Abstract
Self-incompatibility S-locus–encoded F-box (SLF) proteins have been identified in Antirrhinum and several Prunus species. Although they appear to play an important role in self-incompatible reaction, functional evidence is lacking. Here, we provide several lines of evidence directly implicating a role of AhSLF-S2 in self-incompatibility in Antirrhinum. First, a nonallelic physical interaction between AhSLF-S2 and S-RNases was demonstrated by both coimmunoprecipitation and yeast two-hybrid assays. Second, AhSLF-S2 interacts with ASK1- and CULLIN1-like proteins in Antirrhinum, and together, they likely form an Skp1/Cullin or CDC53/F-box (SCF) complex. Third, compatible pollination was specifically blocked after the treatment of the proteasomal inhibitors MG115 and MG132, but they had little effect on incompatible pollination both in vitro and in vivo, indicating that the ubiquitin/26S proteasome activity is involved in compatible pollination. Fourth, the ubiquitination level of style proteins was increased substantially after compatible pollination compared with incompatible pollination, and coimmunoprecipitation revealed that S-RNases were ubiquitinated after incubating pollen proteins with compatible but not with incompatible style proteins, suggesting that non-self S-RNases are possibly degraded by the ubiquitin/26S proteasome pathway. Fifth, the S-RNase level appeared to be reduced after 36 h of compatible pollination. Taken together, these results show that AhSLF-S2 interacts with S-RNases likely through a proposed SCFAhSLF-S2 complex that targets S-RNase destruction during compatible rather than incompatible pollination, thus providing a biochemical basis for the inhibition of pollen tube growth as observed in self-incompatible response in Antirrhinum.
INTRODUCTION
Self-incompatibility (SI) is a widespread mechanism that allows the pistil of a flower to reject self (genetically related) pollen but to accept non-self (genetically unrelated) pollen for fertilization in many families of flowering plants (de Nettancourt, 2001). In most cases, self-incompatible interaction is controlled by the highly polymorphic S-locus with multiple alleles. In self-incompatible species of the Solanaceae, Scrophulariaceae, and Rosaceae, allelic extracellular RNases called S-RNases have been shown to be necessary and sufficient for pollen rejection (for reviews, see McCubbin and Kao, 2000; Wang et al., 2003), and it is generally accepted that S-RNases act as S-allele–specific cytotoxins that inhibit the growth of pollen bearing a matching S-allele (McClure et al., 1990; Gray et al., 1991).
However, it is unclear how S-RNases act during both compatible and incompatible pollinations. Studies on pollen self-compatible mutants (Thompson et al., 1991; Golz et al., 1999) and transformation experiments (Lee et al., 1994; Murfett et al., 1994; Dodds et al., 1999) have shown that knocking out S-RNases or expressing them in pollen had no effect on the expression of the pollen SI phenotype, indicating that a distinct gene controlling the specificity exists in pollen and is often referred to as pollen S (Sp) (Golz et al., 1999). It appears that a direct interaction between the S-RNase and Sp is required for SI (McCubbin and Kao, 2000). Recently, similar candidates for Sp have been identified in the Scrophulariaceae (Lai et al., 2002; Zhou et al., 2003) and Rosaceae (Entani et al., 2003; Ushijima et al., 2003; Yamane et al., 2003). In Antirrhinum, a member of the Scrophulariaceae, clusters of F-box genes known as SLF (S-locus F-box) have been found close to S-RNases and exist as allelic copies (Lai et al., 2002; Zhou et al., 2003). Among them, AhSLF-S2 is the gene nearest to S2-RNase. AhSLF-S2 and its allelic products have ∼95% identity at amino acid levels, whereas paralogous AhSLF products have only 42 to 48% identity either between themselves or with AhSLF-S1, AhSLF-S2, AhSLF-S4, or AhSLF-S5 (Zhou et al., 2003). In several Prunus species, similar pollen-specific F-box genes also have been detected, which include PdSFBs (S haplotype–specific F-box) from P. dulcis (almond) (Ushijima et al., 2003), P. cerasus (cherry), and P. avium (cherry) (Yamane et al., 2003) and PmSLFs from P. mume (Japanese apricot) (Entani et al., 2003). The Prunus sequences have ∼20% amino acid identities with AhSLF-S2 and show a relatively higher level of S haplotype–specific sequence polymorphisms comparable to that of their S-RNases.
The SLF/SFB proteins contain a conserved N-terminal F-box domain, which is usually 40 to 50 amino acids in length (Elledge and Harper, 1998) and plays an essential role in recruiting a protein substrate for degradation through interactions with Skp1 or Skp1 analogs to form the so-called SCF (Skp1/Cullin or CDC53/F-box) complex in yeast, human, and plants (Bai et al., 1996; Cenciarelli et al., 1999). SCF complexes facilitate interaction between the substrates and ubiquitin conjugation enzymes, which then covalently transfer the ubiquitin onto the substrates. Polyubiquitinated substrates are subsequently degraded by the 26S proteasome (Hershko and Ciechanover, 1998). To date, several known protein–protein interaction domains, such as LRRs, WD40 repeats, or Klech repeats (Ruegger et al., 1998; Xie et al., 1998; Xiao and Jang, 2000), occur downstream the F-box domain and confer the substrate specificity for ubiquitination. By contrast, there is no known domain for protein interaction found in the predicted SLF polypeptides, though a domain named CRFB (Kuroda et al., 2002) is present. Nevertheless, these findings raise an interesting possibility that the ubiquitin/26S proteasome proteolytic system plays a role in the discrimination of self/non-self pollen in these species.
In this study, we analyzed the interaction of AhSLF-S2 with S-RNases using both immunoprecipitation and yeast two-hybrid assays. We found that AhSLF-S2 physically interacts with S2-RNase as well as with two known SCF subunits, CULLIN1 (CUL1)-like and ASK1-like proteins in Antirrhinum. We also provided evidence showing that S-RNases are inhibited through the ubiquitin/26S proteasome pathway of protein degradation during compatible but not incompatible pollination. These results suggest that AhSLF-S2 more likely mediates the selective destruction of S-RNase during compatible response but leaves S-RNase active in incompatible response via the ubiquitin/26S proteasome pathway, thus providing a biochemical basis for the inhibition of pollen tube growth during self-incompatible response in Antirrhinum.
RESULTS
Physical Interaction of the AhSLF-S2 C-Terminal Region and S-RNases
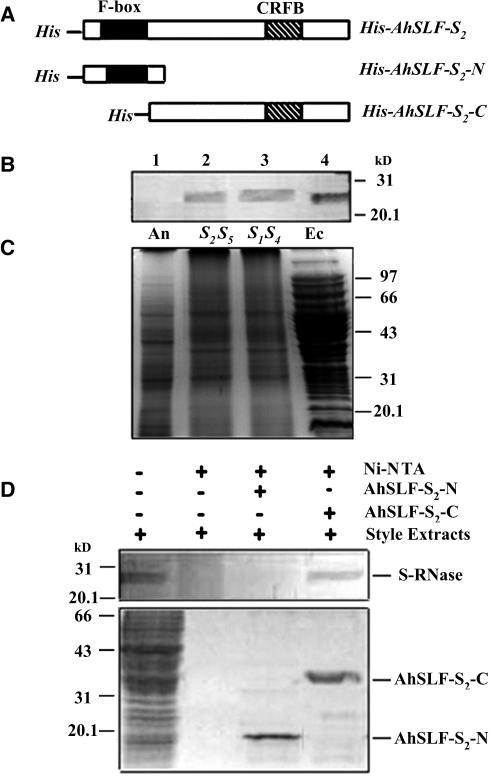
AhSLF-S2 and its allelic products have been proposed as candidates for Sp (Lai et al., 2002; Zhou et al., 2003). This would imply that they directly interact with S-RNases. To examine such a possibility, a pull-down experiment was performed using a recombinant AhSLF-S2 protein fused with an N-terminal His tag. The His-tagged recombinant portions of the full-length AhSLF-S2 of 376 amino acids (His-AhSLF-S2) as well as its N-terminal (101 amino acids, His-AhSLF-S2-N) and C-terminal (275 amino acids, His-AhSLF-S2-C) portions with or without the F-box domain, respectively (Figure 1A), were produced in bacteria and purified by nickel-nitrilotriacetic acid (Ni-NTA) resin. A recombinant S2-RNase without signal peptide of 21 amino acids (Xue et al., 1996) (Figure 2A) also was produced in bacteria and purified. A polyclonal antibody against the recombinant S2-RNase was raised in rabbit and used for detection of its presence in style, anther, and bacteria extracts (Figures 1B and 1C). The antibody detected similar bands with a molecular mass of ∼27 kD from style extracts of self-incompatible lines of S1S4 and S2S5 as well as in bacteria expressing the S2-RNase construct but not in anther total proteins (Figure 1B). Two proteins appeared to be detected in SI lines, consistent with the expression of two heteroallelic S-RNases (lanes 2 and 3 in Figure 1B), indicating that the antibody detected not only S2-RNase but also other allelic S-RNases. Thus, the antibody against S2-RNase detected S-RNases but without allelic specificity. Despite its lack of allelic specificity, the antibody could be used for S-RNase detection and titration experiments (see below).
