Abstract
Most of the previously reported loci for total immunoglobulin E (IgE) levels are related to Th2 cell-dependent pathways. We undertook a genome-wide association study (GWAS) to identify genetic loci responsible for IgE regulation. A total of 479,940 single nucleotide polymorphisms (SNPs) were tested for association with total serum IgE levels in 1180 Japanese adults. Fine-mapping with SNP imputation demonstrated 6 candidate regions: the PYHIN1/IFI16, MHC classes I and II, LEMD2, GRAMD1B, and chr13∶60576338 regions. Replication of these candidate loci in each region was assessed in 2 independent Japanese cohorts (n = 1110 and 1364, respectively). SNP rs3130941 in the HLA-C region was consistently associated with total IgE levels in 3 independent populations, and the meta-analysis yielded genome-wide significance (P = 1.07×10−10). Using our GWAS results, we also assessed the reproducibility of previously reported gene associations with total IgE levels. Nine of 32 candidate genes identified by a literature search were associated with total IgE levels after correction for multiple testing. Our findings demonstrate that SNPs in the HLA-C region are strongly associated with total serum IgE levels in the Japanese population and that some of the previously reported genetic associations are replicated across ethnic groups.
Introduction
Immunoglobulin E (IgE) is a class of antibodies that has an important role in the development of Th2 cell-mediated allergic inflammatory diseases such as asthma, allergic rhinitis, and atopic dermatitis. In atopic individuals, exposure to allergens results in Th2 cell-dependent stimulation of the immune response that causes production of IgE. Recent advances in the understanding of allergen sensitization have also revealed the sentinel role of innate immune mechanisms involved in the development of allergic diseases [1], [2].
Twin and family studies have shown that genetic factors are important for total serum IgE levels [3], [4] and account for about 36% to 78% heritability of its levels [3], [5]. Furthermore, it has been demonstrated that total serum IgE levels are mainly determined by genetic factors that are independent of antigen-specific IgE levels or atopic status [4], [6], [7]. Asthma affection status is known to be related to total serum IgE levels even after adjustment for atopic status [8], [9].
Thus far, a number of candidate gene association studies for total serum IgE levels have demonstrated many polymorphisms in genetic regions related to the Th2 cell-dependent pathways. Recently, 4 genome-wide association studies (GWASs) of total serum IgE levels in independent populations have revealed additional genetic loci, such as TBX18 and SOBP, which seem to be unrelated to the Th2 cell-dependent pathways [10]–[13]. Thus, because GWASs are unbiased by investigator preconceptions, they have the potential of providing new insights into the mechanism of IgE regulation and may be able to clarify unexpected IgE-related genetic loci.
Three of the 4 GWASs of total IgE levels reported so far were conducted solely in populations of European ancestry, and the fourth of those studies also included African-American and Latino populations. In contrast, 2 GWASs recently performed in Asian populations did not identify any loci significantly associated with total serum IgE levels [14], [15]. In genetic association studies, replication of the initial findings in different ethnic groups is important to clarify the relevance of the findings.
Here, we performed a GWAS of total serum IgE levels in a Japanese population and a replication analysis in 2 independent Japanese cohorts that was followed by a meta-analysis. In addition, we validated previously reported gene associations with total serum IgE levels using our GWAS data.
Results
Study Flow Chart
A flow chart outlining the steps of this study is shown in Figure S1.
Characteristics of the Study Cohorts
The characteristics of the original GWAS cohort and the replication cohorts are provided in Table 1. The ratio of female participants was higher in the Fukui cohort than in the Tsukuba and Hokkaido cohorts. The Tsukuba and Hokkaido cohorts included more asthmatic patients than did the Fukui cohort. Age, sex, asthma affection status, atopic status, and IgE levels differed significantly among the cohorts.
Table 1. Characteristics of the study cohorts.
| Tsukuba cohort (n = 1180) | Hokkaido cohort (n = 1110) | Fukui cohort (n = 1364) | |
| Age, y (SD) | 50.3 (10.4) | 44.6 (16.0) | 32.2 (9.8) |
| Female sex | 55.3% | 49.3% | 67.0% |
| Smoking status | |||
| Current smoker | 16.2% | 26.1% | NA |
| Ex-smoker | 17.5% | 16.7% | NA |
| Never smoker | 66.2% | 57.2% | NA |
| Asthma | 18.1% | 44.2% | 6.5% |
| Atopy | |||
| Atopic | 56.6% | 53.7% | 67.2% |
| Nonatopic | 40.5% | 29.5% | 30.5% |
| Unknown | 2.9% | 16.8% | 2.3% |
| Log10 [total serum IgE] (SD) | 1.82 (0.60) | 2.08 (0.69) | 1.91 (0.62) |
NA = not applicable; SD = standard deviation.
