Abstract
Monoclonal antibodies constitute a robust class of therapeutic proteins. Their stability, resistance to stress conditions and high solubility have allowed the successful development and commercialization of over 40 antibody-based drugs. Although mAbs enjoy a relatively high probability of success compared with other therapeutic proteins, examples of projects that are suspended due to the instability of the molecule are not uncommon. Developability assessment studies have therefore been devised to identify early during process development problems associated with stability, solubility that is insufficient to meet expected dosing or sensitivity to stress. This set of experiments includes short-term stability studies at 2−8 þC, 25 þC and 40 þC, freeze-thaw studies, limited forced degradation studies and determination of the viscosity of high concentration samples. We present here three case studies reflecting three typical outcomes: (1) no major or unexpected degradation is found and the study results are used to inform early identification of degradation pathways and potential critical quality attributes within the Quality by Design framework defined by US Food and Drug Administration guidance documents; (2) identification of specific degradation pathway(s) that do not affect potency of the molecule, with subsequent definition of proper process control and formulation strategies; and (3) identification of degradation that affects potency, resulting in program termination and reallocation of resources.
Keywords: developability, discovery, stability, analytics, process control, development, forced degradation, CQA, QbD
Introduction
Development of a therapeutic protein is a long and costly process that can take over a decade from discovery to commercialization. Although therapeutic biologics generally have a higher probability of success than their small molecule counterparts,1 the rate of attrition remains substantial. While decisions to terminate projects can be based on the competitive landscape and commercial opportunities, projects are also terminated for technical reasons, including unsuitable safety profile, lack of efficacy in human, instability of the molecule or formulation, poor expression and purification issues. To reduce the risk associated with a given project, the sooner these technical challenges are identified, the sooner appropriate control strategies can be put in place. When irreducible challenges are identified, the project can be terminated and resources diverted to the development of a more promising molecule. Derisking process development early enough is critical to success because challenges not identified early enough may lead to expensive and time-consuming remediation steps later in the development of a given program. Determination of the safety profile is usually performed during preclinical studies and Phase 1 clinical studies in human, which is well into the development path. Likewise, the true assessment of a molecule efficacy is obtained during Phase 2/3 clinical studies.
In contrast with safety and efficacy assessments, a number of technical hurdles can be evaluated early during development programs. To identify technical challenges, we devised a comprehensive set of developability experiments and applied the strategy to a number of monoclonal antibodies (mAbs) currently in our pipeline. This strategy includes a series of in silico and experimental approaches that we grouped under the Developability Assessment term. In silico work includes structure sequence analysis based on primary sequence alignment and molecular model; prediction of possible degradation pathways is based on prior experience and on published literature. This constitutes the first step toward establishing potential critical attributes within the Quality by Design (QbD) paradigm encouraged by the US Food and Drug Administration.2 The experimental part of the developability assessment includes short-term stability studies at various temperatures, freeze-thaw studies and limited forced degradation studies. This series of studies determine the biochemical (e.g., sensitivity to oxidation and deamidation) and biophysical (e.g., unfolding and formation of large molecular weight species) stability profiles of the molecule of interest. Determination of maximum solubility and associated viscosity are also essential for molecules destined to be formulated at high concentrations for subcutaneous injections. Developability studies offer the first opportunity to asses the technical viability of a project at the industrial scale and function as a bridge between discovery and process development activities. We present in this report three case studies and include data generated during developability assessment of antibody molecules.
Results
General strategy for developability studies
Developability studies are considered a derisking activity designed to provide understanding of the colloidal properties and prevalent degradation pathways of a molecule early in the development process, thereby allowing the analytical strategy to be tailored to the molecule of interest. To be relevant, developability studies should be performed on material produced with a stable cell line of the same host as the one that will be used later for process development; when possible, use of the final stable clone is preferable.
The most widespread host for expression of mAbs is the Chinese hamster ovary (CHO) cell line; however, a developability strategy can be applied to material produced by any expression system. The recombinant protein is produced via an upstream early cell culture process and purified by a downstream, 2–3 columns standard process. It is important to note that, at this stage of development, the formulation has not been optimized and the protein matrix might be suboptimal. Keeping these limitations in mind, developability studies are aimed at detecting gross changes in the protein that could hinder the stability or potency of the molecule.
