Abstract
The Linum usitatissimum (flax) L gene alleles, which encode nucleotide binding site–Leu rich repeat class intracellular receptor proteins, confer resistance against the Melampsora lini (flax rust) fungus. At least 11 different L resistance specificities are known, and the corresponding avirulence genes in M. lini map to eight independent loci, some of which are complex and encode multiple specificities. We identified an M. lini cDNA marker that cosegregates in an F2 rust family with a complex locus determining avirulence on the L5, L6, and L7 resistance genes. Two related avirulence gene candidates, designated AvrL567-A and AvrL567-B, were identified in a genomic DNA contig from the avirulence allele, whereas the corresponding virulence allele contained a single copy of a related gene, AvrL567-C. Agrobacterium tumefaciens–mediated transient expression of the mature AvrL567-A or AvrL567-B (but not AvrL567-C) proteins as intracellular products in L. usitatissimum and Nicotiana tabacum (tobacco) induced a hypersensitive response–like necrosis that was dependent on coexpression of the L5, L6, or L7 resistance gene. An F1 seedling lethal or stunted growth phenotype also was observed when transgenic L. usitatissimum plants expressing AvrL567-A or AvrL567-B (but not AvrL567-C) were crossed to resistant lines containing L5, L6, or L7. The AvrL567 genes are expressed in rust haustoria and encode 127 amino acid secreted proteins. Intracellular recognition of these rust avirulence proteins implies that they are delivered into host cells across the plant membrane. Differences in the three AvrL567 protein sequences result from diversifying selection, which is consistent with a coevolutionary arms race.
INTRODUCTION
The gene-for-gene model of plant disease resistance (Flor, 1971) predicts that plant resistance (R) gene products recognize the direct or indirect product of pathogen avirulence (Avr) genes. This recognition event activates host defense responses, which include a localized host cell death or hypersensitive response (HR). The majority of R genes identified to date encode proteins containing a predicted nucleotide binding site (NBS) followed by a series of Leu-rich repeats (LRR) at their C termini and are predicted to be intracellular (Dangl and Jones, 2001; Staskawicz et al., 2001). Different genes in this group confer host resistance to viruses, bacteria, oomycetes, fungi, nematodes, and sucking insects. NBS-LRR resistance proteins also may contain N-terminal TIR (Toll and Interleukin-1 receptor homology) or coiled coil domains. The highly variable LRR domains are implicated in controlling recognition specificity (Ellis et al., 1999; Jia et al., 2000; Dodds et al., 2001b), whereas intramolecular interactions involving the TIR-NBS or coiled coil NBS regions may control propagation of the perceived signal (Moffet et al., 2002; Hwang and Williamson, 2003). A second major class of R genes encode membrane-bound receptor proteins with an extracellular LRR domain, sometimes coupled to an intracellular kinase signaling domain (Song et al., 1995; Dixon et al., 1996, 1998; Parniske et al., 1997).
In contrast with the conserved structure of most R proteins, pathogen Avr proteins are extremely varied, reflecting the diverse array of pathogen molecules to which plants are exposed and which could be used as recognition targets. For instance, bacterial Avr products are members of a suite of disease effector proteins that are delivered directly into the plant cytoplasm via a type III secretion system (Lahaye and Bonas, 2001; Casper-Lindley et al., 2002). The bacterial type III secretion system is encoded by the hypersensitive response and pathogenicity (Hrp) genes and is essential for infection because hrp mutants are no longer pathogenic on their host plants. Whereas the biochemical functions of most bacterial effector proteins are unknown, some are targeted to the host nucleus and may act as transcription factors (Lahaye and Bonas, 2001; Deslandes et al., 2003). Other bacterial effectors are proteases (Orth et al., 2000), and some of these are known to cleave specific host proteins (Axtell et al., 2003; Shao et al., 2003).
Colocalization of corresponding R and Avr products is probably a prerequisite for recognition. Resistance proteins corresponding to bacterial Avr products targeted to the host cytoplasm all belong to the intracellular NBS-LRR class (Staskawicz et al., 2001), and resistance proteins against intracellular viral pathogens are also NBS-LRR proteins (Whitham et al., 1996; Bendahmane et al., 1999; Cooley et al., 2000). By contrast, the Lycopersicon esculentum (tomato) Cf resistance proteins are members of the extracellular receptor family and recognize Avr products secreted by the fungus Cladosporium fulvum into the apoplastic space (Dixon et al., 1996, 1998; Parniske et al., 1997). In some cases, recognition involves direct protein–protein interactions between corresponding R and Avr proteins (Jia et al., 2000; Deslandes et al., 2003). However, the RPS2 (resistance to Pseudomonas syringae) and RPM1 (resistance to P. syringae pv maculicola) resistance proteins in Arabidopsis (Arabidopsis thaliana) apparently recognize their corresponding Avr products indirectly by detecting changes induced in a host protein that is modified by these Avr products (Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003). Nevertheless, all of the components of the RPM1/RPS2 recognition complexes colocalize and are peripherally associated with the plasma membrane (Boyes et al., 1998; Axtell and Staskawicz, 2003). Similarly, the Arabidopsis RRS1 (resistance to Ralstonia solanacearum) protein and its corresponding Avr protein, PopP2, also colocalize, in this case to the host cell nucleus (Deslandes et al., 2003).
Rust fungi (order Uredinales) cause disease on many important crop plants and are obligate biotrophs that grow only in living plant tissue. During infection, rusts form haustoria, specialized structures that penetrate the plant cell wall but remain separated from the host cytoplasm by the host cell membrane. One of the primary functions of haustoria seems to be to facilitate nutrient uptake from the host (Hahn and Mendgen, 2001). Haustoria formation also induces structural changes in the host cell, including cytoskeletal rearrangements, nuclear migration, and chromatin condensation (Kobayashi et al., 1994; Heath, 1997), and there is some evidence that compatible rusts can suppress host defense responses and influence host cell metabolism (Mellersh and Heath, 2001; Ayliffe et al., 2002; Voegele and Mendgen, 2003). However, the signals leading to these changes are not known. In resistant plants, haustoria formation also seems to be the trigger for HR induction (Kobayashi et al., 1994; Heath, 1997), suggesting that rust Avr products are detected at these sites.
In Linum usitatissimum (flax), at least 30 rust resistance specificities have been distinguished by their recognition of different Melampsora lini (flax rust) strains, and these are distributed among five polymorphic loci, K, L, M, N, and P (Islam and Mayo, 1990). R proteins encoded by genes at the L, M, N, and P loci are members of the intracellular NBS-LRR class (Lawrence et al., 1995; Anderson et al., 1997; Dodds et al., 2001a, 2001b). NBS-LRR proteins also confer resistance to rusts in maize (Zea mays) (Collins et al., 1999), and similar proteins in Arabidopsis, Lactuca sativa (lettuce), and Hordeum vulgare (barley) confer resistance to downy mildew and powdery mildew pathogens (Noel et al., 1999; Halterman et al., 2001; Zhou et al., 2001; Shen et al., 2002), which are also haustoria-producing obligate biotrophs. Although Avr genes have been described at a genetic level in rusts and mildews (Flor, 1971; Lawrence et al., 1981; Zambino et al., 2000; Pedersen et al., 2002; Rehmany et al., 2003), no Avr products have been identified in these pathogens, so the conundrum of how intracellular R proteins recognize these extracellular pathogens remains unresolved.
In this article, we describe the isolation of three members of a family of avirulence genes from M. lini that are recognized by the L5, L6, and L7 resistance genes. A cDNA marker cosegregating with the avirulence phenotype in an F2 rust family was used to identify a genomic DNA region from the avirulence locus. We show that in planta expression of avirulence gene candidates encoded at this locus causes R gene–dependent cell death that is specific to the L5, L6, and L7 genes. The M. lini avirulence genes are expressed in haustoria and encode small secreted proteins. Recognition of these proteins occurs when they are expressed inside the plant cell, suggesting that they are delivered into host cells during rust infection.
RESULTS
Isolation of a cDNA Marker Cosegregating with an M. lini Avirulence Locus
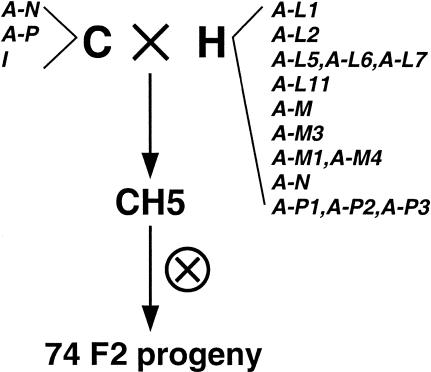
To isolate Avr genes from M. lini, we undertook a map-based cloning approach using an F2 family of 74 individuals produced by crossing the rust strains C and H and then self-fertilizing the F1 hybrid rust strain CH5 (Figure 1; Lawrence et al., 1981). Avr specificities corresponding to sixteen different resistance genes segregate in this family and map to 10 independent loci. Several markers linked to Avr loci were identified using random amplified polymorphic DNA, amplified fragment length polymorphism, and cDNA amplified fragment length polymorphism techniques (P.N. Dodds, M.A. Ayliffe, G.J. Lawrence, and J.G. Ellis, unpublished results) as well as by mapping in planta–expressed rust cDNA clones as restriction fragment length polymorphisms (RFLPs). For the latter approach, we used suppression subtractive hybridization (Diatchenko et al., 1996) to prepare a cDNA library enriched for transcripts present in L. usitatissimum leaves infected with rust H but not in uninfected leaves (see Methods). A second subtracted cDNA library was enriched for transcripts more abundant during infection with rust H than with rust C because most of the Avr genes segregating in the F2 family were derived from rust H. A total of 181 randomly selected cDNA clones from these libraries were sequenced and represented 112 unique cDNAs. DNA gel blot hybridization confirmed that 80 clones were derived from the rust genome, of which 45 detected RFLPs between the parental rust strains C and H. These markers were scored in the 74 F2 individuals.
