Abstract
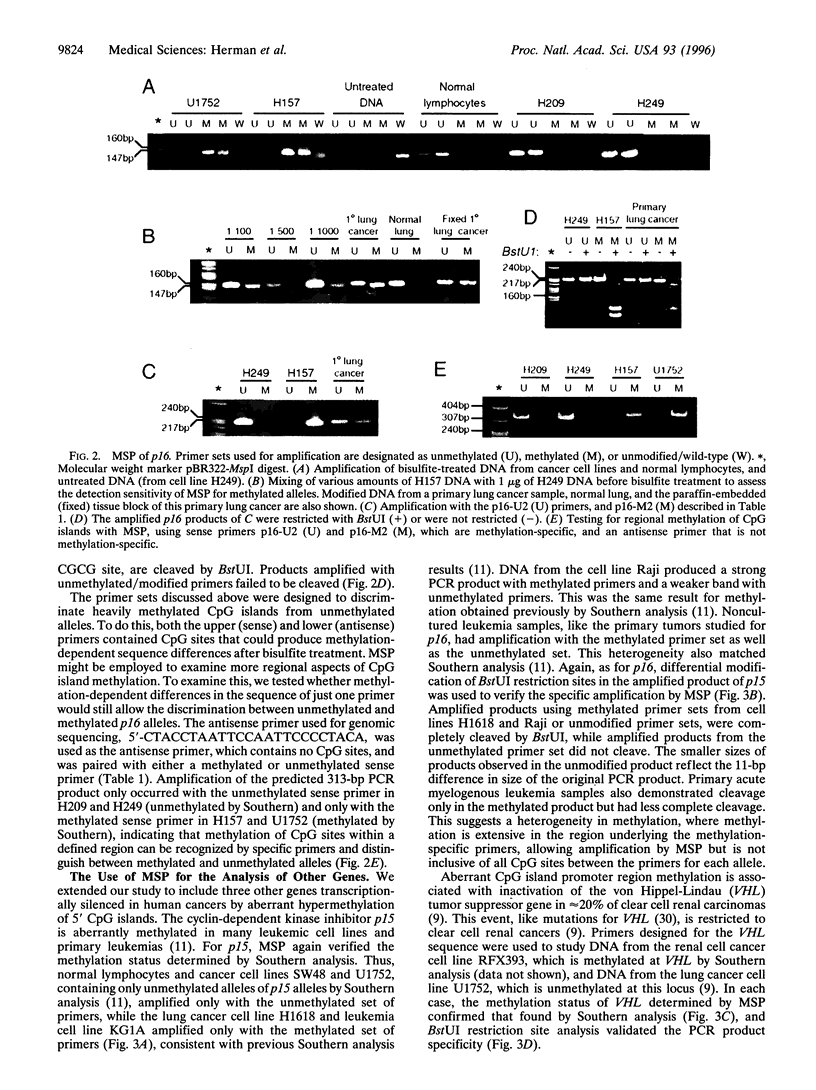
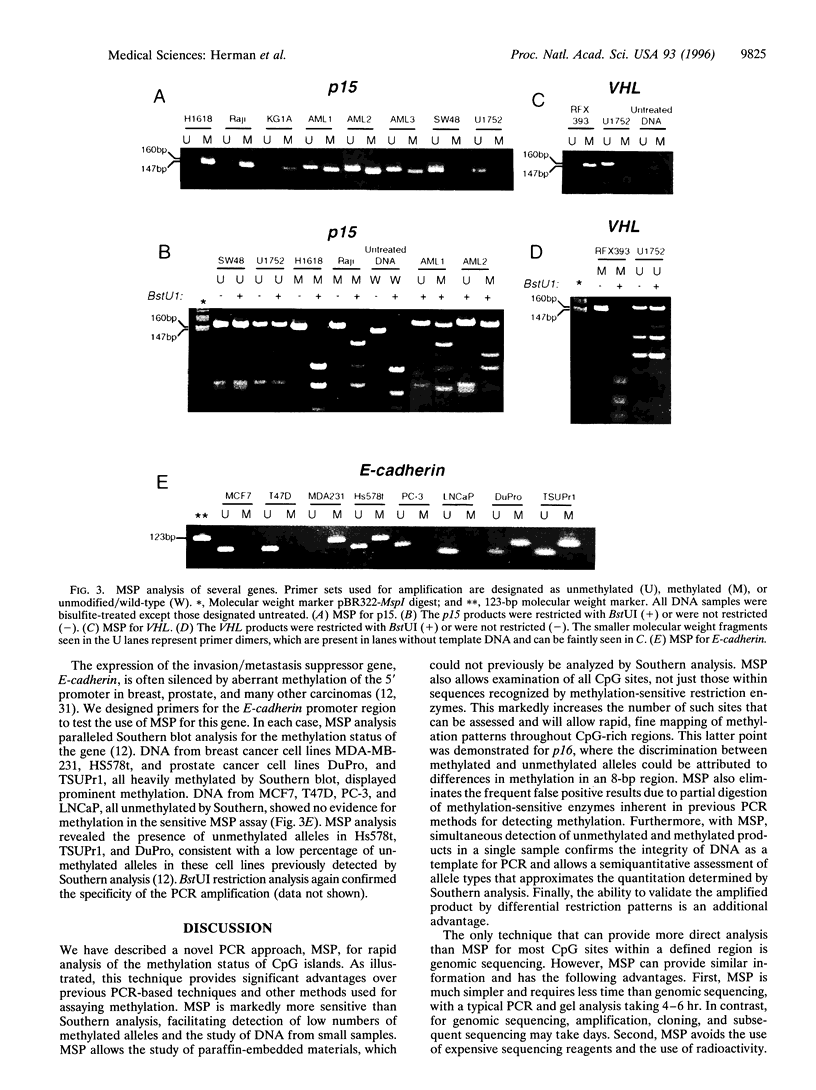
Precise mapping of DNA methylation patterns in CpG islands has become essential for understanding diverse biological processes such as the regulation of imprinted genes, X chromosome inactivation, and tumor suppressor gene silencing in human cancer. We describe a new method, MSP (methylation-specific PCR), which can rapidly assess the methylation status of virtually any group of CpG sites within a CpG island, independent of the use of methylation-sensitive restriction enzymes. This assay entails initial modification of DNA by sodium bisulfite, converting all unmethylated, but not methylated, cytosines to uracil, and subsequent amplification with primers specific for methylated versus unmethylated DNA. MSP requires only small quantities of DNA, is sensitive to 0.1% methylated alleles of a given CpG island locus, and can be performed on DNA extracted from paraffin-embedded samples. MSP eliminates the false positive results inherent to previous PCR-based approaches which relied on differential restriction enzyme cleavage to distinguish methylated from unmethylated DNA. In this study, we demonstrate the use of MSP to identify promoter region hypermethylation changes associated with transcriptional inactivation in four important tumor suppressor genes (p16, p15, E-cadherin, and von Hippel-Lindau) in human cancer.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antequera F., Boyes J., Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990 Aug 10;62(3):503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Bussemakers M. J., Giroldi L. A., van Bokhoven A., Schalken J. A. Transcriptional regulation of the human E-cadherin gene in human prostate cancer cell lines: characterization of the human E-cadherin gene promoter. Biochem Biophys Res Commun. 1994 Sep 15;203(2):1284–1290. doi: 10.1006/bbrc.1994.2321. [DOI] [PubMed] [Google Scholar]
- Clark S. J., Harrison J., Paul C. L., Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994 Aug 11;22(15):2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M., McDonald L. E., Millar D. S., Collis C. M., Watt F., Grigg G. W., Molloy P. L., Paul C. L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnarra J. R., Tory K., Weng Y., Schmidt L., Wei M. H., Li H., Latif F., Liu S., Chen F., Duh F. M. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994 May;7(1):85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Zulueta M., Bender C. M., Yang A. S., Nguyen T., Beart R. W., Van Tornout J. M., Jones P. A. Methylation of the 5' CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995 Oct 15;55(20):4531–4535. [PubMed] [Google Scholar]