Sequence alignment and molecular modeling
Developability assessment starts with analysis of the sequence of the protein entering the development phase. Some of this work has likely been done in the discovery space. For example, potentially problematic residues such as methionine, asparagine or aspartic acid residues localized in exposed region of the mAb, including the complementarity-determining regions (CDRs), may have been removed. Relevant knowledge and experience generated during the discovery phase, such as whether the molecule was generated through phage display or hybridoma cells, should be taken into account. The first examination of the sequence during developability assessment is therefore aimed at identifying potential hot spots for degradation that remain in the sequence. Sequence alignment can be performed by any software available on the market. To complete the analysis of the primary sequence, building a molecular model may help to visualize residues exposed to the solvent, which are susceptible to degradation. The software we currently use for this exercise is MOE (CCG, Montreal, Canada).
Short-term research stability studies and freeze/thaw cycles
To gain a first impression about how the molecule will degrade, short-term stability studies are performed. Material is staged at -80°C, 2–8°C, 25°C and 40°C for up to 6 mo (Table 1). Samples are removed at different time points and analyzed by size exclusion (SEC) HPLC to detect the formation of high molecular weight species and by ion exchange (IEX) HPLC to measure the effect of stress on distribution of charges at the surface of the molecule due to the deamidation and isomerization of asparagine and aspartic acid residues. Samples are also analyzed by reducing and non-reducing sodium dodecyl sulfate capillary electrophoresis (CE-SDS) to detect proteolytic cleavage of the heavy or light chains, peptide mapping to quantify the formation of post-translational modifications formed during stress and a potency test, which is most commonly in a binding ELISA format in the very early stage of the program development.
Table 1. Developability stability schedule.
| Conditions | Initial | t = 0.5 mo | t = 1 mo | t = 2 mo | t = 3 mo | t = 6 mo |
|---|---|---|---|---|---|---|
| -80°C |
X |
/ |
/ |
/ |
/ |
X |
| 2–8°C |
|
X |
X |
X |
X |
X |
| 25°C |
|
X |
X |
X |
X |
X |
| 40°C | X | X | X | X | X |
/ No samples staged at these conditions/timepoints. X Samples staged and analyzed at these conditions/timepoints. Details of the assays used during stability are reported in Table S1
Propensity of the recombinant protein of interest to form aggregates is gauged during freeze/thaw cycles. The protein is subjected to five cycles. Aliquots are analyzed by SEC-HPLC, SEC-multi-angle laser light scatter and dynamic light scattering at each of the five cycles.
Limited stress conditions
To complete the initial assessment of the stability of the molecule, a series of stress conditions are applied (Table 2). These conditions include standard stresses suggested by regulatory agencies, such as exposure to low and high pHs, light and oxidative reagents.3 More emphasis may be put on some of these stress conditions if a potential degradation pathway has been identified by molecular modeling or during early experience with the material. The stresses are likely to trigger deamidation, isomerization, aggregation and oxidation of amino acid side chains. These degradation pathways are the most commonly described for mAbs.4 A direct measurement of the affect of the degradation on potency is also taken.
Table 2. Typical limited forced degradation conditions.
| Condition | Step | Duration | Tests |
|---|---|---|---|
| Low pH |
Adjust pH with HCl to 3.0 |
48hr |
UV, SEC, IEX, peptide mapping, potency |
| High pH |
Adjust pH with NaOH to 9.0 |
48hr |
UV, SEC, IEX, peptide mapping, potency |
| Light stress |
1X ICH (UV, Visible) |
N/A |
UV, SEC, IEX, peptide mapping, potency |
| Methionine oxidation |
0.05% tBHP |
2–5 h |
UV, SEC, IEX, peptide mapping, potency |
| pH jump | Coordinate with purification area | N/A | Measure A280, A320 and DLS before and after pH adjustment to pH5.5 |
Determination of viscosity and ability to concentrate formulated materials (surrogate for solubility studies)
High concentrations of protein are usually required when mAb therapeutics are administered via subcutaneous injection. To verify whether the drug candidate can achieve a target concentration, the protein is concentrated to 100–200 mg/mL in standard buffers. Viscosity of the high concentration solutions is measured to evaluate purification and delivery device strategies.