Figure 1.
An M. lini Family Segregating for 16 Avirulence Specificities.
The rust strains C and H were crossed together to produce the F1 rust CH5, which was then selfed to produce 74 F2 progeny (Lawrence et al., 1981). Avirulence specificities segregating in the F2 family are indicated next to the parental rust from which they were inherited. Some avirulence specificities, such as A-L5, A-L6, and A-L7, cosegregate in this family and are listed next to each other. The A-P gene from rust C segregates as an alternative allele at the same locus as A-P1, A-P2, and A-P3 from rust H. The A-N gene was present in both parents but segregates in the F2. An inhibitor gene (I) that suppresses resistance reactions triggered by some avirulence genes was inherited from rust C.
One cDNA probe, isolated from the infected versus uninfected cDNA library and designated IU2F2, detected RFLPs that cosegregated with an avirulence locus corresponding to the L5, L6, and L7 resistance genes in L. usitatissimum. These three avirulence specificities cosegregate in several independent rust families (Lawrence et al., 1981) and have not been separated by mutation (Flor, 1956). In digests with numerous restriction enzymes, the 180-bp IU2F2 cDNA probe consistently detected two hybridizing fragments that cosegregated with the avirulence allele derived from rust strain H (avirulent on L5, L6, and L7), indicating that this allele contains a duplication of this sequence (data not shown). A single restriction fragment hybridizing to IU2F2 segregated at the alternative allele derived from rust strain C (virulent on L5, L6, and L7).
A Candidate Avirulence Gene Encodes a Small Secreted Protein
Because the IU2F2 marker cosegregated with the avirulence locus, we used this probe to isolate further sequences from this gene and surrounding DNA. A sequence contig of 26.5 kb (Figure 2A) was assembled from several overlapping λ clones isolated from a genomic DNA library prepared from the F1 rust CH5 (Ayliffe et al., 2001). By comparison with the restriction fragment sizes detected by genomic DNA gel blot hybridization, this sequence corresponded to the rust H–derived allele of the IU2F2 RFLP locus. Because the alternative allele was not recovered from the genomic DNA library, the corresponding sequences were amplified by long PCR from an F2 rust strain homozygous for this allele. The restriction map of the resulting 11.5-kb contig was consistent with the IU2F2-detected restriction fragments derived from the virulent rust strain C. An RFLP detected using a probe from the 3′ end of these contigs was separated from the IU2F2 marker and avirulence genes by three recombination events in the F2 family (data not shown). Amplification and sequence analysis of this genomic DNA region from the three recombinants confirmed that the crossovers occurred within 1 kb of the 3′ end of the IU2F2 contig (Figure 2A; data not shown). This region therefore delineated one end of the genetic interval containing the avirulence genes. Although the physical contig did not encompass the other end of this genetic interval, the high frequency of recombination suggested that this region might have a low physical distance to map distance ratio, and we were encouraged to examine the sequences for candidate genes.
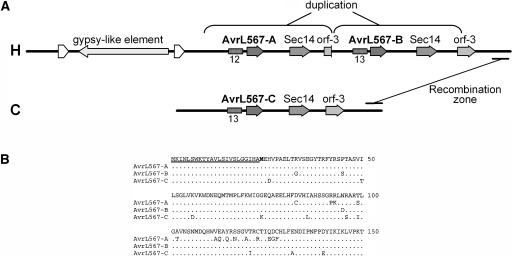
Figure 2.
Characterization of the M. lini AvrL567 Locus.
(A) Schematic diagram of the AvrL567 locus. A 26.5-kb region from the avirulence allele (H) contains the AvrL567-A and AvrL567-B genes, whereas the AvrL567-C gene occurs in an 11.5-kb region at the virulence allele (C). A second gene related to yeast Sec14 (Bankaitis et al., 1989) also occurs in this region along with a third predicted gene (orf-3; no similarity to database sequences). The position of a 7.4-kb duplication in the H allele is indicated, as is a 6.3-kb inserted sequence similar to gypsy class retrotransposons, with 174-bp identical direct repeats flanking a long open reading frame encoding a putative polyprotein. A repeat region containing 12 or 13 copies of an 82-bp sequence, which occurs close to the 5′ ends of the AvrL567 genes, is shown as a closed box. An ∼1-kb region in which three recombination events were resolved is indicated at the 3′ end of the contigs.
(B) Amino acid sequence alignment of the predicted AvrL567-A, AvrL567-B, and AvrL567-C proteins. Only those amino acids that differ from the consensus (top line) are shown, with identical residues indicated by a dotted line. The signal peptide is underlined, and the first amino acid of the predicted mature protein (Met-24) is shown in bold.
Three distinct classes of genes were detected in the DNA sequences derived from the IU2F2 locus, each with two copies present at the H allele and a single copy at the C allele (Figure 2A). One of these corresponded to the IU2F2 cDNA clone and appeared to be a likely candidate to encode avirulence because there was a high level of sequence variation between the virulence and avirulence alleles. In planta expression assays (see below) showed that these genes triggered R gene–dependent cell death responses with specificity for the L5, L6, and L7 resistance genes. We have therefore designated these genes as AvrL567-A, AvrL567-B (both from the avirulence allele), and AvrL567-C (from the virulence allele). Reverse transcriptase (RT)-PCR products amplified from rust-infected leaf RNA indicated that the AvrL567 transcripts contain 450-bp open reading frames. Two introns of 85 and 215 bp, both within the 5′ untranslated leader, were present in the genomic sequence but were spliced from the cDNA-derived PCR products. The 150 amino acid translations of the AvrL567 genes include predicted 23–amino acid cleavable secretion signal peptides (PSORT, http://psort.nibb.ac.jp/), suggesting that they encode secreted proteins of 127 amino acids. The amino acid sequences are highly variable, with 27 polymorphic sites in the 127 amino acid mature products (Figure 2B). No related sequences were detected in public databases, and no other recognizable functional motifs were detected.
A second gene family encoded in this region is closely related to the yeast gene Sec14 (Bankaitis et al., 1989), which encodes a phosphatidylinositol-phosphatidylcholine transfer protein involved in membrane fusion (Lopez et al., 1994). A third gene (orf-3) with no related sequences in the public sequence databases was predicted based on the presence of two open reading frames of 333 and 342 bp separated by a short putative intron (87 bp). However, we did not consider these genes to be likely candidates to encode the avirulence specificities that cosegregate with this locus because there was very little sequence variation between homologs encoded at the two alleles. The three Sec14 homologs are almost identical, with only a single conservative amino acid substitution (glutamate to aspartate) in their predicted products, whereas the orf-3 homologs are identical except for the truncation of one copy at a duplication boundary in the H allele (Figure 2A). In addition, Sec14 and orf-3 transcripts were detected at similar levels in leaves infected with either virulent or avirulent rust strains (data not shown).
Transient Expression of AvrL567 Genes in L. usitatissimum Causes Resistance Gene–Dependent Cell Death
Because no transformation procedure exists for rust fungi, it was not possible to directly test the avirulence phenotype conferred by the AvrL567 genes in transgenic rust. However, one of the primary outcomes triggered by R-Avr recognition is the HR, and Agrobacterium tumefaciens–mediated transient expression of corresponding R and Avr genes in plants has been shown to induce HR-like cell death (Scofield et al., 1996; Tang et al., 1996; van den Ackervecken et al., 1996; Bendahmane et al., 1999; Erickson et al., 1999; Tai et al., 1999; van der Hoorn et al., 2000). We therefore generated T-DNA constructs encoding the 127–amino acid mature AvrL567 proteins beginning at Met-24 (without the 23–amino acid signal peptides) driven by the 35S promoter of Cauliflower mosaic virus for transient expression assays. A. tumefaciens strains containing the AvrL567-A127 construct or an L6 gene construct were infiltrated into leaves of rust-susceptible L. usitatissimum var Hoshangabad (Figure 3A). L. usitatissimum leaves infiltrated with an A. tumefaciens strain containing an empty binary vector, or with the L6 or AvrL567-A127 constructs alone, did not cause any visible effects. However, coexpression of AvrL567-A127 and L6 in A. tumefaciens–infiltrated leaves induced a necrotic response, indicating that the AvrL567-A127 product is recognized by L6.
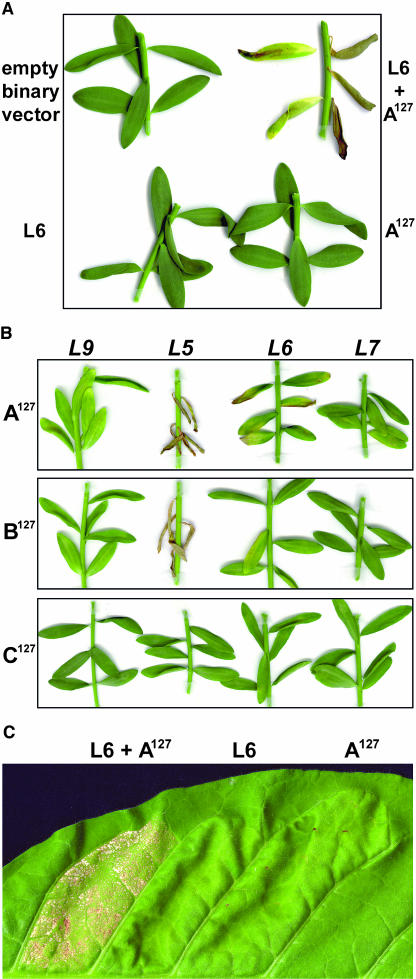
Figure 3.