- Graff J. R., Herman J. G., Lapidus R. G., Chopra H., Xu R., Jarrard D. F., Isaacs W. B., Pitha P. M., Davidson N. E., Baylin S. B. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995 Nov 15;55(22):5195–5199. [PubMed] [Google Scholar]
- Hara E., Smith R., Parry D., Tahara H., Stone S., Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996 Mar;16(3):859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J. G., Jen J., Merlo A., Baylin S. B. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996 Feb 15;56(4):722–727. [PubMed] [Google Scholar]
- Herman J. G., Latif F., Weng Y., Lerman M. I., Zbar B., Liu S., Samid D., Duan D. S., Gnarra J. R., Linehan W. M. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J. G., Merlo A., Mao L., Lapidus R. G., Issa J. P., Davidson N. E., Sidransky D., Baylin S. B. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995 Oct 15;55(20):4525–4530. [PubMed] [Google Scholar]
- Holliday R., Grigg G. W. DNA methylation and mutation. Mutat Res. 1993 Jan;285(1):61–67. doi: 10.1016/0027-5107(93)90052-h. [DOI] [PubMed] [Google Scholar]
- Hussussian C. J., Struewing J. P., Goldstein A. M., Higgins P. A., Ally D. S., Sheahan M. D., Clark W. H., Jr, Tucker M. A., Dracopoli N. C. Germline p16 mutations in familial melanoma. Nat Genet. 1994 Sep;8(1):15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- Issa J. P., Ottaviano Y. L., Celano P., Hamilton S. R., Davidson N. E., Baylin S. B. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994 Aug;7(4):536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- Jen J., Harper J. W., Bigner S. H., Bigner D. D., Papadopoulos N., Markowitz S., Willson J. K., Kinzler K. W., Vogelstein B. Deletion of p16 and p15 genes in brain tumors. Cancer Res. 1994 Dec 15;54(24):6353–6358. [PubMed] [Google Scholar]
- Kuzmin I., Duh F. M., Latif F., Geil L., Zbar B., Lerman M. I. Identification of the promoter of the human von Hippel-Lindau disease tumor suppressor gene. Oncogene. 1995 Jun 1;10(11):2185–2194. [PubMed] [Google Scholar]
- Li E., Beard C., Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993 Nov 25;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
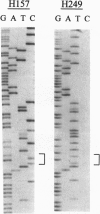
