Abstract
Background
Drosophila melanogaster adult males perform an elaborate courtship ritual to entice females to mate. fruitless (fru), a gene that is one of the key regulators of male courtship behavior, encodes multiple male-specific isoforms (FruM). These isoforms vary in their carboxy-terminal zinc finger domains, which are predicted to facilitate DNA binding.
Results
By over-expressing individual FruM isoforms in fru-expressing neurons in either males or females and assaying the global transcriptional response by RNA-sequencing, we show that three FruM isoforms have different regulatory activities that depend on the sex of the fly. We identified several sets of genes regulated downstream of FruM isoforms, including many annotated with neuronal functions. By determining the binding sites of individual FruM isoforms using SELEX we demonstrate that the distinct zinc finger domain of each FruM isoforms confers different DNA binding specificities. A genome-wide search for these binding site sequences finds that the gene sets identified as induced by over-expression of FruM isoforms in males are enriched for genes that contain the binding sites. An analysis of the chromosomal distribution of genes downstream of FruM shows that those that are induced and repressed in males are highly enriched and depleted on the X chromosome, respectively.
Conclusions
This study elucidates the different regulatory and DNA binding activities of three FruM isoforms on a genome-wide scale and identifies genes regulated by these isoforms. These results add to our understanding of sex chromosome biology and further support the hypothesis that in some cell-types genes with male-biased expression are enriched on the X chromosome.
Keywords: Fruitless, Sex hierarchy, Drosophila, Behavior, Genomics, RNA-seq
Background
In Drosophila melanogaster differences in adult reproductive behaviors are specified by the somatic sex determination hierarchy (hereafter called sex hierarchy), a multi-branched hierarchy with functions in directing both sexual development and dosage compensation (Figure 1) reviewed in [1,2]. The branch of the sex hierarchy critical for specifying adult behaviors consists of a pre-mRNA splicing cascade that regulates the sex-specific splicing of transcripts from doublesex (dsx) and fruitless (fru) (Figure 1) reviewed in [3,4]. fru was initially shown to be important for male courtship behavior based on the phenotypes of mutant males that displayed high levels of male-male courtship behaviors [5]. This was distinct from the phenotypic observations with respect to other mutants that impacted courtship behaviors, in that the phenotype of the fru mutant was specific to courtship behaviors. Later, molecular-genetic analyses of fru demonstrated the position of fru in the sex hierarchy, and showed that it was required for all aspects of male courtship behaviors, providing strong evidence that fru is a key regulator of male courtship behavior [6-10].
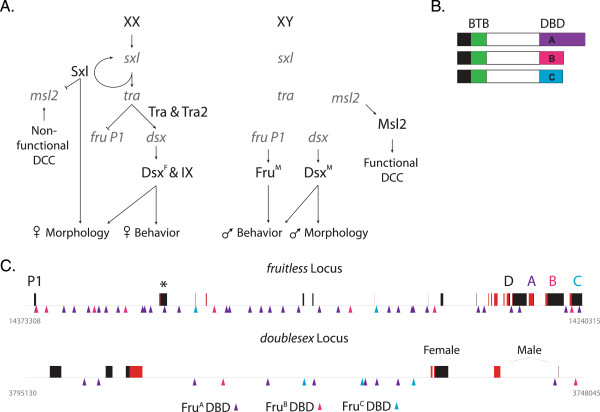
Figure 1.
Fruitless, a gene in the sex-determination hierarchy, encodes multiple male-specific isoforms with distinct zinc finger domains. (A) The Drosophila somatic sex determination hierarchy has two branches--one regulates dosage compensation and the other somatic sexual development. In females (chromosomally XX) Sxl is produced and so dosage compensation is not active. In males there is no Sxl and dosage compensation is active, resulting in increased expression of the single male X chromosome. For somatic sexual development, Sxl regulates the splicing of its own pre-mRNA and the transformer (tra). The product of tra (Tra) along with Transformer-2 (Tra2), coordinate to regulate splicing of transcripts produced from doublesex (dsx) and the P1 promoter of fruitless (fru P1). This branch of the hierarchy culminates in the production of sex-specific transcription factors (DsxF, DsxM, and FruM) that specify sex-specific morphology and behaviors. Grey indicates transcripts; black indicates proteins. (B) Schematic of FruM proteins. Male-specific 101 amino acid region (Black), a bric-a-brac, tramtrack, broad-complex domain (BTB), and distinct zinc finger domains (A, B, or C) are indicated. (C)fruitless and doublesex locus. Coding exons (red bars), non-coding exons (black bars), sex-specifically spliced exon of fru (asterisks), first fru promoter (P1), exons encoding the zinc-finger DNA binding domains (A-D), and male- and female-specific exons for dsx are indicated. The DNA binding motifs (triangles) for A (purple), B (pink) or C (cyan) DNA binding domains of FruM are indicated.
fru is a complex locus that encodes both sex-specific and non-sex-specific proteins through the production of transcripts from at least four different promoters (P1-4) [6,7]. Transcripts expressed from the fru P1 promoter are critical for male courtship behaviors, and are the only fru pre-mRNAs that are spliced by the sex hierarchy (Figure 1). fru P1 transcripts produce multiple male-specific isoforms (FruM) in ~ 2-5% of all central nervous system (CNS) neurons and these neurons have been shown to be important for courtship behaviors [11-14]. fru P1 expressing neurons are present in both males and females [6,11,13,14], but the FruM protein isoforms are produced only in males where they contribute to building the potential for male courtship into the nervous system during development [15-18]. Conversely, fru P1 transcripts in females are not translated [19,20]. All Fru isoforms are members of a family of conserved proteins that contain a BTB (BTB for bric-a-brac, tramtrack, broad-complex) domain and a zinc finger domain (Figure 1). FruM isoforms contain an amino-terminal 101 amino acid region of unknown function that is not present in Fru isoforms common to both sexes. fru P1 transcripts are alternatively spliced at their 3′ ends into one of five exons that encode different zinc finger domains, which are predicted DNA binding domains (DBD; named A-E) [6,19,21,22]. Thus, fru is predicted to encode transcription factors. However, there is no direct evidence of FruM transcriptional activities, other than association with known chromatin modifying proteins [20].
Three of the five predicted FruM isoforms have been shown to be the predominate isoforms in adult head and central nervous system tissues (FruMA, FruMB and FruMC) [22]. These isoforms display differences in their expression patterns and in their ability to rescue male courtship defects [22]. As a first step to mechanistically understanding how FruM isoforms specify the potential for male courtship behaviors, the DNA binding specificities of each FruM isoform needs to be determined and the sets of genes that are regulated downstream of each FruM isoform identified. The identification of genes regulated by each FruM isoform will also contribute to our understanding of how fru functions to establish the potential for sex-specific behaviors.
Here, we identify genes that are induced or repressed by FruM by examining gene expression in adult head tissues where we over-express individual FruM isoforms (FruMA, FruMB and FruMC) in fru P1-expressing neurons of either males or females. We show that each isoform has different regulatory activities and that the sex of the fly impacts which genes change expression as a consequence of FruM over-expression. Similar over-expression conditions were previously used to demonstrate that FruM is sufficient, when expressed in female fru P1-expressing neurons, to specify the potential for nearly all aspects of male courtship behavior [4,12,14]. We used an in vitro binding site selection technique (SELEX) to identify the sequence motifs bound by each of three FruM isoforms and show that each isoform has different binding specificity [reviewed in [23,24]. For each gene, the coding sequence and the regulatory region (defined as 2 kb upstream and 2 kb downstream of the coding sequence) was examined for the presence of these binding sites. Genes containing these binding sites are enriched in the gene sets induced by over-expression of the respective FruM isoform in males, and in the genes identified as induced by FruM in loss-of-function mutant analyses. Additionally, genes induced by FruM, are enriched on the X chromosome, whereas those that are repressed by FruM are under represented on the X chromosome.