GWAS and Replication Analyses
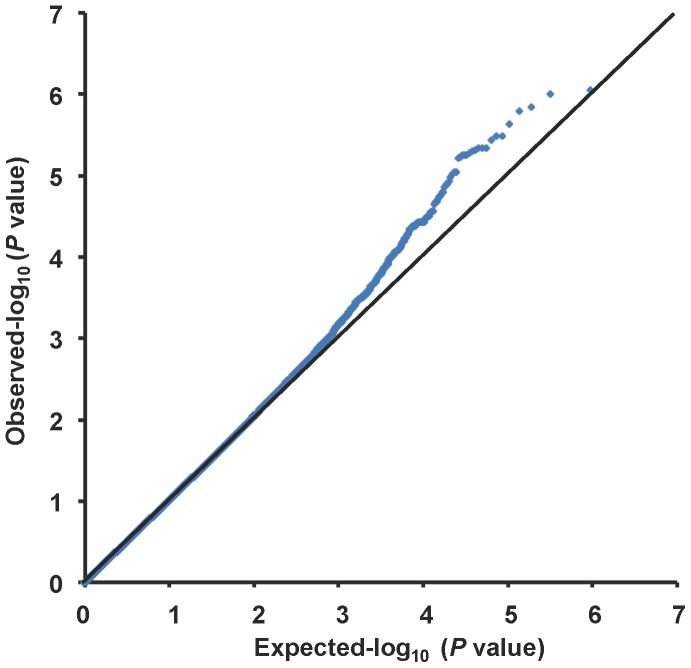
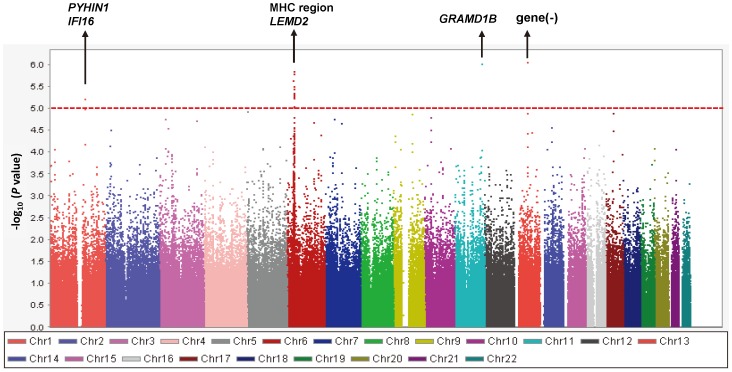
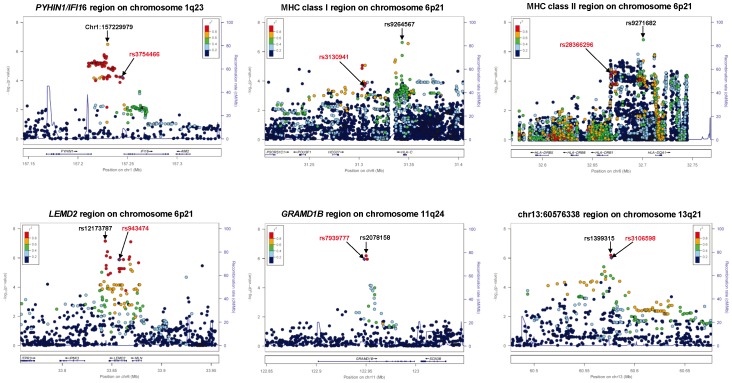
A quantile-quantile plot is shown in Figure 1. The genomic inflation factor of 1.018 indicated a low possibility of false-positive associations resulting from population stratification. A Manhattan plot of the GWAS (Figure 2) showed no SNPs reaching the genome-wide significance threshold of 5.0×10−8. We focused on 4 distinct chromosomal regions in which P values were less than 1.0×10−5: chromosomes 1q23, 6p21, 11q24, and 13q21. Genotypes were imputed to determine the contribution of untyped SNPs to total IgE levels in these regions. Fine-mapping coupled with the imputed SNPs identified 6 candidate genomic regions (Figure 3): the PYHIN1/IFI16 region on chromosome 1q23.1 (chr1∶157229979; P = 3.19×10−7), the MHC class I and II regions on chromosome 6p21.3 (rs9264567, P = 2.33×10−7 and rs9271682, P = 1.55×10−7, respectively), the LEMD2 region on 6p21.31 (rs12173787, P = 7.03×10−8), the GRAMD1B region on chromosome 11q24.1 (rs2078158, P = 6.57×10−7), and the chr13∶60576338 region on chromosome 13q21.31 (rs1399315, P = 6.40×10−7). For each candidate region, we conducted a replication study using validated ready-to-use TaqMan® SNP Genotyping assays. The SNPs available for the replication studies are shown in Figure 3. Table 2 shows the association of these SNPs with total serum IgE levels in the primary Tsukuba cohort and the 2 replication cohorts. The association of rs3130941 in the MHC class I region was consistently replicated in both the Hokkaido and the Fukui cohort. Meta-analysis of the primary and the 2 replication cohorts demonstrated that rs3130941 in the MHC class I region reached the level of genome-wide significance at 1.07×10−10. Rs28366296 in the MHC class II region was replicated in the Hokkaido cohort only. Rs7939777 in the GRAMD1B region was replicated in the Fukui cohort only. As for rs7939777, a meta-analysis using the Tsukuba and Fukui cohorts yielded a P value of 3.35×10−10.
Figure 1. Quantile-quantile (Q-Q) plot of observed versus expected P values of the GWAS results.
The straight line in the Q-Q plot indicates the distribution of SNPs under the null hypothesis.
Figure 2. Manhattan plots of −log10 (P value) for association of 479,940 SNPs with total IgE levels.
Linear regression models adjusted for age, sex, smoking status (never, ex-, or current smoker), pack-year group (0, 0–10, or >10), and asthma affection status were performed. The red line shows the threshold (P = 1×10−5) for selection of genomic regions for further analysis including SNP imputation.
Figure 3. Fine-mapping identification of 6 candidate genomic regions.
Plots show the association results of both genotyped and imputed SNPs in the primary GWAS cohort. The most strongly associated SNPs (black letters) in the GWAS and the SNPs used for the replication studies (red letters) are indicated by arrows. The color of each circle reflects the LD (r2) between a particular SNP and the SNP used for the replication studies.