mAb A case study: No major or unexpected degradation
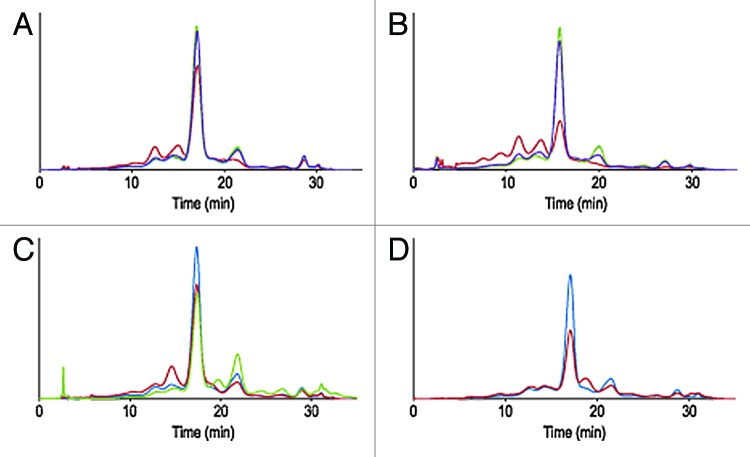
The first case study concerns an IgG1 antibody, termed mAb A. Sequence structure analysis of the CDRs did not reveal any particularly exposed asparagine or aspartic residues in an amino acid context prone to deamidation or isomerization (i.e., located directly upstream from a glycine, serine or proline residues).7 CDRs did not contain exposed methionine or unpaired cysteine residues. The short-term stability of early representative material was evaluated for up to three months at 2−8 þC, 25 þC and 40 þC via a battery of assays. IEX-HPLC profiles after one and three months are shown in Figure 1A and 1B. Decrease of relative peak area of the main and basic species (eluting after the main) and increase of the relative peak area of the acidic variants (eluting before the main) was noted for samples stored at 40 þC. A decrease of basic variants is typical for mAbs; temperature accelerates the cyclisation of N-terminus glutamine into pyroglutamic acid, a post-translational modification common to many therapeutic and endogenous human mAbs.8 Likewise, an increase of acidic variants upon exposure to elevated temperature is a common occurrence that may reflect deamidation of asparagine residue, isomerization of aspartic residues into isoaspartic or structural changes affecting the overall surface charge distribution of the molecule.8 In the present case, deamidation was only detected at conserved position 385 within the Fc portion of the mAb and did not exceed 10% (Table 3). Limited forced degradation studies at low and high pH (3.0 and 9.0) after 48 h at 40 þC, resulted in pH dependant degradation routes: exposure to high pH resulted in an increase of acidic variant (and deamidation of position 385), while low pH resulted in the formation of basic species not identified by peptide mapping (Fig. 1C, Table 3). Exposure to other forms of stress was performed and did not result in unexpected alterations of the primary, secondary or tertiary structure of the molecule. Exposure to UV/visible light following ICH guidelines, for instance, resulted in the formation of a basic species reflecting oxidation of conserved Fc methionines 253 and 429 (Fig. 1D, Table 3). Oxidation of the conserved Fc methionine residues in these conditions was expected and documented in the literature.4 While developability studies of mAb A did not identify major chemistry, manufacturing and control (CMC) challenges, the outcome of these studies provided information about primary degradation pathways and potential critical quality attributes (CQA) for the molecule.
Figure 1. Overlay of IEX-HPLC chromatograms of mAb A samples stored for one month (A) and three months (B) at -80°C, 2–8°C, 25°C and 40°C. Overlay of IEX-HPLC chromatograms of mAb A samples challenged with high and low pHs for 48 h at 40°C (C) and light induced (0.2xICH) stress (D).
Table 3. Relative amounts of PTMs in mAb A by LC-MS peptide mapping.