Results
The goal of our study was to identify genes whose expression was modulated (either up or down) by fru P1 in the nervous system. To this end, we carried out a set of parallel experiments in which the GAL4/UAS system was used to overexpress each of the three best-characterized FruM isoforms (FruMA, FruMB and FruMC; Figure 1) [6,7,22], in just the fru P1-expressing neurons [14]. RNA-sequencing (RNA-seq) analysis was performed on mRNA extracted from the heads of such flies to identify differential expression on a genome-wide level, with exon-level resolution. We compared expression in heads in either male or female flies over-expressing one of the FruM isoforms, as compared to wild type adult male or female heads, respectively. This allows us to determine if there are differences in individual FruM isoform activities and if there are sex-specific factors that function in conjunction with FruM. In addition, we examined gene expression differences in males mutant for fru P1 (two different allele combinations; see Materials and Methods) compared to wild type males.
One of the strengths of RNA-seq analysis is the ability to detect differences in isoform expression levels. This is because exons are used for estimating expression, and thus the presence and difference in amount of transcript from alternative exon cassettes can be used directly to make inferences about isoforms. We focused on exon level expression and used existing models of transcript isoforms to identify alternative exon structures. Next, we tested for differential expression in our comparisons for each exon separately [see Materials and Methods and [25], and then used existing gene models to make inferences about differential expression of isoforms.
FruM isoforms have different activities
To determine if FruMA, FruMB and FruMC had different effects on gene expression in fru P1-expressing neurons we determined the gene sets that have exons that are both (1) statistically significantly induced or repressed and (2) have a ≥2 fold change in expression level when the data from animals overexpressing each FruM isoform are compared separately to the data from both CS and Berlin males (Additional file 1: Table S1). We found that over-expression of each FruM isoform leads to different subsets of genes with exons whose transcription is either induced or repressed. As expected, when we over-express each FruM isoform we found that the exon encoding the respective Fru DBD is significantly up regulated.
Over-expression of FruMA, FruMB and FruMC leads to 752, 739 and 927 genes with higher expression than wild type males, respectively. There were 460 genes with higher expression in all three conditions (Figure 2A and Additional file 1: Table S1; Additional file 2: Figure S1; FruM-induced genes). We found that substantially more genes are up-regulated, than down regulated, relative to wild type expression. Over-expression of FruMA, FruMB and FruMC lead to 204, 259 and 295 genes with lower expression than in wild type males, respectively. There are 55 genes with lower expression in all three conditions (Figure 2B and Additional file 1: Table S1; FruM-repressed genes).
Figure 2.
FruM isoforms have both distinct and overlapping sets of regulated genes. (A-F) Venn diagrams displaying number of distinct and mutual genes between different sets of differentially expressed genes. (A-D) Numbers of genes induced or repressed in males or females by over-expression of FruMA, FruMB or FruMC in fru P1-expressing neurons of the head. (E-F) Numbers of all genes induced or repressed by over-expression of any of the FruM isoforms.
FruM isoforms have different activities in males as compared to females
Using the same criteria as above, we examined the numbers of genes differentially expressed when FruM isoforms are over-expressed in female fru P1-expressing neurons as compared to wild type females of CS and Berlin strains. The rationale for these comparisons was to determine if there are additional sex-specific differences in fru P1-expressing neurons that might influence FruM activities. Over-expression of FruMA, FruMB and FruMC in females leads to 111, 117 and 167 genes with higher expression than wild type females, respectively. There are 42 genes having higher expression in all three conditions, which included the exon encoding the respective Fru DBD (Figure 2C; Additional file 2: Figure S1 and Additional file 3: Table S2). Over-expression of FruMA, FruMB or FruMC lead to 183, 237 and 198 genes with lower expression than wild type females, respectively. There are 42 genes with lower expression in all three conditions (Figure 2D).
These results further demonstrate that each FruM isoform has different activities with respect to regulating gene expression. In female fru P1-expressing neurons, more genes are repressed rather than induced when each isoform is expressed. This is in contrast to our observations in males. Taken together these findings suggest that the activity of each FruM isoform is influenced by the sex in which it is produced. This suggests that there may be other factors that are present in a sex-specific manner in fru P1-expressing neurons to influence FruM isoform activities.
Next, we determined if there are differences in the sets of genes that are induced or repressed by the over-expression of FruM isoforms in males and females. We determined the union of the genes that are induced by any of the three FruM isoforms in either males or females (Figure 2E and F). There are 1217 genes and 267 genes induced by any of the three FruM isoforms in males and females, respectively. The intersection of the induced genes in males and females is 128 genes (Figure 2E). We also determined the union of the genes that are repressed by any of the three FruM isoforms in either males or females. There are 554 genes and 462 genes repressed by one of the three FruM isoforms in males and females, respectively. The intersection of the repressed genes in males and females is 135 genes (Figure 2F). The genes induced in both males and females include dsx, Gustatory receptor 93a, serotonin receptor 1A, semaphorin-5c (Additional file 1: Table S1 and Additional file 3: Table S2). The genes repressed in males and females include genderblind, methuselah-like 8 (Additional file 1: Table S1 and Additional file 3: Table S2). These genes are those for which over-expression of FruM isoforms in fru P1-expressing cells can influence gene expression independent of the sex of the fly.
Overall, many more genes are induced by overexpression of each FruM isoform in males as compared to females (Additional file 2: Figure S1), whereas there is not as large a difference in numbers of genes that are repressed by each isoform in males and females, suggesting that there is a sex-specific factor(s) that functions with FruM in males to facilitate gene induction.
Fru isoforms have different DNA binding specificity
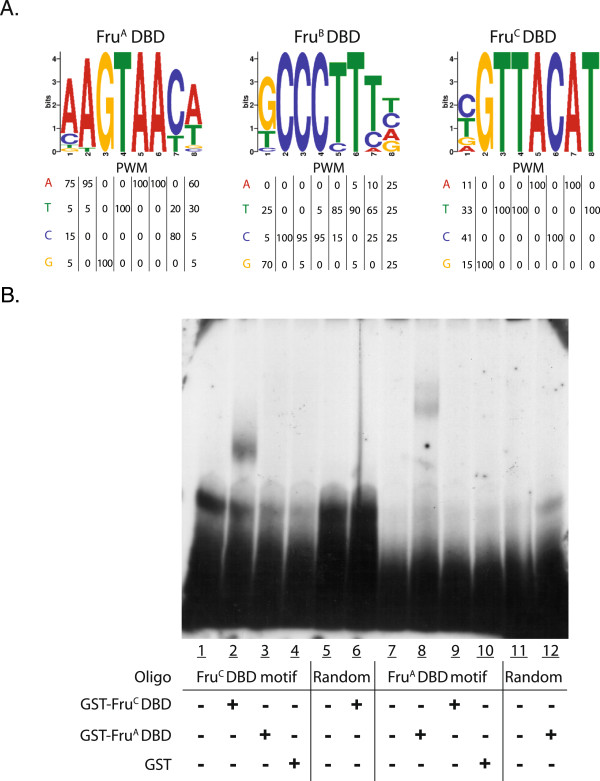
Our results demonstrate that FruM isoforms have different activities with respect to the gene sets that are induced or repressed in response to their expression. One possibility is that this is through differential DNA binding properties of the three isoforms. To address this question, we determined the binding site sequence for FruA, FruB and FruC DBDs using an in vitro selection technique called SELEX (see Materials and Methods). From this analysis, we found that each of the DBDs examined bind different sequence motifs. The consensus motifs identified for the DBDs in the FruA, FruB and FruC proteins are AGTAAC, GCCCTTT, and TGTTACATCA, respectively (Figure 3A).
Figure 3.
Identification of DNA binding motifs for the A, B and C zinc finger domains of FruM. (A) Consensus DNA binding sequences of the zinc finger DNA binding domains (DBD) of FruMA, FruMB or FruMC identified after ten rounds of SELEX. (B) Oligonucleotides containing the consensus sequences identified by SELEX, or sequences of the same nucleotide content but randomized in order, for DBDA or DBDC were incubated with GST-FruA DBD or GST-FruC DBD, or GST alone and assayed by gel mobility shift assay. GST-FruA DBD and GST-FruC DBD bound to oligonucleotides containing their respective consensus sequence (Lane 2 and 8) have slower mobility than GST (Lane 4 and 10), GST-FruA DBD (Lane 3 and 12), or GST-FruC DBD (Lane 6 and 9) incubated with oligonucleotides containing random sequences or the consensus sequence from other zinc finger domains or free oligonucleotides (no protein added in Lanes 1, 5, 7,11).