Figure 1.
Pull-Down Assay for the Physical Interaction between AhSLF-S2 and S-RNases.
(A) Constructs used for expressing the His-tagged products from the full-length AhSLF-S2 (His-AhSLF-S2) and the N-terminal (His-AhSLF-S2-N) and C-terminal (His-AhSLF-S2-C) regions. CRFB represents a conserved domain found among similar F-box proteins to AhSLF-S2 in Arabidopsis (Kuroda et al., 2002).
(B) Immunoblot detection of S-RNases from total proteins of Antirrhinum anther (lane 1), styles of S2S5 (lane 2) and S1S4 (lane 3), and E. coli expressing S2-RNase (lane 4) by a polyclonal antibody against S2-RNase. Fifty micrograms of total proteins were loaded in each lane.
(C) The proteins also were stained by Coomassie blue for loading control. An, anther; Ec, proteins from bacteria expressing S2-RNase; S2S5, style of S2S5; S1S4, style of S1S4.
(D) A pull-down assay for AhSLF-S2 interaction with S-RNase. Ni-NTA resin and the purified fusion proteins of His-AhSLF-S2-N and His-AhSLF-S2-C were incubated with the extracts of Antirrhinum styles of S2S5. Bound proteins were pulled down with His-NAT resin, eluted with lysis buffer, separated by 12% SDS-PAGE, transferred to membranes, and analyzed by immunoblotting using the anti-S-RNase antibody (top panel). Input proteins after washing also were stained by Coomassie blue (bottom panel). Style total proteins were used as a positive control. Molecular mass markers are indicated in kilodaltons.
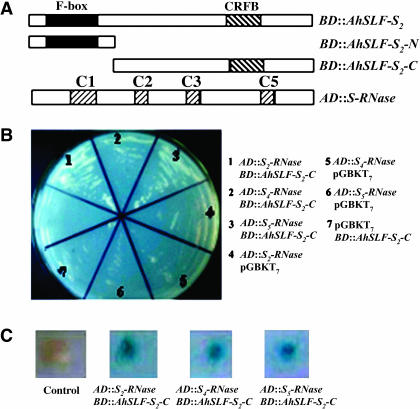
Figure 2.
Physical Interaction between AhSLF-S2 and S-RNases in Yeast Cells.
(A) Constructs of AD::S-RNases and BD::AhSLF-S2, BD::AhSLF-S2-N, and BD::AhSLF-S2-C. S-RNase denotes S2-, S4- or S5-RNases. C1 to C3 and C5 indicate conserved domains present in Antirrhinum S-RNases (Xue et al., 1996). Plasmids containing fusions to the GAL4 DNA binding domain are indicated by BD fusion to GAL4. Transcriptional activation domain is denoted by AD. Plasmid pGBKT7 with various AD:: constructs and plasmid pGADT7 with various BD:: constructs were used as negative controls. Yeast strain AH109 containing various combinations of two-hybrid plasmids were tested for His/Ade-dependent growth and β-galactosidase activity.
(B) Yeast cells containing various combinations of BD and AD fusions were tested for their growth on -Leu/-Trp/-His/-Ade dropout media.
(C) The strains were grown further to test for expression of the β-galactosidase reporter gene.
Initially, we intended to use the full-length product of His-AhSLF-S2 for the pull-down assay, but it was unable to be expressed after our repeated efforts (data not shown). Subsequently, we examined whether part of the protein would interact with S-RNases based on the possibility that AhSLF-S2 encodes an F-box protein, which is expected to possess a domain(s) for protein interaction. His-AhSLF-S2-N and His-AhSLF-S2-C fusion proteins (Figure 1A) were used separately for the pull-down assay (Figure 1D). The purified His-AhSLF-S2-N and His-AhSLF-S2-C fusion proteins were incubated with style extracts of an S2S5 self-incompatible line. After washing with buffer, the Ni-NTA resin–bound proteins were assayed by SDS-PAGE and examined by immunoblot analysis with the S-RNase antibody. Similar to that detected in the style, a specific protein with ∼27 kD was detected by the antibody when using His-AhSLF-S2-C fusion protein, and no protein was detected when only using the style extracts and the resin (Figure 1D), indicating that the C-terminal part of AhSLF-S2 specifically interacts with S-RNases. In addition, no protein was detected for His-AhSLF-S2-N containing only the F-box domain, showing that this part of the protein does not interact with S-RNases. However, it was not clear whether AhSLF-S2 interacts with S2-RNases, S5-RNases, or both. Similar results were obtained when the style extracts of S1S4 were used, indicating that AhSLF-S2 could interact with all of the S-RNases with no allelic specificity (data not shown). Taken together, these data suggest that the C-terminal portion of AhSLF-S2 physically interacts with S-RNases in vitro, but this interaction displayed no allelic specificity.
The C-Terminal Region of AhSLF-S2 Interacts with S-RNases in Yeast
To further examine the interaction of AhSLF-S2 with S-RNases, we used a yeast two-hybrid screening procedure. AhSLF-S2 and S2-, S4-, and S5-RNases were used as prey and bait, respectively. As in the pull-down assay, three versions of AhSLF-S2 were used (Figure 2A). The N-terminal, C-terminal, and full-length AhSLF-S2 were introduced into pGBKT7 vector and expressed as a fusion to GAL4 DNA binding domain (BD), whereas S2-, S4-, and S5-RNases were introduced into pGADT7 plasmid and expressed as a fusion to transcriptional activating domain (AD) (Figure 2A). Three AD::S-RNases were transformed into yeast AH109 in combination with various BD::AhSLF-S2 constructs (Figure 2B). Transformed yeast cells by AD::S-RNases and BD::AhSLF-S2 could grow on -Trp/-Leu media, but no growth of the transformed yeast cells occurred on selective -Trp/-Leu/-His/-Ade media (data not shown). Similar results were obtained when BD::AhSLF-S2-N and AD::S-RNases were cotransformed (data not shown), consistent with the pull-down data. By contrast, yeast cells cotransformed with AhSLF-S2-C and S2-, S4-, or S5-RNase constructs grew well on both Leu-/Trp- and Leu-/Trp-/His-/Ade- media (Figure 2B), showing that a physical interaction had occurred between AhSLF-S2-C and S-RNases. Yeast transformed with the control plasmids AD::S-RNase and pGBKT7 or BD::AhSLF-S2C and pGADT7 did not grow (Figure 2B). Furthermore, the β-galactosidase reporter gene activity was detected and appeared to be positive in yeast cells cotransformed with BD::AhSLF-S2-C and AD::S-RNases (Figure 2C), indicating that these interactions also likely occur in yeast cells in a non-allele-specific manner.