| Storage Temperature | Met 253 ox | Met 429 ox | N385D | |
|---|---|---|---|---|
| Initial |
7% |
4% |
4% |
|
|
1 mo time point | ||||
| 5°C |
8% |
4% |
5% |
|
| 25°C |
8% |
4% |
5% |
|
| 40°C |
9% |
4% |
6% |
|
|
3 mo time point | ||||
| 5°C |
8% |
4% |
8% |
|
| 25°C |
7% |
5% |
8% |
|
| 40°C |
21% |
4% |
8% |
|
|
Limited forced degradation | ||||
| pH 3.0/40°C |
8% |
4% |
14% |
|
| pH 9.0/40°C |
8% |
4% |
7% |
|
| Photostress |
22% |
17% |
5% |
|
| Dark Ctrl | 7% | 4% | 6% | |
Met253ox and Met429 ox refers to methionine oxidation position 253 and 429, respectively. N385D refer to relative amount of aspartic acid at position 385. Photostress conditions were 0.2x ICH, Dark Ctrl is the corresponding control
mAb B case study: Identification of degradation pathways informs appropriate control strategy
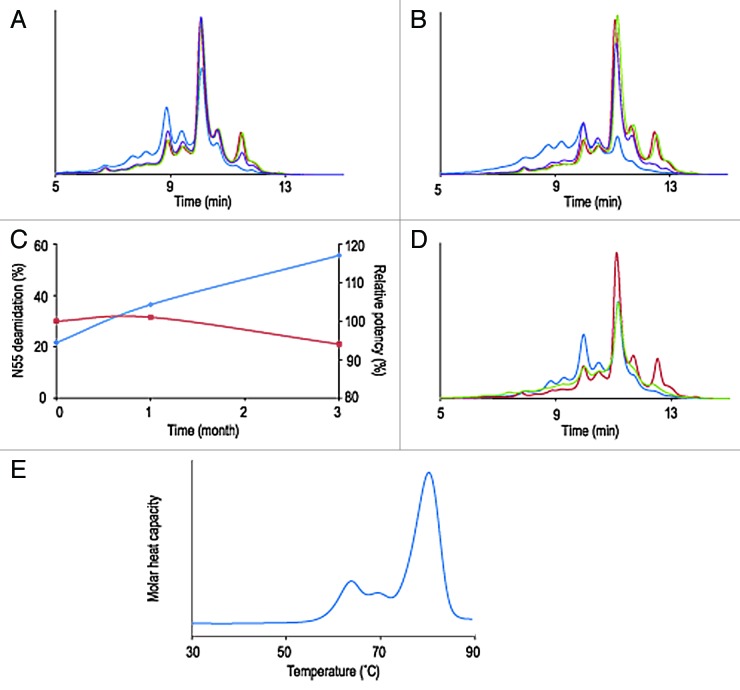
The second example of developability studies involves a S228P hinge-modified IgG4 molecule, termed mAb B. Scanning of the primary sequence for exposed asparagine, aspartic acid, tyrosine, tryptophan and methionine residues did not predict the presence of particularly sensitive hot spots. Results from the short-term stability studies, however, shows a collapsed IEX-HPLC profile after three months at 40°C and a significant increase of acidic variants at 25°C and essentially no changes when stored at 2–8°C or frozen (Fig. 2A and B). No increase of aggregates or fragments resulting from chemical proteolysis was detected by SEC-HPLC (data not shown). Peptide mapping and LC-MS analysis indicated that deamidation of heavy chain Asn55 exceeded 50% and thus was largely responsible for the instability of the molecule (Table 4). Other expected post-translational modifications, such as oxidation of the conserved Fc methionine residues were also detected, albeit to low levels. Testing of relative potency by binding ELISA showed that, in spite of the chemical degradation, mAb B retained its complete ability to bind its target antigen (Table 4, Fig. 2C). Lowering the pH of the mAb B formulation from 6.0 to 4.8 reduced greatly the amount of deamidation after one month at 40°C (Fig. 2D). For this particular molecule, deamidation at Asn55 was recognized as a potential CQA. A proper control strategy involving monitoring deamidation during upstream and down stream process development and manufacturing was put in place. Efforts toward defining an adequate formulation and storage conditions of the drug substance were made to ensure protein stability and the longest possible product shelf-life. mAb B’s desirable commercial drug product form is a highly concentrated solution for subcutaneous injection. Part of the developability exercise is to verify that the protein can remain in solution at concentrations above 100 mg/mL and such a solution has viscosity that would allow development of prefilled syringe image and autoinjector devices. mAb B was successfully concentrated to 190 mg/mL and remained in solution. At these concentrations, viscosity remained under 19 cP, which allows development of purification processes and prefilled syringe image (Table 5). Additionally, mAb B has demonstrated good thermodynamic stability (onset of the first transition above 50°C) and thermal profile with three unfolding transitions at 63, 68 and 80°C, respectively (Fig. 2E, Table 5), which is typical for multidomain proteins. Developability studies of mAb B highlighted primary degradation pathways and potential critical attributes. It also identified CMC issues addressable with an appropriate manufacturing and analytical control strategy.
Figure 2. Overlay of IEX-HPLC chromatograms of mAb B samples stored for one month (A) and three months (B) at -80°C, 2–8°C, 25°C and 40°C. Evolution during stability studies of relative amount of deamidation at HCAsn55 (N55, blue trace) and mAb B potency (red trace) (C). Overlay of IEX-HPLC chromatograms of mAb B samples stored for one month at 40°C at pH 4.5 and pH 6.0 (D). Thermal unfolding profile of high concentration solution (190 mg/ml) of mAb B (E).