Transient Expression of AvrL567 Genes in L. usitatissimum and N. tabacum Causes R Gene–Dependent Necrosis.
(A) Leaves of rust-susceptible L. usitatissimum var Hoshangabad were infiltrated with A. tumefaciens cultures containing an empty binary vector, an L6 T-DNA vector, a T-DNA vector encoding AvrL567-A127, or a mixture of two cultures containing the L6 and AvrL567-A127 T-DNAs.
(B) Leaves of near-isogenic L. usitatissimum lines containing the L9, L5, L6, or L7 resistance gene were infiltrated with A. tumefaciens cultures containing T-DNA expression vectors encoding AvrL567-A127, AvrL567-B127, or AvrL567-C127. Images were prepared 12 d after infiltration.
(C) W38 N. tabacum leaves were infiltrated with A. tumefaciens cultures containing either an L6 T-DNA vector, a T-DNA vector encoding AvrL567-A127, or a mixture of two cultures containing the L6 and AvrL567-A127 T-DNAs. Each A. tumefaciens culture was infiltrated into separate regions of the leaf bounded by the major veins, and the leaf was photographed 11 d after infiltration.
To further test the recognition specificity of the avirulence genes, A. tumefaciens cultures containing T-DNA constructs encoding AvrL567-A127, AvrL567-B127, or AvrL567-C127 were infiltrated into leaves of near-isogenic L. usitatissimum lines carrying different L locus rust resistance alleles (Figure 3B). Expression of AvrL567-A127 induced a strong necrotic response in L5 and L6 plants and a weak response in L7 plants, whereas expression of AvrL567-B127 induced a strong response in L5 plants, a weak response in L6 plants, and no response on L7. Thus, the AvrL567-A and AvrL567-B genes are sufficient to account for the three recognition specificities that cosegregate at this avirulence locus. No responses were observed when these constructs were tested in L. usitatissimum lines containing L, L1, L2, L3, L4, L8, L9, or L10 (Figure 3, Table 1), indicating that recognition was specific to the L5, L6, and L7 resistance genes. Importantly, transient expression of AvrL567-C127, encoded by the homologous gene from the virulent parental rust strain C, did not induce necrosis in any L. usitatissimum line (Figure 3B, Table 1). RNA gel blot analysis confirmed that the AvrL567127 constructs were each expressed at similar levels in A. tumefaciens–infiltrated leaves (data not shown).
Table 1.
Response of Near-Isogenic L. usitatissimum Lines to Avr Gene Transient Expressiona
|
L Gene Allele Present in L. usitatissimum Lines
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Construct | L | L1 | L2 | L3 | L4 | L5 | L6 | L7 | L8 | L9 | L10 |
| AvrL567-A127 | − | − | − | − | − | + | + | +/− | − | − | − |
| AvrL567-B127 | − | − | − | − | − | + | +/− | − | − | − | − |
| AvrL567-C127 | − | − | − | − | − | − | − | − | − | − | − |
Leaves were infiltrated with A. tumefaciens strains containing T-DNA constructs encoding the AvrL567127 proteins and scored for the induction of necrotic responses 12 d after infiltration. A plus sign indicates necrosis observed, a minus sign indicates no necrosis, and a plus and minus sign together indicates a weak necrotic response.
Infiltration with A. tumefaciens strains containing the AvrL567-A127 or AvrL567-B127 T-DNA construct, but no Ti plasmid to enable T-DNA transfer, did not induce necrosis, and AvrL567 transcripts were not detected by RNA gel blot analysis (data not shown). Thus, the necrotic reactions were dependent on production of the AvrL567127 proteins in transformed plant cells rather than recognition of A. tumefaciens–expressed proteins. Infiltration with A. tumefaciens strains containing T-DNA constructs encoding the full-length AvrL567150 proteins (including the secretion signal) induced much weaker reactions than the truncated versions, although they maintained a similar relative specificity (data not shown). These responses took longer to develop (10 to 12 d compared with 4 to 5 d) and were characterized by chlorosis rather than necrosis.
Transient Coexpression of L6 and AvrL567 Induces Cell Death in Nicotiana tabacum
To determine whether coexpression of the L6 and AvrL567 genes could induce an HR in other species, these genes were transiently expressed in Nicotiana tabacum (tobacco). A. tumefaciens–mediated transient coexpression of AvrL567-A127 and L6 produced a necrotic response in N. tabacum (Figure 3C). Again this was dependent on the expression of both transgenes because transient expression of either L6 or AvrL567-A127 alone did not produce a necrotic response. In a similar assay, coexpression of L6 and AvrL567-B127 gave a weak response in N. tabacum, but no response was observed with coexpression of L6 and AvrL567-C127 (data not shown). Similar results also were observed when the AvrL567 genes were expressed in N. tabacum lines carrying an L6 transgene (data not shown). Thus, the responses in N. tabacum are consistent with the recognition specificities observed in L. usitatissimum. Also, as observed in L. usitatissimum, constructs encoding the full-length AvrL567-A150 and AvrL567-B150 proteins, including the predicted signal peptides, produced much weaker responses than the truncated versions (data not shown).
Coexpression of Resistance and Avirulence Genes in Transgenic Plants Causes Seedling Death
Specific HR induction also has been observed when stably transformed plants expressing Avr products are crossed to plants containing the corresponding R genes, with the progeny showing seedling death or stunted growth phenotypes (Jones et al., 1994; Gopalan et al., 1996; Hammond-Kosack et al., 1998; Erickson et al., 1999). We therefore generated transgenic plants containing the AvrL567127 or AvrL567150 construct in L. usitatissimum var Ward, which contains L9. No visible phenotypes were observed among the primary T0 transgenic plants. For each construct, at least three independent T0 plants were crossed to lines homozygous for the L5, L6, L7, or Lx resistance gene (Lx has an identical specificity to L7 but confers a stronger resistance phenotype; Luck et al., 2000). As a control, each T0 plant was crossed to the universally susceptible line Hoshangabad, and self-fertilized T1 seed were collected. All progeny from these control fertilizations showed normal growth (data not shown). However, progeny resulting from some crosses involving L5, L6, L7, or Lx segregated for seedling lethal or stunted growth phenotypes. For example, Figure 4A shows progeny resulting from crosses involving one T0 plant containing a single (hemizygous) copy of the AvrL567-A150 transgene. No abnormal phenotypes were observed among the self-progeny or the outcross progeny involving the Hoshangabad, L7, or Lx parent. However, in the crosses to the L5 or L6 parent, the progeny segregate for wild-type and stunted phenotypes. Table 2 summarizes the seedling phenotypes observed in progeny of crosses between each of the T0 transgenic plants and the panel of resistant lines. The severity of these phenotypes varied between different outcross families but was consistent within each family. In the most severe cases, thin incompletely filled seeds were produced, and these failed to germinate when placed on wet filter paper (score = 0; Figure 4B). In other cases, apparently normal seeds were produced that germinated in vitro but gave rise to stunted seedlings. When germinated in soil, these seedlings either failed to emerge (score = 1; Figure 4A, L5 cross), were arrested in growth after cotyledon emergence (score = 2; Figure 4A, L6 cross), or gave rise to extremely stunted (score = 3; Figure 4C) or moderately stunted (score = 4; data not shown) dwarf plants. In some crosses, only wild-type progeny (score = 5) were observed. Segregation of wild-type and stunted phenotypes was generally consistent with the 1:1 ratio expected for single locus transgenes, although the sample sizes were small (12 seeds for each cross were assayed).
Figure 4.
Phenotypes Resulting from Crossing AvrL567 Transgenes into L. usitatissimum Lines with Different L Genes.
(A) A single T0 transgenic plant containing one copy of an AvrL567-A150 transgene was crossed to L. usitatissimum lines homozygous for the L5, L6, L7, or Lx resistance gene as well as to the rust susceptible line Hoshangabad (Hosh). Twelve seeds from each cross as well as self-fertilized seeds were planted in soil. Photographs were taken 2 weeks after planting.
(B) Seeds resulting from a cross between a T0 transgenic plant containing one copy of the AvrL567-A127 transgene and a L. usitatissimum line homozygous for L5. Approximately half of the seeds have a shrunken appearance (left) and are unable to germinate, whereas the phenotypically wild-type seeds (right) give rise to normal plants.
(C) Segregation of wild-type and dwarf phenotypes among 3-week-old seedlings resulting from a cross between a T0 transgenic plant containing one copy of the AvrL567-B127 transgene and a L. usitatissimum line homozygous for L6.
Table 2.
Abnormal Growth Phenotypes of Progeny from Crosses between AvrL567-Transformed L. usitatissimum and Various L Gene Lines
| Phenotypesb of Progeny from Crosses to:
|
|||||
|---|---|---|---|---|---|
| Transgene | # T0 Plantsa | L5 | L6 | L7 | Lx |
| AvrL567-A127 | 5 | 0 | 0 | 3 to 4 | 0 |
| AvrL567-A150 | 9 | 1 to 3 | 1 to 5 | 5 | 4 to 5 |
| AvrL567-B127 | 7 | 0 to 3 | 2 to 5 | 5 | 5 |
| AvrL567-B150 | 7 | 1 to 3 | 4 to 5 | 5 | 5 |
| AvrL567-C127 | 3 | 5 | 5 | 5 | 5 |
| AvrL567-C150 | 7 | 5 | 5 | 5 | 5 |
The number of independent transgenic T0 plants used in crosses to L gene lines.