Table 2. Replication studies and meta-analysis.
| Tsukuba cohort | Hokkaidocohort | FukuiCohort | Meta-analysis | |||||||
| Top SNP | ||||||||||
| Chromosome | Gene | SNP forreplicationstudy | P value | LD withtop SNP(r2) | MAF | Minorallele | β | P value | P value | P value |
| 1q23.1 | PYHIN1/IFI16 | chr1∶157229979 | 3.19E-07 | 1 | 0.16 | G | −0.168 | |||
| rs3754466 | 5.65E-05 | 0.637 | 0.20 | C | −0.119 | 0.244 | 0.222 | 0.378 | ||
| 6p21.3 | MHC class I | rs9264567 | 2.33E-07 | 1 | 0.28 | A | 0.132 | |||
| rs3130941 | 2.28E-04 | 0.549 | 0.25 | C | 0.098 | 5.03E-04 | 6.12E-05 | 1.07E-10 | ||
| 6p21.3 | MHC class II | rs9271682 | 1.55E-07 | 1 | 0.48 | A | 0.127 | |||
| rs28366296 | 2.67E-05 | 0.608 | 0.39 | A | −0.098 | 4.15E-02 | 5.44E-02 | 2.17E-05 | ||
| 6p21.31 | LEMD2 | rs12173787 | 7.03E-08 | 1 | 0.17 | G | 0.161 | |||
| rs943474 | 1.30E-06 | 0.786 | 0.16 | G | 0.157 | 5.56E-02 | 0.308 | 2.29E-02 | ||
| 11q24.1 | GRAMD1B | rs2078158 | 6.57E-07 | 1 | 0.41 | A | −0.118 | |||
| rs7939777 | 1.13E-06 | 0.943 | 0.42 | C | −0.114 | 7.63E-02* | 7.58E-05 | 0.263 | ||
| 13q21.31 | none | rs1399315 | 6.40E-07 | 1 | 0.48 | C | 0.114 | |||
| rs3106598 | 9.51E-07 | 0.997 | 0.48 | G | 0.112 | 0.817 | 0.813 | 0.326 | ||
The direction of the effect was opposite to that of the Tsukuba cohort.
LD = linkage disequilibrium; MAF = minor allele frequency.
When we repeated the meta-analyses by studying nonasthmatic healthy individuals only (n = 2861) or by adding atopic sensitization as an additional covariate, we confirmed the association between rs3130941 in the MHC class I region and levels of total serum IgE at genome-wide significance (Table S1 and Table S2).
Validation of Previous Genetic Associations
The PubMed search identified 448 related publications. After screening of the titles, abstracts, and text, 156 eligible publications were selected. Screening of the references of those publications identified 33 additional relevant publications. From the 189 selected publications, we found 32 candidate genes associated with total serum IgE levels; 25 of those genes were reported in 3 or more candidate gene association studies, and 7 (DARC, HLA-A, HLA-DQA2, HLA-G, RAD50, SOBP, and TBX18) were reported in at least 1 GWAS.
Table 3 shows the SNP most significantly associated with total IgE levels in our GWAS data of each candidate gene on an autosomal chromosome. Nominal P values were less than 0.05 for 17 of the 32 candidate genes, including HLA-C. After corrections for multiple testing using SNPSpD software [16], the associations of 9 genes with total IgE levels remained significant. The strongest associations were detected at the genes in the MHC class I/II regions, including LTA on chromosome 6p21.3, although none of the top SNPs reached genome-wide significance. Previously reported polymorphisms associated with total serum IgE levels are shown in Table S3.
Table 3. Top SNPs with the strongest statistical evidence of association with total serum IgE levels.
| Gene | Chromosome | Number of SNPs | SNP | Nominal P value | Corrected P value* |
| ADAM33 | 20 | 12 | rs2853215 | 1.66E-02 | 0.150 |
| ADRB2 | 5 | 17 | rs17640574 | 7.27E-02 | 0.728 |
| CCL11 | 17 | 7 | rs6505403 | 0.225 | 0.898 |
| CD14 | 5 | 8 | rs3822356 | 0.719 | 1 |
| CMA1 | 14 | 9 | rs1956932 | 0.179 | 1 |
| CTLA4 | 2 | 3 | rs231726 | 1.21E-02 | 2.31E-02 |
| DARC | 1 | 3 | rs863002 | 0.394 | 0.789 |
| FCER1A | 1 | 9 | rs2427824 | 3.06E-02 | 0.184 |
| FLG | 1 | 3 | rs3126085 | 0.288 | 0.577 |
| GSTP1 | 11 | 3 | rs614080 | 0.766 | 1 |
| HLA-A | 6 | 2 | rs2734959 | 6.64E-03 | 1.33E-02 |
| HLA-C | 6 | 26 | rs3132486 | 1.50E-04 | 1.88E-03 |
| HLA-DQA2 | 6 | 24 | rs17500468 | 7.94E-05 | 8.06E-04 |
| HLA-DQB1 | 6 | 11 | rs2647025 | 7.48E-05 | 3.74E-04 |
| HLA-DRB1 | 6 | 66 | rs35465556 | 8.51E-05 | 1.83E-03 |
| HLA-G | 6 | 21 | rs1633053 | 7.58E-03 | 4.56E-02 |
| IFNG | 12 | 4 | rs3181032 | 3.56E-02 | 0.104 |
| IL10 | 1 | 8 | rs1800896 | 0.110 | 0.660 |
| IL13 | 5 | 4 | rs1295686‡ | 7.41E-02 | 0.216 |
| IL4 | 5 | 5 | rs2243288 | 2.08E-03 | 8.34E-03 |
| IL4R | 16 | 23 | rs4787948 | 0.219 | 1 |
| LTA | 6 | 12 | rs2857709 | 4.24E-05 | 2.97E-04 |
| MS4A2 | 11 | 4 | rs574700 | 0.201 | 0.602 |
| NOD2 | 16 | 6 | rs7194886 | 0.156 | 0.468 |
| NOS1 | 12 | 45 | rs4767535 | 8.60E-03 | 0.235 |
| NPSR1 | 7 | 66 | rs323928 | 3.29E-02 | 0.867 |
| RAD50 | 5 | 7 | rs17772583 | 1.82E-02 | 7.29E-02 |
| SOBP | 6 | 41 | rs3734747 | 5.43E-03 | 0.135 |
| STAT6 | 12 | 7 | rs841718 | 0.171 | 0.854 |
| TBX18 | 6 | 5 | rs2015519 | 3.71E-02 | 0.146 |
| TLR2 | 4 | 4 | rs7656411 | 0.328 | 0.983 |
| TNF | 6 | 11 | rs2844484 | 7.96E-02 | 0.478 |
The significance level was corrected for multiple testing using the SNPSpD program [16].