Gel shift analysis, using purified GST-Fru DBD fusion proteins and the identified binding sites for FruA and FruC, demonstrate that they each bind specifically to the sequence identified in the SELEX experiment, but do not bind to an oligonucleotide (oligo) containing a randomized sequence with the same nucleotide content (Figure 3B). Furthermore, FruA and FruC did not bind the oligos containing the motifs identified for FruC and FruA, respectively (Figure 3B).
When gel shifts were performed using the DBD in the FruB protein, high levels of binding to DNA sequences that are present in each oligo used in these assays, flanking the SELEX identified motif, were observed in the gel shift assay (see Materials and Methods). This could be because the FruB protein has a poly-glutamine tract, which is not observed in the other binding domains and this facilitates non-specific association with DNA in the gel shift assay. To ensure that the correct sequence motif was identified for FruB, the full SELEX technique was performed two independent times and identified nearly the same consensus sequence.
Fru DNA binding site motifs are significantly enriched in genes that are induced by FruM isoforms
Next, we determined the frequency of each Fru binding site motif on a genome-wide level, in annotated gene regions, including introns and regions 2 kb upstream and downstream from each gene’s annotated start and stop site. We found that genes containing at least one sequence motif for FruA DBD are significantly enriched in the sets of genes that are induced downstream of FruMA, and similar enrichments were observed for FruB and FruC in males, respectively (Table 1 and Additional file 4: Table S3). We do not see a similar enrichment when we examine the genes repressed downstream FruA, FruB and FruC in males. A very different pattern is observed for genes induced downstream of FruA, FruB and FruC in females, with only the gene sets induced downstream FruA showing enrichment of genes containing at least one FruA binding site (Table 1). These results suggest that the sets of genes that are induced in males by FruM over-expression includes genes that may be direct targets of FruM, whereas those that are induced in females, or repressed in either males or females by FruM over-expression are not as likely to be direct targets.
Table 1.
Genes containing DNA binding motifs are significantly enriched in all FruM -induced sets of genes in males
| Total Genes | Number of Genes with Motif | Expected Number of Genes | Chi-Square Value | Degrees of Freedom | Exact P-value for Chi-Square | Fisher raw P-value (2-tail) | |||
|---|---|---|---|---|---|---|---|---|---|
|
FruA DBD Motif |
Male |
Induced by over-expression of FruMA |
14903 |
644 |
473.4123331 |
174.7423677 |
1 |
9.87E-39 |
2.39E-45 |
| Repressed by over-expression of FruMA |
14903 |
134 |
128.4256861 |
0.66217505 |
1 |
0.42340488 |
0.465483604 |
||
| Induced by FruM (As determined by fru P1 mutant analysis) |
14903 |
575 |
444.4536 |
108.65168 |
1 |
5.99946E-25 |
8.5546E-28 |
||
| Repressed by FruM (As determined by fru P1 mutant analysis) |
14903 |
276 |
274.4784272 |
0.023454623 |
1 |
0.880224856 |
0.919841345 |
||
| Female |
Induced by over-expression of FruMA |
14903 |
80 |
69.87868215 |
3.98687096 |
1 |
0.04846709 |
0.04846709 |
|
| Repressed by over-expression of FruMA |
14903 |
116 |
115.2053949 |
0.014977928 |
1 |
0.9387153 |
0.938715296 |
||
|
FruB DBD Motif |
Male |
Induced by over-expression of FruMB |
14903 |
378 |
183.7706502 |
287.4837937 |
1 |
3.99E-56 |
6.65E-56 |
| Repressed by over-expression of FruMB |
14903 |
50 |
64.40676374 |
4.36503163 |
1 |
0.041977653 |
0.035594233 |
||
| Induced by FruM (As determined by fru P1 mutant analysis) |
14903 |
309 |
175.564383 |
141.6955672 |
1 |
2.20046E-29 |
3.27561E-29 |
||
| Repressed by FruM (As determined by fru P1 mutant analysis) |
14903 |
128 |
108.4221969 |
4.847043913 |
1 |
0.028457111 |
0.032441531 |
||
| Female |
Induced by over-expression of FruMB |
14903 |
35 |
29.09494733 |
1.607773993 |
1 |
0.236941316 |
0.199286144 |
|
| Repressed by over-expression of FruMB |
14903 |
64 |
58.93591894 |
0.588511944 |
1 |
0.448926212 |
0.44892621 |
||
| FruC DBD Motif | Male |
Induced by over-expression of FruMC |
14903 |
296 |
131.8687513 |
253.964127 |
1 |
3.55E-46 |
4.53E-46 |
| Repressed by over-expression of FruMC |
14903 |
33 |
28.16614105 |
0.261120798 |
1 |
0.613295999 |
0.613295997 |
||
| Induced by FruM (As determined by fru P1 mutant analysis) |
14903 |
209 |
100.4307858 |
143.6364162 |
1 |
2.29794E-27 |
2.78422E-27 |
||
| Repressed by FruM (As determined by fru P1 mutant analysis) |
14903 |
62 |
62.0224116 |
9.72597E-06 |
1 |
1 |
1 |
||
| Female | Induced by over-expression of FruMC |
14903 |
30 |
23.75629068 |
1.934825144 |
1 |
0.180129952 |
0.180129951 |
|
| Repressed by over-expression of FruMC | 14903 | 33 | 28.08642555 | 0.980189811 | 1 | 0.355875025 | 0.306776226 |
FruA, FruB and FruC sites are found in overlapping sets of genes and the presence of a motif for one significantly increases the likelihood of finding a binding site for at least one of the other. This result may explain why there were several genes that were induced by each of the three Fru isoforms in both males and females (see above).
fru has many binding sites with 32, 8 and 2 binding sites motifs for each FruA, FruB and FruC DBD, respectively (Figure 1C). Additionally, dsx has a large number of binding sites with 9, 2 and 3 binding sites motifs for FruA, FruB and FruC DBD, respectively (Figure 1C), although it should be noted that both fru and dsx are large genes. FruM may regulate dsx expression directly, consistent with the overlap observed between Dsx and FruM in the CNS [26,27], it may also regulate its own expression. This regulation of both dsx and fru transcript levels by FruM isoforms could ensure sufficient dsx and fru expression in neurons important for male courtship behaviors.
Genes with roles in neuronal patterning and physiology are enriched among genes regulated by FruM isoforms in males and not in females
fru P1-expressing neurons are present in very similar positions and numbers in adult males and females. To gain insight into the processes FruM regulates, we examined gene ontology (GO) enrichment of protein domains [28] and biological processes, molecular functions and cellular processes [29] for FruM regulated genes (Additional file 5: Table S4). For the 1,217 genes up-regulated by over-expression of any of the FruM isoforms in males, there is an enrichment of genes that contain protein domains that function in neuronal patterning and physiology. Among the enriched categories for protein domains are immunoglobulin-like fold, pleckstrin homology domain, PDZ domain, Ion transport domain, epidermal growth factor-like domain, and voltage dependent potassium channel. Many proteins with these domains function at the plasma membrane and mediate neuronal projection patterns, form complexes with channels, and make junctions, including synaptic and neuromuscular junctions, which are consistent with functions ascribed to FruM isoforms (Additional file 5: Table S4).
Among the enriched GO terms for those 1,217 up-regulated genes are those that underlie diverse functions in development of the nervous system and adult physiological functions. The Biological Process GO terms include axon guidance, regulation of response to stimulus, regulation of neuron differentiation and behavior, among many others. The Cellular Component GO terms include synapse, ion channel and neuromuscular junction, among many others (for a complete list see Additional file 5: Table S4).
In contrast, an analysis of the 554 genes down-regulated by over-expression of any of the FruM isoforms in males revealed an enrichment of genes that contain protein domains that function in lipid and triglyceride metabolism, consistent with previous studies [30-32] (Additional file 5: Table S4).
