Abstract
RANTES (CCL5) is a chemokine that recruits immune cells to inflammatory sites by interacting with the G-protein coupled receptor CCR5, which is also the primary coreceptor used together with CD4 by HIV to enter and infect target cells. Ligands of CCR5, including chemokines and chemokine analogs, are capable of blocking HIV entry, and studies of their structures and interactions with CCR5 will be key to understanding and optimizing HIV inhibition. The RANTES derivative 5P12-RANTES is a highly potent HIV entry inhibitor that is being developed as a topical HIV prevention agent (microbicide). We have characterized the structure and dynamics of 5P12-RANTES by solution NMR. With the exception of the nine flexible N-terminal residues, 5P12-RANTES has the same structure as wild-type RANTES but unlike the wild-type, does not dimerize via its N-terminus. To prepare the ground for interaction studies with detergent-solubilized CCR5, we have also investigated the interaction of RANTES and 5P12-RANTES with various commonly used detergents. Both RANTES variants are stable in Cymal-5, DHPC, Anzergent-3-12, dodecyltrimethylammonium chloride, and a DDM/CHAPS/CHS mixture. Fos-Cholines, dodecyldimethylglycine, and sodium dodecyl-sulfate denature both RANTES variants at low pH, whereas at neutral pH the stability is considerably higher. The onset of Fos-Choline-12-induced denaturation and the denatured state were characterized by circular dichroism and NMR. The detergent interaction starts below the critical micelle concentration at a well-defined mixed hydrophobic/positive surface region of the chemokine, which overlaps with the dimer interface. An increase of Fos-Choline-12 concentration above the critical micelle concentration causes a transition to a denatured state with a high α-helical content.
Introduction
RANTES (CCL5) belongs to the class of chemokines (chemotactic cytokines), i.e., small (8–10 kDa) soluble secreted proteins that regulate immune responses by attracting various immune cell types to sites of inflammation. Based on the arrangement of conserved N-terminal cysteines, chemokines are divided into four subclasses: C, CC, CXC, and CX3C (1). Chemokines signal by interacting with G protein-coupled receptors, which are named according to the chemokine subclass they interact with and a number, e.g., CCR5, CXCR4, etc. (2).
The fact that the CC chemokines RANTES, MIP-1α, and MIP-1β can suppress HIV infection, with RANTES being the most effective (3), has made RANTES an interesting candidate as an HIV entry inhibitor. This discovery was followed by the identification of the respective chemokine receptor CCR5, which binds all three of these ligands, and also functions as the principal HIV-1 coreceptor (4). Depending on concentration, RANTES is known to signal via two distinct pathways (5): at low RANTES concentrations (≤50 nM), a Gαi-type GPCR pathway regulates chemotaxis and transient calcium mobilization, whereas higher RANTES concentrations (≥1 μM) cause general T cell activation via protein tyrosine kinases (6,7). Interestingly, high concentrations of RANTES enhance viral infection instead of inhibiting it (8).
RANTES and other chemokines can form dimers, higher-order oligomers, and large molecular-weight aggregates (9,10). Wild-type RANTES is particularly aggregation-prone (10), and this property correlates with inflammation (11). Key residues involved in oligomerization are E26 and E66, because the charge-neutralizing E26A and E66S mutations suppress formation of very high molecular weight aggregates and lead predominantly to tetrameric and dimeric RANTES, respectively (12). Although such disaggregated RANTES mutants retain CCR5-binding and Gαi-dependent signaling properties, they fail to activate the protein tyrosine kinase pathway and do not activate T lymphocytes, neutrophils, and monocytes (11).
Structures of numerous chemokines including RANTES, MIP-1α, MIP-1β, vMIP-II, and SDF-1α have been solved by x-ray crystallography and NMR (12–20). Their tertiary structures are highly conserved (Fig. 1): the first two cysteines (C9 and C10 for RANTES) delimit the often flexible N-terminus and form disulphide bonds with cysteines in the center (30s loop) and the C-terminus (50s loop). The N-terminal cysteines are followed by a single-turn 310-helix, three β-strands, and a C-terminal α-helix (22). Although the tertiary structures are highly conserved, the quaternary homo-oligomeric structures vary strongly (22). Thus the CCR5 ligands, i.e., RANTES, MIP-1α, MIP-1β, and vMIP-II dimerize via an intermolecular N-terminal antiparallel β-sheet involving the first cysteine and two preceding residues (PDB:1EQT, PDB:2X6G, PDB:2X6L, and PDB:1CM9), whereas the CXCR4 ligand SDF-1α dimerizes via an extended structural motif comprising the C-terminal α-helix and the first β-strand (PDB:1A15).
Figure 1.
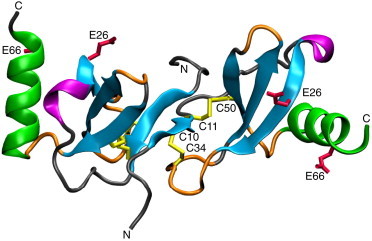
Cartoon representation of the crystal structure of the RANTES dimer (PDB:1EQT) highlighting secondary structure elements: α-helix (green), 310-helix (purple), β-strand (cyan), β-turn (orange), and random coil (gray). The four cysteines are shown (in yellow), and the residues involved in RANTES aggregation (E26 and E66, in red). The RANTES dimer is stabilized by an intermolecular β-sheet formed by the N-termini. The figure was generated using the software VMD (75). To see this figure in color, go online.
Dimer formation and interactions with peptides from the N-terminal tail of CCR5 have been studied in detail by NMR (23). RANTES monomer and dimer are in slow exchange and can be observed as separate sets of 1H-15N HSQC resonances, yielding a dissociation constant of the RANTES monomer-dimer equilibrium of 17.6 μM at pH 3.8 and of 2.9 μM at pH 6.0. This indicates that the dimeric RANTES form is more stable at higher pH. It was also shown that an N-terminal CCR5 peptide sulfated at Y10 and Y14 predominantly binds to monomeric RANTES at pH 3.8 and that the interaction interface overlaps with the interface of RANTES dimerization observed in the crystal structure PDB:1EQT. A more recent study using a slightly longer N-terminal CCR5 peptide at pH 4.8 showed strong binding to the RANTES monomer (KD = 16 μM), but also weak binding (KD = 100 μM) to the dimer (24).
Based on NMR, mass spectrometry, and small-angle x-ray scattering data, a structural model of higher RANTES oligomers has been proposed (25). In this model, tetramers form from dimers by interactions between the second β-strand and the C-terminal α-helix of one monomer within a dimer with corresponding locations in a second dimer. Because these interactions are limited to one monomer in each dimer, the structure can propagate into long, linear oligomers. The interdimer contacts involve hydrophobic (residues Y27, F28, I62, and L65) and electrostatic interactions (K25, E26, E66, and R44), which provides a rationale for the reduced aggregation properties of the RANTES E26A and E66S mutants (11,12).
Since the discovery of its HIV entry-inhibiting properties, numerous strategies, e.g., peptide derivatives (26,27), N-terminal truncations (28–34), chemical modifications of the N-terminus (35–38), or mutagenesis (39–41) have been pursued to develop a topical HIV prevention agent (microbicide) based on RANTES. The variant 5P12 (5P12-RANTES) was obtained using a phage display of N-terminal RANTES mutants on living cells expressing CCR5, with isolated candidates assessed not only for high anti-HIV potency, but also low CCR5 signaling activity (42). In 5P12-RANTES, the wild-type N-terminus S1PYSSDTTP9 has been replaced by the more hydrophobic amino acid sequence Q0GPPLMATQS9. 5P12-RANTES shows very high anti-HIV potency (28 nM) and does not activate CCR5. Hence, it carries a lower risk of unwanted inflammatory side effects that could be elicited by wild-type RANTES (wt-RANTES) or highly potent analogs with agonist activity such as PSC-RANTES (43). As a fully recombinant chemokine analog, 5P12-RANTES has the potential to be produced economically by using low-cost fermentation approaches (42), and shows suitable stability for use as a microbicide (44). Based on its promising profile, 5P12-RANTES has been taken forward into clinical development (Oliver Hartley, personal communication, 2013).
Here, we have characterized the structure, dynamics, and oligomeric state of 5P12-RANTES (in the reduced aggregation form 5P12-RANTES-E66S) by solution NMR and circular dichroism (CD) spectroscopy and compared these properties to wt-RANTES and RANTES-E66S. In addition, we have screened various commonly used detergents for interactions with 5P12-RANTES-E66S and RANTES-E66S to prepare the ground for in vitro interaction studies with detergent-solubilized receptors.