AhSLF-S2 Interacts with S-RNases in Vivo
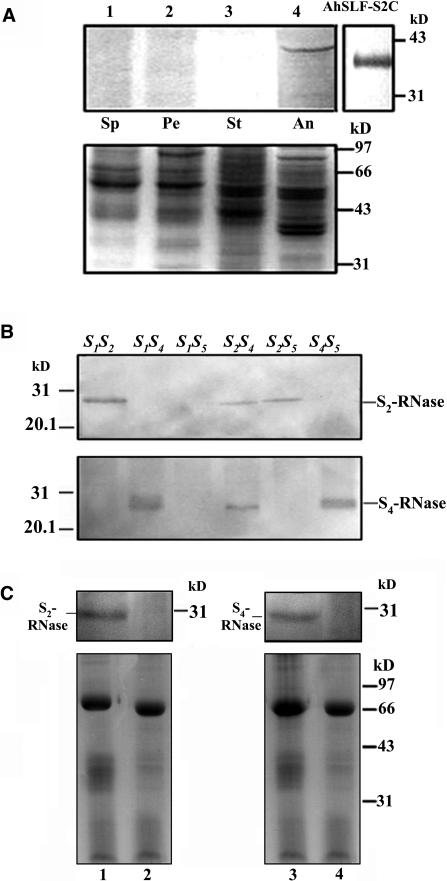
To determine whether the interaction between AhSLF-S2 and S-RNases occurs in planta, we performed a coimmunoprecipitation experiment with an antibody against the C-terminal part of AhSLF-S2. The antibody specifically detected a protein with a similar size to that of the predicted AhSLF-S2 polypeptide (41.4 kD) in anther but none in other tissues (Figure 3A). Meanwhile, two peptide antibodies against S2- and S4-RNases were developed, and immunoblot analysis showed that they detected them in an allele-specific manner (Figure 3B). To perform the coimmunoprecipitation, equal amounts of the mixtures of style extracts (50 μg) from S2S5 or S1S4 and total pollen extracts (50 μg) from S2S5 were incubated together with anti-AhSLF-S2-C antibody or preimmune serum at 4°C. The reason why we used the style and pollen extracts was that we had been unable to express a full-length AhSLF-S2 (see above), and it was difficult to recover functional S-RNases after SDS-PAGE. The bound proteins to the anti-AhSLF-S2-C antibody were analyzed by immunoblotting with the allele-specific anti-S-RNases antibody (Figure 3C). A protein similar to S2-RNase was specifically precipitated by the anti-AhSLF-S2-C antibody (lane 1 in Figure 3C) but not by control preimmune serum (lane 2 in Figure 3C), suggesting that an association of AhSLF-S2 with S2-RNase occurs in vivo. In addition, a protein similar to S4-RNase also was precipitated by anti-AhSLF-S2-C antibody (lane 3 in Figure 3C) but not by control preimmune serum (lane 4 in Figure 3C), indicating that AhSLF-S2 also interacts with S4-RNases in vivo, consistent with the findings of the pull-down and yeast two-hybrid assays.
Figure 3.
Coimmunoprecipitation of AhSLF-S2 and S-RNases.
(A) Detection of AhSLF-S2 in several tissues and E. coli expressing AhSLF-S2C by a polyclonal antibody against the C-terminal region of AhSLF-S2. Bottom panel: Coomassie blue–stained gel before protein gel blot analysis showing control loading of proteins. Lanes 1 to 4 represent protein extracts from sepal (Sp), petal (Pe), style (St), and anther (An) from an S2S5 line. AhSLF-S2C indicates the detection of AhSLF-S2C in E. coli expressing AhSLF-S2C.
(B) Immunoblot analysis of style proteins from several lines of an S-allele–segregating family using allele-specific antibodies against peptides derived from S2- and S4-RNases. Genotypes are indicated at the top.
(C) Extracts of pollinated styles of S1S4 (lane 1) or S2S5 (lane 3) lines (50 μg) and pollen of an S2S5 line (50 μg) were immunoprecipitated with anti-AhSLF-S2-C antibody (3 μL) (lanes 1 and 3) or control preimmune serum (3 μL) (lanes 2 and 4), respectively. After immunoprecipitation, the proteins were subjected to 12% SDS-PAGE followed by protein gel blotting with specific antibodies against S2- and S4-RNases, respectively (top panels). S-RNases detected are indicated. Bottom panels: Coomassie blue–stained gels before protein gel blot analysis showing equal loading of proteins. The strongest bands represent immunoglobulin heavy chains. Molecular mass markers are indicated in kilodaltons.
Paralogous Copies of AhSLF-S2 Show No Physical Interaction with S-RNases
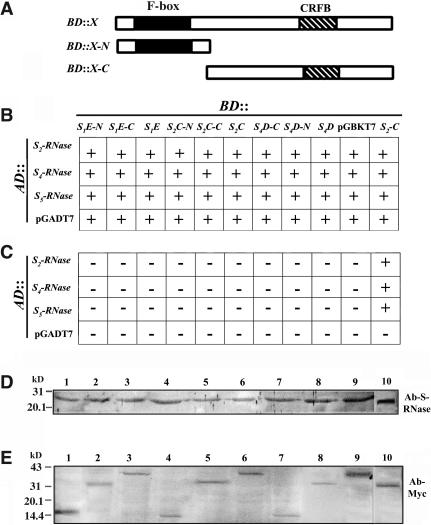
Several duplicated genes similar to AhSLF-S alleles have been found in the S-locus of Antirrhinum, and most of them also are present as allelic copies (Zhou et al., 2003). Interestingly, three of them (AhSLF-S1E, AhSLF-S2C, and AhSLF-S4D) are expressed specifically in pollen, similar to AhSLF-S2 (Zhou et al., 2003). To examine if they interact with S-RNases, we performed the test using the yeast two-hybrid screening procedure described above. The BD plasmids of AhSLF-S1E, AhSLF-S2C, and AhSLF-S4D similar to AhSLF-S2 were constructed (Figure 4A) and transformed into yeast together with AD::S-RNase constructs (Figures 4B and 4C). Yeast cells transformed with all of the plasmid combinations grew normally on -Trp/-Leu media (Figure 4B), showing that both plasmids were present in the transformed yeast and produced no growth defect. By contrast, no yeast growth occurred for all of the plasmid combinations except for AhSLF-S2-C on -Trp/-Leu/-His/-Ade media (Figure 4C), indicating that the three duplicated SLF proteins are not capable of physically interacting with S-RNases. Immunoblot analysis of transformed yeast cells showed that both S-RNases and AhSLF paralogs were expressed (Figures 4D and 4E). These results further imply that the interaction of AhSLF-S2 and possibly its allelic products with S-RNases is of specificity.
Figure 4.
Proteins Encoded by Parologous Copies of the Pollen-Expressed AhSLF Genes Do Not Interact with S-RNases in Yeast.
(A) Constructs made for the full-length, N-terminal, and C-terminal regions of AhSLF-S1E, AhSLF-S2C, and AhSLF-S4D in pGBTK7 plasmid and expressed as fusions to GAL4 DNA binding domains, which are represented by an X. AD::S-RNases were as described in Figure 2.
(B) Growth of yeast strains transformed with various combinations of BD and AD plasmids on -Leu/-Ade media. S1E, AhSLF-S1E; S2C, AhSLF-S2C; S4D, AhSLF-S4D; S2, AhSLF-S2. Plus signs indicate the normal growth of the transformed yeast strains.
(C) Growth of AH109 yeast strains transformed with various combinations of BD and AD plasmids on -Trp/-His/-Leu/-Ade media. No growth occurred except BD::AhSLF-S2 and AD::S-RNases. Minus signs indicate no growth of the transformed yeast strains.
(D) Expression of the S-RNase fusion proteins were assayed by immunoblot analysis with the anti-S-RNase antibody in transformed yeast cells. The S2-RNase (lanes 1, 4, and 7), S4-RNase (lanes 2, 5, and 8), and S5-RNase (lanes 3, 6, and 9) fusion proteins were expressed properly in the transformed yeast cells. Lane 10 represents style proteins of S2S5.
(E) Expression of the AhSLF fusion proteins were assayed by immunoblot analysis with the anti-Myc antibody in transformed yeast cells. Lanes 1 to 10 represent AhSLF-S1E-N, -S1E-C, -S1E-F, -S2C-N, -S2C-C, -S2C-F, -S4D-N, -S4D-C, -S4D-F, and -S2-C fusion proteins expressed in the transformed yeast cells, respectively. Suffixes -C, -N, and -F denote the C-terminal, N-terminal, and full-length products, respectively.