An analysis of the genes induced by overexpression of FruM in females identified fewer significantly enriched GO Biological Process categories, as compared to our observations in males. These include response to caffeine, response to purines, and potassium ion transport. The GO category male sex differentiation is included, but only contains dsx and fru. Nearly all the enriched GO Biological Process categories identified in the genes repressed by FruM in females include defense response genes (Additional file 5: Table S4).
Genes that are up regulated in response to over-expression of FruM isoforms include those that were previously implicated in playing a role in fru P1 expressing neurons, including the ecdysone receptor gene EcR and the ecdysone hierarchy gene broad[33]. Several neuronally-expressed genes, not previously known to be regulated by FruM isoforms, were identified that play critical roles in axon target recognition and attraction, axon guidance, dendrite guidance axon defasiculation, and sensory perception. These genes include roundabout (1 and 3), Dscam (1,2,3 and 4), prospero, semaphorin (1a, 2a), Netrin A and B, fasiclin (I and II), Notch, Cadherin N, Gustatory receptor 93a and abnormal chemosensory jump 6, Gaba receptor, serotonin receptor, nicotinic Acetylcholine Receptor alpha 7E, Neuroligin 1, foxo, Target of rapamycin, cacophony, muscarinic Acetylcholine Receptor 60C, spinster, and Dopamine receptor 2, among many genes.
An analysis of the genes induced in response to the expression of FruM isoforms reveals that a large fraction of these genes had previously been shown to have high expression in nervous system tissues. Thus of the genes with induced expression in response to FruM isoform expression, 649, 645 and 592 genes were previously shown to be significantly highly expressed in the adult brain, larval brain and adult ventral nerve cord, respectively, when compared to Flyatlas data using the Flymine portal [28,34]. Of the tissues examined in the Flyatlas study, these three tissues had the largest overlap of genes with significantly high expression with the genes induced by FruM from this study.
Comparison of differential gene expression in response to FruM over-expression vs. FruM loss of function
We have also analyzed gene expression differences in head tissues of fru P1 mutant males as compared to wild type males to further confirm our over-expression analysis. Over-expression of FruM is likely to yield higher fold-differences in gene expression than observed in the loss-of-function mutants, because the absolute difference in fru P1 mRNA amounts is greater between over-expressor flies and wild type flies than between loss-of-function fru P1 flies and wild type flies based on RNA-seq data. Thus, we identified genes based on significant differences in expression between fru P1 mutants and wild type males, but did not require a ≥2 fold change.
Based on the loss-of-function analyses, of the 706 genes that are induced by FruM, 209 genes were also induced by at least one of the FruM isoforms in the over-expression experiments. If we do not restrict the list of genes to those with ≥ 2-fold induction by overexpression of at least one of the FruM isoforms, 360 genes were identified as induced by FruM in both the loss-of-function and overexpression experiment. Of the 436 genes that are repressed by FruM, 19 genes were also repressed by at least one of the FruM isoforms in the overexpression experiments (Additional file 2: Figure S1 and Additional file 6: Table S5). There is a significant association between the lists of genes that we identified as regulated downstream of fru P1 in the loss-of-function analyses to those identified in the overexpression analyses (p <0.05).
The genes induced by fru P1 in the fru P1 loss-of-function analysis have a significant enrichment of genes that contained either the FruA, FruB or FruC binding sites motif, whereas those that are repressed downstream of fru P1 show an enrichment of only the FruB binding site motif (Table 1). Taken together, these results further support the idea that genes that are induced downstream of FruM isoform are likely direct targets.
Genes that are regulated downstream of FruM isoforms do not have the expected chromosome distribution
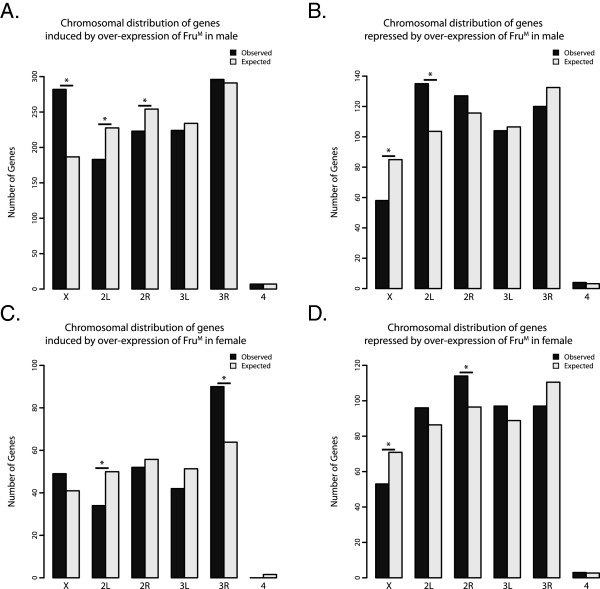
An examination of the chromosomal distribution of the genes with exons that are either induced or repressed downstream of FruM isoforms in the male over-expression experiments revealed a significant enrichment and depletion on the X chromosome, respectively. The genes with up-regulated expression downstream of FruM isoforms in males are enriched on the X chromosome and the second chromosome (Figure 4 and Additional file 7: Table S6). The genes with reduced expression downstream of FruM isoforms are significantly depleted from the X chromosome. In addition, if we examine the genes that are induced by FruM that were previously shown to be significantly highly expressed in the adult brain, larval brain or adult ventral nerve cord these genes sets are present on the X chromosome at a higher level than expected, based on Flyatlas data using the Flymine portal [28,34]. These results suggest that the X chromosome has properties distinct from the autosomes with respect to genes important for the potential for male courtship behaviors.
Figure 4.
Chromosomal distributions of genes regulated by FruM. (A-D) Observed (black) and expected (grey) number of genes on each chromosome for the sets of genes induced and repressed by over-expression of FruMA, FruMB, and FruMC in males or females. Asterisks indicate significant enrichment or depletion between observed and expected (Fisher’s exact test, p-value < 0.05).
Discussion
In this study we identified hundreds of genes regulated downstream of FruM isoform activity. FruMA, FruMB and FruMC have differences in the gene sets induced or repressed when they are over-expressed, demonstrating that each isoform has distinct biochemical activities (Figure 2). Consistent with this observation is that each FruM isoform has different DNA binding specificity (Figure 3). Our results suggest that there are sex-specific factors that influence FruM isoform activity, as over-expression of FruMA, FruMB and FruMC isoforms in males and females resulted in different genes that are induced and repressed by each isoform (Figure 2 and Additional file 2: Figure S1). The gene sets identified as induced downstream of FruM isoforms in males are enriched with genes with nervous system function, based on GO annotations (Additional file 5: Table S4).
Additionally, it is worth noting that there may be other possible sources for the differences observed in the gene expression levels in these experiments. First, the FruM proteins contain a BTB domain that in previous work has been shown to contain a dimerization domain that can mediate homodimeric or heterodimeric interactions. Thus, some of the effects we observe could be due to 1) differences in the stoichiometric ratios of FruM with each other, and/or 2) differences in the stoichiometric ratios of FruM with other potential dimerization partners. However, based on immunofluorescence results we do not observe substantially different levels of over-expression of each isoform, nor is there substantial expression, if any, outside of the normal fru P1 expression pattern (see Additional file 8: Figure S2). Second, there is a significant association between the genes that are either induced or repressed when FruM is over-expressed, with those genes identified in loss-of-function fruM mutant analyses, demonstrating the physiological relevance of the genes identified by over-expression. Third, there is significant enrichment of the binding site sequences identified for each isoform within the genes that are induced by each isoform. Fourth, while our criteria were stringent (significant and substantial differences from two different wild type strains), strain differences may account for some of the differences between wild type male and female and FruM over-expressor male and female strains, respectively. However, such strain differences are not likely to account for the differences we observe between the FruM isoforms, which are in the same genetic background, nor can strain differences account for differences observed across sex. Taken together, these results demonstrate that in context of the over-expression experiments each of the three FruM isoforms examined has different activities with respect to genes that are induced or repressed in males, with many more genes having induced rather than repressed expression in males.