Material and Methods
Cloning, expression, and purification
The DNA sequence of 5P12-RANTES-E66S DNA was obtained by mutating the previously described GB1-RANTES-E66S construct inside the pGEV2 plasmid (23) to a GB1-5P12-RANTES-E66S construct. The GB1-5P12-RANTES-E66S and GB1-RANTES-E66S fusions are cleavable by the blunt-cutting enterokinase, releasing 5P12-RANTES-E66S and RANTES-E66S, respectively, without additional amino acids at their N-termini. The cloning strategy (see Fig. S1 in the Supporting Material) used three consecutive site-directed mutagenesis PCR steps with the following primer pairs:
-
1.
5′-GATCCGACGACGACGACAAGCAG-GGCCCATATTCCTCGGACACC-3′ and 5′-GGTGTCCGAGGAATATGGGCCCTGCTTGTCGTCGTCGTCGGATC-3′;
-
2.
5′-CCCATATTCCTCGGACACCCAATCCTGCTGCTTTGCCTACATTG-3′ and 5′-CAATGTAGGCAAAGCAGCAGGA-TTGGGTGTCCGAGGAATATGGG-3′;
-
3.
5′-GACGACAAGCAGGGCCCACCTTTAATGGCCACCCAATCCTGCTGC-3′ and 5′-GCAGCAGGATTGGGTGGCCATTAAAGGTGGGCCCTGCTTGTCGTC-3′.
Each PCR step changed 3–7 nucleotides resulting in a mutation of 2–4 amino acids. Before proceeding to the next step, the plasmid was amplified in self-prepared TOP10 cells followed by gene sequencing to screen for the correctly mutated clones. Isotopically labeled RANTES-E66S and 5P12-RANTES-E66S were expressed and purified as described in Duma et al. (23).
CD spectroscopy
CD spectra were recorded on 25–60 μM RANTES-E66S/5P12-RANTES-E66S samples under various conditions specified in the figure legends. Measurements were performed on a Chirascan CD spectrometer (Applied Photophysics, Leatherhead, UK) at 9–94°C in a 1-mm quartz Suprasil cuvette (Hellma, Müllheim, Germany). Data points in a wavelength range of 204–260 nm were collected every 1 nm (bandwidth 1 nm) and averaged for 3 s. For thermal denaturation, the temperature was increased in 2°C increments, allowing 60 s equilibration after reaching the set value (0.5°C tolerance). Accurate temperature readings were obtained from a sensor placed in the sample cuvette. After baseline (buffer spectra) subtraction, the mean residue molar ellipticity ΘMRM was calculated as ΘMRM = Θ/(C × n × l), where Θ is the ellipticity, C is the concentration, n is the number of residues, and l is the optical path length. For thermal melts, sigmoidal two-state denaturation curves that included sloping baselines (45,46) and ΔG expressed in terms of ΔH, ΔCp, and melting temperature Tm (47) were fitted using PRO FIT 6.2.9 (Quantum Soft, Uetikon Am See, Switzerland).
Sample preparation for NMR
Samples without detergent contained 0.6 mM 15N/13C-labeled 5P12-RANTES-E66S, 50 mM KH2PO4, 0.02% NaN3, pH 3.8, 5% D2O. Unless noted otherwise, samples containing the detergent Fos-Choline-12 (FC-12, dodecylphosphocholine) contained 0.25 mM 15N/13C-labeled 5P12-RANTES-E66S or 50 μM 2H/15N/13C-labeled RANTES-E66S, 50 mM DCOONa, 0.02% NaN3, 1% (28 mM) FC-12, pH 3.8, 5% D2O. Samples for screening of other detergents by 1H-15N HSQC spectra typically contained 50 μM 15N-labeled protein, 1% detergent, and 50 mM DCOONa pH 3.8 (5P12-RANTES-E66S/RANTES-E66S) or 50 mM CD3COONa pH 4.7 (ubiquitin). Detergents were obtained from Anatrace, Maumee, OH. A full list of the used detergents together with their chemical structures and several physicochemical properties is given in Table S1 in the Supporting Material and a full list of tested conditions is given in Table 1.
Table 1.
Protein state perturbation by detergents
| No. | Proteina | pHb | Temp [°C] | Detergentc | [Detergent]/[CMC] | Protein state |
|---|---|---|---|---|---|---|
| 1 | RANTES-E66S | 3.8 | 25 | — | — | Folded, ID < IMd |
| 2 | RANTES-E66S | 3.8 | 60 | — | — | Onset of thermal unfolding |
| 3 | RANTES-E66S | 3.8 | 25 | 1% FC-8 | 0.3 | Folded, ID > IM |
| 4 | RANTES-E66S | 3.8 | 25 | 5% FC-8 | 1.5 | Folded and denatured in slow exchange |
| 5 | RANTES-E66S | 3.8 | 25 | 1% FC-10 | 2.9 | Denatured, only terminal residues visible |
| 6 | RANTES-E66S | 3.8 | 25 | 1% FC-12 | 21 | Denatured, only terminal residues visible |
| 7 | RANTES-E66S | 3.8 | 60 | 1% FC-12 | 21 | Denatured, most residues visible |
| 8 | RANTES-E66S | 3.8 | 25 | 1% FC-16 | 1881 | Denatured, only terminal residues visible |
| 9 | RANTES-E66S | 3.8 | 25 | 5% DHPC | 75 | Folded, ID > IM |
| 10 | RANTES-E66S | 3.8 | 25 | 1% ANZ 3-12 | 11 | Folded, ID > IM |
| 11 | RANTES-E66S | 3.8 | 25 | 1% TMA-12 | 1.9 | Folded, ID > IM |
| 12 | RANTES-E66S | 3.8 | 25 | 1% DMG-12 | 24 | Folded and (mostly) denatured in slow exchange |
| 13 | RANTES-E66S | 3.8 | 49 | 1% DMG-12 | 24 | Denatured, most residues visible |
| 14 | RANTES-E66S | 3.8 | 25 | 1% SDS | 4.3 | Denatured, only terminal residues visible |
| 15 | RANTES-E66S | 3.8 | 25 | 1% Cymal-5 | 8.3 | Folded, ID ≈ IM |
| 16 | RANTES-E66S | 3.8 | 25 | 1% DDM, | 12 | Folded, ID ≈ IM |
| 1% CHAPS, | 2.0 | |||||
| 0.2% CHS | N/A | |||||
| 17 | RANTES-E66S | 6.3 | 25 | 1% FC-12 | 21 | Folded and denatured in slow exchange |
| 18 | RANTES-E66S | 8.8 | 25 | — | — | Folded, only dimer resonances visible |
| 19 | RANTES-E66S | 8.8 | 25 | 1% FC-12 | 21 | Folded, only dimer resonances visible |
| 20 | 5P12-RANTES-E66S | 3.8 | 25 | 1% FC-12 | 21 | Denatured, only terminal residues visible |
| 21 | 5P12-RANTES-E66S | 3.8 | 60 | 1% FC-12 | 21 | Denatured, most residues visible |
| 22 | 5P12-RANTES-E66S | 3.8 | 25 | 1% DDM, | 12 | Folded |
| 1% CHAPS, | 2.0 | |||||
| 0.2% CHS | N/A | |||||
| 23 | Ubiquitin | 4.7 | 25 | — | — | Folded |
| 24 | Ubiquitin | 4.7 | 25 | 1% FC-12 | 21 | Folded |
| 25 | Ubiquitin | 4.7 | 25 | 1% SDS | 21 | Denatured, most resonances visible |
Effects of detergents on RANTES-E66S, 5P12-RANTES-E66S, and ubiquitin at various buffer, pH, and temperature conditions.
Protein concentrations were 50 μM.
Buffers used: 50 mM DCOONa (pH 3.8), 50 mM CD3COONa (pH 4.7), 50 mM DCOONa (pH 6.3), and 50 mM Tris-HCl (pH 8.8).
For nomenclature, chemical structures of detergent, and CMC, see Table S1.
Intensity of monomer (Im) and dimer (Id) resonances in HSQC spectra.