Each T0 plant was crossed to the L5, L6, L7, and Lx lines. Each cross was scored according to the severity of any stunted growth phenotype segregating among the progeny. For each AvrL567 transgene/L gene combination, the range of scores obtained for crosses involving independent T0 plants is shown. Numerical scores represent the following phenotypes: 0, incompletely filled seed, no germination in vitro; 1, seeds germinate in vitro, but seedlings do not emerge from soil; 2, seedling growth arrested after cotyledon emergence; 3, extreme dwarf seedlings; 4, moderate dwarf seedlings; and 5, all progeny have wild-type growth.
The specificity of the interactions observed in this experiment were very similar to those observed in the agroinfiltration transient expression assays (Figure 3, Table 1). Five independent T0 plants containing the AvrL567-A127 transgene each gave rise to nonviable seed when crossed to the L5, L6, or Lx line. A milder stunted phenotype was observed when these plants were crossed to the L7 line (score = 3 to 4). Of seven T0 plants expressing AvrL567-B127, four gave rise to nonviable seed when crossed to L5, whereas the remainder gave rise to extremely stunted progeny (one plant each with a score of 1, 2, or 3). Progeny of these seven T0 plants crossed to L6 showed a range of weaker phenotypes from seedlings arrested after cotyledon emergence (score = 2, one T0 plant) to dwarf seedlings (four T0 plants with a score of 3 and one T0 plant with a score of 4), and one T0 plant gave only wild-type progeny (score = 5). This weaker phenotype in the L6 background compared with L5 was consistently observed for each of the individual T0 plants, with seedling phenotype scores 2 to 3 points higher in the crosses to L6 than in the crosses to L5. No effects were observed when the AvrL567-B127 plants were crossed to L7 or Lx (Table 2). DNA gel blot hybridization and/or PCR analysis of genomic DNA from individual progeny of several crosses showed that the AvrL567 transgene cosegregated with the abnormal growth phenotypes (data not shown).
Just as in the transient assays, constructs encoding the AvrL567-A150 and AvrL567-B150 full-length proteins showed weaker responses than the truncated versions but with a similar relative specificity (Table 2). For instance, nine independent AvrL567-A150 T0 plants gave rise to progeny with a range of severe to mild stunted phenotypes when crossed to L5 or L6, a very mild phenotype when crossed to Lx, and only wild-type progeny when crossed to L7. This weaker interaction was not because of lower transgene expression because we detected equivalent transcript levels in transgenic plants with the full-length and truncated constructs (data not shown). No abnormal phenotypes were observed in crosses involving AvrL567-C127 or AvrL567-C150, although DNA gel blot hybridization showed that these transgenes were inherited in crosses to the L5 and L6 lines. RNA gel blot analysis also showed that the AvrL567-C150 construct was expressed at similar levels to the avirulence allele constructs in the transgenic plants. However, the three AvrL567-C127 transgenic plants showed low expression of this transgene, so the lack of any response in these crosses is not conclusive.
AvrL567 Genes Are Expressed in Haustoria
We examined the expression pattern of the AvrL567 genes in rust by RNA gel blot analysis (Figure 5A). AvrL567 transcripts were detected in RNA samples derived from L. usitatissimum leaves infected with rust strain CH5 but not in RNA from germ tubes of rust spores germinated in vitro. This suggested that AvrL567 expression was induced during infection. On the other hand, the linked Sec14 and orf-3 rust genes and an M. lini β-tubulin gene (Ayliffe et al., 2001) were expressed in germinated spores as well as during infection. AvrL567 transcripts were detected in RNA from leaves infected with rust strains homozygous for either the virulence or avirulence allele (Figure 5A), indicating that the AvrL567-C gene is expressed in rust. The slightly lower amount of transcript detected from the virulent rust is consistent with the presence of only a single AvrL567 gene at this allele compared with the two copies present at the avirulence allele. Because of the low levels of rust RNA present in infected leaves at early time points after infection, we used RT-PCR to examine AvrL567 gene expression at these early stages (Figure 5B). Using this more sensitive assay, we still did not detect any AvrL567 transcript in RNA from rust spores germinated in vitro, but it was detected in leaves 24 h after inoculation. Expression of the rust tubulin and Sec14 genes also was detected by RT-PCR at 24 h after inoculation.
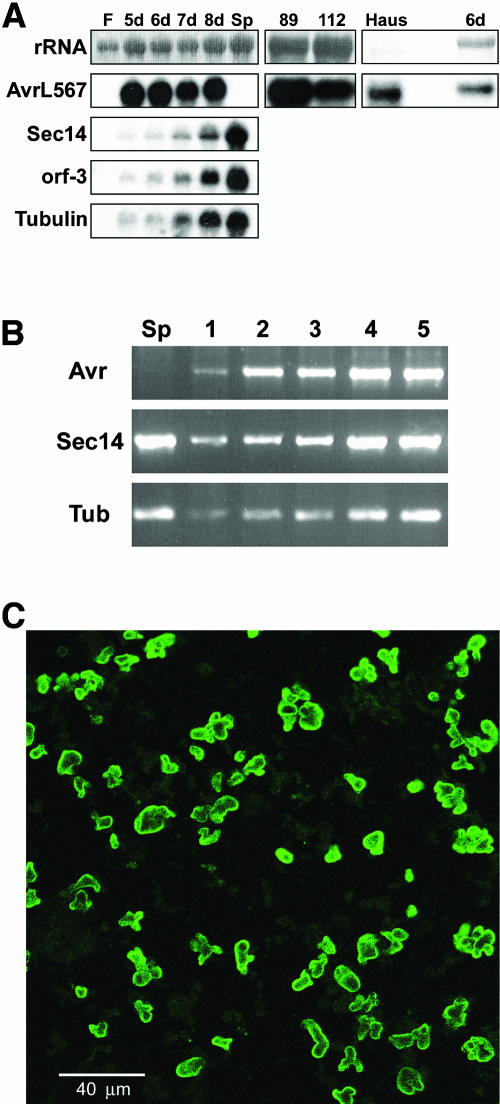
Figure 5.
AvrL567 Transcripts Are Induced during Infection and Are Expressed in Haustoria.
(A) RNA samples (5 μg) from uninfected L. usitatissimum leaves (F), rust CH5–infected L. usitatissimum leaves 5 to 8 d after infection (5d to 8d), and in vitro–germinated CH5 rust spores (Sp) were separated on a 1.5% agarose gel, transferred to nylon membranes, and hybridized with probes for the AvrL567, Sec14, orf-3, and tubulin genes of M. lini. Note that the infected leaf samples contain largely L. usitatissimum RNA with an increasing proportion of rust RNA as the fungal biomass increases during infection. The AvrL567 probe also was hybridized to RNA from leaves infected with rust strain CH5-F2-89 (homozygous for the avirulence allele) or CH5-F2-112 (homozygous for the virulence allele). RNA extracted from 50 mg of haustoria purified from L. usitatissimum leaves 6 d after infection with rust CH5 (Haus) was run alongside 2.5 μg of RNA from rust CH5 infected leaves (6d) and hybridized to the AvrL567 probe. RNA filters were stained with methylene blue to detect rRNA loading (top panel). Given the low amount of haustorial RNA, the AvrL567 signal in haustoria reflects a considerable enrichment compared with the infected leaf RNA.
(B) RNA from in vitro–germinated rust spores (Sp) and infected L. usitatissimum leaves after 1 to 5 d was reversed transcribed and amplified by PCR using primers specific for the AvrL567 (Avr), Sec14, or tubulin (Tub) gene. RT-PCR products were separated on a 2% low melting point agarose gel and visualized by ethidium bromide staining and UV irradiation.
(C) Haustoria isolated from L. usitatissimum leaves 6 d after infection were fixed on glass slides and labeled with monoclonal antibody mL1 (Murdoch et al., 1998), followed by a goat anti-mouse secondary antibody coupled to the fluorophore Alexa-488 and visualized by fluorescence under illumination by 488-nm light.
The HR induced by avirulent M. lini strains on L. usitatissimum plants containing L6 first occurs in plant cells that contain a haustorium (Kobayashi et al., 1994), suggesting that avirulence genes are likely to be expressed in these structures. We used affinity chromatography with a Sepharose concanavalin A column (Hahn and Mendgen, 1992) to purify haustoria from L. usitatissimum leaves infected with rust CH5. The presence of intact haustoria in these samples was confirmed by microscopy and immunolabeling with a monoclonal antibody that detects polysaccharide epitopes present on the haustorial surface (Figure 5C; Murdoch et al., 1998). No contaminating plant cells or fungal mycelia were observed in the purified haustorial samples, although some chloroplasts were present. The AvrL567 transcript was highly abundant in RNA isolated from the purified haustoria (Figure 5A). A cDNA library prepared from the purified M. lini haustorial RNA was screened with an AvrL567 probe, which identified 4, 2, and 13 cDNA clones derived from AvrL567-A, AvrL567-B, and AvrL567-C, respectively, indicating that each of the three genes is expressed in haustoria.
Evidence for Diversifying Selection at the AvrL567 Locus
Because the three AvrL567 proteins were highly polymorphic (Figure 2B), we examined the patterns of nucleotide variation at the AvrL567 locus for evidence of positive selection. We aligned the 7357-bp DNA sequences surrounding each of the three AvrL567 gene homologs, which correspond to the region duplicated in the AvrL567-A and AvrL567-B alleles (Figure 2A). The nucleotide variation in this region was highly concentrated within the 450-bp AvrL567 coding sequence. A total of 30 single nucleotide differences, giving rise to 27 amino acid differences, occur in the coding region (average pairwise distance = 0.049). By contrast, only 25 single nucleotide changes occur in the 6907 bp of flanking sequence (average pairwise distance = 0.0022). This difference is highly significant (t test; P < 0.001) and suggests that selection has contributed to the accumulation of sequence differences within the coding sequences. There is also an excess of nucleotide changes at nonsynonymous sites (average distance = 0.061) over synonymous sites (average distance = 0.007) in the AvrL567 genes (ratio = 8.6). A G-test showed that this difference was significant for the pairwise comparisons between AvrL567-A and AvrL567-B (P < 0.04; 19 substitutions in 341 nonsynonymous sites and 1 substitution in 109 synonymous sites) and between AvrL567-A and AvrL567-C (P < 0.002; 27/341 and 0/109) but not between AvrL567-B and AvrL567-C (P < 0.08; 15/342 and 1/108). (Zhang et al., 1997 recommended a G-test for significance when there are fewer than 10 substitutions at one class of site because the t test overestimates the significance value under these conditions.) This indicates that selection has favored the accumulation of amino acid variation in these genes (Hughes and Nei, 1988).