AhSLF-S2 Interacts with ASK1- and CUL1-Like Proteins
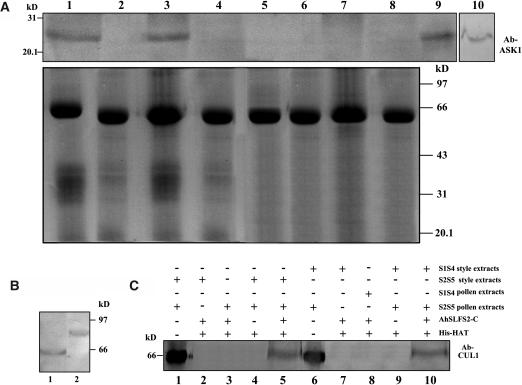
Based on the facts that AhSLF-S2 encodes an F-box protein and interacts with S-RNases, it is likely that it is a subunit of an SCF complex. To examine this possibility, we performed coimmunoprecipitation and pull-down assays using antibodies against AtASK1 and AtCUL1 (Liu et al., 2002), which are the two known subunits of several SCF complexes (Bai et al., 1996; Deshaies, 1999). To test if these antibodies detect similar proteins in Antirrhinum, an immunoblot analysis was performed (Figure 5). A similar protein of ∼23 kD was detected in Antirrhinum using the AtASK1 antibody (lanes 9 and 10 in Figure 5A). However, a smaller protein (∼67 kD) than that of Arabidopsis (Arabidopsis thaliana) (∼81.2 kD) was detected in Antirrhinum using the AtCUL1 antibody (Figure 5B). To examine if AhSLF-S2 interacts with an Skp1-like protein, equal amounts of the mixtures of the total style extracts (50 μg) from S2S5 or S1S4 and total pollen extracts (50 μg) from S2S5 were incubated with the AhSLF-S2-C antibody at 4°C as described above. As controls, the preimmune serum, the style extracts (50 μg) from S2S5 or S1S4, or total pollen extracts (50 μg) from S2S5 or S1S4 plants were incubated with the AhSLF-S2-C antibody. The proteins bound to the anti-AhSLF-S2-C antibody were analyzed by immunoblotting with the AtAsk1 antibody (Figure 5A). A protein similar to AtASK1 was specifically precipitated by the anti-AhSLF-S2-C antibody using a mixture of the style and pollen proteins (lanes 1 and 3 in Figure 5A) but not by control preimmune serum (lanes 2 and 4 in Figure 5A) or by only the pollen extracts or style extracts (lanes 5 to 8 in Figure 5A), suggesting that an association of AhSLF-S2 with an Skp-like protein from Antirrhinum occurs in vivo in both compatible and incompatible reactions.
Figure 5.
AhSLF-S2 Interacts with ASK1- and CUL1-Like Proteins.
(A) Extracts of styles of S1S4 or S2S5 lines (50 μg each) and pollen of an S2S5 line (50 μg) were coimmunoprecipitated with anti-AhSLF-S2-C antibody (3 μL) (lanes 1 and 3) or control preimmune serum (3 μL) (lanes 2 and 4), respectively. As additional controls, the extracts of styles of S1S4 (lane 5) and S2S5 (lane 6) lines or pollen of S1S4 (lane 7) and S2S5 (lane 8) lines also were coimmunoprecipitated with anti-AhSLF-S2-C antibody (3 μL). After immunoprecipitation, the proteins were subjected to 12% SDS-PAGE followed by protein gel blotting with specific antibodies against AtASK1 of Arabidopsis (top panel). Total proteins of Antirrhinum pollen (lane 9) and Arabidopsis leaf (lane 10) also were used as positive controls. The bottom panel represents the Coomassie blue–stained gel before protein gel blotting for equal loading of proteins.
(B) Immunoblot detection of CUL1-like protein from total pollen proteins of Antirrhinum pollen (lane 1) and Arabidopsis (lane 2).
(C) A pull-down assay for an interaction between AhSLF-S2 and CUL1-like proteins. His-NAT resin or the purified fusion proteins of His-AhSLF-S2-C were incubated with the extracts of Antirrhinum styles from S2S5 and pollen of S2S5 or S1S4 (lanes 5 and 10), respectively. The extracts of the Antirrhinum styles from S2S5 and pollen of S2S5 or S1S4 were incubated without His-AhSLF-S2-C as negative controls (lanes 4 and 9), and style and pollen extracts were used as positive controls (lanes 1 and 6). As additional controls, the extracts of the styles of S2S5 (lane 2) and S1S4 (lane 7) lines or pollen of S2S5 (lane 3) and S1S4 (lane 8) lines were incubated with anti-AhSLF-S2-C antibody (3 μL). Proteins bound were pulled down with His-NAT resin, eluted with lysis buffer, separated by 12% SDS-PAGE, transferred to membranes, and analyzed by immunoblotting with Arabidopsis anti-CUL1 antibody.
To further confirm this association, a pull-down experiment was performed using the AtCUL1 antibody (Figure 5C). The purified His-AhSLF-S2-C fusion protein was incubated with the mixtures of the style extracts from S2S5 or S1S4 plants and pollen extracts from an S2S5 plant. The bound proteins were assayed by SDS-PAGE and examined by immunoblot analysis with the AtCul1 antibody. Similar to that detected in pollen, a specific protein of ∼65 kD was detected by the antibody when using the His-AhSLF-S2-C fusion protein in the mixed style and pollen extracts, whereas no protein was detected when only using the style or pollen extracts and the resin (Figure 5C). Taken together, these results indicate that AhSLF-S2 is associated with the two known subunits of SCF complex, and together, they likely form an SCFAhSLF-S2 complex in Antirrhinum pollen.
The 26S Proteasome Pathway of Protein Degradation Is Required for Pollen Tube Growth through an Inhibition of S-RNase Activity
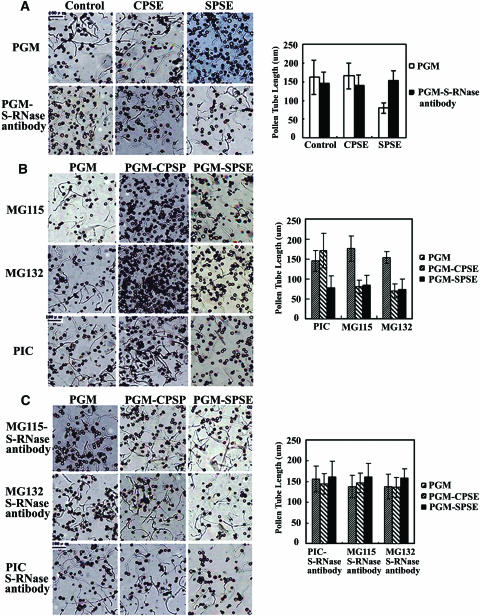
Because F-box proteins are mostly known to be involved in the 26S proteasome–mediated protein degradation pathway, we postulated that such a pathway also likely plays a role in self-incompatible reaction in Antirrhinum. To investigate this possibility, we performed the following experiments. First, pollen tube growth was assayed in vitro in a pollen germination medium (PGM) containing either incompatible or compatible style extracts and pollen tubes incubated with the germination medium containing 0.1% DMSO as control (Figure 6). In this system, Antirrhinum pollen germination rate was >80%, and the pollen growth occurred normally in PGM containing 0.1% DMSO as untreated controls (data not shown). Using this in vitro bioassay system, inhibition of the pollen tube growth was observed using incompatible style extracts, whereas the growth occurred normally in compatible style extracts after 2 h (Figure 6A). On average, we measured ∼100 pollen tubes per treatment and repeated four times. The pollen tube length treated by incompatible style extracts was reduced to approximately one-half (47%) of that treated by compatible style extracts. Addition of the S-RNase antibody into the control media reduced the pollen tube growth by ∼10%, which was not statistically significant as judged by standard deviations of the means. However, addition of the S-RNase antibody into the medium containing the incompatible style extracts reversed the inhibition of the pollen tube growth (Figure 6A), indicating that the in vitro inhibition of the pollen tube growth was directly related to S-RNase activities.
Figure 6.
Effects of the Proteasome-Specific Inhibitors on Pollen Tube Growth in Vitro.
Pollen tube growth is shown at left, and the mean length of the pollen tubes under various conditions is shown at right.
(A) Pollen was incubated in PGM (control) or in medium containing 3 μL of S-RNase antibody together with the cross-pollinated style extracts (CPSE) or self-pollinated style extracts (SPSE), whose final concentration was 25 mg/mL.
(B) Pollen was incubated in PGM or in PGM with CPSE or SPSE with 50 μM MG115 or MG132. As a control, a recommended concentration of protease inhibitors cocktail (PIC) and 0.1% DMSO (control) also were used.
(C) Effects of the S-RNase antibody on the pollen tube growth in combination with the proteasomal inhibitors. The pollen tube length was measured microscopically at 2 h after incubation.