Backbone assignment
All NMR experiments were recorded on either a DRX600 spectrometer equipped with a triple resonance, triple-axis gradient TXI probe or a DRX800 spectrometer with a triple resonance, Z-gradient TCI cryoprobe (Bruker, Billerica, MA). The following three-dimensional spectra were used for assignment of backbone resonances in the different samples:
CBCANH, HNCO (5P12-RANTES-E66S, 37°C);
CBCANH (5P12-RANTES-E66S, 1% FC-12, 25°C);
CBCANH, CBCACONH, HNCO (5P12-RANTES-E66S, 1% FC-12, 60°C);
HNCACB (RANTES-E66S, 1% FC-12, 25°C).
NMR spectra were processed using the software NMRPIPE (48) and evaluated with the software SPARKY (49).
Chemical-shift analysis
The 1H chemical shift was referenced relative to DSS using the frequency of the water resonance as described in Vajpai et al. (50), while the chemical shifts of 13C (relative to DSS) and 15N (relative to NH3) were referenced indirectly from the obtained 1H DSS frequency (51). Chemical shifts of the RANTES-E66S dimer were taken from BMRB entry No. 16803. Secondary shifts were calculated by subtraction of random coil shifts from the experimental shifts. Random coil shifts (except for cystines) were obtained from the web server of the University of Kopenhagen (http://www1.bio.ku.dk/english/research/pv/sbin_lab/staff/MAK/randomcoil/script/), which uses protein sequence, pH, and temperature corrections (52–54), while cystine random coil shifts were obtained from the web server of the Vendruscolo laboratory (http://www-vendruscolo.ch.cam.ac.uk/camcoil.php) using the CAMCOIL software program (55).
Relaxation experiments
Standard 15N relaxation measurements (R1 = 1/T1, R2 = 1/T2, {1H}-15N nuclear Overhauser effect (NOE)) were recorded on uniformly 15N/13C-labeled 5P12-RANTES-E66S at 600 MHz at 37°C. The R1, R2 decay curves were fitted by an in-house written routine implemented in MATLAB (The MathWorks, Natick, MA) providing Monte Carlo estimation of errors. NMR spectra were processed using the software NMRPIPE (48) and evaluated with the software PIPP (56).
Results and Discussion
5P12-RANTES-E66S heteronuclear backbone assignment and aggregation state
Similarly to other chemokines, RANTES forms dimers as well as higher-order oligomers, which makes it precipitation-prone and challenging to handle in vitro (10). For these reasons, we used the less-aggregation-prone E66S mutation in 5P12-RANTES (11). 5P12-RANTES-E66S gives a well-dispersed 1H-15N HSQC spectrum characteristic for folded proteins containing β-sheets (Fig. 2). Unlike wt-RANTES and RANTES-E66S, where monomer and dimer resonances can be observed in slow exchange in the micro- to millimolar range (23), 5P12-RANTES-E66S displays only a single set of concentration-independent resonances in this concentration range. Furthermore, the 5P12-RANTES-E66S resonances agree much better with the monomer than with the dimer resonances of RANTES-E66S (see Fig. S2). This indicates that 5P12-RANTES-E66S exists in a single, i.e., monomeric state. This is also inferred from the analysis of the retention volume on size-exclusion chromatography, where RANTES-E66S elutes as 15.7 ± 0.1 (standard deviation) kDa, while 5P12-RANTES-E66S elutes as 8.5 ± 0.4 kDa globular protein (see Fig. S3). Apparently, 5P12-RANTES-E66S does not form dimers even at millimolar concentrations, which can be explained by the fact that residues S4-D6, T8, and P9 of the wild-type N-terminus, which are strongly involved in intermolecular β-sheet contacts (Fig. 1), are all mutated in 5P12-RANTES-E66S. Accordingly, also 5P12-RANTES does not show any indication of dimer formation in one-dimensional 1H spectra (Lydia Nisius, University of Basel and Oliver Hartley, personal communication, 2011).
Figure 2.
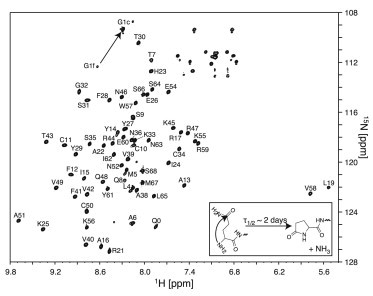
1H-15N HSQC spectrum of 0.6 mM 15N/13C-labeled 5P12-RANTES-E66S (50 mM KH2PO4, 0.02% NaN3, pH 3.8, 5% D2O) recorded at 37°C on a 600-MHz spectrometer. Backbone amide resonances are labeled with the assignments. The N-terminal glutamine Q0 undergoes a spontaneous cyclization to pyroglutamate (mechanism shown in the inset) resulting in the appearance of Q0 resonance and a shift of the G1 resonance from G1f to G1c (marked by arrow) corresponding to the free and the cyclic N-terminus, respectively.
Backbone assignments for 1HN, 15N, 13C′, 13Cα, and 13Cβ resonances were obtained for all nonproline residues of 5P12-RANTES-E66S from three-dimensional CBCANH and HNCO spectra. Interestingly, the 1H-15N correlation of the N-terminal Q0 is also detected in the HSQC spectrum. Due to fast hydrogen exchange, this correlation of an N-terminal NH3+ group is usually not observable. The observation of this correlation for Q0 is consistent with the expected cyclization of the N-terminal 5P12-RANTES glutamine residue (42) via a nucleophilic attack of the backbone nitrogen lone pair on the side-chain carbonyl carbon that results in the formation of a stable pentacyclic pyroglutamate ring (57) with an uncharged NH group (Fig. 2, inset). The cyclization also strongly affects the chemical shift of the second residue G1 (Fig. 2). The reaction requires deprotonated amine and therefore takes days to complete at low pH. At the tested conditions (pH 3.8, set of measurements at 37°C interleaved with storage at 4°C), the approximate half-life of the reaction was approximately two days. Presumably, the increase in hydrophobicity by the N-terminal cyclization is advantageous for HIV inhibition (42), since also the efficient anti-HIV variant PSC-RANTES (Nα-(n-nonanoyl)-des-Ser1-[L-thioproline2, L-α-cyclohexyl-glycine3]) contains highly hydrophobic N-terminal modifications (43).
Secondary structure of 5P12-RANTES-E66S
Secondary chemical shifts ΔδCα, ΔδCβ (Fig. 3), ΔδC′, and ΔδN (see Fig. S4) were obtained from the measured 13Cα, 13Cβ, 13C′, and 15N shifts by subtraction of respective random coil values. From residue F12 to the C-terminal S66, the (secondary) chemical shifts of 5P12-RANTES-E66S closely follow those of RANTES-E66S, which indicates that in this region 5P12-RANTES-E66S should have a very similar structure to RANTES-E66S. However, larger deviations are found in the N-terminal region before residue F12, where RANTES-E66S dimerizes via the N-terminal β-sheet, but 5P12-RANTES-E66S adopts a random structure as evident from close to zero secondary chemical shifts. Furthermore, reduced secondary chemical shifts in the region L65-M67 indicate that the C-terminal helix α1 of both RANTES-E66S and 5P12-RANTES-E66S is three residues shorter in solution than in the PDB:1EQT crystal structure (Fig. 3, top). This can be explained by crystal contacts between the end of helix α1 (residues E66–S68) and residues P20, H23, and R47 of neighboring molecules, which apparently stabilize the helix end by electrostatic and hydrophobic interactions.
Figure 3.
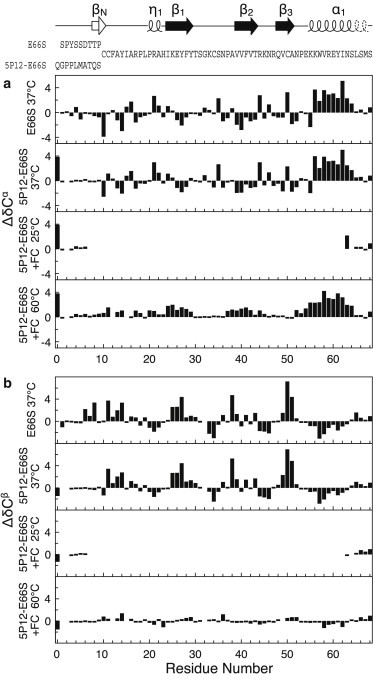
Secondary 13Cα (a) and 13Cβ (b) chemical shifts analysis of RANTES-E66S dimer (37°C), 5P12-RANTES-E66S (37°C), and 5P12-RANTES-E66S in the presence of 1% FC-12 (at 25 and 60°C). Secondary structure elements according to the crystal structure PDB:1EQT and the amino acid sequence of RANTES-E66S as well as of 5P12-RANTES-E66S are drawn at the top. (Note that the sequence used in PDB:1EQT differs from human wt-RANTES by the mutations S1M, P2G, and L19M.) The extraordinary large secondary shifts for Q0 are the result of the pyroglutamate formation.