SNP previously reported for association with total serum IgE (the direction of the effect was the same).
Discussion
To the best of our knowledge, ours is the first GWAS that demonstrates positive results for levels of total serum IgE in an Asian population. In our primary GWAS cohort, fine-mapping using the imputed SNPs on chromosome 6p revealed 3 independent peaks: the MHC class I, MHC class II, and LEMD2 regions (Figure S2). In the meta-analysis, rs3130941 in the MHC class I region reached levels of genome-wide significance. This finding was not significantly influenced by the presence or absence of asthma or atopy. Rs3130941 is located between HLA-C and HCG27 (HLA complex group 27) (Figure 3). In the MHC class I region, 4 genes have been previously reported to be associated with total serum IgE levels: HLA-A (rs2517754, rs2571391), HLA-G (rs2523809), LTA (rs909253), and TNF (rs1800629, rs361525, rs1800630) [12], [17]–[22]. Linkage disequilibrium (LD) between rs3130941 and each of these SNPs estimated by r2 in our population was very weak (Table S4, Figure S3). Furthermore, the association of rs3130941 with total IgE levels was not influenced by inclusion of each of these SNPs in the statistical model as a covariate (Table S4). Therefore, rs3130941 in the HLA-C region is associated with total serum IgE levels independent of the genetic influence of HLA-A, HLA-G, LTA, or TNF. With respect to the functional consequence of rs3130941, the GENEVAR database (http://www.sanger.ac.uk/humgen/genevar/) [23] revealed that rs3094609 and rs3130931, both of which are in weak LD with rs3130941 (r2 = 0.566 and 0.324, respectively), are significantly associated with expression levels of HLA-C mRNA (P = 0.0495 and 0.00521, respectively), suggesting that rs3130941 may also be related to HLA-C expression.
The MHC class II region is another candidate for regulation of IgE levels because MHC class II molecules are importantly involved in antigen-specific IgE synthesis [24]. Three genes in the MHC class II region have been reported to be associated with total IgE levels: HLA-DQA2, HLA-DQB1, and HLA-DRB1. In our primary GWAS, associations of SNPs in these genes were replicated (Table 3). Because the MHC class I and II regions are in very close proximity on chromosome 6p21.3, we examined whether the genetic impact of rs3130941 in the MHC class I region was influenced by rs28366296 in the MHC class II region. Although both rs3130941 and rs28366296 showed a strong association with total IgE levels in our study, the LD between these 2 SNPs was weak (Table S4) and a linear regression model including these 2 SNPs showed that each genetic association maintained significance after controlling for the effect of each of the remaining SNPs (Table S4). Therefore, we believe that the association of rs3130941 with total serum IgE levels is independent of the effects of MHC class II genes.
A set of specific infections that strongly promote Th1 and natural killer (NK) cells likely has the potential to inhibit atopic disorder by repression of Th2 immunity. Among Japanese schoolchildren, positive tuberculin responses predicted a lower incidence of asthma, lower serum IgE levels, and biased Th1 cytokine profiles [25]. MHC class I-restricted CD8 T cells collaborate with CD4 Th1 cells to invoke Th1-type immunity, thereby counteracting CD4 Th2 cells, which results in inhibition of IgE production [26], [27]. HLA-C molecules also modulate NK cell function [28]. NK cytotoxicity is negatively controlled by inhibitory receptors, such as human killer cell Ig-like receptors (KIRs) specific for HLA-B and HLA-C [29]. Accordingly, as an intrinsic abnormality, impaired HLA-C-mediated triggering of protective immunity to microbial exposure would predispose an individual to increased levels of total serum IgE as a principal determinant of allergy.
Although localizing the causal effects of the genes within the MHC region has been limited by the complexity and strong LD of this region, the genetic association between the MHC region and several immune and inflammatory diseases has been among the most robust. The HLA-C region has been associated particularly with Behçet disease, psoriasis, and sarcoidosis [30]–[32]. Of note, dysregulation of IgE has been reported in these diseases [33]–[35], which may also imply that HLA-C is involved in the genetic regulation of total IgE levels. Finally, it is also interesting to note that 1 locus (rs9266772) near HLA-C and MICA has been recently identified as one of the loci of allergy-specific susceptibility [36].
The meta-analysis of rs7939777 in GRAMD1B using the Tsukuba and Fukui cohorts yielded a P value of 3.35×10−10. Interestingly, although no studies demonstrated a relationship between GRAMD1B and serum IgE levels, a GWAS has identified a SNP near GRAMD1B on chromosome 11q24.1 associated with chronic lymphocytic leukemia (CLL) [37]. CLL is characterized by coexpression of CD19 and CD23 coupled with low levels of surface immunoglobulins [38]. CD23, also known as FCER2, is a low-affinity receptor for IgE and important for regulation of serum IgE levels. In addition, total serum IgE levels are inversely associated with risk of CLL [39]. Accordingly, polymorphisms of GRAMD1B could be related to regulation of total serum IgE levels.
Although the SNP in the PYHIN1/IFI16 region did not reach genome-wide significance in the primary GWAS (P = 3.19×10−7), a recent meta-analysis of GWASs of asthma has identified a SNP in PYHIN1 in populations of African descent [40]. PYHIN1 and IFI16 have recently emerged as sensors of microbial DNA [41], and the innate immune response relies on the ability of immune cells to detect the presence of infection through these germline-encoded pattern recognition receptors. Given that environments with a wide range of microbial exposures are associated with protection from childhood asthma and atopy in proportion to their level of exposure to bacterial and fungal microbes [42], PYHIN1 and IFI16 deserve further attention as candidate genes for association with asthma and atopy.