The only other significant variation between these sequences occurs in the AvrL567 promoter region. Each gene contains either 12 (AvrL567-A) or 13 (AvrL567-B and AvrL567-C) copies of an 82-bp repeated sequence (Figure 2A). The last of these repeats ends ∼150 bp from the putative transcription start site. There is also one 13-bp insertion/deletion polymorphism that occurs 750 bp downstream of the AvrL567 coding sequence. The DNA sequences flanking the 7357-bp duplication also are highly conserved between the two alleles, with only 10 nucleotide differences in the 3200-bp 5′ region and five differences in the 2460-bp 3′ region (distance = 0.0026). The only major difference is the insertion of a 6270-bp sequence in the 5′ flanking region of the avirulence allele (Figure 2A), which includes 173-bp identical direct repeats and contains an open reading frame that is closely related to gypsy class retrotransposon pol protein sequences from Tricholoma matsutake, Magnaporthe grisea, and Fusarium oxysporum.
DISCUSSION
We have isolated genes previously from four major rust resistance loci in L. usitatissimum (Lawrence et al., 1995; Anderson et al., 1997; Dodds et al., 2001a, 2001b), but the corresponding avirulence genes in M. lini had not been identified. We now have identified M. lini avirulence genes corresponding to the L5, L6, and L7 alleles of the L resistance locus in L. usitatissimum. The AvrL567-A and AvrL567-B genes cosegregate with the avirulence phenotype in an F2 family of 74 individuals and cause R gene–dependent cell death when expressed in transiently transformed leaves of L. usitatissimum (Figures 3A and 3B, Table 1) and N. tabacum (Figure 3C). Induction of cell death occurred when the AvrL567 genes were coexpressed with L5, L6, or L7 but not other L locus resistance genes. Importantly, the AvrL567-C gene, which segregates with the virulence allele, did not cause necrosis with any of the L resistance genes. In addition, a stunted growth phenotype was observed when the corresponding R and Avr genes were brought together by crossing transgenic L. usitatissimum expressing AvrL567 genes to resistant lines (Figure 4, Table 2). Although we have not been able to directly test the phenotype conferred by these genes in rust, these results provide compelling evidence that the AvrL567-A and AvrL567-B genes are responsible for recognition by the L5, L6, and L7 resistance genes.
AvrL567 Proteins Are Secreted from Rust Haustoria and Recognized in Plant Cells
Haustoria are the primary site of contact between rust pathogens and host mesophyll cells. Rust infection involves germination of the rust spore, growth of the germ tube on the leaf surface, and then appresorium formation over a stoma to allow penetration of the leaf. Inside the leaf, a substomatal vesicle forms and growth of infection hyphae leads to differentiation of a haustorial mother cell that extends a haustorium into a leaf mesophyll cell. In rust-infected L. usitatissimum, the first haustoria appear ∼12 h after infection, and by 24 h, haustoria have formed at almost all penetration sites (Kobayashi et al., 1994). In L6-mediated resistance, hypersensitive cell death is induced in cells containing developing haustoria by 24 h after infection (Kobayashi et al., 1994). The AvrL567 transcripts were not detected in germ tubes produced by in vitro germinated rust spores but were induced during infection within 24 h (Figures 5A and 5B). The relative enrichment of the AvrL567 transcript in the isolated haustoria samples (Figure 5A) suggests that these genes may be predominantly expressed in haustoria, although we do not know whether there is expression in other rust cell types present in infected leaves. These observations are consistent with the timing and location of HR induction in resistant L6 plants.
The AvrL567 protein sequences contain predicted signal peptides (Figure 2B), which can direct secretion of a reporter protein in yeast (A.-M. Catanzariti, unpublished results). Thus, we predict that these avirulence products are secreted from M. lini haustoria into the surrounding extrahaustorial matrix. In planta expression of the AvrL567-A127 and AvrL567-B127 proteins, which lack the signal peptide, induced strong necrotic responses in conjunction with the corresponding L resistance genes (Figures 3 and 4, Tables 1 and 2). Thus, recognition of the AvrL567 products occurs when they are expressed within plant cells, which is consistent with the predicted location of the L resistance proteins. However, intracellular recognition during infection would require translocation of the secreted AvrL567 proteins across the plant plasma membrane, which separates the extrahaustorial matrix from the host cytoplasm. Similar observations for bacterial Avr proteins provided the first indication that these products were translocated directly into plant cells (Gopalan et al., 1996; Leister et al., 1996; Scofield et al., 1996; Tang et al., 1996; van den Ackervecken et al., 1996). Other known rust resistance genes in L. usitatissimum (Lawrence et al., 1995; Anderson et al., 1997; Dodds et al., 2001a, 2001b), maize (Collins et al., 1999), and H. vulgare (Brueggeman et al., 2002) also encode predicted cytoplasmic proteins, so it is possible that a variety of rust Avr products are targeted to the host cytoplasm. The AvrL567 proteins may represent a class of rust effector proteins that are translocated into plant cells to facilitate infection during a compatible infection.
No specific translocation mechanism to direct effector proteins into host cells has been described in fungal pathogens. Although Avr products identified in other plant pathogenic fungi also are secreted proteins (Lauge and de Wit, 1998), the Avr2, Avr4, and Avr9 products of Cladosporium fulvum (Parniske et al., 1997; Luderer et al., 2002) and the Rhyncosporium secalis NIP1 protein (Knogge, 1996) are recognized extracellularly and probably do not enter plant cells. Delivery of the Avr1b protein from the oomycete pathogen Phytophthora sojae to the extracellular space of Glycine max (soybean) leaves induced R gene–dependent cell death (Tyler 2002), suggesting that it also may be recognized extracellularly. On the other hand, the M. grisea–secreted protein AVR-PITA is recognized via a direct interaction with a putatively cytoplasmic R protein from rice (Oryza sativa) (Jia et al., 2000). However, infection hyphae of M. grisea penetrate plant cells directly, and no host membrane separating the fungus from the plant cytoplasm has been described.
AvrL567 Proteins Have Overlapping Recognition Specificities
The cosegregation of avirulence on L5, L6, and L7 has suggested that multiple avirulence genes may occur at this locus. However, although there are two related avirulence genes at this locus in the CH5-F2 family, these genes show overlapping recognition specificities. AvrL567-A is recognized by all three resistance genes, whereas AvrL567-B is recognized most strongly by L5 but only weakly by L6 and not at all by L7 (Tables 1 and 2, Figure 3B). Similar overlapping recognition specificities have been observed in other gene-for-gene resistance systems. For instance, the RPM1 resistance protein in Arabidopsis recognizes two unrelated avirulence products from P. syringae, AvrRpm1 and AvrB (Grant et al., 1995), and these two Avr proteins are recognized by distinct R genes in G. max (Ashfield et al., 1995).
We speculated previously that the L6 and L7 resistance proteins may recognize the same or very similar avirulence products because there are only 11 amino acid differences between these proteins, all in the N-terminal TIR domain, and just three of these changes, in the recombinant Lx protein, are sufficient to alter L6 to express the L7 specificity (Ellis et al., 1999; Luck et al., 2000). Furthermore, these specificities are only distinguished by a separate inhibitor gene (I) from M. lini, which prevents L7- and Lx-mediated resistance but has no effect on L6-mediated resistance (Lawrence et al., 1981). The difference between the L6 and L7 specificities in our assays with the AvrL567 genes seems to be largely one of degree. L6 interacts strongly with AvrL567-A but weakly with AvrL567-B, whereas L7 interacts weakly with AvrL567-A but not with AvrL567-B (Tables 1 and 2, Figure 3B). This is consistent with the weaker resistance phenotype conferred by L7, which allows the formation of small pustules, relative to L6, which completely restricts rust growth (Islam and Mayo, 1990). The recombinant Lx gene gives a strong resistance phenotype in infected plants and also gives stronger recognition of AvrL567-A in transgenic plants than L7 (Table 2) but is still completely inhibited by the I gene. It is not clear how the I gene product may inhibit L7 and Lx resistance, although it apparently does not affect the expression of AvrL567 genes because similar transcript levels accumulate in rust strains with or without I (P.N. Dodds, unpublished results). One intriguing possibility is that the I gene product may interfere directly with the L7 and Lx proteins through their TIR domains to block signaling. Alternatively, the AvrL567 products may be altered in a way that prevents recognition by L7 and Lx but not L6.
The overlap in recognition properties between L5 and L6 was more surprising because there are 104 amino acid differences between these proteins, which are among the most highly diverged of the L protein family. There are seven other rust resistance specificities encoded by the known alleles of the L. usitatissimum L gene, and the corresponding Avr genes in M. lini map to seven independent loci. However, hybridization of AvrL567 probes to M. lini genomic DNA did not detect any related sequences even at low stringency (data not shown). Therefore, despite the high sequence similarity of these L gene alleles, the corresponding Avr genes are not closely related to the AvrL567 genes. The overlap between the L5 and L6 recognition specificities may be a result of convergent evolution. Although all of the rusts in our collection show the same reaction on both L5 and L6, Flor (1955) described a rust strain that was avirulent on plants containing L6 but virulent on L5 plants. Thus, some rust strains may carry variants of AvrL567 that can distinguish between L5 and L6. The weaker recognition of AvrL567-B by L6 compared with L5 in our expression assays provides some evidence for differential recognition. In fact, one AvrL567 gene variant, isolated from a rust strain unrelated to strains C and H, is recognized by L6 but not L5 in the transient expression assay (P.N. Dodds, unpublished results).