5P12-RANTES-E66S dynamics
The dynamics of 5P12-RANTES-E66S in solution was characterized by 15N relaxation experiments (R1, R2, {1H}-15N NOE, Fig. 4). The core of 5P12-RANTES-E66S (residues C10-N63, excluding Y14 and G32, which are affected by exchange broadening) has very uniform R1, R2, and {1H}-15N NOE values of 2.26 ± 0.13 s−1, 6.08 ± 0.78 s−1, and 0.71 ± 0.04, respectively. These values correspond to a well-folded protein core tumbling with an isotropic rotational correlation time of 4.0 ns, which is somewhat larger than the 3.1 ns expected for a rigid isotropic rotator of 7.9 kDa at 37°C. Possibly, this increase is caused by the onset of aggregation at the used 0.6 mM concentration.
Figure 4.
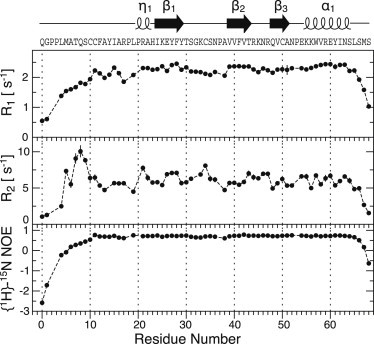
15N R1, R2, and {1H}-15N NOE relaxation data recorded on 0.6 mM 15N/13C-labeled 5P12-RANTES-E66S at 37°C on a 600-MHz spectrometer. The data are an average of two (R1 and {1H}-15N NOE) or three (R2) independent experiments with error bars representing standard deviations. Secondary structure elements and the amino acid sequence of 5P12-RANTES-E66S are drawn at the top.
In contrast to the core, the N-terminal residues Q0–S9 and C-terminal residues beyond approximately N63 show strongly reduced R1 and {1H}-15N NOE values indicative of large amplitude motions in the pico- to nanosecond range from terminal fraying. In addition, increased R2 values for residues M5-S9 give evidence of further slow micro- to millisecond motion in this region. The high mobility of the N-terminal region preceding cysteines (Q0–S9) is in agreement with the monomeric state of 5P12-RANTES-E66S, which, in contrast to the wild-type, does not form an intermolecular β-sheet via its N-terminus.
Interactions of RANTES with Fos-Choline-12
In vitro studies of GPCR ligand interactions are often carried out with detergent-solubilized receptors. The suitability of a detergent is usually judged based on the activity of the solubilized receptor, e.g., efficiency of binding a soluble ligand. Absence of activity is then often ascribed to unfavorable effects of detergent on the receptor and less attention is given to effects on the ligands. In an effort to find a suitable detergent for RANTES CCR5 interaction studies, we have screened several detergents for their effects on RANTES. A list of all investigated detergents together with their chemical structure and other physiochemical parameters like the critical micelle concentration (CMC) is shown in Table S1.
FC-12 solubilizes CCR5 very efficiently (58,59). However, only very little or no ligand binding has been observed in surface plasmon resonance experiments (60). Thus we sought to test its interactions with RANTES. Size exclusion chromatography (Superdex 200 10/300 GL; GE Healthcare, Little Chalfont, UK) shows that 5P12-RANTES-E66S in the presence of 0.5% FC-12 elutes much earlier (Fig. 5 a, 10.7 mL) than in its absence (16.2 mL). This indicates a major increase in molecular mass (to ≥36 kDa as judged based on the column calibration in Fig. S3), suggesting a direct interaction of 5P12-RANTES-E66S and FC-12.
Figure 5.
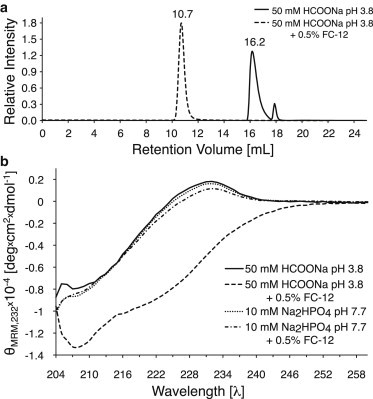
(a) Size exclusion chromatogram of 5P12-RANTES-E66S with (dashed line) and without (solid line) 0.5% FC-12 performed on a Superdex 75 10/300 GL column (GE Healthcare) in 50 mM HCOOH pH 3.8 at room temperature and 0.5 mL/min flow rate. The large decrease of the retention volume (from 16.2 to 10.7 mL) resulting from a major increase of mass (to ≥36 kDa) suggests a direct interaction between 5P12-RANTES-E66S and FC-12. (b) pH-dependent effect of detergent on RANTES-E66S secondary structure measured by CD spectroscopy at 25°C. The spectrum of RANTES-E66S at pH 3.8 without detergent (solid line) changes dramatically after addition of 0.5% FC-12 (dashed line). However, at pH 7.7, the spectra without detergent (dotted line) and after addition of 0.5% FC-12 (dashed-dotted line) are very similar.
Structural analysis of RANTES in Fos-Choline-12
Possible effects of FC-12 on the RANTES structure were first examined by CD spectroscopy (Fig. 5 b). Without detergent, the RANTES-E66S CD spectrum has a maximum at 232 nm and a shallow minimum at 207 nm, which reflects its mixed secondary structure consisting of α/310 helices, β-sheet, and the flexible N-terminus (Fig. 4). Upon addition of 0.5% FC-12 at pH 3.8, the ellipticity strongly decreases, consistent with a conversion to a more helical secondary structure. Similar observations were made for 5P12-RANTES-E66S (see Fig. S5, d and f). At pH 7.7, however, the spectra with and without detergent are very similar, indicating that at the higher pH, FC-12 does not have a pronounced effect on the RANTES-E66S secondary structure (Fig. 5 b).
Subsequently, a more detailed NMR structural analysis was carried out on 5P12-RANTES-E66S in FC-12. Only about a dozen backbone resonances are visible in the 1H-15N HSQC spectrum of 5P12-RANTES-E66S in 0.5% FC-12 at 25°C and pH 3.8 (Fig. 6 a), most of which could be assigned by a CBCANH experiment. The resonances correspond to the apparently flexible N- (Q0, G1, L4, M5, A6, T7) and C-terminal (I62, N63, S64, L65, S66, M67, S68) residues. Other residues from the core of the molecule are undetectable. This can be caused by strong interactions of the core residues with the large detergent micelle or also by conformational exchange on the intermediate timescale. An increase in temperature from 25 to 60°C (Fig. 6 b) leads to the reappearance of most resonances from the core residues. Analogous observations were made for RANTES-E66S at both 25 and 60°C (see Fig. S6, f and g).
Figure 6.
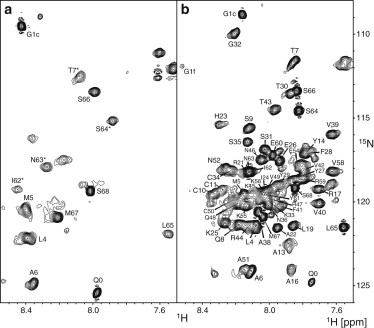
1H-15N HSQC spectra of 0.25 mM 15N/13C-labeled 5P12-RANTES-E66S in the presence of 1% FC-12 (50 mM KH2PO4, 0.02% NaN3, pH 3.8, 5% D2O) recorded at 25°C (a) and at 60°C (b) on a 600-MHz spectrometer. Backbone amide resonances are labeled with assignment information. At 25°C, only terminal residues are visible. An increase to 60°C causes most of the resonances to reappear. Compared to the spectrum in Fig. 2, the presence of FC-12 reduces 5P12-RANTES-E66S peak dispersion from >4 to only ∼1 ppm, suggesting a significant change in the secondary structure.
