lu YH, Xi T. Knockdown of SMYD3 by RNA interference inhibits cervical carcinoma cell growth and invasion in vitro. BMB Rep. 2008;41:294–9. doi: 10.5483/BMBRep.2008.41.4.294. [DOI] [PubMed] [Google Scholar]
- 17.Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–40. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 18.Hamamoto R, Silva FP, Tsuge M, Nishidate T, Katagiri T, Nakamura Y, et al. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–8. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SYR, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–95. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, et al. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–34. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–22. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, et al. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–19. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–7. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, et al. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–5. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 25.Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–26. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 27.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 28.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fingerman IM, Wu C-L, Wilson BD, Briggs SD. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J Biol Chem. 2005;280:28761–5. doi: 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venkatasubrahmanyam S, Hwang WW, Meneghini MD, Tong AHY, Madhani HD. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci U S A. 2007;104:16609–14. doi: 10.1073/pnas.0700914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 32.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–33. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 33.Millar CB, Kurdistani SK, Grunstein M. Acetylation of yeast histone H4 lysine 16: a switch for protein interactions in heterochromatin and euchromatin. Cold Spring Harb Symp Quant Biol. 2004;69:193–200. doi: 10.1101/sqb.2004.69.193. [DOI] [PubMed] [Google Scholar]
- 34.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–92. doi: 10.1016/0092-8674(95)90512-X. [DOI] [PubMed] [Google Scholar]
- 35.Park EC, Szostak JW. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol. 1990;10:4932–4. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–7. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 37.Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30:3582–95. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci U S A. 2005;102:5501–6. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/S0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 40.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–5. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 41.Ma XJ, Wu J, Altheim BA, Schultz MC, Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc Natl Acad Sci U S A. 1998;95:6693–8. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou BO, Wang S-S, Zhang Y, Fu X-H, Dang W, Lenzmeier BA, et al. Histone H4 lysine 12 acetylation regulates telomeric heterochromatin plasticity in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1001272. doi: 10.1371/journal.pgen.1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Y, Song C, Zhang Q, Dimaggio PA, Garcia BA, York A, et al. Histone H3 Lysine 56 Methylation Regulates DNA Replication through Its Interaction with PCNA. Mol Cell. 2012;46:7–17. doi: 10.1016/j.molcel.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guillemette B, Drogaris P, Lin H-HS, Armstrong H, Hiragami-Hamada K, Imhof A, et al. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 2011;7:e1001354. doi: 10.1371/journal.pgen.1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal Biochem. 2003;316:23–33. doi: 10.1016/S0003-2697(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 47.Nourani A, Doyon Y, Utley RT, Allard S, Lane WS, Côté J. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol Cell Biol. 2001;21:7629–40. doi: 10.1128/MCB.21.22.7629-7640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]