Another question we have considered is how the virulent rust strains carrying AvrL567-C escape recognition. This does not appear to be because of differences in gene expression because AvrL567 transcripts were observed at similar levels in both virulent and avirulent rust strains (Figure 5A), and a cDNA library from rust CH5 haustoria contained representatives of all three AvrL567 genes. The lack of any significant sequence divergence outside the coding sequences of the AvrL567 genes also would argue against differential expression of these genes. The AvrL567-C expression constructs in our transient expression assays were identical to the AvrL567-A and AvrL567-B constructs outside of the coding sequence and generated similar amounts of mRNA. Thus, the lack of recognition of AvrL567-C in these assays suggests that the virulence phenotype is conferred by the differences in the coding sequence rather than differences in gene expression or protein modifications occurring in the rust. There are 10 polymorphic amino acids unique to the AvrL567-C protein (Figure 2B) that may explain its lack of recognition by any known R gene in L. usitatissimum. One possibility is that these changes may result in reduced protein stability. However, both the AvrL567-A and AvrL567-C proteins can accumulate to high levels in Escherichia coli when expressed with a 6xHis tag (A.-M. Catanzariti, unpublished results). An alternative possibility is that the AvrL567-C protein is stable, but the amino acid changes interfere with recognition by the resistance proteins. In this scenario, AvrL567-C may retain the presumed pathogenicity function performed by this family of proteins in M. lini.
Adaptive Evolution of the AvrL567 Genes
Evolution of the AvrL567 genes has been characterized by strong positive selection for amino acid variation between the encoded products. This is shown by the significant excess of nucleotide variation within the AvrL567 coding sequence relative to the flanking DNA as well as the evidence for diversifying selection acting in these genes. This could be explained by selective pressure for the pathogen to avoid recognition-triggered resistance in the host and is consistent with our interpretation that the lack of AvrL567-C recognition is attributable to the amino acid differences from AvrL567-A and AvrL567-B. The observation that diversifying selection acts on the AvrL567 genes as well as on the L. usitatissimum L genes (Dodds et al., 2000) is consistent with an arms race model of coevolution between R and Avr genes driven by selection for resistance in the host and virulence in the pathogen (Bergelson et al., 2001). This system provides the first example in which evidence for such adaptive evolution has been found in both components of an R/Avr gene pair. Although R genes impose an obvious selective pressure on pathogen Avr genes and host resistance strongly influences pathogen virulence in natural populations of M. lini on the wild flax species L. marginale (Thrall and Burdon, 2003), it is possible that other factors have driven the AvrL567 gene diversification. For instance, if, like some bacterial Avr proteins (Lahaye and Bonas, 2001; Axtell et al., 2003; Mackey et al., 2003; Shao et al., 2003), the AvrL567 proteins have an effector role in rust infection that involves targeting a specific host protein(s), an alternative arms race independent of R genes could occur between AvrL567 and the host gene(s) encoding this target. In this case, variation in the host gene that prevents targeting may select for changes in the pathogen Avr gene to re-engage the host target.
Conservation of L6/AvrL567-A Recognition and HR Induction in N. tabacum
Many resistance proteins have been shown to function in heterologous plant species to trigger a defense response that is dependent on their corresponding Avr products (Thilmony et al., 1995; Whitham et al., 1996; Hammond-Kosack et al., 1998; Tai et al., 1999; van der Hoorn et al., 2000). However, most of these examples involve R proteins transferred to closely related species, and transfer to species outside the original family has generally not been successful, with a few exceptions. For instance, the L. esculentum Cf resistance proteins can function in some more distantly related species because Cf-4/Avr4 recognition induces an HR in L. sativa (van der Hoorn et al., 2000), and Cf-9/Avr9 recognition induces an HR in Brassica (Hennin et al., 2001). The Arabidopsis RPW8.1 and RPW8.2 powdery mildew resistance genes also confer resistance to powdery mildew pathogens when expressed in N. tabacum (Xiao et al., 2003). We have found that the recognition interaction between the L6 and AvrL567 proteins can occur in N. tabacum cells and results in HR induction. Thus, if any other host proteins are involved in this recognition interaction as suggested by the guard hypothesis (Dangl and Jones, 2001), then these proteins must be sufficiently conserved between L. usitatissimum and N. tabacum to support the same recognition events. Alternatively, the L6 and AvrL567 products may interact directly. Furthermore, this observation indicates that the L6 protein can interact productively with the N. tabacum resistance-signaling pathway.
METHODS
Rust and Plant Material
An M. lini F2 family derived from selfing rust strain CH5, the F1 hybrid of a cross between parental strains C and H, was described by Lawrence et al. (1981). Near-isogenic L. usitatissimum lines containing the L, L1, L2, L3, L4, L5, L6, L7, L8, or L10 allele backcrossed for 6 to 12 generations into L. usitatissimum var Bison, which contains L9, were described by Flor (1954). Rust inoculations were performed as described by Lawrence et al. (1981). Rust spores were germinated overnight on water, and genomic DNA was isolated from the resulting mycelial mat as described (Anderson et al., 1997).
Isolation of Rust cDNAs
RNA was isolated from leaves of rust-susceptible L. usitatissimum var Hoshangabad 6 d after inoculation with urediospores of rust strain C or H or a mock treatment. Poly(A)+ RNA was isolated from total RNA using a PolyAtract kit (Promega, Madison, WI) according to the manufacturer's instructions. Double-stranded cDNA was synthesized using a Superscript Choice cDNA synthesis kit (Gibco BRL, Rockville, MD). Subtracted cDNA libraries enriched for cDNAs more abundant in the tester than driver samples were prepared using suppression subtractive hybridization as described by Diatchenko et al. (1996) using an Advantage cDNA polymerase kit (Clontech Laboratories, Palo Alto, CA) and cloned into pGEMT-Easy (Promega). For the infection-specific library, the tester cDNA was prepared from leaves infected with rust stain H for 6 d, and the driver was from uninfected L. usitatissimum leaves. For the differential library, the driver was prepared from leaves infected with rust strain C for 6 d.
Gel Blot Analysis
Restriction enzyme–digested genomic DNA was separated on 1.0% agarose gels and transferred to Hybond N+ nylon membranes (Amersham, Buckinghamshire, UK). RNA samples were separated on 1.5% agarose gels and transferred to Hybond N+. Prehybridization and hybridization with 32P-dCTP–labeled DNA probes were performed in 7% (w/v) SDS, 1% (w/v) BSA, 0.5 M sodium phosphate, pH 7.2, and 1 mM EDTA at 65°C, and washing was in 1× SSC and 0.1% SDS at 65°C.
Isolation of Genomic Sequences
Lambda clones hybridizing to the IU2F2 cDNA probe were isolated from a λEMBL3 genomic DNA library prepared from rust strain CH5 (Ayliffe et al., 2001). A sequence contig of 26.5 kb was assembled by shotgun cloning and direct sequencing of overlapping λ clones. A second contig of 11 kb was assembled by direct sequencing of PCR products amplified from the F2 rust CH5F2-112 (homozygous for the virulence allele). The Perkin-Elmer/Applied Biosystems GeneAmp XL PCR kit was used (Roche Molecular Systems, Branchburg, NJ) with thermal cycling conditions as follows: 94°C for 2 min, 40 cycles of 94°C for 20 s, 55°C for 30 s, 72°C for 5 min, and then a final 10 min at 72°C. The primer pairs 10-1.8 (5′-TGCCTTGAGCCGGTGATTC-3′) and 10-1.16 (5′-TAATCCTCGTTGACATCAGTC-3′), 10-1.15 (5′-AAGCTTGAGAGCTCCGCTC-3′) and 10-1.9 (5′-GTCTCTTCGTCCTTCCAAG-3′), and 10-1.4 (5′-GTCGACCGATCTATATCGAG-3′) and 10-1.37 (5′-CGAGCATTTGTAGGCATTTGTC-3′) were used to generate overlapping PCR fragments of 4.5, 3.3, and 4.5 kb, respectively. These were purified using the QIAquick PCR purification kit (Qiagen, Clifton Hill, Australia) and sequenced directly. An additional 500-bp region at the 3′ end of this contig was identified by inverse PCR. Genomic DNA from the rust CH5F2-78 (homozygous for the virulent allele) was digested with EcoRV (MBI Fermentas, Vilnius, Lithuania) and religated with T4 DNA ligase (2 units; MBI Fermentas). The circularized DNA was amplified by PCR (94°C for 2 min, 40 cycles of 94°C for 20 s, 55°C for 30 s, 72°C for 2 min, and 72°C for 10 min) with the primer pairs 10-1.16 and 10-1.40 (5′-AGCGATTATAAAATGAGACAAG-3′) and then reamplified in a nested PCR with the primers 10-1.6 (5′-TTGCATGGATCCCACCAAG-3′) and 10-1.39 (5′-CACTGGAATTGATTTCAATCAC-3′). An amplified fragment of ∼800 bp was cloned into pGEM T-Easy (Promega), and four independent clones were sequenced.