The HSQC spectrum of 5P12-RANTES-E66S/RANTES-E66S in 0.5% FC-12 at 60°C shows a strongly reduced dispersion as compared to the spectra without detergent at 25°C (Fig. 2) and at 60°C (see Fig. S6 b) representing the folded protein. This is consistent with the loss of native β secondary structure in favor of α and/or random coil conformations and hence with the CD data. The majority of backbone resonances of 5P12-RANTES-E66S in FC-12 at 60°C could be assigned by a set of CBCANH, CBCACONH, and HNCO experiments. Their secondary chemical shift analysis (Figs. 3 and see Fig. S4) reveals a severe structural perturbation by FC-12. The only preserved secondary structure segment is helix α1, which, however, is four residues longer (52–64 instead of 56–64) in FC-12. All indications of β-structure (negative ΔδCα and ΔδC′) are lost for all β-strands. Instead, positive ΔδCα and ΔδC′ of ∼1 ppm in the regions of residues P20-Y29 and P37-R44 indicate the appearance of ∼25% α-helical populations. Analysis of the secondary chemical shifts at 25°C confirms a similar secondary structure as at 60°C for the few detectable terminal residues for 5P12-RANTES-E66S (Fig. 3) and RANTES-E66S (see Fig. S7).
Interactions of RANTES with other detergents
To investigate the specificity and mechanism of the RANTES detergent interaction, we have tested a series of detergents for their effects on 1H-15N HSQC spectra of RANTES-E66S and 5P12-RANTES-E66S at several temperatures and pH values. The results are summarized in Table 1 and corresponding spectra are shown in Fig. S6.
Members of the Fos-Choline family with a shorter (FC-10) and with a longer (FC-16) hydrocarbon tail caused a very similar effect to FC-12 (see Fig. S6, e and h): RANTES is in an NMR dark state with only a dozen resonances visible at 25°C. Interestingly, the C-terminus experienced larger chemical shift changes by the variation of the hydrocarbon tail length than the N-terminus (see Fig. S8). This may indicate that the RANTES C-terminus interacts more strongly with the hydrophobic part of the detergent. However, the addition of 1% FC-8 did not change the RANTES spectrum. Only at 5% FC-8 were the RANTES resonances partially affected, suggesting the coexistence of a nativelike and a detergent-perturbed RANTES-E66S state (see Fig. S6 d). The reported CMC (see Table S1) of FC-8 is 3.4% and thus considerably higher than that of FC-10, -12, and -16 (≤0.35%). This dependence on concentration shows that, similar to findings on sodium dodecyl-sulfate (SDS) (61), Fos-Choline micelle formation is required for effective RANTES denaturation.
To study the dependence on Fos-Choline concentration in more detail, we have carried out a titration of 5P12-RANTES-E66S with FC-12 (37°C, pH 3.8). HSQC spectra show a monotonous decrease of native-state 5P12-RANTES-E66S resonances with increasing FC-12 concentration (Fig. 7 b and see Fig. S9). The midpoint occurs at ∼3 mM (0.1%, i.e., in the vicinity of the CMC). In addition to the decrease in intensity, small but significant chemical shift changes are observable (Fig. 7 a), which start well below the CMC and are monotonous with FC-12 concentration. A chemical-shift map reveals that the interaction surface spans an elongated, mostly hydrophobic crevice between residues C10/C11 and a more polar (positive) region around residues R22/H23 (Fig. 7, c and d). This surface appears suited to accommodate the FC-12 hydrocarbon chain within the hydrophobic crevice and the phosphocholine headgroup at its polar end. Interestingly, the hydrophobic crevice also coincides in part with the RANTES dimer interface harboring residues S4-P9 of the interacting monomer (Fig. 7 d). The mixed hydrophobic/polar RANTES surface perturbed by FC-12 is somewhat reminiscent of the ubiquitin surface affected by SDS at sub-CMC concentration (61). In the latter case, many interacting residues are also hydrophobic, but surface regions with a positive potential dominate.
Figure 7.
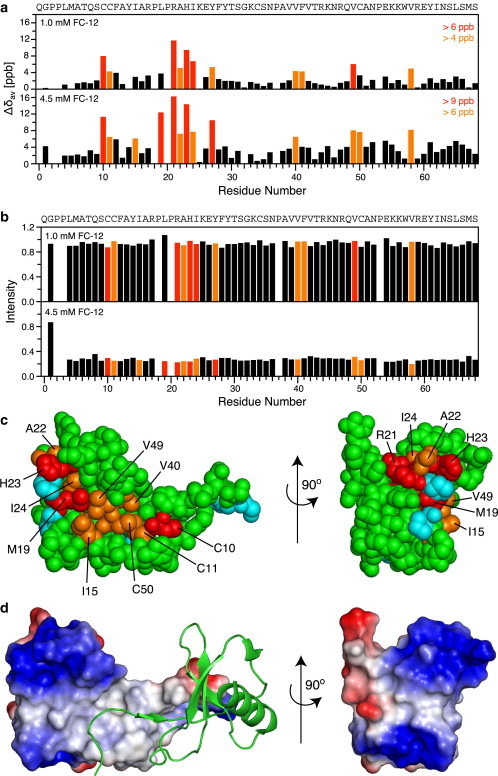
Residue-specific interaction of 5P12-RANTES-E66S with FC-12 from chemical-shift mapping. A titration of 0.10 mM 15N-labeled 5P12-RANTES-E66S (50 mM DCOONa pH 3.8) was carried out with FC-12 at 37°C. (a) Weighted average chemical-shift differences (Δδav = ((ΔδN2/25+ΔδHN2)/2)1/2) for 1.0 mM and 4.5 mM FC-12 relative to no FC-12. Significantly perturbed residues are marked (>6/4 ppb, orange; and >9/6 ppb, red). The amino acid sequence of 5P12-RANTES-E66S is given at the top. (b) Intensities of 5P12-RANTES-E66S backbone 1H-15N resonances in the presence of 1.0 and 4.5 mM FC-12 relative to the intensity without detergent. Residues are colored as in panel a. (c) Space-filling representation of the crystal structure PDB:1EQT (one of two monomers) with chemical-shift map of 5P12-RANTES-E66S residues significantly perturbed by FC-12 (colored in red and orange according to panel a). (Cyan) Positions of proline residues, for which no 1H-15N resonances can be detected. Residues perturbed by FC-12 form a continuous stretch across the molecule. (d) Electrostatic surface representation of panel c. Positive (blue) and negative (red) surface potential is shown. To reflect the charge distribution at pH 3.8, residue H23 was assumed in the protonated state. The second RANTES dimer (green cartoon) occupies a part of the hydrophobic crevice where FC-12 binds. Panels c and d were generated using the software PyMOL (76). To see this figure in color, go online.
All Fos-Choline detergents have a similar denaturing effect on RANTES above their respective CMC regardless of the hydrocarbon tail length. However, a change from Fos-Choline with a single fatty acid chain to DHPC (1,2-diheptanoyl-sn-glycero-3-phosphocholine) with two fatty acids chains abolished the denaturing effect. Even at 5% DHPC (CMC 0.067%) no RANTES denaturation could be observed (see Fig. S6 i). This indicates that the particular topology of the hydrocarbon tail and/or its connection to the choline headgroup are crucial for the interaction with RANTES.
In addition to Fos-Cholines, the zwitterionic surfactant DMG-12 (dodecyl-n,n-dimethylglycine, CMC 0.041%) also shows a strong effect on RANTES and puts it into a dark state similar to that of the Fos-Cholines (see Fig. S6 l). Analogously to FC-12, again many more resonances become visible at higher temperature (see Fig. S6 m). However, the zwitterionic surfactant ANZ-3-12 (n-dodecyl-n,n-dimethyl-3-ammonio-1-propanesulfonate, CMC 0.094%), containing an identical dodecyl hydrocarbon chain, but an ammonium-sulfonate headgroup and also the positively charged TMA-12 (n-dodecyltrimethylammonium chloride, CMC 0.03–0.5% (62)), did not cause RANTES denaturation (see Fig. S6, j and k). Not surprisingly, the strong, anionic detergent SDS (CMC 0.2%) very efficiently denatures RANTES and bleaches most resonances (see Fig. S6 n). In contrast, the mild nonionic detergent Cymal-5 (CMC 0.1%) did not affect the position and linewidth of the RANTES resonances, but interestingly shifted the RANTES-E66S monomer-dimer equilibrium toward the monomeric state (see Fig. S6 o). The same was observed for the frequently used (60) GPCR detergent mixture 1% DDM/1% CHAPS/0.2% CHS (see Fig. S6 p). This shift may be rationalized by assuming that these detergents interact with RANTES via the same hydrophobic crevice as FC-12, and thus block the dimer interaction site.