To examine if the ubiquitin/26S proteasome pathway of protein degradation is involved in the pollen tube growth, two proteasomal inhibitors (MG115 and MG132) were added into the media and 0.1% DMSO as control. The results showed that both inhibitors had little effect on the inhibition of the pollen tube growth after being added in the media containing incompatible style extracts but inhibited the pollen tube growth significantly (45.2 and 48.65%) when the compatible style extracts were added into media (Figure 6B), indicating that the ubiquitin/26S proteasome pathway is required for the pollen tube growth during compatible pollination. As a control, protease inhibitor cocktails were used, and no significant inhibition was observed on the pollen tube growth (Figure 6B).
To further investigate if the proteasome pathway is involved in the S-RNase–mediated pollen tube growth inhibition, the proteasomal inhibitors and style extracts were added together with the S-RNase antibody in the media, and the pollen tube growth recovered normally, indicating that the inhibition of the proteasomal pathway specifically reversed the inhibitory effect of the self S-RNases on the pollen tube growth, and such an inhibition is mediated by the self S-RNase activity (Figure 6C). Taken together, these results suggest that the pollen tube growth likely requires the inhibition of the S-RNase activity by the 26S proteasome activity during compatible pollination.
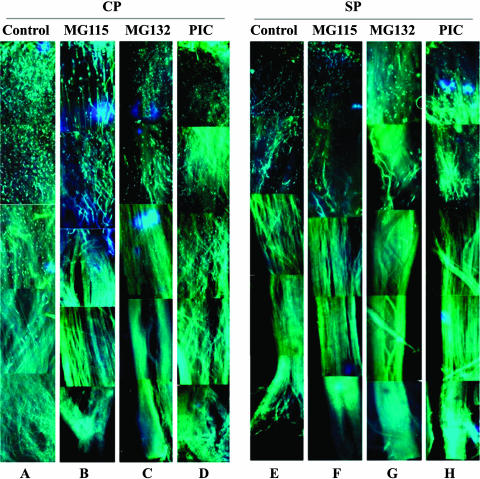
To confirm this observation in vivo, we treated pollen with the two proteasomal inhibitors (MG115 and MG132), the protease inhibitors cocktail, and the 0.1% DMSO as a control. They were subsequently used to pollinate either incompatible or compatible styles. We observed ∼20 styles per treatment, and 80 to 90% of pollen germinated normally (Figure 7). After 60 h of compatible pollination, most of the control pollen tubes reached the ovary (Figure 7A), but ∼80 to 85% of the pollen treated with the proteasomal inhibitors reached about the top one-third of the style length, and no further growth occurred (Figures 7B and 7C). As a control, the pollen treated with the protease inhibitors cocktail grew as well as that of the untreated pollen (Figure 7D), consistent with the in vitro bioassay. After 60 h of incompatible pollination, both of the control and the treated pollen tubes reached approximately the top one-third length of the style (Figures 7E to 7H), also similar to what we had observed in the in vitro experiments. These results further support the possibility that the ubiquitin/26S proteasome pathway is required for pollen tube growth during compatible pollination.
Figure 7.
Effects of the Proteasomal Inhibitors on Compatible and Self-Incompatible Pollination in Vivo.
The pollen tube growth was observed by fluorescence microscopic analyses in the style pollinated by non-self pollen treated by 0.1% DMSO (A), by non-self pollen treated by the proteasomal inbitors MG115 (B), MG132 (C), and the protease inhibitors cocktail (D), by self-pollen 0.1% DMSO (E), and by self-pollen treated by the proteasomal inhibitors MG115 (F), MG132 (G), and the protease inhibitors cocktail (H). The pollen tube growth was assayed at 60 h after pollination. CP, compatible pollination; SP, self-incompatible pollination.
Compatible Pollination Is Associated with an Increased Ubiquitination of Proteins, Including Non-Self S-RNase
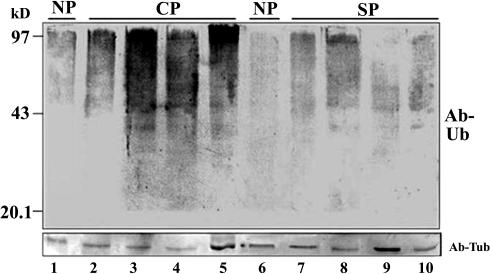
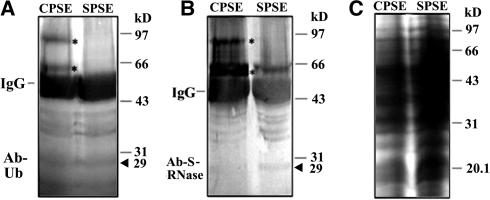
To examine if the involvement of the proteasome activity in the pollen tube growth is related to protein ubiquitination, an equal amount of style proteins (50 μg) from compatible and incompatible pollination were analyzed by immunoblotting with an anti-ubiquitin antibody. Figure 8 shows that a substantial increase of protein ubiquitination was detected in the styles pollinated by compatible pollen compared with that by incompatible pollen. It was not clear whether a specific protein or a group of proteins were ubiquinated based on this analysis. However, the treatment of the proteasomal inhibitors on the styles from compatible or incompatible pollination did not reveal an accumulation of ubiquinated S-RNases (data not shown). S-RNases are abundant extracellular proteins. It is possible that only a small amount of S-RNases are uptaken into pollen tubes, and the ubiquitinated S-RNases were below the detection limit of this assay. To increase the detection sensitivity, a coimmunoprecipitation experiment was performed on the total proteins from pollen incubated with compatible and incompatible style extracts using the anti-S-RNase antibody. In the pollen proteins treated with the compatible style proteins, two proteins of higher molecular mass (∼63 and ∼95 kD) were specifically detected by both the anti-S-RNase and the anti-ubiquitin antibodies, but no equivalent proteins were found in the pollen proteins treated with the incompatible style proteins (Figure 9), indicating that the S-RNases are likely ubiquitinated during compatible pollination.
Figure 8.
Immunoblot Analysis of the Style Proteins after Compatible and Self-Incompatible Pollination for Ubiquitination.
Fifty micrograms of proteins from the unpollinated (lanes 1 and 6), cross-pollinated (lanes 2 to 5: 8, 24, 48, and 60 h), and self-pollinated (lanes 7 to 10: 8, 24, 48, and 60 h) styles were separated by 12% SDS-PAGE and blotted for immunodetection by the anti-ubiquitin antibody (Ab-Ub) (top panel). The proteins also were analyzed by protein gel blotting using anti-tubulin antibody (Ab-Tub) for equal loading in each lane (bottom panel). CP, compatible pollination; SP, self-incompatible pollination.
Figure 9.
Detection of Ubiquitinated S-RNases.
The pollen total proteins (50 μg) after incubation with CPSE or SPSE were immunoprecipitated with anti-S-RNase antibody (3 μL). The precipitated proteins were subjected to 12% SDS-PAGE followed by protein gel blot analysis with the anti-ubiquitin antibody (A) and the anti-S-RNase antibody (B). The 25 μg of total proteins also were subjected to 12% SDS-PAGE and stained by Coomassie blue for loading control (C). Immunoglobulin heavy chains are indicated by IgG. Asterisks indicate two extra protein bands detected by both the anti-ubiquitin antibody and the anti-S-RNase antibody after the incubation with CPSE. By contrast, no similar proteins were detected after the incubation with SPSE. Arrowheads indicate the predicted size of S-RNases.
The S-RNase Level Is Reduced during Compatible Pollination
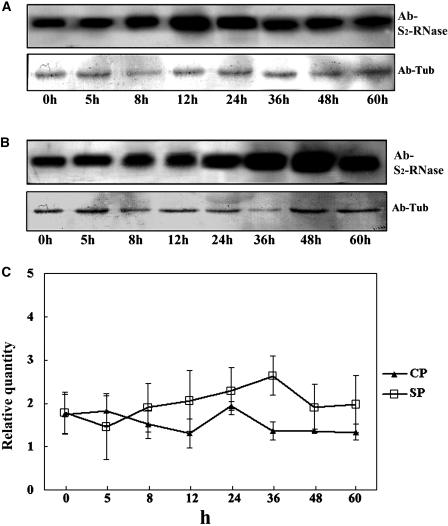
To examine if S-RNases are degraded, we determined their levels after either compatible or incompatible pollinations (Figure 10). The S2S5 styles were collected after being pollinated with compatible or incompatible pollen at different time points. Equal amounts of the total pollinated style proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-S-RNase antibody. The S-RNases in both the compatible and incompatible pollinated styles increased with time during the first 24 h of pollination, although a reduction of S-RNases was detected at 12 h after compatible pollination (Figures 10A and 10B). However, a noticeable decrease of the S-RNase level appeared to occur after 36 h of the compatible pollination but not in the incompatible pollination (Figure 10C). In fact, the levels of the S-RNases during incompatible pollination were slightly higher than those during compatible pollination. These results suggest that the level of the S-RNases is specifically reduced during compatible pollination, consistent with the action of the proteasome in this process.