We have here tried to validate previously reported gene associations with IgE regulation. Among 32 autosomal genes, which were previously identified mainly in European populations, we found that 9 (28.1%) were replicated in a Japanese population after correction for multiple testing, indicating that some of the candidate genes for association with total serum IgE levels are effective across ethnic groups. Heterogeneity seems to exist in the genetic factors for total serum IgE levels among different ethnic groups [13], [43]. For SNPs that were not replicated in our study, causal SNPs for IgE regulation or SNPs tightly in LD with the causal SNPs may not exist in the regions analyzed in the Japanese population. Alternatively, our study sample size may have not have provided a significant power to detect the associations due to low minor allele frequencies of the true causal SNPs.
In terms of the limitations of this study, because we chose only 1 SNP for each region to replicate the original findings in the discovery cohort, we cannot exclude the possibility that we have missed true functional genetic variants in the replication cohorts. In addition, we observed many differences in the population characteristics of the discovery cohort and of the replication cohorts, including in the proportion of asthmatic patients and levels of total serum IgE. These differences might have affected our results, especially because the genetic background for increased levels of total IgE may differ in nonasthmatic healthy individuals and in asthmatic patients [44]. Nevertheless, analyses excluding asthmatic patients produced similar results with genome-wide significance for the HLA-C region, indicating the robustness of our findings.
As the mechanisms mediating the risk conferred by the HLA-C region remains to be found, future studies will identify the causal genes/variants within the susceptibility loci associated with levels of total serum IgE by fine-mapping and by investigating the biological link between rs3130941/HLA-C and regulation of IgE production.
In summary, we performed a GWAS showing positive results for total serum IgE levels for the first time in an Asian population. Association of a SNP in the HLA-C region with total serum IgE levels reached genome-wide significance in our meta-analysis involving a total of 3654 Japanese adults. We also demonstrated that some of the previously reported genetic associations with total serum IgE levels were replicated across ethnicities.
Materials and Methods
Ethical Statement
This study was approved by the Human Genome Analysis and Epidemiology Research Ethics Committee of the University of Tsukuba and by the Human Genome/Gene Analysis Research Ethics Review Committees of the Tsukuba Medical Center, RIKEN, the Hokkaido University School of Medicine, and the University of Fukui. Written informed consent was obtained from each participant in accordance with institutional requirements and the principles of the Declaration of Helsinki.
Study Participants
The discovery cohort (Tsukuba cohort) consisted of 1180 individuals of Japanese ethnicity (967 healthy volunteers and 213 patients with asthma). The healthy volunteers without pulmonary diseases such as asthma and COPD were originally recruited for a genetic study of pulmonary function from the general population who visited the Tsukuba Medical Center for an annual health checkup [45]. All the participants were asked about their respiratory health, medical history, lifestyle, and exposure to environmental irritants (eg, cigarette smoke, allergens, and air pollution) and underwent heart and lung auscultation. The patients with asthma were recruited for genetic analysis of asthma from the Tsukuba University Hospital and its affiliated hospitals [46]. Asthma was diagnosed by pulmonary physicians according to the American Thoracic Society criteria as previously described [47]. Specific serum IgE antibody was measured for both the healthy and the asthmatic groups with the multiple allergen simultaneous test (MAST)-26 chemiluminescent assay systems (Hitachi Chemical Company, Tokyo, Japan) [48]. Atopy was assessed by measurement of specific IgE responsiveness to 14 common inhaled allergens including Dermatophagoides farinae, grass pollens, animal dander, and molds. We defined atopy as a positive response (>4.40 lumicount) to at least 1 of the 14 allergens.
To replicate our findings in the discovery cohort, we analyzed 2 independent Japanese cohorts. The first replication cohort (Hokkaido cohort) comprised 619 healthy volunteers and 491 asthmatic patients from the Hokkaido University Hospital and its affiliated hospitals. This population was originally recruited for a case-control genetic association study searching for susceptibility genes to asthma and atopy [49]. Serum-specific IgE to Dermatophagoides species, molds, pollen, and animal dander was measured by a radioallergosorbent test (RAST). Atopy was defined as a positive response (>0.70 UA/mL) to at least 1 of these allergens.
The second replication cohort (Fukui cohort) comprised 1275 healthy volunteers and 89 asthmatic patients. This population was originally recruited from workers and students of the University of Fukui for a study of the genetic epidemiology of allergic rhinitis [50]. Serum IgE antibody specific to Japanese cedar, house dust, orchard grass, ragweed mix, Candida species, or Aspergillus species was measured by RAST. Atopy was defined as a positive response (>0.70 UA/mL) to at least 1 of these allergens.
Genotyping
Genomic DNA was extracted from peripheral blood samples of all participants by an automated DNA extraction system (QuickGene-610L; Fujifilm, Tokyo, Japan). Genotyping of the Tsukuba cohort was carried out using the Illumina HumanHap550v3 BeadChip (Illumina, San Diego, CA, USA) for the healthy volunteers and the HumanHap610-Quad BeadChip for the asthmatic patients. The concordance rate between the genotypes determined by the Illumina HumanHap550v3 BeadChip and the Illumina HumanHap610-Quad BeadChip among 182 duplicated samples was 0.99998 [46]. Quality control checks for the SNPs were performed separately using PLINK version 1.07 software [51]. None of the healthy volunteers or the asthmatic patients were removed owing to a call rate for autosomal SNPs of <0.02. SNPs with a missing genotype rate >0.01, minor allele frequency <0.01, or Hardy-Weinberg equilibrium P value <1.0×10−6 were excluded, leaving 479,940 SNPs that were common to the 2 arrays for analysis. Raw data is available upon request.
Genotyping accuracy on the X chromosome is often lower than that on other chromosomes because of difficulties involving clustering algorithms, higher frequencies of chromosome anomalies, and more missing data on X chromosome variants [52]. Genotyping of the pseudoautosomal region shared with the Y chromosome and hemizygous males can also be problematic. These analytic complexities could reduce the power of X chromosome analyses, making detection of reliable associations difficult. Therefore, in the current study, we decided to exclude the X chromosome from the analysis.