Transcript Analysis by RT-PCR
Total RNA was reverse transcribed using Superscript reverse transcriptase (Gibco BRL) with an oligo(dT)25 primer and then amplified by Taq polymerase with the following thermal profile: 94°C for 2 min, 38 cycles of 94°C for 20 s, 55°C for 30 s, 72°C for 1 min, and 72°C for 5 min. AvrL567 transcripts were amplified with the primers 10-1.15 and 10-1.16, Sec14 transcripts with 10-1.4 and 10-1.9, and M. lini tubulin transcripts with tub10 (5′-AAACACTAAATCAAACATGAGGG-3′) and tub12 (5′-ACAAAGAACCAAAAGGACCCGA-3′). Each primer set spans one or more introns to distinguish cDNA and genomic DNA derived products. AvrL567 RT-PCR products from rust H–infected leaves were cloned into pGEMT-Easy, and 21 independent clones were sequenced to confirm the predicted coding regions and splicing sites.
Transient Expression Assays
Gene expression constructs contained the AvrL567 coding sequences inserted between a 35S promoter of Cauliflower mosaic virus and a nopaline synthase terminator in the binary vector pTNotTReg (Anderson et al., 1997). Constructs encoding the truncated AvrL567127 proteins, lacking the signal peptide, begin at a BamHI site at position 59 relative to the first ATG, have an A-to-T alteration at position 65 to remove an out-of-frame ATG codon, and include 45 nucleotides 3′ to the stop codon. The open reading frame starts at Met-24, the first amino acid of the expected mature AvrL567 proteins. Constructs encoding the full-length AvrL567150 proteins contain the entire coding sequences as well as 105 nucleotides 5′ to the first ATG. All constructs were fully sequenced to confirm their integrity. A. tumefaciens strains GV3101-pMP90 or C58C1 (which lacks the Ti plasmid) containing the binary vector expression constructs were prepared at an OD600 of 1.0 in liquid MS medium containing 200 μM acetosyringone and infiltrated into L. usitatissimum or N. tabacum leaves. For coinfiltration, equal volumes of two A. tumefaciens cultures containing the AvrL567 gene constructs (at OD600 = 1.0) or an L6 gene construct (Lawrence et al., 1995; at OD600 = 1.0 for L. usitatissimum or 0.5 for N. tabacum) were mixed together before infiltration. For control infiltrations of the AvrL567 or L6 constructs alone, these cultures were mixed with an A. tumefaciens culture containing the empty binary vector in similar proportions.
L. usitatissimum Transformation and Crosses
Transformation of the L. usitatissimum line Ward was as described by Anderson et al. (1997), except that the selective agent spectinomycin was used at 50 μg/mL. Genomic DNA was extracted from T0 plants using DNAzol reagent (Molecular Research Center, Cincinnati, OH) and analyzed by gel blot hybridization to determine transgene copy number. Selected plants containing one to three intact copies of the transgene were used as the female parents in crosses to L. usitatissimum lines carrying various L resistance genes. Flowers were emasculated before maturity and then hand pollinated. Twelve seeds from each cross were planted in soil to examine the phenotypes of the resultant progeny. Seeds from some crosses also were germinated in Petri dishes on wet filter paper.
Isolation of Haustoria
Haustoria were isolated from L. usitatissimum leaves 6 d after inoculation with rust strain CH5 by affinity chromatography as described by Hahn and Mendgen (1992), except that an 11-μM pore size nylon mesh was used to remove plant cell material from the crude preparation. An affinity column was prepared by covalently attaching concanavalin A (Pharmacia Biotech, Uppsala, Sweden) to cyanogen bromide–activated Sepharose 6MB (Pharmacia Biotech) as described in the manufacturer's protocol. Samples of purified haustoria were examined by immunofluorescence labeling with monoclonal antibody mL1 as described by Murdoch et al. (1998), except that samples were fixed to slides in 0.5% gelatin, 0.05% chromic potassium sulfate, and 0.02% NaN3, and the secondary antibody used was the goat anti-mouse coupled to the fluorophore Alexa 488 (Molecular Probes, Eugene, OR). Slides were examined on a Leica TCS SP2 confocal microscope (Leica, Mannheim, Germany) under light or under excitation by an argon ion laser at a wavelength of 488 nm. Images were collected at 500 to 530 nm with pseudocolor green. RNA was prepared from purified haustoria using the Qiagen Plant RNeasy kit. A cDNA library was prepared from 2 μg total RNA from haustoria using the SMART cDNA library kit (Clontech) according to the manufacturer's instructions.
Sequence Analysis
The AvrL567 coding sequence or flanking sequences were aligned, and nucleotide sequence distances were calculated across all sites or specifically for nonsynonymous and synonymous sites using the Jukes-Cantor algorithm of the Molecular Evolutionary Genetics Analysis software version 1.02 (Kumar et al., 1993). The significance of differences between average pairwise nucleotide distances was assessed by a t test, whereas a G-test was used to test the significance of differences in the number of nucleotide changes at nonsynonymous and synonymous sites in individual pairwise comparisons.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY510102 and AY510103.
Acknowledgments
Valerie Ryle, Patricia Atkinson, and Diana Hall provided excellent technical assistance. The ML1 mAb was kindly provided by Adrienne Hardham (Research School of Biological Science, Australian National University, Canberra, Australia). This research was supported by grants from the Grains Research and Development Corporation. A.-M.C. was supported by an Australian postgraduate award.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Peter N. Dodds (peter.dodds@csiro.au).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.020040.
References
- Anderson, P.A., Lawrence, G.J., Morrish, B.C., Ayliffe, M.A., Finnegan, E.J., and Ellis, J.G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield, T., Keen, N.T., Buzzell, R.I., and Innes, R.W. (1995). Soybean resistance genes specific for different Pseudomonas syringae avirulence genes are allelic, or closely linked. Genetics 141, 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., Chisholm, S.T., Dahlbeck, D., and Staskawicz, B.J. (2003). Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 49, 1537–1546. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance is coupled to AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. [DOI] [PubMed] [Google Scholar]
- Ayliffe, M.A., Dodds, P.N., and Lawrence, G.J. (2001). Characterisation of the beta-tubulin genes from Melampsora lini and comparison of fungal beta-tubulin genes. Mycol. Res. 105, 818–826. [Google Scholar]
- Ayliffe, M.A., Roberts, J.K., Mitchell, H.J., Zhang, R., Lawrence, G.J., Ellis, J.G., and Pryor, T.J. (2002). A plant gene upregulated at rust-infection sites. Plant Physiol. 129, 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis, V.A., Malehorn, D.E., Emr, S.D., and Greene, R. (1989). The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for the transport of secretory proteins from the yeast Golgi complex. J. Cell Biol. 108, 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane, A., Kanyuka, K., and Baulcombe, D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson, J., Kreitman, M., Stahl, E.A., and Tian, D. (2001). Evolutionary dynamics of plant R-genes. Science 292, 2281–2284. [DOI] [PubMed] [Google Scholar]
- Boyes, D.C., Nam, J., and Dangl, J.L. (1998). The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 95, 15849–15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggeman, R., Rostoks, N., Kudrna, D., Kilian, A., Chen, J., Druka, A., Steffenson, B., and Kleinhofs, A. (2002). The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc. Natl. Acad. Sci. USA 99, 9328–9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper-Lindley, C., Dahlbeck, D., Clark, E.T., and Staskawicz, B.J. (2002). Direct biochemical evidence for type III secretion-dependent translocation of the AvrBs2 effector protein into plant cells. Proc. Natl. Acad. Sci. USA 99, 8336–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, N., Drake, J., Ayliffe, M., Sun, Q., Ellis, J., Hulbert, S., and Pryor, T. (1999). Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11, 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley, M.B., Pathirana, S., Wu, H.-J., Kachroo, P., and Klessig, D.F. (2000). Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Deslandes, L., Olivier, J., Peeters, N., Feng, D.X., Khounlotham, M., Boucher, C., Somssich, I., Genin, S., and Marco, Y. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100, 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko, L., Lau, Y.F., Campbell, A.P., Chenchik, A., Moqadam, F., Huang, B., Lukyanov, S., Lukyanov, K., Gurskaya, N., Svberdlov, E.D., and Siebert, P.D. (1996). Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93, 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, M.S., Hatzixanthis, K., Jones, D.A., Harrison, K., and Jones, J.D.G. (1998). The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10, 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, M.S., Jones, D.A., Keddie, J.S., Thomas, C.M., Harrison, K., and Jones, J.D.G. (1996). The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84, 451–459. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Pryor, A.J., and Ellis, J.G. (2000). Genetic analysis and evolution of plant disease resistance genes. In Molecular Plant Pathology, M. Dickinson and J. Beynon, eds (Sheffield, UK: Sheffield Academic Press), pp. 88–107.