Figure 10.
S-RNase Levels Change after Compatible or Incompatible Pollination.
(A) Detection of the S-RNase level during compatible pollination. h, hours after pollination.
(B) Detection of the S-RNase level during incompatible pollination. The proteins also were analyzed by immunoblotting using the anti-tubulin antibody for loading control in both cases.
(C) Quantification of the RNase level differences between compatible and incompatible pollination. Values are means of relative quantity ± se (n = 3). CP, compatible pollination; SP, self-incompatible pollination.
DISCUSSION
Although several SLF proteins with allelic polymorphisms have been identified from Antirrhinum and Prunus, their roles in self-incompatible reactions are not clear. In this study, we have provided several lines of evidence implicating that both AhSLF-S2 and the ubiquitin/26S pathway play important roles in SI in Antirrhinum.
AhSLF-S2 Physically Interacts with S-RNases Likely through the Formation of an SCF Complex
If AhSLF-S2 acts as an Sp or an S-RNase inhibitor as proposed (Lai et al., 2002; Entani et al., 2003; Ushijima et al., 2003; Zhou et al., 2003), its most probable target would be S-RNases that are possibly recruited as substrates for destruction through the 26S proteasome pathway, based on the findings of several known F-box proteins (Hershko and Ciechanover, 1998; Vierstra, 2003). Both yeast two-hybrid assays and coimmunoprecipitation indicated that AhSLF-S2 physically interacts with S-RNases but in a nonallelic manner. Importantly, no physical interaction was detected between three paralogous proteins (AhSLF-S1E, AhSLF-S2C, and AhSLF-S4D) and S-RNases (Figure 4), suggesting that they play no direct role(s) in self-incompatible reactions, and the interaction between AhSLF-S2 and S-RNases is of functional relevance.
The well-characterized SCF complex consists of CUL1, the Regulator of Cullins 1 ring finger protein, the SKP1 adaptor, and an F-box protein that binds directly to substrates (Weissman, 2001). The facts that AhSLF-S2 encodes an F-box protein (Lai et al., 2002) and interacts with S-RNases (Figures 1 to 3) together with CULl- and ASK1-like proteins in Antirrhinum (Figure 5) suggest that they form a multifactor SCF complex in pollen. However, other additional subunits of this complex still remain to be identified. Recently, a stylar Asn-rich protein called HT has been demonstrated to play a role in self-incompatible reactions in Nicotiana alata, Lycopersicon esculentum, and Solanum tuberosum (McClure et al., 1999, 2000; Kondo et al., 2002a, 2002b). Although HT genes are developmentally regulated during anthesis similar to S-RNases after compatible and incompatible pollination, HT is encoded by a gene unlinked to the S-locus and does not interact directly with S-RNases (McClure et al., 1999; Kondo et al., 2002a; O'Brien et al., 2002). Furthermore, a protein named PhSBP1 from Petunia hybrida (petunia) has been shown to interact with S-RNases in vitro, but it is not known if this also occurs in vivo (Sims and Ordanic, 2001). Thus, it will be interesting to test whether these proteins or other proteins also interact with AhSLF-S2 to form the complex together with S-RNases.
Inhibition of S-RNase Activity Likely Occurs during Compatible Pollination by the 26S Proteasome Pathway
Although S-RNases are thought to function as highly specific cytotoxins that inhibit the growth of incompatible pollen (McClure et al., 1990; Gray et al., 1991), it is unclear how the inhibition is accomplished. The specific block of compatible pollen tube growth by the proteasomal inhibitors and the reverse of such an inhibition by S-RNase antibody indicate that S-RNase is possibly degraded through the ubiquitin/26S proteolytic pathway during compatible pollination. These findings are consistent with genetic and molecular evidence that S-RNases are responsible for pollen rejection (Huang et al., 1994; Lee et al., 1994; Murfett et al., 1994) and provide a biochemical basis of the inhibition of S-RNase activity. Nevertheless, this appears to be different from what Scoccianti et al. (2003) have found in Actinidia deliciosa (kiwifruit), in which the concentration of MG132 of 50 μM strongly affected the A. deliciosa pollen tube elongation. By contrast, we found that the MG132 or MG115 of 50 μM had little effect on the growth of pollen tube, although the tips of some pollen tubes appeared to be swollen in Antirrhinum (Figure 6).
The detection of ubiquitinated RNases and their reduction during compatible pollination but not incompatible pollination further supports the possibility that this pathway is involved in S-RNase destruction during compatible pollination despite the findings that both non-self and self S-RNases interact with AhSLF-S2. This implies that either there is an additional factor(s) protecting or attenuating self S-RNases from their ubiquitination or that a subtle difference occurs between AhSLF-S2 and self or non-self RNase interactions in vivo, resulting in the ubiquitination of only non-self RNases. Recently, Luu et al. (2001) proposed a model for Sp consisting of two components: a general S-RNase inhibitor (RI) capable of inhibiting any S-RNases and S-allele–specific products that maintain the activity of a specific S-RNase inside pollen tubes by blocking RI biding. It is possible that AhSLF-S2 functions as a RI because of their direct interaction with S-RNases in a nonallelic fashion. In this case, AhSLF-S2 is unlikely to encode Sp.
In the rosaceous species, both predicted PdSFB and PmSLF proteins have been found to display haplotype-specific sequence polymorphisms as postulated for Sp genes (Entani et al., 2003; Ushijima et al., 2003; Yamane et al., 2003). Whether they are orthologous of AhSLF-S2 requires further functional tests. Nevertheless, it appears that the genomic structures of the S-locus of Antirrhinum and the rosaceous species are different from each other. In fact, the Antirrhinum S-locus appears to be a more diversified and larger genomic structure than that in the rosaceous species (Ushijima et al., 2001, 2003; Lai et al., 2002; Entani et al., 2003; Yamane et al., 2003; Zhou et al., 2003). The implication of this structural diversity in operation of SI remains to be tested. AhSLF-S2 and its allelic products only have a few amino acid variations among them (Zhou et al., 2003). Whether such a low level of differences could be related to allelic specificity postulated for Sp needs further investigation. If they are capable of showing specificity in vivo, they could encode Sp as a single component combining both S-RNase inhibition and recognition.
An Emerging Active Role of Protein Degradation in SI
The ubiquitin/26S proteasome system of intracellular protein degradation regulates abundance of proteins that control many developmental processes (Vierstra, 2003). Recent studies have revealed that it also is important for pollen recognition and rejection. In Brassica, ARM-repeat-containing protein 1 (ARC1), an S receptor kinase domain–interacting protein (Gu et al., 1998), has been demonstrated to be an E3 ubiquitin ligase (Stone et al., 2003). Furthermore, the level of ubiquitinated proteins increases in pistils pollinated by self-pollen but does not change in ARC1 antisense-suppressed pistils. More importantly, the inhibition of the proteasomal proteolytic activity disrupts the SI response. Thus, ARC1 has been proposed to promote the ubiquitination and proteasomal degradation of a compatibility factor(s) in the pistil and its destruction during self-incompatible reaction, which in turn leads to pollen rejection.
Our data appear to support that AhSLF-S2 is a component of an SCF complex to recruit substrates for destruction just like several known F-box proteins (Woo et al., 2001; Grima et al., 2002). Thus, the putative SCFAhSLF-S2 complex would target non-self S-RNases for destruction but leave self S-RNases active during self-incompatible response. Both types of S-RNases enter pollen tube after pollination (Luu et al., 2000). It appears that AhSLF-S2 together with ASK1- and CUL1-like molecules form a complex that targets S-RNases for ubiquitination and destruction by the 26S proteasome during compatible pollination. On the other hand, the interaction of AhSLF-S2 with self S-RNases is somehow ineffective for the target ubiquitination and degradation. As a result, they function as a cytotoxin that degrades RNA, causing the cessation of pollen tube growth. In addition, most known substrates of the SCF complexes in animals and plants need to be phosphorylated before being recognized by the F-box protein (Hershko and Ciechanover, 1998; Jackson et al., 2000). Kunz et al. (1996) found that S-RNases could be phosphorylated in vitro by a CDPK from pollen tube without allelic specificity. Thus, it needs testing for phosphorylation of S-RNases in vivo.