Table 4. Potency and Relative amounts of PTMs in mAb B by LC-MS peptide mapping.
| Storage Temperature | N55D | M250ox | M426ox | Potency |
|---|---|---|---|---|
|
1 mo time point | ||||
| -80°C |
21.6% |
10.1% |
3.7% |
100% 100% 103% 101% |
| 5°C |
21.7% |
10.0% |
3.4% |
|
| 25°C |
23.9% |
10.8% |
3.8% |
|
| 40°C |
36.4% |
11.7% |
4.1% |
|
|
3 mo time point | ||||
| 5°C |
21.1% |
14.7% |
3.1% |
113% 108% 94% |
| 25°C |
28.1% |
16.7% |
2.7% |
|
| 40°C | 55.6% | 18.6% | 3.5% | |
N55D refers to relative amount of aspartic acid at position 55 on the heavy chain. Met248ox and Met250ox refer to methionine oxidation position 248 and 250. Potency by Binding ELISA is expressed as % of reference standard
Table 5. Viscosity and thermal melt of mAb B.
| Concentration | Viscosity (cP) | Thermal transitions (°C) | |||
|---|---|---|---|---|---|
| Onset |
Tm1 |
Tm2 |
Tm3 |
||
| 190 | 19 | 56 | 63 | 68 | 80 |
60
2
Tm1, Tm2, Tm3 refers to melting temperature transition 1, 2, 3 shown in thermal melt curves (Fig. 2E)
mAb C case study: Identification of degradation pathways leads to project termination and reallocation of resources
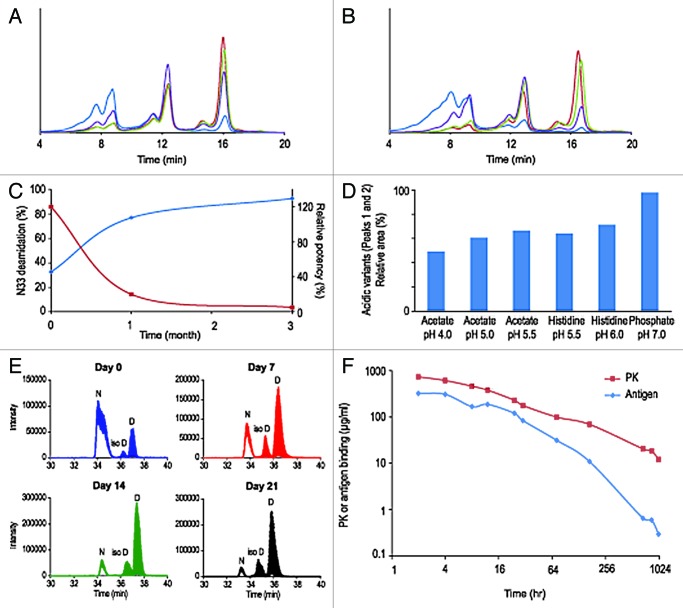
The third case study concerns an IgG2 molecule, termed mAb C. Analysis of the primary sequence of the molecule showed the presence of an asparagine-glycine (Asn33-Gly34) sequence in one of the CDRs of the antibody light chain. Analysis of the structural context of these residues in the molecular model showed that Asn33 is exposed to the solvent and located in a flexible loop. The Asn-Gly sequence has been documented as a site particularly sensitive to facile deamidation to aspartic acid and isomerization to isoaspartic acid.7 IEX-HPLC chromatograms of mAb C samples showed a series of three charge variant peaks eluting at about 8, 12 and 16 min (Fig. 3A and B). These peaks were first believed to be associated with the three different hinge region disulfide isomers commonly found in IgG2s with kappa light chain.9 Isolation and characterization of the charge variants demonstrated, however, that the two acidic variants (eluting at 8 and 12 min) are mAb C molecules with one or both of the Asn33 deamidated (data not shown). Analysis by IEX-HPLC of the short-term stability at one and three months time points shows a rapid increase of acidic variants when the molecule is stored at 25°C and 40°C (Fig. 3A and B). In the same storage conditions and at the same time points, the molecule showed only minimal increase of high molecular weight species by SEC-HPLC (data not shown). Consistent with the increase of the acidic variants peaks, analysis of the stability samples by peptide mapping shows dramatic formation of aspartic acid and isoaspartic acid at light chain position 33 (Table 6). Deamidation of light chain Asn33 reaches 92.6% at 40°C after three months and correlates with mAb C loss of potency (Table 6, Fig. 3C). Other post-translational modifications were also detected: oxidation of the conserved methionine 248 in the Fc domain and limited cyclization of the glutamic acid residue located at heavy chain position 1 (Table 6). These later modifications are well-documented and shown to have no effect on the potency of mAbs.4,8 Asn33 deamidation can be controlled by optimizing the formulation; lowering of the pH reduced deamidation at elevated temperature over time (Fig. 3D). However, stabilization of the molecule in vitro is not sufficient. Ex vivo and in vivo rhesus monkey plasma stability studies indicate that degradation of mAb C drug candidate occurs in plasma (Fig. 3E and F). Titration of mAb C by anti-kappa light chain and antigen ligand-capture ELISAs, in serum samples drawn from rhesus monkeys, shows a time-dependent separation of the PK and antigen binding curves, suggesting a loss of ligand binding capacity consistent with our in vitro observations (Fig. 3F). To remain viable as a product candidate, mAb C had to be reengineered by replacing Asn33 by an alternative amino acid without altering affinity for the target antigen. Developability studies of mAb C led to the identification of major CMC challenges that could be addressed only via re-engineering of the molecule.