- Merlo A., Herman J. G., Mao L., Lee D. J., Gabrielson E., Burger P. C., Baylin S. B., Sidransky D. 5' CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995 Jul;1(7):686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- Myöhänen S., Wahlfors J., Jänne J. Automated fluorescent genomic sequencing as applied to the methylation analysis of the human ornithine decarboxylase gene. DNA Seq. 1994;5(1):1–8. doi: 10.3109/10425179409039698. [DOI] [PubMed] [Google Scholar]
- Otterson G. A., Khleif S. N., Chen W., Coxon A. B., Kaye F. J. CDKN2 gene silencing in lung cancer by DNA hypermethylation and kinetics of p16INK4 protein induction by 5-aza 2'deoxycytidine. Oncogene. 1995 Sep 21;11(6):1211–1216. [PubMed] [Google Scholar]
- Park J. G., Chapman V. M. CpG island promoter region methylation patterns of the inactive-X-chromosome hypoxanthine phosphoribosyltransferase (Hprt) gene. Mol Cell Biol. 1994 Dec;14(12):7975–7983. doi: 10.1128/mcb.14.12.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Razin A., Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991 Sep;55(3):451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeben M., Myöhänen S., Saarma M., Prydz H. Sequencing of the rat light neurofilament promoter reveals differences in methylation between expressing and non-expressing cell lines, but not tissues. Gene. 1995 May 19;157(1-2):325–329. doi: 10.1016/0378-1119(95)00084-j. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Pfeifer G. P. X-chromosome inactivation and cell memory. Trends Genet. 1992 May;8(5):169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- Singer-Sam J., Grant M., LeBon J. M., Okuyama K., Chapman V., Monk M., Riggs A. D. Use of a HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol Cell Biol. 1990 Sep;10(9):4987–4989. doi: 10.1128/mcb.10.9.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöger R., Kubicka P., Liu C. G., Kafri T., Razin A., Cedar H., Barlow D. P. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993 Apr 9;73(1):61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- Tremblay K. D., Saam J. R., Ingram R. S., Tilghman S. M., Bartolomei M. S. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995 Apr;9(4):407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- Wang R. Y., Gehrke C. W., Ehrlich M. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res. 1980 Oct 24;8(20):4777–4790. doi: 10.1093/nar/8.20.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiura K., Kanai Y., Ochiai A., Shimoyama Y., Sugimura T., Hirohashi S. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci U S A. 1995 Aug 1;92(16):7416–7419. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]