In summary, all investigated detergents, which denature RANTES, are fully active only above their CMC and have a single fatty acid chain in their hydrophobic part. There is no clear correlation to the type of headgroup: some (phosphocholine and dimethylglycine), but not all zwitterionic, and the investigated negatively charged (SDS) detergent, were denaturing. The positively charged detergent (n-dodecyltrimethylammonium) was not denaturing, which may be explained by the partially positive interaction surface found in the FC-12 titration. A further common feature is that all denaturing detergents (FC-12, DMG-12, SDS) caused rather similar changes in the HSQC spectra of RANTES, which may indicate a similar denatured conformational ensemble in these detergents.
This unfolded state in FC-12 is characterized by a shift to a more α-helical secondary structure content. The fact that detergents may affect the conformation of soluble proteins has been known for over 50 years (63–65). If detergents have an effect, often all-helical proteins lose, while nonhelical proteins gain helicity (65,66). The changes in secondary structure usually result from hydrophobic as well as charged interactions between the polypeptide and the detergent at concentrations above the CMC (61,67,68). However, for certain detergents like the anionic SDS, shifts to helical structure can also be observed below the CMC (61,67,69). In agreement with our observation, several unfolded or water-insoluble polypeptides have been shown to increase their α-helical content in FC-12 (67,69–71). However, for RANTES the structural transition occurs from a well-folded, soluble protein.
pH dependence of FC-12-induced RANTES denaturation and thermal stability
The perturbation of RANTES by FC-12 is most pronounced at pH 3.8 (see Fig. S6 f), whereas it is only partial at pH 6.3 (see Fig. S6 q), and no perturbation is observed at pH 8.8 (see Fig. S6 s). To exclude that this behavior was caused by a pH-induced change of FC-12 surface charges, a pH titration of FC-12 was carried out by adding NaOH (see Fig. S10). The titration curve is completely flat between pH 4 and 10, corresponding to a normalized buffering capacity
of 0.0065, which indicates that there is no titrable surface charge on FC-12 in this pH range (72). Hence, the differing interactions of RANTES and FC-12 at low and high pH should be attributed to RANTES itself, i.e., to its titratable surface groups such as the side chains of aspartic and glutamic acid residues (total 4/3 in RANTES-E66/5P12-RANTES-E66S) or of its single histidine (H23), which have pK values in the respective pH range. Indeed, the influence of the protonation state of H23 on the FC-12 interaction is highly likely due to its location within the interaction surface (Fig. 7, c and d).
Because the interaction with FC-12 destabilizes the native structure of RANTES, we asked whether there is also a correlation with the (thermal) stability of RANTES at the different pH values. A CD melting experiment (see Fig. S11 a and Table 2) revealed that RANTES-E66S has a lower melting temperature (Tm = 64.5°C) and melting enthalpy (ΔH = 26.9 kcal/mol) at pH 3.8 than at pH 6.3 and 7.7 (75.8°C/38.4 kcal/mol and 77.5°C/37.9 kcal/mol, respectively). Melting temperatures and enthalpies at the even higher pH 8.8 had considerable errors, because Tris significantly changes its pK (and therefore the pH of the solution) with temperature, while the measurement in borate was jeopardized by protein precipitation. Nevertheless, the determined values are still higher than at pH 3.8. Similar increases in melting temperature and enthalpy at higher pH were observed for 5P12-RANTES-E66S (see Fig. S11 b and Table 2). Thus both the decrease in stability and the change to a more positive surface charge may contribute to the increased susceptibility of RANTES to Fos-Choline-induced denaturation at low pH values.
Table 2.
Summary of 5P12-RANTES-E66S and RANTES-E66S CD thermal denaturation experiments
| RANTES | Buffer | pH at 20°C | ΔH [kcal/mol] | Tm [°C] |
|---|---|---|---|---|
| E66S | 50 mM HCOONa | 3.8 | 26.9 | 64.5 |
| E66S | 10 mM Na2HPO4 | 6.3 | 38.4 | 75.8 |
| E66S | 10 mM Na2HPO4 | 7.7 | 37.9 | 77.5 |
| E66S | 50 mM Tris-HCl | 8.8a | 53.6 | 73.3 |
| E66S | 10 mM NaH2BO3 | 8.8 | 48.3 | 70.4 |
| 5P12-E66S | 50 mM HCOONa | 3.8 | 24.5 | 65.5 |
| 5P12-E66S | 10 mM Na2HPO4 | 6.3 | 33.9 | 77.0 |
Parameters ΔH and Tm were derived from fits in Fig. S11.
Because for Tris d(pKa)/dT = −0.028, the real pH value at Tm is ∼1.5 units lower than the value measured at room temperature.
Interactions of ubiquitin with FC-12 and SDS
To check whether other proteins are also denatured by Fos-Choline, we tested the thermostable protein ubiquitin (Tm > 100°C (73)). No sign of chemical shift perturbation was observed in 1% FC-12 (see Fig. S6, w and x), indicating that this detergent is not strong enough to denature this extraordinary stable biomolecule. However, consistent with previous studies (61,74), the stronger detergent SDS caused denaturation at 1% concentration, as evident by a strong reduction of dispersion in the backbone 1HN resonances (see Fig. S6 y).
Conclusion
This study presents a biophysical characterization of the nonaggregating E66S mutant of the microbicide candidate 5P12-RANTES. In contrast, to wt-RANTES, 5P12-RANTES-E66S (and also 5P12-RANTES) is unable to dimerize up to millimolar concentrations, apparently as a result of its mutated N-terminus, which is highly flexible on the nanosecond timescale and unable to form an intermolecular β-sheet. The remainder of the tertiary structure beyond residue C10 is very similar to wt-RANTES.
We have also analyzed the behavior of 5P12-RANTES-E66S and RANTES-E66S in the presence of various detergents. The zwitterionic surfactants, i.e., Fos-Cholines and DMG-12, as well as the anionic SDS, denature RANTES above their CMC at low pH. The nonionic Cymal-5, the zwitterionic ANZ-3-12, and DHPC, the positively charged TMA-12, and the lipid/detergent mixture DDM/CHAPS/CHS cause only moderate (shift in monomer/dimer equilibrium) or no effects. A more detailed analysis of the FC-12-induced denaturation showed that the interaction starts below CMC within a hydrophobic crevice that is delimited on one side by a positively charged surface patch. This hydrophobic crevice overlaps in part with the dimerization interface. Higher FC-12 concentrations induce RANTES denaturation to a state with a slightly extended native α-helix and two further regions with α-helical propensities of ∼25%. We have also observed that at neutral pH, RANTES is more stable than at low pH against thermal as well as FC-12-induced denaturation. This pH dependence must be linked to changes in the RANTES surface charge and may in particular be connected to the titration of residue H23, which is part of the FC-12 interaction surface.
In summary, we have characterized the structure, dynamics, detergent interactions, and other properties of the anti-HIV microbicide 5P12-RANTES. These data should help to better understand its physicochemical behavior, choose suitable detergents for interaction studies with CCR5, and provide general insights into the mechanism of detergent denaturation of soluble proteins.
Acknowledgments
We thank Marco Rogowski and Klara Rathgeb-Szabo for protein preparation and Dr. Timothy Sharpe for help in CD data collection and evaluation.
This work was supported by the EU grant No. FP7 Combined Highly Active Anti-Retroviral Microbicides (CHAARM) and Swiss National Science Foundation grant No. 31-132857.