We imputed the genotypes of missing SNPs by using MACH version 1.0 software [53] to improve the resolution of candidate regions identified as associated with total IgE levels at P values <1×10−5. MACH employs a Markov chain algorithm and imputes missing genotypes by taking phased haplotypes as templates. We used 1000 Genomes Project data of Asian origin (JPT+CHB) (http://www.sph.umich.edu/csg/abecasis/MACH/download/1000G-2010-06.html) as the reference panel. To evaluate missing genotypes, we used 50 iterations of the Markov sampler to ensure reliable results.
To obtain the genomic inflation factor, we performed the multidimensional scaling (MDS) method using PLINK version 1.07 software. The MDS method is widely used in stratification methods, matching cases to controls based on genotype information (identity-by-state), resulting in discrete strata of individuals that can be analyzed using the Cochran–Mantel–Haenszel test [51].
For replication analyses of the original GWAS data, to obtain high-confidence results, we selected SNPs that are available in the ready-to-use predesigned TaqMan® SNP Genotyping assays (Applied Biosystems, Foster City, CA, USA) in each candidate region that satisfied the following conditions in the discovery Tsukuba cohort: (1) in strongest LD with the SNP most significantly associated with total IgE levels and (2) minor allele frequency >0.15. All assays are quality control tested using a mass spectrophotometer to verify sequence and yield. All assays have 1 VIC® and 1 FAM™ dye-labeled probe and 2 target-specific primers and undergo bioinformatics evaluation of target SNP sequences.
Validation of Association of Previously Reported Genes
A literature search was conducted in PubMed of publications up to June 1, 2013 on genetic association studies of total serum IgE levels. Keywords in the search strategy were (“IgE level” or “IgE concentration” or “serum IgE”) and (“polymorphism” or “SNP” or “genetics”) and (“association”). The search was restricted to human studies written in English. We reviewed the titles, abstracts, and texts of the publications to identify positive genetic association studies. Review articles and studies analyzing antigen-specific IgE production were excluded, as were studies using linkage analysis and transmission disequilibrium tests. We selected only genetic association studies. The references of the collected articles were also screened to find additional matching studies. From the retrieved publications, we selected eligible genes that were reported in 3 or more independent association studies or demonstrated by at least 1 GWAS so that we could as far as possible exclude potentially false-positive findings.
Because LD structures may be quite different between Japanese and Caucasian populations, we attempted gene-level replication instead of SNP-level replication. From the primary GWAS data, we chose the SNP with the strongest statistical evidence in a region extending +/−10 kilobases (kb) from each literature-selected candidate gene. The significance level was corrected for multiple testing using the SNPSpD program [16], which corrects for multiple testing of SNPs in LD with each other on the basis of the spectral decomposition of matrices of pairwise LD between SNPs. This method provides a useful alternative to more computationally intensive permutation tests.
Statistical Analysis
In the primary GWAS cohort, associations of genotypes of all the SNPs with log-transformed (base 10) levels of total serum IgE were analyzed by multiple linear regression models in PLINK version 1.07. Because total serum IgE levels are influenced by age, sex, smoking status, and asthma affection status [8], [54], the original GWAS of the total serum IgE levels in the current study was adjusted according to these variables. Quantile-quantile plots and genomic inflation factors were calculated in PLINK version 1.07. In the replication studies, the associations were examined by the same methods in the Hokkaido cohort. As smoking behavior was not available in the Fukui cohort, the associations were adjusted only for age, sex, and asthma affection status in this cohort. Replication was declared only if P<0.05 and the direction of the effect was the same as in the primary GWAS. Combined analysis of the primary GWAS with the replication studies was performed by the basic meta-analysis function in PLINK version 1.07. Random-effect meta-analysis P values were estimated. We used the Haploview 4.2 program [55] to analyze the LD values between SNPs.
Supporting Information
Study flow chart. GWAS for total IgE levels was performed, followed by replication studies and meta-analysis. Validation of previously reported genes for IgE was also conducted using the GWAS data.
(TIFF)
Fine-mapping association plots on chromosome 6p21. Three peaks are identified: the MHC class I, MHC class II, and LEMD2 regions.
(TIFF)
Fine-mapping association plots in the MHC class I region. The color of each circle reflects the LD (r2) between a particular SNP and rs3130941 indicated as a purple diamond.
(TIFF)
Results of meta-analysis for nonasthmatic healthy individuals only.
(DOCX)
Results of meta-analysis after inclusion of atopic status as a covariate.
(DOCX)
Previously reported polymorphisms significantly associated with total serum IgE.
(DOCX)
Genetic influences of SNPs in the MHC class I/II regions on the association between rs3130941 and total IgE levels.
(DOCX)
Acknowledgments
We would like to thank Ms Flaminia Miyamasu, an associate professor at the Faculty of Medicine, University of Tsukuba, who proofread and commented on this paper. We also thank Ms Takako Nakamura for technical assistance with genotyping.
Funding Statement
This study was partly supported by a Grant-in-Aid for Scientific Research (B), No. 24390206, from the Japan Society for the Promotion of Science. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding received for this study.