Figure 3. Overlay of IEX-HPLC chromatograms of mAb C samples stored for one month (A) and three months (B) at -80°C, 2–8°C, 25°C and 40°C. Evolution during stability studies of relative amount of deamidation at HCAsn33 (N33, blue trace) and mAb B potency (red trace) (C). The bar graph represents the relative area of acidic variants after one month at 40°C in six different formulations (D). Deamidation at Asn33 in rhesus monkey serum stability studies (E): TIC of the doubly charged ion of peptide 25–39 at day 0, 7, 14 and 21: The peak at ~33–34 min is the early eluting non deamidated parent peptide (Asn33), the two peaks eluting at 34–36 and 35–37 min are the deamidated product peptides (iso-Asp33 and Asp33, respectively). At all-time points the ratio of deamidated product peptides IsoAsp/Asp is the same, ~1:5 throughout the whole experiment. Titration of mAb C in rhesus monkeys serum by an anti-kappa light chain (PK curve) and antigen ligand-capture (Antigen curve) ELISAs after second injection (F).
Table 6. Potency and Relative amounts of PTMs in mAb C by LC-MS peptide mapping.
| Storage Temperature | N33D | N33isoD | N33D+N33isoD | Met248 ox | HC pE1 | Potency |
|---|---|---|---|---|---|---|
| Initial |
3.5% |
29.2% |
32.6% |
3.2% |
2.2% |
N/A |
|
1 mo time point | ||||||
| 5°C |
3.4% |
30.3% |
33.6% |
1.4% |
2.5% |
108% |
| 25°C |
5.2% |
44.1% |
49.2% |
1.5% |
2.9% |
101% |
| 40°C |
12.4% |
64.5% |
76.9% |
2.4% |
5.2% |
20% |
|
3 mo time point | ||||||
| 5°C |
3.5% |
26.6% |
30.1% |
3.2% |
1.5% |
96% |
| 25°C |
6.4% |
56.2% |
62.6% |
5.8% |
2.2% |
28% |
| 40°C | 24.1% | 68.5% | 92.6% | 21.7% | 6.1% | 5% |
N33D and N33IsoD refer to relative amount of aspartic acid and isoaspartic acid at position 33, respectively. Total deamidation at position 33 is indicated as the sum of (N33D and N33IsoD). Met248ox refers to methionine oxidation position 248. HC pE1 refers to the proportion of pyroglutamic acid at HC position1. Potency by Binding ELISA is expressed as % of control (Initial, indicated as N/A)
Discussion
Developability studies are an important link between drug discovery and process development activities. After expression of a proposed therapeutic biologic in the expected host organism, early assessment of the molecule’s stability is the first occasion to gather knowledge on it to determine potential CQAs as encouraged by the QbD paradigm. We presented three cases of developability studies, one for each of the typical therapeutic mAb frameworks, that correspond essentially to three scenarios encountered during developability studies. For the first example, short-term stabilities, freeze-thaw and limited forced degradation studies did not uncover unexpected sources of concerns about the stability of the molecule. In this particular case, developability studies showed that the molecule’s degradation pathways conformed to those of standard IgGs. The second case study provides an example of a therapeutic mAb undergoing degradation without affecting potency. In this case, a proper analytical strategy is put in place to monitor the degradation detected. Proper process controls are also instituted to limit the degradation during manufacturing. Lastly, formulation studies are put in place to ensure that the degradation does not occur during storage and does not compromise the shelf-life of the drug product. The last case study corresponds to the case where stability studies detect a major problem with the molecule’s stability and efficacy, leading to termination of the project and reallocation of resources.