Supporting Material
References
- 1.Allen S.J., Crown S.E., Handel T.M. Chemokine: receptor structure, interactions, and antagonism. Annu. Rev. Immunol. 2007;25:787–820. doi: 10.1146/annurev.immunol.24.021605.090529. [DOI] [PubMed] [Google Scholar]
- 2.Murphy P.M., Baggiolini M., Power C.A. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol. Rev. 2000;52:145–176. [PubMed] [Google Scholar]
- 3.Cocchi F., DeVico A.L., Lusso P. Identification of RANTES, MIP-1 α, and MIP-1 β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 4.Feng Y., Broder C.C., Berger E.A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 5.Bacon K.B., Premack B.A., Schall T.J. Activation of dual T cell signaling pathways by the chemokine RANTES. Science. 1995;269:1727–1730. doi: 10.1126/science.7569902. [DOI] [PubMed] [Google Scholar]
- 6.Bacon K.B., Szabo M.C., Schall T.J. RANTES induces tyrosine kinase activity of stably complexed p125FAK and ZAP-70 in human T cells. J. Exp. Med. 1996;184:873–882. doi: 10.1084/jem.184.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szabo M.C., Butcher E.C., Bacon K.B. RANTES stimulation of T lymphocyte adhesion and activation: role for LFA-1 and ICAM-3. Eur. J. Immunol. 1997;27:1061–1068. doi: 10.1002/eji.1830270504. [DOI] [PubMed] [Google Scholar]
- 8.Gordon C.J., Muesing M.A., Trkola A. Enhancement of human immunodeficiency virus type 1 infection by the CC-chemokine RANTES is independent of the mechanism of virus-cell fusion. J. Virol. 1999;73:684–694. doi: 10.1128/jvi.73.1.684-694.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel S.R., Evans S., Craig S. Characterization of the quaternary structure and conformational properties of the human stem cell inhibitor protein LD78 in solution. Biochemistry. 1993;32:5466–5471. doi: 10.1021/bi00071a024. [DOI] [PubMed] [Google Scholar]
- 10.Skelton N.J., Aspiras F., Schall T.J. Proton NMR assignments and solution conformation of RANTES, a chemokine of the C-C type. Biochemistry. 1995;34:5329–5342. doi: 10.1021/bi00016a004. [DOI] [PubMed] [Google Scholar]
- 11.Appay V., Brown A., Czaplewski L.G. Aggregation of RANTES is responsible for its inflammatory properties. Characterization of nonaggregating, noninflammatory RANTES mutants. J. Biol. Chem. 1999;274:27505–27512. doi: 10.1074/jbc.274.39.27505. [DOI] [PubMed] [Google Scholar]
- 12.Czaplewski L.G., McKeating J., Hunter M.G. Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1α, MIP-1β, and RANTES. Characterization of active disaggregated chemokine variants. J. Biol. Chem. 1999;274:16077–16084. doi: 10.1074/jbc.274.23.16077. [DOI] [PubMed] [Google Scholar]
- 13.Chung C.W., Cooke R.M., Wells T.N. The three-dimensional solution structure of RANTES. Biochemistry. 1995;34:9307–9314. doi: 10.1021/bi00029a005. [DOI] [PubMed] [Google Scholar]
- 14.Crump M.P., Gong J.H., Clark-Lewis I. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dealwis C., Fernandez E.J., Lolis E. Crystal structure of chemically synthesized [N33A] stromal cell-derived factor 1α, a potent ligand for the HIV-1 “fusin” coreceptor. Proc. Natl. Acad. Sci. USA. 1998;95:6941–6946. doi: 10.1073/pnas.95.12.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez E.J., Wilken J., Lolis E. Comparison of the structure of vMIP-II with eotaxin-1, RANTES, and MCP-3 suggests a unique mechanism for CCR3 activation. Biochemistry. 2000;39:12837–12844. doi: 10.1021/bi001166f. [DOI] [PubMed] [Google Scholar]
- 17.Hoover D.M., Shaw J., Wells T.N. The crystal structure of MET-RANTES: comparison with native RANTES and AOP-RANTES. Protein Pept. Lett. 2000;7:73–82. [Google Scholar]
- 18.Liwang A.C., Wang Z.X., Liwang P.J. The solution structure of the anti-HIV chemokine vMIP-II. Protein Sci. 1999;8:2270–2280. doi: 10.1110/ps.8.11.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lodi P.J., Garrett D.S., Clore G.M. High-resolution solution structure of the β chemokine hMIP-1 β by multidimensional NMR. Science. 1994;263:1762–1767. doi: 10.1126/science.8134838. [DOI] [PubMed] [Google Scholar]
- 20.Ren M., Guo Q., Tang W.J. Polymerization of MIP-1 chemokine (CCL3 and CCL4) and clearance of MIP-1 by insulin-degrading enzyme. EMBO J. 2010;29:3952–3966. doi: 10.1038/emboj.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reference deleted in proof.
- 22.Fernandez E.J., Lolis E. Structure, function, and inhibition of chemokines. Annu. Rev. Pharmacol. Toxicol. 2002;42:469–499. doi: 10.1146/annurev.pharmtox.42.091901.115838. [DOI] [PubMed] [Google Scholar]
- 23.Duma L., Häussinger D., Grzesiek S. Recognition of RANTES by extracellular parts of the CCR5 receptor. J. Mol. Biol. 2007;365:1063–1075. doi: 10.1016/j.jmb.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 24.Schnur E., Kessler N., Anglister J. NMR mapping of RANTES surfaces interacting with CCR5 using linked extracellular domains. FEBS J. 2013;280:2068–2084. doi: 10.1111/febs.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Watson C., Prestegard J.H. Oligomeric structure of the chemokine CCL5/RANTES from NMR, MS, and SAXS data. Structure. 2011;19:1138–1148. doi: 10.1016/j.str.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lusso P., Vangelista L., Pavone V. Molecular engineering of RANTES peptide mimetics with potent anti-HIV-1 activity. FASEB J. 2011;25:1230–1243. doi: 10.1096/fj.10-167627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Secchi M., Longhi R., Vangelista L. Enhancement of anti-HIV-1 activity by hot spot evolution of RANTES-derived peptides. Chem. Biol. 2012;19:1579–1588. doi: 10.1016/j.chembiol.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Arenzana-Seisdedos F., Virelizier J.L., Baggiolini M. HIV blocked by chemokine antagonist. Nature. 1996;383:400. doi: 10.1038/383400a0. [DOI] [PubMed] [Google Scholar]
- 29.Oravecz T., Pall M., Norcross M.A. Regulation of the receptor specificity and function of the chemokine RANTES (regulated on activation, normal T cell expressed and secreted) by dipeptidyl peptidase IV (CD26)-mediated cleavage. J. Exp. Med. 1997;186:1865–1872. doi: 10.1084/jem.186.11.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ylisastigui L., Vizzavona J., Benjouad A. Synthetic full-length and truncated RANTES inhibit HIV-1 infection of primary macrophages. AIDS. 1998;12:977–984. [PubMed] [Google Scholar]
- 31.Struyf S., De Meester I., van Damme J. Natural truncation of RANTES abolishes signaling through the CC chemokine receptors CCR1 and CCR3, impairs its chemotactic potency and generates a CC chemokine inhibitor. Eur. J. Immunol. 1998;28:1262–1271. doi: 10.1002/(SICI)1521-4141(199804)28:04<1262::AID-IMMU1262>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 32.Proost P., De Meester I., van Damme J. Amino-terminal truncation of chemokines by CD26/dipeptidyl-peptidase IV. Conversion of RANTES into a potent inhibitor of monocyte chemotaxis and HIV-1-infection. J. Biol. Chem. 1998;273:7222–7227. doi: 10.1074/jbc.273.13.7222. [DOI] [PubMed] [Google Scholar]
- 33.Schols D., Proost P., De Clercq E. CD26-processed RANTES(3-68), but not intact RANTES, has potent anti-HIV-1 activity. Antiviral Res. 1998;39:175–187. doi: 10.1016/s0166-3542(98)00039-4. [DOI] [PubMed] [Google Scholar]
- 34.Lim J.K., Lu W., DeVico A.L. N-terminal proteolytic processing by cathepsin G converts RANTES/CCL5 and related analogs into a truncated 4-68 variant. J. Leukoc. Biol. 2006;80:1395–1404. doi: 10.1189/jlb.0406290. [DOI] [PubMed] [Google Scholar]
- 35.Simmons G., Clapham P.R., Proudfoot A.E. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 36.Mack M., Luckow B., Proudfoot A.E. Aminooxypentane-RANTES induces CCR5 internalization but inhibits recycling: a novel inhibitory mechanism of HIV infectivity. J. Exp. Med. 1998;187:1215–1224. doi: 10.1084/jem.187.8.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosier D.E., Picchio G.R., Wilken J. Highly potent RANTES analogues either prevent CCR5-using human immunodeficiency virus type 1 infection in vivo or rapidly select for CXCR4-using variants. J. Virol. 1999;73:3544–3550. doi: 10.1128/jvi.73.5.3544-3550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartley O., Gaertner H., Offord R. Medicinal chemistry applied to a synthetic protein: development of highly potent HIV entry inhibitors. Proc. Natl. Acad. Sci. USA. 2004;101:16460–16465. doi: 10.1073/pnas.0404802101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proudfoot A.E., Power C.A., Wells T.N. Extension of recombinant human RANTES by the retention of the initiating methionine produces a potent antagonist. J. Biol. Chem. 1996;271:2599–2603. doi: 10.1074/jbc.271.5.2599. [DOI] [PubMed] [Google Scholar]
- 40.Polo S., Nardese V., Lusso P. Enhancement of the HIV-1 inhibitory activity of RANTES by modification of the N-terminal region: dissociation from CCR5 activation. Eur. J. Immunol. 2000;30:3190–3198. doi: 10.1002/1521-4141(200011)30:11<3190::AID-IMMU3190>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 41.Hartley O., Dorgham K., Gorochov G. Human immunodeficiency virus type 1 entry inhibitors selected on living cells from a library of phage chemokines. J. Virol. 2003;77:6637–6644. doi: 10.1128/JVI.77.12.6637-6644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaertner H., Cerini F., Hartley O. Highly potent, fully recombinant anti-HIV chemokines: reengineering a low-cost microbicide. Proc. Natl. Acad. Sci. USA. 2008;105:17706–17711. doi: 10.1073/pnas.0805098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lederman M.M., Veazey R.S., Hartley O. Prevention of vaginal SHIV transmission in rhesus macaques through inhibition of CCR5. Science. 2004;306:485–487. doi: 10.1126/science.1099288. [DOI] [PubMed] [Google Scholar]
- 44.Cerini F., Landay A., Hartley O. Chemokine analogues show suitable stability for development as microbicides. J. Acquir. Immune Defic. Syndr. 2008;49:472–476. doi: 10.1097/QAI.0b013e31818c953f. [DOI] [PubMed] [Google Scholar]
- 45.Clarke J., Fersht A.R. Engineered disulfide bonds as probes of the folding pathway of barnase: increasing the stability of proteins against the rate of denaturation. Biochemistry. 1993;32:4322–4329. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]
- 46.Santoro M.M., Bolen D.W. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 47.Nicholson E.M., Scholtz J.M. Conformational stability of the Escherichia coli HPr protein: test of the linear extrapolation method and a thermodynamic characterization of cold denaturation. Biochemistry. 1996;35:11369–11378. doi: 10.1021/bi960863y. [DOI] [PubMed] [Google Scholar]
- 48.Delaglio F., Grzesiek S., Bax A. NMRPIPE: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 49.Goddard, T. D., and D. G. Kneller. SPARKY 3. University of California at San Francisco, San Francisco, CA.