References
- 1. Holgate ST (2012) Innate and adaptive immune responses in asthma. Nat Med 18: 673–683. [DOI] [PubMed] [Google Scholar]
- 2. Jacquet A (2011) The role of innate immunity activation in house dust mite allergy. Trends Mol Med 17: 604–611. [DOI] [PubMed] [Google Scholar]
- 3. Jacobsen HP, Herskind AM, Nielsen BW, Husby S (2001) IgE in unselected like-sexed monozygotic and dizygotic twins at birth and at 6 to 9 years of age: high but dissimilar genetic influence on IgE levels. J Allergy Clin Immunol 107: 659–663. [DOI] [PubMed] [Google Scholar]
- 4. Lebowitz MD, Barbee R, Burrows B (1984) Family concordance of IgE, atopy, and disease. J Allergy Clin Immunol 73: 259–264. [DOI] [PubMed] [Google Scholar]
- 5. Meyers DA, Beaty TH, Freidhoff LR, Marsh DG (1987) Inheritance of total serum IgE (basal levels) in man. Am J Hum Genet 41: 51–62. [PMC free article] [PubMed] [Google Scholar]
- 6. Dizier MH, Hill M, James A, Faux J, Ryan G, et al. (1995) Detection of a recessive major gene for high IgE levels acting independently of specific response to allergens. Genet Epidemiol 12: 93–105. [DOI] [PubMed] [Google Scholar]
- 7. Palmer LJ, Burton PR, Faux JA, James AL, Musk AW, et al. (2000) Independent inheritance of serum immunoglobulin E concentrations and airway responsiveness. Am J Respir Crit Care Med 161: 1836–1843. [DOI] [PubMed] [Google Scholar]
- 8. Sears MR, Burrows B, Flannery EM, Herbison GP, Hewitt CJ, et al. (1991) Relation between airway responsiveness and serum IgE in children with asthma and in apparently normal children. N Engl J Med 325: 1067–1071. [DOI] [PubMed] [Google Scholar]
- 9. Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG (1989) Association of asthma with serum IgE levels and skin-test reactivity to allergens. N Engl J Med 320: 271–277. [DOI] [PubMed] [Google Scholar]
- 10. Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, et al. (2008) Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet 4: e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, et al. (2010) A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 363: 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granada M, Wilk JB, Tuzova M, Strachan DP, Weidinger S, et al.. (2012) A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J Allergy Clin Immunol 129: 840–845 e821. [DOI] [PMC free article] [PubMed]
- 13. Levin AM, Mathias RA, Huang L, Roth LA, Daley D, et al. (2013) A meta-analysis of genome-wide association studies for serum total IgE in diverse study populations. J Allergy Clin Immunol 131: 1176–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liao M, Shi D, Wang Y, Zhang K, Chen X, et al. (2013) Genome-wide scan on total serum IgE levels identifies no common variants in a healthy Chinese male population. Immunogenetics 65: 561–568. [DOI] [PubMed] [Google Scholar]
- 15. Kim JH, Cheong HS, Park JS, Jang AS, Uh ST, et al. (2013) A Genome-Wide Association Study of Total Serum and Mite-Specific IgEs in Asthma Patients. PLoS One 8: e71958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nyholt DR (2004) A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 74: 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shin HD, Park BL, Kim LH, Jung JH, Wang HJ, et al. (2004) Association of tumor necrosis factor polymorphisms with asthma and serum total IgE. Hum Mol Genet 13: 397–403. [DOI] [PubMed] [Google Scholar]
- 18. Trabetti E, Patuzzo C, Malerba G, Galavotti R, Martinati LC, et al. (1999) Association of a lymphotoxin alpha gene polymorphism and atopy in Italian families. J Med Genet 36: 323–325. [PMC free article] [PubMed] [Google Scholar]
- 19. Malerba G, Trabetti E, Patuzzo C, Lauciello MC, Galavotti R, et al. (1999) Candidate genes and a genome-wide search in Italian families with atopic asthmatic children. Clin Exp Allergy 29 Suppl 427–30. [PubMed] [Google Scholar]
- 20. Krasznai M, Szaniszlo K, Kraxner H, Vargha E, Kovacs M, et al. (2011) Association of TLR-4 and TNF-alpha polymorphisms with clinical symptoms and cytokine levels in patients with allergic rhinitis. Eur Arch Otorhinolaryngol 268: 561–567. [DOI] [PubMed] [Google Scholar]
- 21. Gueant-Rodriguez RM, Gueant JL, Viola M, Tramoy D, Gaeta F, et al. (2008) Association of tumor necrosis factor-alpha −308G>A polymorphism with IgE-mediated allergy to betalactams in an Italian population. Pharmacogenomics J 8: 162–168. [DOI] [PubMed] [Google Scholar]
- 22. Sharma S, Sharma A, Kumar S, Sharma SK, Ghosh B (2006) Association of TNF haplotypes with asthma, serum IgE levels, and correlation with serum TNF-alpha levels. Am J Respir Cell Mol Biol 35: 488–495. [DOI] [PubMed] [Google Scholar]
- 23. Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, et al. (2010) Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 26: 2474–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marsh DG, Meyers DA, Bias WB (1981) The epidemiology and genetics of atopic allergy. N Engl J Med 305: 1551–1559. [DOI] [PubMed] [Google Scholar]
- 25. Shirakawa T, Enomoto T, Shimazu S, Hopkin JM (1997) The inverse association between tuberculin responses and atopic disorder. Science 275: 77–79. [DOI] [PubMed] [Google Scholar]
- 26. Thomas MJ, Noble A, Sawicka E, Askenase PW, Kemeny DM (2002) CD8 T cells inhibit IgE via dendritic cell IL-12 induction that promotes Th1 T cell counter-regulation. J Immunol 168: 216–223. [DOI] [PubMed] [Google Scholar]
- 27. Kalinski P, Moser M (2005) Consensual immunity: success-driven development of T-helper-1 and T-helper-2 responses. Nat Rev Immunol 5: 251–260. [DOI] [PubMed] [Google Scholar]
- 28. Boyton RJ, Smith J, Ward R, Jones M, Ozerovitch L, et al. (2006) HLA-C and killer cell immunoglobulin-like receptor genes in idiopathic bronchiectasis. Am J Respir Crit Care Med 173: 327–333. [DOI] [PubMed] [Google Scholar]
- 29. Das A, Long EO (2010) Lytic granule polarization, rather than degranulation, is the preferred target of inhibitory receptors in NK cells. J Immunol 185: 4698–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hughes T, Coit P, Adler A, Yilmaz V, Aksu K, et al. (2013) Identification of multiple independent susceptibility loci in the HLA region in Behcet’s disease. Nat Genet 45: 319–324. [DOI] [PubMed] [Google Scholar]