Nevertheless, it is unclear how the nonubiquitination of self-RNases is achieved. It is possible that the expression of AhSLF-S2 as an extra allele would lead to the breakdown of SI because of its action on non-self S-RNases, similar to the proposed inhibitor model of Sp (Golz et al., 2001). Thus, genetic transformation of AhSLF-S2 and detailed characterization of the proposed SCFAhSLF-S2 complex will help to understand better how pollen rejection occurs during self-incompatible reactions in species such as Antirrhinum.
METHODS
Plant Materials
Antirrhinum self-incompatible lines derived from interspecific crosses between A. majus and A. hispanicum have been described previously (Xue et al., 1996; Lai et al., 2002).
Pollen Culture Conditions
Fresh mature pollen was collected and incubated in a standard medium for pollen germination and growth (PGM). PGM contained 0.44 M sucrose, 1 mM CaCl2, 1 mM H3BO3, 1 mM MgSO4, and 2 mM citrate–phosphate buffer, pH 5.8. Self-pollinated style extracts, non-self pollinated style extracts, MG132, MG115, protease inhibitor cocktail, or anti-RNase antibody were added directly into the medium for various treatments, as indicated in Figure 6. For germination, pollen was incubated in small Petri dishes at 25°C ± 1°C in a saturated atmosphere (100% relative humidity) for 2 h. The measurements of pollen tube length were determined by Image-Pro software (Media Cybernetics, Silver Spring, MD) under a microscope.
Proteasomal Inhibitor Studies and Immunofluorescence Microscopy
Antirrhinum pollen were collected 1 d before anthesis and incubated 20 min in PGM with 50 μM proteasome inhibitor MG132 or MG115 (10 mM stock in DMSO; Sigma-Aldrich, St. Louis, MO). For controls, the Sigma-Aldrich plant protease inhibitor cocktail was used at a concentration of 30 g of plant tissue in a total volume of 100 mL/mL of protease inhibitor cocktail, or only DMSO was added. Styles were pollinated by the treated self or non-self pollen and harvested after 60 h, stained with decolorized aniline blue, and examined by epifluorescence (Kho and Baer, 1968).
Protein Gel Blot Analysis
To generate S-RNase and AhSLF-S2 antisera, the open reading frame (ORF) of S2-RNase lacking signal peptide and AhSLF-S2 missing the F-box domain were cloned into pET-28b (Clontech, Palo Alto, CA) separately, and the proteins were expressed in Escherichia coli and purified by SDS-PAGE. Purified protein was injected into a rabbit to raise an anti-S-RNase or AhSLF-S2 serum. To generate peptide antibodies against S2- and S4-RNases, the peptides from hypervariable regions of S2-RNase (NCRDTSADVLKIT) and S4-RNase (QAPQSCTTNFQRFT) (Xue et al., 1996) were synthesized, and the antibodies were generated by Genemed Synthesis (San Francisco, CA). For protein gel blot analysis, styles were weighed, homogenized in SDS-loading buffer (0.2 M Tris-HCl, pH 6.8, 0.5 M DTT, 4% SDS, and 25% glycerol), boiled, and centrifuged. Proteins were separated in 12% Tris-Tricine gels (Schägger and von Jagow, 1987) and blotted onto Nitrobind (Micron Separations, Westborough, MA) using a Bio-Rad Transblot sd wet electroblotting apparatus (Hercules, CA). Blots were treated with the rabbit S-RNase (1:1000) or AhSLF-S2 antiserum (1:1000), a mouse monoclonal anti-tubulin Ab or anti-Myc (1:100000) (Sigma-Aldrich), or rabbit anti-ubiquitin Ab (1:100000) (Sigma-Aldrich), and immune complexes were detected using alkaline phosphatase-conjugated secondary antibodies and nitroblue tetrazolumy 5-bromo-4-chloro-3-indolyl phosphate (Harlow and Lane, 1988). Antibodies against AtASK1 and AtCUL1 were kindly provided by William Crosby and Xingwang Deng, respectively. S-RNase levels during incompatible or compatible pollination were measured by calculating signal density after immunoblot analysis using Image-Pro and tubulin signal as a control.
Immunoprecipitation
Immunoprecipitation was performed following the manufacturer's recommendation of an immunoprecipitation kit (IP-50; Sigma-Aldrich). Approximately 50 μg of proteins from style or pollen or 100 μg of proteins of their mixtures were incubated for 8 h with constant rotation at 4°C with 3 μL of antibody or preimmune serum. At the end of the incubation, 40 μL of Protein G–Sepharose (Amersham Biosciences, Uppsala, Sweden) was added to immunoprecipitate the proteins, and the precipitate was washed five times with 50 mM Tris-HCl and 0.5 M NaCl, pH 8.0, and the proteins were loaded on a 12% polyacrylamide gel. After gel electrophoresis, proteins were assayed by anti-S-RNase antibody or anti-ubiquitin antibody.
Expression of His Fusion Proteins and Pull-Down Assay
Recombinant fusion proteins His-AhSLF-S2C were expressed in E. coli BL21 (DE3) strain (Promega, Madison, WI) after induction by 0.1 mM isopropyl-d-thio-galactopyranoside (Sigma-Aldrich) for 3 h at 37°C and purified on Ni-NTA (Qiagen, Valencia, CA) following the manufacturer's recommendation (P6611; Sigma-Aldrich). Ten micrograms of each protein was coupled to 100 μL of a 50% suspension (v/v) of beads in equilibration buffer for 20 min at 4°C. After prewashing, style extracts were transferred to a clean tube containing His-AhSLF-S2C coupled to the Ni-NAT resin and incubated overnight with constant rotation at 4°C. Subsequently, the beads were washed more than five times with ice-cold 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 1 mM EDTA buffer, and then 50 μL of SDS-PAGE sample buffer was added to each sample. Bound proteins were dissociated by heating at 37°C for 1 h and resolved by SDS-PAGE in 12% gels as described above.
Yeast Two-Hybrid Cotransformation Assay
S2-RNase ORF lacking signal peptide was cloned into pGBKT7 (BD) (Clontech), and the C-terminal region, the N-terminal region, and the full ORF of AhSLF-S2, AhSLF-S1E, AhSLF-S2C, and AhSLF-S4D (Lai et al., 2002; Zhou et al., 2003) were cloned into pGADT7 (AD) (Clontech). The BD and AD vectors were cotransformed into AH109 and grown on -Leu/-Trp medium containing 2% agar at 30°C for 4 to 5 d. The clones were further grown on -Leu/-Trp/-Ade/-His medium containing 2% agar at 30°C for 3 to 4 d to test interaction.
For liquid β-galactosidase assay, yeast clones grown for 48 h at 30°C were transferred onto filter paper, and the clones were lysed in liquid nitrogen for 1 min, and then 5 mL of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 1 mM MgSO4, pH 7.0) containing β-mercaptoethanol (27 mL/10 mL) was added and incubated at 30°C or room temperature (for <8 h) and checked periodically for the appearance of blue color.
Acknowledgments
We are grateful to E.S. Coen and R. Carpenter for providing Antirrhinum plants and constant support. We also thank William Crosby and Xingwang Deng for providing AtASK1 and AtCUL1 antibodies, respectively, and Weicai Yang for critically reading the manuscript. The work was supported by Chinese Academy of Sciences and the National Natural Science Foundation of China (39825103 and 30221002).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yongbiao Xue (ybxue@genetics.ac.cn).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.017673.
References
- Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W., and Elledge, S.J. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274. [DOI] [PubMed] [Google Scholar]
- Cenciarelli, C., Chiaur, D.S., Guardavaccaro, D., Parks, W., Vidal, M., and Pagano, M. (1999). Identification of a family of human F-box proteins. Curr. Biol. 9, 1177–1179. [DOI] [PubMed] [Google Scholar]
- De Nettancourt, D. (2001). Incompatibility and Incongruity in Wild and Cultivated Plants. (Berlin: Springer-Verlag).