In previous studies, production of FruM in females, by expression of a tra-2 RNAi transgene in fru P1-expressing neurons, was sufficient to endow females with the potential to perform the first four sub-steps of the male courtship ritual, following, tapping, wing extension and proboscis extension, but not attempted copulation [14]. In contrast, overexpression of FruMA or FruMC in fru P1-expressing neurons, resulted in flies that displayed only following and tapping reviewed in [4], suggesting that overexpression of FruM in females is not sufficient to endow females with the potential to perform courtship behaviors. The 42 genes identified as induced by all three FruM isoforms in females will be interesting to examine, with respect to their role in establishing the potential for these early courtship steps. Interestingly, one of these genes is Ir54a, which encodes a member of a diverse family of ionotropic receptors, some of which are expressed in the adult antenna and underlie chemosensory functions [35]. It is also known that DsxM plays a role in establishing the potential for courtship behaviors [26,27,36-38], which would not have been present in females in which FruM was produced, though DsxM is not present in all fru P1-expressing neruons, so is unlikely to account for all the differences between males and females observed here [26,37]. Our results may further explain why there was not a complete rescue of male courtship behavior. It is clear that the sex of the fly in which FruM is produced has an impact on the genes that are induced and repressed. These results suggest that there are additional sex-specific factors that influence FruM activity, which may include DsxM. Further biochemical characterization of FruM protein interactions will be important to understand FruM activities.
While fru has been predicted to be a transcription factor based on the observation that fru encodes BTB-zinc finger products, no direct transcriptional targets of fru have been identified, leaving this an open question. A recent study has shown that FruM associates with a cofactor, Bonus, and subsequently associates with two chromatin modifying proteins, HP1a and HDAC1, however it was not clear if the association of FruM with chromatin was direct [20]. The results presented here demonstrate that FruM can bind DNA and that three FruM isoforms examined have different binding activities. Given our observation that the binding sites are significantly enriched in all gene sets identified as induced, but not repressed by FruM, suggests that FruM may function by binding enhancer DNA directly, but acts in an indirect manner to repress gene expression (Table 1).
In previous studies we and others have show that genes with male-biased expression were enriched on the X chromosome in the adult head [31,39] and brain [40]. There was also a significant enrichment of genes with male-biased expression that reside near dosage compensation entry sites [39,40]. Here, we observe significant enrichment of genes that reside on the X chromosome that are induced by FruM in males. This observation supports the idea that over evolutionary time there may have been a selection for genes with male-specific functions to reside on the X chromosome and in particular those regulated by FruM. Perhaps, FruM isoforms and their gene targets have evolved to take advantage of the unique properties of the male nucleus. These differences include the dosage compensation complex that is bound to the male X chromosome that leads to less compact chromatin reviewed in [41], the presence of the Y chromosome that affects chromatin architecture throughout the nucleus [42], or other differences in the chromatin and three-dimensional architecture of the nucleus in males [for example see [43]. It is possible that there are more interconnections in the sex hierarchy model between chromosomal sex, the sex hierarchy branches and sexual development that is downstream of FruM than shown in the model (Figure 1).
Conclusions
The results in this study add to the information regarding FruM function with the identification of hundreds of genes regulated by FruM, many of which have known roles in nervous system development and physiology. One of the next exciting challenges to our understanding of how complex behaviors are specified at a molecular-genetic level will be to develop tools to interrogate the functions of specific transcript isoforms in a cell-specific manner.
Methods
Flies
Flies were raised on standard cornmeal food medium at 25°C on a 12 hour light and 12 hour dark cycle. Wild type flies were the Canton-S (CS) and Berlin strains. Transheterozygous mutants for fru P1 are Df(3R)P14/Df(3R)fru4-40 and fruw12/Df(3R)ChaM5, which are phenotypically void of all male courtship behaviors [10]. In addition, no detectable fru P1 transcript or FruM protein is present in Df(3R)P14/Df(3R)fru4-40[11], and no full length fru P1 mRNAs are present in fruw12/Df(3R)ChaM5[10]. Df(3R)P14 contains a breakpoint in fru extending proximally removing common fru coding exons and ending in the cytological location 90C2-D1 [5]; Df(3R)fru4-40 contains a breakpoint in fru extends distally thus removing the fru P1 promoter [11]; fruw12 is an inversion-cum-translocation that removes P1-3 from common fru coding exons [10]; and Df(3R)ChaM5 contains a breakpoint between P1 and P2 that extends distally removing P1 ending in the cytological location 91D [5,10]. The fru P1-Gal4, UAS-FruMA, UAS-FruMB, and UAS-FruMC were described previously [14,44]. y1w1118; P(UAS-Gal4.H)12B stock was obtained from Bloomington stock center.
Flies that ectopically expressed FruM isoforms were of the genotypes y w/(w or Y); P(w+mC, UAS-Gal4)/P(w+mC, UAS-FruMA,B, orC); fru P1-Gal4/+. While each UAS-FruM transgene is inserted at a different location, they are homozygous viable and each respective DNA binding domain (A, B, and C) encoding exon is > four-fold induced compared to wild type in our over-expression assay conditions, by examining the RNA-seq expression data. Immunofluorescence using an antibody specific to the male-specific region common to all FruM proteins demonstrates relatively similar levels of the male-specific proteins in fru P1-Gal4 expressing neurons and is not readily detectable in other regions of the CNS (Additional file 8: Figure S2). Additionally, examination of male-female courtship of the above transgenic strains demonstrated that over-expression is not changing male behaviors significantly (Additional file 9: Figure S3).
Tissue collection
Adult head cDNA libraries were prepared from three independent biological replicates from each of the following genotypes: 1) from males and females: Canton S, Berlin, y w/(w or Y); P(w+mC, UAS-Gal4)/P(w+mC, UAS-FruMA); fru P1-Gal4/+, y w/(w or Y); P(w+mC, UAS-Gal4)/P(w+mC, UAS-FruMB); fru P1-Gal4/+, y w/(w or Y); P(w+mC, UAS-Gal4)/P(w+mC, UAS-FruMC); fru P1-Gal4/+ and 2) in males only: Df(3R)P14/Df(3R)fru4-40 and, fruw12/Df(3R)ChaM5 males. For each experimental condition, approximately 200 flies that were 8 to 24 hours post-eclosion were used. All flies were collected 0 to 16 hours post-eclosion under anesthetization and allowed to recover for 8 hours before being snap frozen in liquid nitrogen. Snap frozen whole animals were stored at -80°C until heads were collected. Adult heads were separated from bodies by mechanical tapping of the cryovial. A piece of plastic was cooled on dry ice, on which the frozen heads were separated from the bodies and immediately transferred and homogenized in 1 mL of TRIzol® (Invitrogen).
Illumina sequencing library preparation
Total RNA was extracted using TRIzol® Reagent (Invitrogen), and RNA was precipitated by addition of 250 μL 100% isopropanol and 250 μL 1.2 M NaCitrate, 0.8 M NaCl in DEPC-treated H2O. Approximately 25 μg total RNA was DNase treated to remove any trace amounts of DNA, following Zymo Research RNA Clean & Concentrator™-25 In-Column DNase Digestion protocol, using 10 units Ambion® TURBO™ DNase. Poly(A) + transcripts were subsequently isolated from total RNA using Ambion® MicroPoly(A)Purist™ Kit. 100 ng mRNA was chemically fragmented to a range of approximately 200-500 base pairs using the Ambion® RNA Fragmentation Reagent, and the reaction was cleaned using Zymo Research RNA Clean & Concentrator™-5. First strand cDNA was synthesized using SuperScript® II Reverse Transcriptase (Invitrogen™) and a combination of 3 μg random hexamers and 0.15 μg oligo(dT)20 primers. Following first strand synthesis, the second strand of the cDNA was synthesized by addition of DNA polymerase I (Invitrogen™), RNase H (New England Biolabs® Inc.), dNTPs and second strand buffer (Invitrogen™). This reaction and all subsequent reactions were cleaned using Zymo Research DNA Clean & Concentrator™-5 kit. Double stranded cDNA templates were blunt ended using End-It™ Repair Kit (Epicentre®). Next, A-overhangs were then added to both ends with Klenow fragment (3′ → 5′ exo-minus) (New England Biolabs® Inc.). Illumina sequencing adapters were then ligated to both ends of the cDNA templates using Fast-Link™ DNA Ligation Kit (Epicentre®). cDNA templates were then amplified by performing polymerase chain reaction (PCR; 18 cycles) that extended the adapter and incorporated a different six base pair index into each sample. The product was then isolated by gel purification of 250-550 base pair fragments. Samples were then pooled and sequenced on the Illumina Genome Analyzer GAII platform with 72 base pair single end reads, and the reads were matched to their corresponding sample via the index.