Materials and Methods
Materials
mAb A, mAb B and mAb C are humanized IgG1, IgG4 and IgG2, respectively. All molecules have λ light chains. These Merck proprietary mAbs are expressed in CHO cells and purified using three chromatographic steps (protein A, anion exchange and cation exchange). Formulations included histidine, acetate and typical excipients such as sucrose and PS80; pHs ranged from 5.0 to 6.0.
Ion exchange HPLC (IEX-HPLC)
Ion exchange HPLC for mAbs A, B and C is run on the Dionex WCX-10 column and uses either a citrate or Mes-based mobile phase A and a phosphate buffer with or without added NaCl as mobile phase B. Gradient is adjusted to suit the needs of each molecule.
Peptide mapping
A typical peptide mapping procedure was followed. Samples were diluted in reducing buffer (50 mM TRIS-HCl at pH 8.0, 6 M guanidine-HCl, 5 mM EDTA and 20 mM DTT) and were incubated at 56°C for 30 min. Each sample was cooled at room temperature for 5 min prior to alkylation with 50 mM iodoacetamide at room temperature for 30 min in the dark. Each sample was then buffer-exchanged to digestion buffer containing 50 mM Tris buffer at pH 8.0, 2 M urea and treated with an aliquot of trypsin. The mixture was incubated at 37°C. The digestion was quenched by the addition of 20% trifluoroacetic acid (TFA). Agilent 6538 Q-TOF or Xevo G2 Q-TOF system mass spectrometers were set up with Agilent 1290 Infinity HPLC system or Waters Acquity UPLC. The samples were loaded on a C18 column held at 75°C. Mobile phase A was 0.05% TFA in water and mobile phase B was 0.05% TFA in acetonitrile. Gradient is adjusted to suit the needs of each molecule Peptide mapping analysis was performed by using Agilent MassHunter Qualitative Analysis software.
Quantitation of mAb B deamidated peptide in serum matrix by multiple reaction monitoring
mAb B was spiked in rhesus monkey plasma and incubated for up to 21 d at 37 þC. Blank plasma was prepared and incubated along with the spiked samples serving as a negative control for the rhesus monkey plasma’s IgG2. mAb B was subsequently affinity-purified from the biomatrices using a goat anti-human IgG-heavy and light chain monkey adsorbed antibody conjugated to an agarose based Affi-Gel hydrazine gel, by coupling the sugar residues of the antibody to the hydrazine groups on the gel. The eluates were digested by trypsin followed by endoproteinase V8 to obtain a reasonably sized peptide amenable to mass spectrometry analysis. The eluates were separated by reverse phase chromatography, using a very shallow gradient necessary to separate the peptides containing Asn33, isoAsp33 and Asp33. The separated peptides were analyzed online by multiple reaction monitoring (MRM) targeted analysis on a TSQ Vantage Thermo mass spectrometer. Areas under the chromatogram were subsequently extracted for each species using Skyline software (MacCoss lab, Department of Genome Sciences, UW) as a platform or Xcalibur (Thermo Scientific) and plotted against time.
Potency by direct binding ELISA
Typical direct binding ELISA was used. The 96-well ELISA plates were coated with the appropriate antigen for 1 h at 25°C for mAb B or overnight at 4°C for mAb C. Plates were washed with TBS (mAb B) or PBS (mAb C) containing 0.05% Tween 20 and blocked with a blocking buffer for 1 h at 25°C. For the primary incubations, various concentrations of mAbs were added into the pre-coated plates. The amount of bound mAb was measured by either alkaline phosphatase (mAb B) or horseradish peroxidase (mAb C) labeled anti-human IgG. The enzymatic reaction was developed with either PhosphoGlo AP substrate (mAb B) or Pico SuperSignal (mAb C). The EC50 vales were determined by nonlinear regression.