- Deshaies, R.J. (1999). SCF and cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Ferguson, C., Clarke, A.E., and Newbigin, E. (1999). Pollen-expressed S-RNases are not involved in self-incompatibility in Lycopersicon peruvianum. Sex. Plant Reprod. 12, 286–287. [Google Scholar]
- Elledge, S.J., and Harper, J.W. (1998). The role of protein stability in cell cycle control and cancer. Biochim. Biophys. Acta 1337, M61–M70. [DOI] [PubMed] [Google Scholar]
- Entani, T., Iwano, M., Shiba, H., Che, F.S., Isogai, A., and Takayama, S. (2003). Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: Identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8, 203–213. [DOI] [PubMed] [Google Scholar]
- Golz, J.F., Oh, H.-Y., Su, V., Kusaba, M., and Newbigin, E. (2001). Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-Ribonuclease inhibitor at the S-locus. Proc. Natl. Acad. Sci. USA 98, 15372–15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz, J.F., Su, V., Clarke, A.E., and Newbigin, E. (1999). A molecular description of mutations affecting the pollen component of the Nicotiana alata S locus. Genetics 152, 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, J.E., McClure, B.A., Bonig, I., Anderson, M.A., and Clarke, A.E. (1991). Action of the style product of the self-incompatibility gene of Nicotiana alata (S-RNase) on in vitro-grown pollen tubes. Plant Cell 3, 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima, B., Lamouroux, A., Chelot, E., Papin, C., Limbourg-Bouchon, B., and Rouyer, F. (2002). The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 420, 178–182. [DOI] [PubMed] [Google Scholar]
- Gu, T., Mazzurco, M., Sulaman, W., Matias, D.D., and Goring, D.R. (1998). Binding of a novel arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 95, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), p. 349.
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Huang, S., Lee, H.S., Karunanandaa, B., and Kao, T.H. (1994). Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. Plant Cell 6, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, P.K., Eldridge, A.G., Freed, E., Furstenthal, L., Hsu, J.Y., Kaiser, B.K., and Reimann, J.D. (2000). The lore of the RINGs: Substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10, 429–439. [DOI] [PubMed] [Google Scholar]
- Kho, Y.O., and Baer, J. (1968). Observing pollen tubes by means of fluorescence. Euphytica 17, 298–302. [Google Scholar]
- Kondo, K., Yamamoto, M., Itahashi, R., Sato, T., Egashira, H., Hatori, T., and Kowyama, Y. (2002. a). Insights into the evolution of self-compatibility in Lycopersicon from a study of stylar factors. Plant J. 30, 143–153. [DOI] [PubMed] [Google Scholar]
- Kondo, K., Yamamoto, M., Matton, D.P., Sato, T., Hirai, M., Norioka, S., Hatori, T., and Kowyama, Y. (2002. b). Cultivated tomato has defects in both S-RNase and HT genes required for stylar function of self-incompatibility. Plant J. 29, 627–636. [DOI] [PubMed] [Google Scholar]
- Kunz, C., Chang, A., Faure, J.D., Clarke, A.E., Polya, G.M., and Anderson, M.A. (1996). Phosphorylation of style S-RNases by Ca-[2+]-dependent protein kinases from pollen tubes. Sex. Plant Reprod. 9, 25–34. [Google Scholar]
- Kuroda, H., Takahashi, N., Shimada, H., Seki, M., Shinozaki, K., and Matsui, M. (2002). Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol. 43, 1073–1085. [DOI] [PubMed] [Google Scholar]
- Lai, Z., Ma, W., Han, B., Liang, L., Zhang, Y., Hong, G., and Xue, Y. (2002). An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol. Biol. 50, 29–42. [DOI] [PubMed] [Google Scholar]
- Lee, H.S., Huang, S., and Kao, T.H. (1994). S proteins control rejection of incompatible pollen in Petunia inflata. Nature 367, 560–563. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Serino, G., Deng, X.W., and Dinesh-Kumar, S.P. (2002). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to tobacco mosaic virus. Plant Cell 14, 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu, D.T., Qin, X., Laublin, G., Yang, Q., Morse, D., and Cappadocia, M. (2001). Rejection of S-heteroallelic pollen by a dual-specific s-RNase in Solanum chacoense predicts a multimeric SI pollen component. Genetics 159, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu, D.T., Qin, X., Morse, D., and Cappadocia, M. (2000). S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407, 649–651. [DOI] [PubMed] [Google Scholar]
- McClure, B.A., Cruz-Garcia, F., Beecher, B.S., and Sulaman, W. (2000). Factors affecting inter- and intra-specific pollen rejection in Nicotiana. Ann. Bot. 85, 113–123. [Google Scholar]
- McClure, B.A., Gray, J.E., Anderson, M.A., and Clarke, A.E. (1990). Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature 250, 937–941. [Google Scholar]
- McClure, B.A., Mou, B., Canevascini, S., and Bernatzky, R. (1999). A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proc. Natl. Acad. Sci. USA 96, 13548–13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubbin, A.G., and Kao, T.H. (2000). Molecular recognition and response in pollen and pistil interactions. Annu. Rev. Cell Dev. Biol. 16, 333–364. [DOI] [PubMed] [Google Scholar]
- Murfett, J., Atherton, T.L., Mou, B., Gasser, C.S., and McClure, B.A. (1994). S-RNase expressed in transgenic Nicotiana causes S-allelespecific pollen rejection. Nature 367, 563–566. [DOI] [PubMed] [Google Scholar]
- O'Brien, M., Kapfer, C., Major, G., Laurin, M., Bertrand, C., Kondo, K., Kowyama, Y., and Matton, D.P. (2002). Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. Plant J. 32, 985–996. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arobidopsis functions in auxin response and is related to human SKP2 and yeast grrlp. Genes Dev. 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Scoccianti, V., Ovidi, E., Taddei, A.R., Tiezzi, A., Crinelli, R., Gentilin, L., and Speranza, A. (2003). Involvement of the ubiquitin/proteasome pathway in the organisation and polarised growth of kiwifruit pollen tubes. Sex. Plant Reprod. 16, 123–133. [Google Scholar]
- Sims, T.L., and Ordanic, M. (2001). Identification of a S-ribonuclease-binding protein in Petunia hybrida. Plant Mol. Biol. 47, 771–783. [DOI] [PubMed] [Google Scholar]
- Stone, S.L., Anderson, E.M., Mullen, R.T., and Goring, D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15, 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, R.D., Uhrig, H., Hermsen, J.G.T., Salamini, F., and Kaufmann, H. (1991). Investigation of a self-compatible mutation in Solanum tuberosum clones inhibiting S-allele activity in pollen differentially. Mol. Gen. Genet. 226, 283–288. [DOI] [PubMed] [Google Scholar]
- Ushijima, K., Sassa, H., Dandekar, A.M., Gradziel, T.M., Tao, R., and Hirano, H. (2003). Structural and transcriptional analysis of the self-incompatibility locus of almond: Identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima, K., Sassa, H., Tamura, M., Kusaba, M., Tao, R., Gradziel, T.M., Dandekar, A.M., and Hirano, H. (2001). Characterization of the S-Locus region of almond (Prunus dulcis): Analysis of a somaclonal mutant and a cosmid contig for an S haplotype. Genetics 158, 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8, 135–142. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Wang, X., Skirpan, A.L., and Kao, T.H. (2003). S-RNase-mediated self-incompatibility. J. Exp. Bot. 54, 115–122. [DOI] [PubMed] [Google Scholar]
- Weissman, A.M. (2001). Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2, 169–178. [DOI] [PubMed] [Google Scholar]
- Woo, H.R., Chung, K.M., Park, J.H., Oh, S.A., Ahn, T., Hong, S.H., Jang, S.K., and Nam, H.G. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. 13, 1779–1790. [DOI] [PMC free article] [PubMed]
- Xiao, W., and Jang, J.C. (2000). F-box proteins in Arabidopsis. Trends Plant Sci. 5, 454–457. [DOI] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xue, Y., Carpenter, R., Dickinson, H.G., and Coen, E.S. (1996). Origin of allelic diversity in Antirrhinum S locus RNases. Plant Cell 8, 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane, H., Ikeda, K., Ushijima, K., Sassa, H., and Tao, R. (2003). A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol. 44, 764–769. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Wang, F., Ma, W., Zhang, Y., Han, B., and Xue, Y. (2003). Structural and transcriptional analysis of S-Locus F-box genes in Antirrhinum. Sex. Plant Reprod. 16, 165–177. [Google Scholar]