SELEX
Each of the three fru DNA binding domain (DBD) encoding sequences were PCR amplified from fru cDNAs using the following primer pairs that contain EcoR1 or XhoI restriction sites engineered at their ends: FruMA primers are 5′CCGGAATTCCGC GTCAAGTGTTTTAACATTAAGC and 5′CCGCTCGAGGTTTGCTTGATTCTTGGTTACTTA; FruMB primers are 5′GGC CGGAATTCTCCAAGGCCTGGCACATG and 5′ CCCGCTCGAGTGTGCTG CTGTTGCTGC; FruMC primers are 5′CCGGAATTCCAGCAGCGCCCGCCACC and 5′GCCGCTCGAGCGGGATGGGCTGCACTTGGGC. For each DBD-encoding exon, the first primer has the EcoR1 site and the second primer has the XhoI site. The primers were designed to amplify beginning where the divergent sequence for each Fru isoform begins (see Figure 1), and the amplicon includes each isoforms respective stop codon at the end. Each region was cloned as in-frame fusions with Glutathione S-transferase at the amino terminus (GST-Fru), into the pGEX-4 T1 plasmid for expression in bacteria.
Each DBD containing plasmid was transformed into E.coli BL21 and single colonies were grown to ~ OD600 = 0.8 when IPTG was added to 0.1 mM to induce protein expression. Bacteria were grown for approximately two more hours in the presence of IPTG and then harvested by centrifugation at 8,000 rpm for 10 minutes. Protein extract was made by resuspending a 200 mL culture in 10 mL of ice cold Buffer 1 (100 mM KCl, 50 mM HEPES, pH 7.5, 10% glycerol, 5 mM MgCl2, 1 mM DTT and protease inhibitors). The cells were lysed on ice by sonication and were visually monitored with a compound microscope to assess the efficiency of sonication. Lysates were cleared by centrifugation and the supernatant was retained. Fusion proteins were purified by binding to ~500 μL of a 50% slurry of glutathione Sepharose 4B that had been washed and equilibrated in Buffer 1. The protein was mixed with the Sepharose resin for 2 hour at 4°C and then loaded into a gravity flow column. The protein bound to the Sepharose was washed with 5 column volumes of Buffer 1. For the SELEX, the Sepharose beads with the bound purified protein was resuspended in 250 μL Buffer 2 (100 mM KCl, 50 mM HEPES, pH 7.5, 50% glycerol, 5 mM MgCl2, 1 mM DTT) and stored at 4°C. For gel shifts, the protein was eluted with glutathione by incubating the slurry for two hours in elution buffer (10 mM glutathione in 50 mM Tris, pH 8.0) and dialyzed into Buffer 1. Examination of protein by SDS-PAGE and Commassie staining showed a high degree of purity and proteins of the expected sizes.
The SELEX procedure was performed as previously described with some modification [24]. Here we used oligoR76: CAGGTCAGTTCAGCGGATCCTGTCGN26GAGGCGAATTCAGTGCAACTGCAGC, primer F GCTGCAGTTGCACTGAATTCGCCTC and primer R CAGGTCAGTTCAGCGGATCCTGTCG. For the first round of the SELEX procedure, a complementary strand of OligoR76 was generated by a single cycle of PCR using primer F, 100 ng of OligoR76 in a 20 μL PCR reaction to generate double stranded molecules (1 minute at 94°C, 3 minutes at 62°C, 9 minutes at 72°C). The first round of SELEX was performed with 5 μL of the primer extension oligoR76 DNA reaction, 20 μL of the GST-Fru/bead slurry and 100 μL of Buffer 3 (Buffer 1 with poly dI,dC 4 μg/μL and BSA 40 μg/mL) for 2 hours at 4°C. The protein/bead slurry was washed twice with 800 μL Buffer 1 and then was resuspended in 30 μL of high quality water, boiled for 2 minutes and the supernatant that included the bound DNA retained. For the next nine rounds of SELEX, 10 μL of the eluted DNA from each preceding round was used in a 100 μL PCR reaction for 20 cycles, using primers F and R. The DNA was loaded onto a 2% Nuseive gel and separated by electrophoresis. The DNA band at 75 base pairs was excised and purified using Qiaquick gel extraction columns (Qiagen). For the last rounds of the SELEX, 1 μL of the purified DNA from the previous round (~300 ng) was mixed with 20 μL of the protein/bead slurry in 100 μL Buffer 3. The bound DNA fragments were purified as described for round 1 of the SELEX. After the tenth round of the SELEX, the DNA fragments were cloned by ligation into Bluescript at the EcoRI and BamHI sites and sequenced by Sanger sequencing. For each Fru DBD, at least 20 independent clones were sequenced. A Gibbs sampling algorithm was used to find the consensus motif [45]. For FruMB a sequence of low complexity was identified from the first SELEX experiment and so a second full SELEX experiment was performed, each with 10 rounds of selection. Nearly the same consensus sequence was identified in both rounds, providing confidence in the result.
To determine if the motifs identified in the SELEX bind specifically to the DNA binding domain used in the SELEX procedure, gel shift reactions were performed using annealed phosphorylated oligonucleotides that contain common flanking DNA sequence chosen to facilitate cloning and either the binding sequences identified in the SELEX (in bold below), or oligonucleotides of the same nucleotide content but randomized but in the same position as the identified binding site resides in the sequence (in italic and bold below). For FruMA the oligonucleotide sequences were 5′TCGACCTGCAGAGTAACCTGCAGG and 5′TCGACCTGCAGGTTACTCTGCAGG. The FruMA randomized sequences were 5′TCGACCTGCAGATAGACCTGCAGG and 5′TCGACCTGCAGGTCTATCTGCAGG. For FruMC the oligonucleotide sequences were 5′TCGACCTGCAGTGTTACATCACTGCAGG and 5′TCGACCTGCAGTGATGTAACACTGCAGG. The FruMC randomized sequences were 5′TCGACCTGCAGGCATCTATATCTGCAGG and 5′TCGACCTGCAGATATAGATGCCTGCAGG. Gel shift reactions were performed as previously described [46].
For FruMB, two independent trials with ten rounds of selection for the FruMB binding motif identified very similar sequences. The consensus sequence for FruMB from the two independent trials is GCCCTTT. The GST-FruB protein bound DNA in the invariant region present in all the synthesized oligos (see above). To determine if the binding site identified for FruMB was correct, a modified assay was performed in test tubes. In this assay 5 μL of P32 labeled DNA from the first and tenth round of the SELEX procedure were mixed with 30 μL of the FruMB protein/bead slurry in 150 μL Buffer 3 for two hours at 4°C, washed three times in Buffer 1 and added to 5 mL of scintillation fluid. The percent of the input retained from the first and tenth SELEX round was quantified using a scintillation counter. The P32 labeled DNA was generated by a standard PCR reaction, using Primer F and R, 1 μL of DNA from the SELEX round, and included dATP-P32gamma. Only 0.8% of the labeled input DNA from the first round of SELEX was retained on the FruMB protein/bead slurry, whereas 3.6% was retained from the tenth round of SELEX, demonstrating that the SELEX enriched for a sequence bound by FruMB.