- 50.Vajpai N., Nisius L., Grzesiek S. High-pressure NMR reveals close similarity between cold and alcohol protein denaturation in ubiquitin. Proc. Natl. Acad. Sci. USA. 2013;110:E368–E376. doi: 10.1073/pnas.1212222110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Markley J.L., Bax A., Wüthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids—IUPAC-IUBMB-IUPAB Inter-Union Task Group on the standardization of data bases of protein and nucleic acid structures determined by NMR spectroscopy. Eur. J. Biochem. 1998;256:1–15. doi: 10.1046/j.1432-1327.1998.2560001.x. [DOI] [PubMed] [Google Scholar]
- 52.Kjaergaard M., Poulsen F.M. Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR. 2011;50:157–165. doi: 10.1007/s10858-011-9508-2. [DOI] [PubMed] [Google Scholar]
- 53.Schwarzinger S., Kroon G.J., Dyson H.J. Sequence-dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 54.Kjaergaard M., Brander S., Poulsen F.M. Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J. Biomol. NMR. 2011;49:139–149. doi: 10.1007/s10858-011-9472-x. [DOI] [PubMed] [Google Scholar]
- 55.De Simone A., Cavalli A., Vendruscolo M. Accurate random coil chemical shifts from an analysis of loop regions in native states of proteins. J. Am. Chem. Soc. 2009;131:16332–16333. doi: 10.1021/ja904937a. [DOI] [PubMed] [Google Scholar]
- 56.Garrett D.S., Powers R., Clore G.M. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 2011;213:357–363. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 57.Suchanek G., Kreil G. Translation of melittin messenger RNA in vitro yields a product terminating with glutaminylglycine rather than with glutaminamide. Proc. Natl. Acad. Sci. USA. 1977;74:975–978. doi: 10.1073/pnas.74.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren H., Yu D., Zhang S. High-level production, solubilization and purification of synthetic human GPCR chemokine receptors CCR5, CCR3, CXCR4 and CX3CR1. PLoS ONE. 2009;4:e4509. doi: 10.1371/journal.pone.0004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiktor M., Morin S., Grzesiek S. Biophysical and structural investigation of bacterially expressed and engineered CCR5, a G protein-coupled receptor. J. Biomol. NMR. 2013;55:79–95. doi: 10.1007/s10858-012-9688-4. [DOI] [PubMed] [Google Scholar]
- 60.Navratilova I., Sodroski J., Myszka D.G. Solubilization, stabilization, and purification of chemokine receptors using biosensor technology. Anal. Biochem. 2005;339:271–281. doi: 10.1016/j.ab.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 61.Shaw B.F., Schneider G.F., Whitesides G.M. Complexes of native ubiquitin and dodecyl sulfate illustrate the nature of hydrophobic and electrostatic interactions in the binding of proteins and surfactants. J. Am. Chem. Soc. 2011;133:17681–17695. doi: 10.1021/ja205735q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sarac B., Bester-Rogac M. Temperature and salt-induced micellization of dodecyltrimethylammonium chloride in aqueous solution: a thermodynamic study. J. Colloid Interface Sci. 2009;338:216–221. doi: 10.1016/j.jcis.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 63.Jirgensons B. Effect of detergents on the conformation of proteins. I. An abnormal increase of the optical rotatory dispersion constant. Arch. Biochem. Biophys. 1961;94:59–67. doi: 10.1016/0003-9861(61)90011-x. [DOI] [PubMed] [Google Scholar]
- 64.Jirgensons B. Optical rotatory dispersion and conformation of various globular proteins. J. Biol. Chem. 1963;238:2716–2722. [PubMed] [Google Scholar]
- 65.Jirgensons B. Effect of anionic detergents on the optical rotatory dispersion of proteins. J. Biol. Chem. 1966;241:4855–4860. [PubMed] [Google Scholar]
- 66.Parker W., Song P.S. Protein structures in SDS micelle-protein complexes. Biophys. J. 1992;61:1435–1439. doi: 10.1016/S0006-3495(92)81949-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alvares R.D., Tulumello D.V., Prosser R.S. Effects of a polar amino acid substitution on helix formation and aggregate size along the detergent-induced peptide folding pathway. Biochim. Biophys. Acta. 2013;1828:373–381. doi: 10.1016/j.bbamem.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 68.Kleinschmidt J.H., den Blaauwen T., Tamm L.K. Outer membrane protein A of Escherichia coli inserts and folds into lipid bilayers by a concerted mechanism. Biochemistry. 1999;38:5006–5016. doi: 10.1021/bi982465w. [DOI] [PubMed] [Google Scholar]
- 69.Singarapu K.K., Tonelli M., Markley J.L. Structural characterization of Hsp12, the heat shock protein from Saccharomyces cerevisiae, in aqueous solution where it is intrinsically disordered and in detergent micelles where it is locally α-helical. J. Biol. Chem. 2011;286:43447–43453. doi: 10.1074/jbc.M111.306464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hauge H.H., Mantzilas D., Nissen-Meyer J. Membrane-mimicking entities induce structuring of the two-peptide bacteriocins plantaricin E/F and plantaricin J/K. J. Bacteriol. 1999;181:740–747. doi: 10.1128/jb.181.3.740-747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grace R.C., Chandrashekar I.R., Cowsik S.M. Solution structure of the tachykinin peptide eledoisin. Biophys. J. 2003;84:655–664. doi: 10.1016/S0006-3495(03)74885-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grzesiek S., Dencher N.A. Dependency of ΔpH-relaxation across vesicular membranes on the buffering power of bulk solutions and lipids. Biophys. J. 1986;50:265–276. doi: 10.1016/S0006-3495(86)83460-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Makhatadze G.I., Lopez M.M., Thomas S.T. Anion binding to the ubiquitin molecule. Protein Sci. 1998;7:689–697. doi: 10.1002/pro.5560070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schneider G.F., Shaw B.F., Whitesides G.M. Pathway for unfolding of ubiquitin in sodium dodecyl sulfate, studied by capillary electrophoresis. J. Am. Chem. Soc. 2008;130:17384–17393. doi: 10.1021/ja804736t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
- 76.Schrödinger, LLC. 2010. The PyMOL Molecular Graphics System, Ver. 1.3r1. http://pymol.org/.
- 77.Turro N.J., Yekta A. Luminescent probes for detergent solutions. A simple procedure for determination of the mean aggregation number of micelles. J. Am. Chem. Soc. 1978;100:5951–5952. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