- 31. Strange A, Capon F, Spencer CC, Knight J, Weale ME, et al. (2010) A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet 42: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adrianto I, Lin CP, Hale JJ, Levin AM, Datta I, et al. (2012) Genome-wide association study of African and European Americans implicates multiple shared and ethnic specific loci in sarcoidosis susceptibility. PLoS One 7: e43907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Onat AM, Buyukhatipoglu H, Yilmaz M, Geyik R, Celik A, et al. (2007) Immunoglobulin E: a new diagnostic clue for Behcet’s disease? IgE and Behcet’s disease. Clin Rheumatol 26: 81–83. [DOI] [PubMed] [Google Scholar]
- 34. Ovcina-Kurtovic N, Kasumagic-Halilovic E (2010) Serum levels of total immunoglobulin E in patients with psoriasis: relationship with clinical type of disease. Med Arh 64: 28–29. [PubMed] [Google Scholar]
- 35. Hattori T, Konno S, Shigemura M, Matsuno K, Shimizu C, et al. (2012) Total serum IgE levels and atopic status in patients with sarcoidosis. Allergy Asthma Proc 33: 90–94. [DOI] [PubMed] [Google Scholar]
- 36. Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, et al. (2013) A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet 45: 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Di Bernardo MC, Crowther-Swanepoel D, Broderick P, Webb E, Sellick G, et al. (2008) A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet 40: 1204–1210. [DOI] [PubMed] [Google Scholar]
- 38. Gaidano G, Foa R, Dalla-Favera R (2012) Molecular pathogenesis of chronic lymphocytic leukemia. J Clin Invest 122: 3432–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ellison-Loschmann L, Benavente Y, Douwes J, Buendia E, Font R, et al. (2007) Immunoglobulin E levels and risk of lymphoma in a case-control study in Spain. Cancer Epidemiol Biomarkers Prev 16: 1492–1498. [DOI] [PubMed] [Google Scholar]
- 40. Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, et al. (2011) Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet 43: 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goubau D, Rehwinkel J, Reis e Sousa C (2010) PYHIN proteins: center stage in DNA sensing. Nat Immunol 11: 984–986. [DOI] [PubMed] [Google Scholar]
- 42. Rabinovitch N, Liu AH, Zhang L, Rodes CE, Foarde K, et al. (2005) Importance of the personal endotoxin cloud in school-age children with asthma. J Allergy Clin Immunol 116: 1053–1057. [DOI] [PubMed] [Google Scholar]
- 43. Mathias RA, Freidhoff LR, Blumenthal MN, Meyers DA, Lester L, et al. (2001) Genome-wide linkage analyses of total serum IgE using variance components analysis in asthmatic families. Genet Epidemiol 20: 340–355. [DOI] [PubMed] [Google Scholar]
- 44. Hizawa N, Yamaguchi E, Jinushi E, Kawakami Y (2000) A common FCER1B gene promoter polymorphism influences total serum IgE levels in a Japanese population. Am J Respir Crit Care Med 161: 906–909. [DOI] [PubMed] [Google Scholar]
- 45. Masuko H, Sakamoto T, Kaneko Y, Iijima H, Naito T, et al. (2011) Lower FEV1 in non-COPD, nonasthmatic subjects: association with smoking, annual decline in FEV1, total IgE levels, and TSLP genotypes. Int J Chron Obstruct Pulmon Dis 6: 181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, et al. (2011) Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet 43: 893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harada M, Obara K, Hirota T, Yoshimoto T, Hitomi Y, et al. (2009) A functional polymorphism in IL-18 is associated with severity of bronchial asthma. Am J Respir Crit Care Med 180: 1048–1055. [DOI] [PubMed] [Google Scholar]
- 48. Miller SP, Marinkovich VA, Riege DH, Sell WJ, Baker DL, et al. (1984) Application of the MAST Immunodiagnostic System to the determination of allergen-specific IgE. Clin Chem 30: 1467–1472. [PubMed] [Google Scholar]
- 49. Isada A, Konno S, Hizawa N, Tamari M, Hirota T, et al. (2010) A functional polymorphism (−603A –> G) in the tissue factor gene promoter is associated with adult-onset asthma. J Hum Genet 55: 167–174. [DOI] [PubMed] [Google Scholar]
- 50. Sakashita M, Hirota T, Harada M, Nakamichi R, Tsunoda T, et al. (2010) Prevalence of allergic rhinitis and sensitization to common aeroallergens in a Japanese population. Int Arch Allergy Immunol 151: 255–261. [DOI] [PubMed] [Google Scholar]
- 51. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wise AL, Gyi L, Manolio TA (2013) eXclusion: toward integrating the X chromosome in genome-wide association analyses. Am J Hum Genet 92: 643–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li Y, Willer C, Sanna S, Abecasis G (2009) Genotype imputation. Annu Rev Genomics Hum Genet 10: 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wuthrich B, Schindler C, Medici TC, Zellweger JP, Leuenberger P (1996) IgE levels, atopy markers and hay fever in relation to age, sex and smoking status in a normal adult Swiss population. SAPALDIA (Swiss Study on Air Pollution and Lung Diseases in Adults) Team. Int Arch Allergy Immunol 111: 396–402. [DOI] [PubMed] [Google Scholar]
- 55. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Study flow chart. GWAS for total IgE levels was performed, followed by replication studies and meta-analysis. Validation of previously reported genes for IgE was also conducted using the GWAS data.
(TIFF)
Fine-mapping association plots on chromosome 6p21. Three peaks are identified: the MHC class I, MHC class II, and LEMD2 regions.
(TIFF)
Fine-mapping association plots in the MHC class I region. The color of each circle reflects the LD (r2) between a particular SNP and rs3130941 indicated as a purple diamond.
(TIFF)
Results of meta-analysis for nonasthmatic healthy individuals only.
(DOCX)
Results of meta-analysis after inclusion of atopic status as a covariate.
(DOCX)
Previously reported polymorphisms significantly associated with total serum IgE.
(DOCX)
Genetic influences of SNPs in the MHC class I/II regions on the association between rs3130941 and total IgE levels.
(DOCX)