Surrogate solubility
While not strictly a maximum solubility study determination, viscosity measurements in conjunction with protein concentration experiments in standard buffers are meant to evaluate the feasibility of 100 – 200 mg/mL solutions. Buffer exchanges were performed at 5°C in 50 kDa Amicon® Ultra – 15 centrifugal filter units, with sample to buffer volume ratio of 1:1 and three volume exchanges. mAb C concentration was determined by UV absorption at 280 nm using a calculated extinction coefficient.
Viscosity
Viscosity measurements were performed at 30°C using mVROC instrument.
Thermal melts (DSC)
Differential scanning calorimetry (DSC) was performed with Microcal VP-DSC. 400 μL of each sample and matching buffer were placed in a 96-well plate, and the thermograms were acquired between 10 and 90°C at a scan rate of 90°C/h, with 5 min pre-scan equilibration. The data was processed by subtracting from each sample thermogram, a corresponding placebo–placebo scan, fitting a baseline to the trace using the ORIGIN® software package and determining the onset temperature of the first transition and maximum temperature of observed thermal transitions.
In vivo titration of mAb B
Two injections of mAb B were performed subcutaneously in rhesus monkeys cohorts. Blood samples were drawn at various time points and assayed by two sandwich ELISAs (PK and antigen). Antigen and PK Sandwich ELISAs were performed as follows: Costar plates coated with antigen or mouse anti-human kappa light chain antibody. Bound antibodies were detected either with anti-human IgG-AP or mouse anti-human IgG (Fc)-AP. Standard (mAb B 14-point curve from 100ng/ml) and plasma samples (antigen –1:10000, PK – 1:20000 dilution, respectively). Total antigen and PK (µg/ml) was calculated based on standard curve.
Supplementary Material
Acknowledgments
We would like to express our gratitude to Rebecca Goski and Mussadeq Hussain for their assistance during these studies. We would also like to express our gratitude to Mary Savage, Andrea Webber, Susan Cannon-Carlson, Mohammed Shameem, Steve Farrand and the BioProcess Development (MRL) group.
Submitted
04/28/2013
Revised
05/31/2013
Accepted
06/03/2013
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Supplemental Materials
Supplemental materials can be found here: www.landesbioscience.com/journals/mabs/article/25269
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/25269
References
- 1.Reichert JM. Metrics for antibody therapeutics development. MAbs. 2010;2:695–700. doi: 10.4161/mabs.2.6.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rathore AS, Winkle H. Quality by design for biopharmaceuticals. Nat Biotechnol. 2009;27:26–34. doi: 10.1038/nbt0109-26. [DOI] [PubMed] [Google Scholar]
- 3.Hawe A, Wiggenhorn M, van de Weert M, Garbe JH, Mahler HC, Jiskoot W. Forced degradation of therapeutic proteins. J Pharm Sci. 2012;101:895–913. doi: 10.1002/jps.22812. [DOI] [PubMed] [Google Scholar]
- 4.Yan B, Valliere-Douglass J, Brady L, Steen S, Han M, Pace D, et al. Analysis of post-translational modifications in recombinant monoclonal antibody IgG1 by reversed-phase liquid chromatography/mass spectrometry. J Chromatogr A. 2007;1164:153–61. doi: 10.1016/j.chroma.2007.06.063. [DOI] [PubMed] [Google Scholar]
- 5.Keck RG. The use of t-butyl hydroperoxide as a probe for methionine oxidation in proteins. Anal Biochem. 1996;236:56–62. doi: 10.1006/abio.1996.0131. [DOI] [PubMed] [Google Scholar]
- 6.International Conference on Harmonisation. Guidance for Industry, Q1B “Photostability Testing of New Drug Substances and Product”, ICH-Q1B. 1996 November. http://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Quality/Q1B/Step4/Q1B_Guideline.pdf
- 7.Robinson NE, Robinson AB. Molecular Clocks: Deamidation of Asparaginyl and Glutaminyl Residues in Peptides and Proteins. 2004 Althouse Press, Cave Junction, OR. [Google Scholar]
- 8.Du Y, Walsh A, Ehrick R, Xu W, May K, Liu H. Chromatographic analysis of the acidic and basic species of recombinant monoclonal antibodies. MAbs. 2012;4:578–85. doi: 10.4161/mabs.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wypych J, Li M, Guo A, Zhang Z, Martinez T, Allen MJ, et al. Human IgG2 antibodies display disulfide-mediated structural isoforms. J Biol Chem. 2008;283:16194–205. doi: 10.1074/jbc.M709987200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.