- Dodds, P.N., Lawrence, G.J., and Ellis, J.G. (2001. a). Contrasting modes of evolution acting on the complex N locus for rust resistance in flax. Plant J. 27, 439–453. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Pryor, T., and Ellis, J.G. (2001. b). Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J.G., Lawrence, G.J., Luck, J.E., and Dodds, P.N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, F.L., Holzberg, S., Calderon-Urrea, A., Handley, V., Axtell, M., Corr, C., and Baker, B. (1999). The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 18, 67–75. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1954). Seed-flax improvement. III. Flax rust. Adv. Agron. 6, 152–161. [Google Scholar]
- Flor, H.H. (1955). Host-parasite interaction in flax rust – Its genetic and other implications. Phytopathol. 45, 680–685. [Google Scholar]
- Flor, H.H. (1956). Mutations in flax induced by ultraviolet radiation. Science 124, 888–889. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Gopalan, S., Bauer, D.W., Alfano, F.R., Loniello, A.O., He, S.Y., and Collmer, A. (1996). Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell 8, 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M.R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R.W., and Dangl, J.L. (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269, 843–846. [DOI] [PubMed] [Google Scholar]
- Hahn, M., and Mendgen, K. (1992). Isolation by ConA binding of haustoria from different rust fungi and comparison of their surface qualities. Protoplasma 170, 95–103. [Google Scholar]
- Hahn, M., and Mendgen, K. (2001). Signal and nutrient exchange at biotrophic plant-fungus interfaces. Curr. Opin. Plant Biol. 4, 322–327. [DOI] [PubMed] [Google Scholar]
- Halterman, D., Zhou, F., Wei, F., Wise, R.P., and Schulze-Lefert, P. (2001). The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 25, 335–348. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., Tang, S., Harrison, K., and Jones, J.D.G. (1998). The tomato Cf-9 disease resistance gene functions in tobacco and potato to confer responsiveness to the fungal avirulence gene product Avr9. Plant Cell 10, 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, M.C. (1997). Signalling between pathogenic rust fungi and resistant or susceptible host plants. Ann. Bot. 80, 713–720. [Google Scholar]
- Hennin, C., Hofte, M., and Diedrichsen, E. (2001). Functional expression of Cf9 and Avr9 genes in Brassica napus induces enhanced resistance to Leptospheria maculans. Mol. Plant Microbe Interact. 14, 1075–1085. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L., and Nei, M. (1988). Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335, 167–170. [DOI] [PubMed] [Google Scholar]
- Hwang, C.-F., and Williamson, V.M. (2003). Leucine-rich repeat-mediated intramolecular interactions in nematode recognition and cell death signaling by the tomato resistance protein Mi. Plant J. 34, 585–593. [DOI] [PubMed] [Google Scholar]
- Islam, M.R., and Mayo, G.M.E. (1990). A compendium on host genes in flax conferring resistance to flax rust. Plant Breed. 104, 89–100. [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., Thomas, C.M., Hammond-Kosack, K.E., Balint-Kurti, R.J., and Jones, J.D.G. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793. [DOI] [PubMed] [Google Scholar]
- Knogge, W. (1996). Fungal infection of plants. Plant Cell 8, 1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, I., Kobayashi, Y., and Hardham, A.R. (1994). Dynamic reorganisation of microtubules and microfilaments in flax cells during the resistance response to flax rust infection. Planta 195, 237–247. [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (1993). MEGA: Molecular Evolutionary Genetics Analysis, version 1.02. http://evolgen.biol.metro-u.ac.jp/MEGA/. [DOI] [PubMed]
- Lahaye, T., and Bonas, U. (2001). Molecular secrets of bacterial type III effector proteins. Trends Plant Sci. 6, 479–485. [DOI] [PubMed] [Google Scholar]
- Lauge, R., and de Wit, P.J. (1998). Fungal avirulence genes: Structure and possible functions. Fungal Genet. Biol. 24, 285–297. [DOI] [PubMed] [Google Scholar]
- Lawrence, G.J., Finnegan, E.J., Ayliffe, M.A., and Ellis, J.G. (1995). The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene Rps2 and the tobacco viral resistance gene N. Plant Cell 7, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, G.J., Mayo, G.M.E., and Shepherd, K.W. (1981). Interactions between genes controlling pathogenicity in the flax rust fungus. Phytopathol. 71, 12–19. [Google Scholar]
- Leister, R.T., Ausubel, F.M., and Katagiri, F. (1996). Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc. Natl. Acad. Sci. USA 93, 15497–15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, M.C., Nicaud, J.M., Skinner, H.B., Vergnolle, C., Kader, J.C., Bankaitis, V.A., and Gaillardin, C. (1994). A phosphatidylinositol/phosphatidylcholine transfer protein is required for differentiation of the dimorphic yeast Yarrowia lipolytica from the yeast to the mycelial form. J. Cell Biol. 125, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck, J.E., Lawrence, G.J., Dodds, P.N., Shepherd, K.W., and Ellis, J.G. (2000). Regions outside of the leucine-rich repeats of flax rust resistance proteins have a role in specificity determination. Plant Cell 12, 1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderer, R., Takken, F.L., de Wit, P.J., and Joosten, M.H. (2002). Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 45, 875–884. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonsos, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopisis. Cell 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Mellersh, D.G., and Heath, M.C. (2001). Plasma membrane-cell wall adhesion is required for expression of plant defense responses during fungal penetration. Plant Cell 13, 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffet, P., Farnham, G., Peart, J., and Baulcombe, D.C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21, 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch, L.J., Kobayashi, I., and Hardham, A.R. (1998). Production and characterisation of monoclonal antibodies to cell wall components of the flax rust fungus. Eur. J. Plant Path. 104, 331–346. [Google Scholar]
- Noel, L., Moores, T.L., van der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D.G. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11, 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Orth, K., Xu, Z., Mudgett, M.B., Bao, Z.Q., Palmer, L.E., Bliska, J.B., Mangel, W.F., Staskawicz, B.J., and Dixon, J.E. (2000). Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290, 1594–1597. [DOI] [PubMed] [Google Scholar]
- Parniske, M., Hammond-Kosack, K.E., Golstein, C., Thomas, C.M., Jones, D.A., Harrison, K., Wulff, B.B.H., and Jones, J.D.G. (1997). Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91, 821–832. [DOI] [PubMed] [Google Scholar]
- Pedersen, C., Rasmussen, S.W., and Giese, H. (2002). A genetic map of Blumeria graminis based on functional genes, avirulence genes and molecular markers. Fungal Genet. Biol. 35, 235–246. [DOI] [PubMed] [Google Scholar]
- Rehmany, A.P., Grenville, L.J., Gunn, N.D., Allen, R.L., Paniwnyk, Z., Byrne, J., Whisson, S.C., Birch, P.R.J., and Beynon, J.L. (2003). A genetic interval and physical contig spanning the Peronospora parasitica (At) avirulence gene locus ATR1Nd. Fungal Genet. Biol. 38, 33–42. [DOI] [PubMed] [Google Scholar]
- Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.M., and Staskawicz, B.J. (1996). Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274, 2063–2065. [DOI] [PubMed] [Google Scholar]
- Shao, F., Golstein, C., Ade, J., Stoutemyer, M., Dixon, J.E., and Innes, R.W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301, 1230–1233. [DOI] [PubMed] [Google Scholar]
- Shen, K.A., Chin, D.B., Arroyo-Garcia, R., Ochoa, O.E., Lavelle, D.O., Wroblewski, T., Meyers, B.C., and Michelmore, R.W. (2002). Dm3 is one member of a large constitutively expressed family of nucleotide binding site-leucine-rich repeat encoding genes. Mol. Plant Microbe Interact. 15, 251–261. [DOI] [PubMed] [Google Scholar]
- Song, W.-Y., Wang, G.-L., Chen, L.-L., Kim, H.-S., Pi, L.-Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.-X., Zhu, L.-H., Fauquet, C., and Ronald, P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Staskawicz, B.J., Mudgett, M.B., Dangl, J.L., and Galan, J.E. (2001). Common and contrasting themes of plant and animal diseases. Science 292, 2285–2289. [DOI] [PubMed] [Google Scholar]
- Tai, T.H., Dahlbeck, D., Clark, E.T., Gajiwala, P., Pasion, R., Whalen, M.C., Stall, R.E., and Staskawicz, B.J. (1999). Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA 96, 14153–14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X., Frederick, R.D., Zhou, J., Halterman, D.A., Jia, Y., and Martin, G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274, 2060–2063. [DOI] [PubMed] [Google Scholar]
- Thilmony, R.L., Chen, Z., Bressan, R.A., and Martin, G.B. (1995). Expression of the tomato Pto gene in tobacco enhances resistance to Pseudomonas syringae pv tabaci expressing avrPTO. Plant Cell 7, 1529–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrall, P.H., and Burdon, J.J. (2003). Evolution of virulence in a plant host-pathogen metapopulation. Science 299, 1735–1737. [DOI] [PubMed] [Google Scholar]
- Tyler, B.M. (2002). Molecular basis of recognition between Phytophthora pathogens and their hosts. Annu. Rev. Phytopathol. 40, 137–167. [DOI] [PubMed] [Google Scholar]
- van den Ackervecken, G., Marois, E., and Bonas, U. (1996). Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host cell. Cell 87, 1307–1316. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L., Laurent, F., Roth, R., and de Wit, P.J. (2000). Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13, 439–446. [DOI] [PubMed] [Google Scholar]
- Voegele, R.T., and Mendgen, K. (2003). Rust haustoria: Nutrient uptake and beyond. New Phytol. 159, 93–100. [DOI] [PubMed] [Google Scholar]
- Whitham, S.M., McCormick, S., and Baker, B. (1996). The N gene of tobacco confers resistance to tobacco mosaic virus in transgenic tomato. Proc. Natl. Acad. Sci. USA 93, 8776–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S.Y., Charoenwattana, P., Holcombe, L., and Turner, J.G. (2003). The Arabidopsis genes RPW8.1 and RPW8.2 confer induced resistance to powdery mildew diseases in tobacco. Mol. Plant Microbe Interact. 16, 289–294. [DOI] [PubMed] [Google Scholar]
- Zambino, P.J., Kubelik, A.R., and Szabo, L.J. (2000). Gene action and linkage of avirulence genes to DNA markers in the rust fungus Puccinia graminis. Phytopathol. 90, 819–826. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Kumar, S., and Nei, M. (1997). Small-sample tests of episodic adaptive evolution: A case study of primate lysozymes. Mol. Biol. Evol. 14, 1335–1338. [DOI] [PubMed] [Google Scholar]
- Zhou, F., Kurth, J., Wei, F., Elliott, C., Vale, G., Yahiaoui, N., Keller, B., Somerville, S., Wise, R., and Schulze-Lefert, P. (2001). Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]