Illumina read mapping
We used a sequential mapping pipeline that mapped approximately 95% of all reads to the Drosophila genome. Barcode, primer, and adapter sequences were trimmed. Reads were aligned to the D. melanogaster genome FB5.30 (FlyBase v5.30) using Bowtie (--tryhard, --best, --strata, -m1)[47], unaligned reads were 3′ end quality trimmed and homopolymers (5+) were removed. Quality trimmed reads were mapped as above. Unaligned reads at this step were aligned to junctions estimated by Tophat [48]. Any remaining unaligned reads were mapped allowing for gaps to FB5.30 using LAST [49]. Reads were visualized as wiggle tracks on FB5.30 genome using a custom R script [25].
Within a gene, exons from different isoforms may overlap due to alternative start and end positions (Additional file 10: Figure S4). Exons from different genes may also overlap. Overlapping exons, regardless of strand, were combined into the maximum exonic region see [25]. If there was a single exon in the region it was labeled as S####_SI and if there were multiple overlapping exons, they were combined and labeled as F####_SI (Additional file 10: Figure S4). Exonic regions can be further classified as constitutive (a single exon present in all isoforms), common (exonic region present in all isoforms), and alternative (not present in all isoforms) (Additional file 10: Figure S4). Expression was quantified for each exon in FB5.30 using a perl script. In regions where exons overlap, exons were combined. Of the 60,291 exonic regions 53,459 did not overlap with any other exon. Most overlaps are due to alternate start or end positions (4,503). About 30% (2,329) are due to exonic regions from different genes, these regions were not considered further. We considered an exonic region as detected if at least one read mapped to that region. Exonic regions with no reads mapping for any observed samples were not considered further (1,550 regions). For each exonic region, Reads Per Kilobase per Million mapped reads RPKM [50] was calculated and the natural log taken. If no variation was observed in one condition, or if it was not detected at least once in each treatment group no statistical analysis is possible. These 11,104 regions and the remaining 45,339 exonic regions that were analyzed for quantitative differences in expression are reported in Additional file 11: Table S7.
Differential expression
A linear model was fit for each exonic region separately and models were examined for conformation to assumptions. All comparisons were performed as contrasts in a single model and a single FDR correction was performed for all contrast simultaneously [51]. Results were partitioned into induced or repressed based upon the direction of the observed difference. To declare that an exon was significantly differentially expressed, we required that exons be both (1) statistically significantly different [False Discovery Rate (FDR) p-value < 0.20] and (2) have a ≥2 fold change in expression level (Additional file 1: Table S1 and Additional file 3: Table S2). We also provide the full lists of genes that have exons that are significantly differentially expressed (Additional file 12: Tables S8 and Additional file 13: Table S9). To reduce the chances of identifying exons only due to background differences in strain, we required that the FruM be different from both wild type backgrounds in order to be declared differentially expressed. For the fru null comparisons, we required that both fru allele combinations were each statistically different from CS and Berlin (four statistical comparisons).
Enrichments and motif analysis
Enrichments for chromosomal locations were tested by constructing contingency tables and conducting a Fisher’s exact test [52]. Position weight matrices (PWMs) generated by SELEX enrichment, were used to identify the locations of FruMA, FruMB, or FruMC binding sites. MAST [53] was used to identify putative binding sites in a region that included the gene of interest and 2 kb upstream of the transcription start site, 2 kb downstream of the 3′ UTR and throughout the entire genic region and did not attempt to normalize the counts. Enrichment of Fru binding sites in genes that were significantly induced or repressed was tested using a Fisher’s exact test [52]. Chromosomal enrichment for genes identified as differentially expressed was tested using a Fisher’s exact test for each chromosomal arm.
Gene Ontology (GO) enrichment analysis was performed using Gene Ontology enrichment analysis and visualization tool (GOrilla) [29]. Target list containing FBgns from genes either induced or repressed in the FruM over-expression or FruM loss-of-function were supplied against a background list containing all 14903 FBgns to obtain significantly enriched (p value < 10-3) GO terms for biological processes, cellular components, and molecular functions. Enriched protein domain analysis was implemented with the Holm-Bonferroni correction in the Flymine portal [28].
Abbreviations
Dsx: Doublesex; fru: fruitless; GO: Gene Ontology; RNA-seq: RNA-sequencing; oligo: Oligonucleotide; DBD: DNA binding domain; CNS: Central nervous system.
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
JED, MNA performed experiments. JED, JF, SK, LMM and MNA contributed to data and statistical analyses. JED, JF, SK, BB, LMM and MNA contributed to writing the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Genes induced and repressed by FruM isoforms in males (gene lists are based on the statistical tests and the ≥ 2-fold difference criterion).
Venn diagrams of comparisons.
Genes induced and repressed by FruM isoforms in females. (gene lists are based on the statistical tests and the ≥ 2-fold difference criterion).
Genomic locations for FruM DNA binding motifs.
Gene Ontology analyses.
Genes induced and repressed by FruM, as determined by examination of fru P1 mutants and wild type males. All data described is included.
Statistical analysis of chromosomal distribution of genes regulated by FruM.
FruM is localized in the fru P1-expression pattern in flies over-expressing FruMA,B or C.
Courtship analyses.
Schematic of how exon IDs are determined.
Exon region ID information and all associated data from this study.
Genes induced and repressed by FruM isoforms in males (gene lists are based on the statistical tests).
Genes induced and repressed by FruM isoforms in females (gene lists are based on the statistical tests).
Contributor Information
Justin E Dalton, Email: justin.eric.dalton@gmail.com.
Justin M Fear, Email: jfear@ufl.edu.
Simon Knott, Email: srvknott22@gmail.com.
Bruce S Baker, Email: bakerb@janelia.hhmi.org.
Lauren M McIntyre, Email: mcintyre@ufl.edu.
Michelle N Arbeitman, Email: michelle.arbeitman@med.fsu.edu.
Acknowledgements
This work was supported by research start-up funds from FSU and NIH grants R01GM073039 (PI is MNA), R01MH091561 (PI is S.V. Nuzhdin, MNA and LMM co-PIs). The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. We are grateful to S. Goodwin for UAS-FruM fly stocks, to Scott Greenberg for his experimental assistance, and D. Luo for comments on the manuscript.
The data sets supporting the results of this article are available in the NCBI’s Gene Expression Omnibus and are available through GEO series accession number GSE50515 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50515).
References
- Yamamoto D. The neural and genetic substrates of sexual behavior in Drosophila. Adv Genet. 2007;59:39–66. doi: 10.1016/S0065-2660(07)59002-4. [DOI] [PubMed] [Google Scholar]
- Villella A, Hall JC. Neurogenetics of courtship and mating in Drosophila. Adv Genet. 2008;62:67–184. doi: 10.1016/S0065-2660(08)00603-2. [DOI] [PubMed] [Google Scholar]
- Dauwalder B. The roles of fruitless and doublesex in the control of male courtship. Int Rev Neurobiol. 2011;99:87–105. doi: 10.1016/B978-0-12-387003-2.00004-5. [DOI] [PubMed] [Google Scholar]
- Manoli DS, Meissner GW, Baker BS. Blueprints for behavior: genetic specification of neural circuitry for innate behaviors. Trends Neurosci. 2006;29:444–451. doi: 10.1016/j.tins.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Gailey DA, Hall JC. Behavior and cytogenetics of fruitless in Drosophila melanogaster: different courtship defects caused by separate, closely linked lesions. Genetics. 1989;121:773–785. doi: 10.1093/genetics/121.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, Baker BS, Hall JC, Taylor BJ, Wasserman SA. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–1089. doi: 10.1016/S0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- Ito H, Fujitani K, Usui K, Shimizu-Nishikawa K, Tanaka S, Yamamoto D. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci USA. 1996;93:9687–9692. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SF, Taylor BJ, Villella A, Foss M, Ryner LC, Baker BS, Hall JC. Aberrant splicing and altered spatial expression patterns in fruitless mutants of Drosophila melanogaster. Genetics. 2000;154:725–745. doi: 10.1093/genetics/154.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villella A, Gailey DA, Berwald B, Ohshima S, Barnes PT, Hall JC. Extended reproductive roles of the fruitless gene in Drosophila melanogaster revealed by behavioral analysis of new fru mutants. Genetics. 1997;147:1107–1130. doi: 10.1093/genetics/147.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, Villella A, Ryner LC, Carlo T, Goodwin SF, Song HJ, Gailey DA, Morales A, Hall JC, Baker BS, Taylor BJ. Molecular genetic dissection of the sex-specific and vital functions of the Drosophila melanogaster sex determination gene fruitless. Genetics. 2001;158:1569–1595. doi: 10.1093/genetics/158.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Foss M, Goodwin SF, Carlo T, Taylor BJ, Hall JC. Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J Neurobiol. 2000;43:404–426. doi: 10.1002/1097-4695(20000615)43:4<404::AID-NEU8>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Demir E, Dickson BJ. Fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121:785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- Stockinger P, Kvitsiani D, Rotkopf S, Tirian L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- Manoli DS, Foss M, Villella A, Taylor BJ, Hall JC, Baker BS. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature. 2005;436:395–400. doi: 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- Kimura K, Ote M, Tazawa T, Yamamoto D. Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature. 2005;438:229–233. doi: 10.1038/nature04229. [DOI] [PubMed] [Google Scholar]
- Cachero S, Ostrovsky AD, Yu JY, Dickson BJ, Jefferis GS. Sexual dimorphism in the fly brain. Curr Biol. 2010;20:1589–1601. doi: 10.1016/j.cub.2010.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, Kanai MI, Demir E, Jefferis GS, Dickson BJ. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr Biol. 2010;20:1602–1614. doi: 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- Ruta V, Datta SR, Vasconcelos ML, Freeland J, Looger LL, Axel R. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. 2010;468:686–690. doi: 10.1038/nature09554. [DOI] [PubMed] [Google Scholar]
- Usui-Aoki K, Ito H, Ui-Tei K, Takahashi K, Lukacsovich T, Awano W, Nakata H, Piao ZF, Nilsson EE, Tomida J, Yamamoto D. Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression. Nat Cell Biol. 2000;2:500–506. doi: 10.1038/35019537. [DOI] [PubMed] [Google Scholar]
- Ito H, Sato K, Koganezawa M, Ote M, Matsumoto K, Hama C, Yamamoto D. Fruitless recruits two antagonistic chromatin factors to establish single-neuron sexual dimorphism. Cell. 2012;149:1327–1338. doi: 10.1016/j.cell.2012.04.025. [DOI] [PubMed] [Google Scholar]
- McQuilton P, St Pierre SE, Thurmond J. FlyBase 101–the basics of navigating FlyBase. Nucleic Acids Res. 2012;40:D706–D714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter JC, Villella A, Allendorfer JB, Dornan AJ, Richardson M, Gailey DA, Goodwin SF. Isoform-specific control of male neuronal differentiation and behavior in Drosophila by the fruitless gene. Curr Biol. 2006;16:1063–1076. doi: 10.1016/j.cub.2006.04.039. [DOI] [PubMed] [Google Scholar]
- Klug SJ, Famulok M. All you wanted to know about SELEX. Mol Biol Rep. 1994;20:97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- Ausubel FM. Current protocols in molecular biology. Brooklyn, N.Y. Media, Pa: Greene Pub. Associates; J. Wiley; 1987. [Google Scholar]
- Graze RM, Novelo LL, Amin V, Fear JM, Casella G, Nuzhdin SV, McIntyre LM. Allelic imbalance in Drosophila hybrid heads: exons, isoforms, and evolution. Mol Biol Evol. 2012;29:1521–1532. doi: 10.1093/molbev/msr318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders LE, Arbeitman MN. Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Dev Biol. 2008;320:378–390. doi: 10.1016/j.ydbio.2008.05.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, Janssens H, Ji W, McLaren P, North P. et al. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8:R129. doi: 10.1186/gb-2007-8-7-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Anzi B, Sapin V, Waters C, Zinn K, Wyman RJ, Benzer S. Obesity-Blocking Neurons in Drosophila. Neuron. 2009;63:329–341. doi: 10.1016/j.neuron.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman TD, Arbeitman MN. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 2007;3:e216. doi: 10.1371/journal.pgen.0030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazareva AA, Roman G, Mattox W, Hardin PE, Dauwalder B. A role for the adult fat body in Drosophila male courtship behavior. PLoS Genet. 2007;3:e16. doi: 10.1371/journal.pgen.0030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton JE, Lebo MS, Sanders LE, Sun F, Arbeitman MN. Ecdysone receptor acts in fruitless- expressing neurons to mediate drosophila courtship behaviors. Curr Biol. 2009;19:1447–1452. doi: 10.1016/j.cub.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SW, Herzyk P, Dow JA, Leader DP. FlyAtlas: database of gene expression in the tissues of Drosophila melanogaster. Nucleic Acids Res. 2013;41:D744–D750. doi: 10.1093/nar/gks1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hachiya T, Koganezawa M, Tazawa T, Yamamoto D. Fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron. 2008;59:759–769. doi: 10.1016/j.neuron.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Rideout EJ, Billeter JC, Goodwin SF. The sex-determination genes fruitless and doublesex specify a neural substrate required for courtship song. Curr Biol. 2007;17:1473–1478. doi: 10.1016/j.cub.2007.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BJ, Villella A, Ryner LC, Baker BS, Hall JC. Behavioral and Neurobiological Implications of Sex-Determining Factors in Drosophila. Dev Genet. 1994;15:275–296. doi: 10.1002/dvg.1020150309. [DOI] [PubMed] [Google Scholar]
- Chang PL, Dunham JP, Nuzhdin SV, Arbeitman MN. Somatic sex-specific transcriptome differences in Drosophila revealed by whole transcriptome sequencing. BMC Genom. 2011;12:364. doi: 10.1186/1471-2164-12-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan A, Hutter S, Parsch J. Population and sex differences in Drosophila melanogaster brain gene expression. BMC Genom. 2012;13:654. doi: 10.1186/1471-2164-13-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet. 2011;13:123–134. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- Lemos B, Branco AT, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci USA. 2010;107:15826–15831. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Song HJ, Billeter JC, Reynaud E, Carlo T, Spana EP, Perrimon N, Goodwin SF, Baker BS, Taylor BJ. The fruitless gene is required for the proper formation of axonal tracts in the embryonic central nervous system of Drosophila. Genetics. 2002;162:1703–1724. doi: 10.1093/genetics/162.4.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberg LA, Thompson WA, Conlan S, Smith TM, Mccue LA, Lawrence CE. A phylogenetic Gibbs sampler that yields centroid solutions for cis-regulatory site prediction. Bioinformatics. 2007;23:1718–1727. doi: 10.1093/bioinformatics/btm241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman MN, Hogness DS. Molecular chaperones activate the Drosophila ecdysone receptor, an RXR heterodimer. Cell. 2000;101:67–77. doi: 10.1016/S0092-8674(00)80624-8. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Wan R, Horton P. Incorporating sequence quality data into alignment improves DNA read mapping. Nucleic Acids Res. 2010;38:e100. doi: 10.1093/nar/gkq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Meth. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc B Methodol. 1995;57:289–300. [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Methods and statistics for combining motif match scores. J Comput Biol. 1998;5:211–221. doi: 10.1089/cmb.1998.5.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes induced and repressed by FruM isoforms in males (gene lists are based on the statistical tests and the ≥ 2-fold difference criterion).
Venn diagrams of comparisons.
Genes induced and repressed by FruM isoforms in females. (gene lists are based on the statistical tests and the ≥ 2-fold difference criterion).
Genomic locations for FruM DNA binding motifs.
Gene Ontology analyses.
Genes induced and repressed by FruM, as determined by examination of fru P1 mutants and wild type males. All data described is included.
Statistical analysis of chromosomal distribution of genes regulated by FruM.
FruM is localized in the fru P1-expression pattern in flies over-expressing FruMA,B or C.
Courtship analyses.
Schematic of how exon IDs are determined.
Exon region ID information and all associated data from this study.
Genes induced and repressed by FruM isoforms in males (gene lists are based on the statistical tests).
Genes induced and repressed by FruM isoforms in females (gene lists are based on the statistical tests).