Abstract
Topoisomerases are crucial to solve DNA topological problems, but they have not been linked to RNA metabolism. Here we show that human topoisomerase 3β (Top3β) is an RNA topoisomerase that biochemically and genetically interacts with FMRP, a protein deficient in Fragile X syndrome and known to regulate translation of mRNAs important for neuronal function and autism. Notably, the FMRP-Top3β interaction is abolished by a disease-associated FMRP mutation, suggesting that Top3β may contribute to pathogenesis of mental disorders. Top3β binds multiple mRNAs encoded by genes with neuronal functions related to schizophrenia and autism. Expression of one such gene, ptk2/FAK, is reduced in neuromuscular junctions of Top3β mutant flies. Synapse formation is defective in Top3β mutant flies and mice, as observed in FMRP mutant animals. Our findings suggest that Top3β acts as an RNA topoisomerase and works with FMRP to promote expression of mRNAs critical for neurodevelopment and mental health.
Keywords: schizophrenia, autism, topoisomerase, Fragile X syndrome, Top3β, TDRD3, synapse
Introduction
Topoisomerases are “magicians of the DNA world”1, working their wizardry to solve topological problems in DNA metabolism. A hallmark of these enzymes is the catalysis of strand passage reactions. Type I topoisomerases can create a transient break on one DNA strand, allow the other strand to pass through the break, and rejoin the broken ends. As a result, supercoils generated during DNA metabolism can be relaxed, interlocked DNA rings can be unlinked (decatenation), and knots can be introduced into or removed from DNA circles. Whereas most DNA metabolizing enzymes (polymerases, helicases, nucleases, and ligases) have their counterparts in the RNA world, topoisomerases seem to be an exception. In fact, DNA topoisomerase III (Top3; a type I topoisomerase) from E. coli is capable of catalyzing RNA strand passage reactions2, hinting that other members of this family might possess comparable topoisomerase activity for RNA. However, the relevance of this activity in RNA metabolism is unclear. No protein with RNA topoisomerase activity has been reported in eukaryotes.
Whereas bacteria and yeast have a single Top3 enzyme, metazoans have two, Top3α and Top3β, which have similar sequences and DNA topoisomerase activities but distinct functions. Top3α is essential for viability in mice, Drosophila, and chicken DT40 cells, whereas Top3β is dispensable3-8. Mice ablated for Top3β display reduced life span, aneuploidy, and a defective DNA damage response6,7, but Drosophila and DT40 cells defective in Top3β lack obvious phenotypes5,8. Mechanistically, Top3α is part of the DNA “dissolvasome” that resolves intermediates generated during repair and recombination9. The dissolvasome comprises BLM helicase, Top3α, RMI1 and RMI2, with RMI1 acting as a bridge between the other subunits (Fig. 1a). No comparable Top3β complex has been reported.
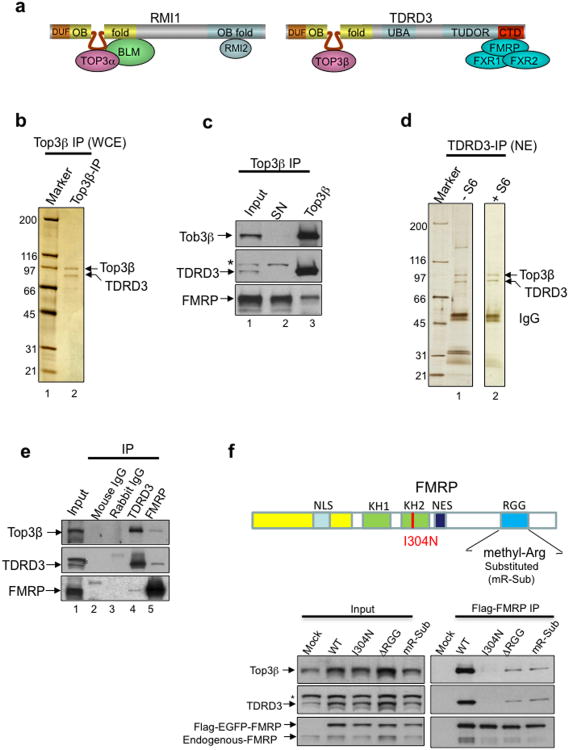
Figure 1. Top3β and TDRD3 form a complex that associates with FMRP; this association is disrupted by a patient-derived point mutation or by substitution of methylated arginine residues in FMRP.
(a) Schematic representations of Top3α and Top3β complexes. TDRD3 and RMI1 are similar to each other in that both contain DUF1767 and OB-fold domains at their N-terminus. The unique intervening region within each OB-fold is indicated by a loop. (b-e) Silver-stained SDS gels (b,d), and immunoblotting (c,e), show the endogenous Top3β-TDRD3 complex immunoprecipitated either from whole-cell extracts by a Top3β antibody (b, c), or from nuclear extract by a TDRD3 antibody (d,e), respectively. In (d), IP was performed from nuclear extract with (+SP6) or without (-SP6) Superose 6 column fractionation (see Supplementary Fig. 1c). The major polypeptides on the gel (marked with arrows) were identified by mass spectrometry. Immunoblotting in (c,e) also shows that Top3β-TDRD3 co-immunoprecipitates with FMRP. (f) IP-Western (bottom panels) using transfected HEK293 cells shows that FMRP variants with a patient-derived I304N mutation, or the RGG-box deletion (ΔRGG), or substitution of methylarginines (mR-sub) within the RGG-box16, are defective in association with Top3β-TDRD3. The mR-sub variant substituted 5 arginine residues within the RGG-box, R527K, R533K, R538K, R543K and R545H; the last 4 residues are methylated16. FMRP is double-tagged with Flag and EGFP; and a Flag antibody was used for IP. A Mock IP was done using untransfected HEK293 cells. Asterisk marks a crossreactive polypeptide. A schematic representation of FMRP domain structure and various mutations was shown on top. Full-length pictures of the blots are in Supplementary Fig. 10. The representative images shown have been repeated at least twice, and the results are reproducible.
Schizophrenia and Fragile X syndrome (FXS) are two mental disorders that occur in worldwide populations. Whereas multiple genes contribute to susceptibility to schizophrenia10, inappropriate silencing or mutation of a single gene, FMR1, causes FXS11. FXS is a leading cause of inherited intellectual disability and autism. The product of FMR1, FMRP, preferentially binds coding regions of mRNAs and can stall ribosomal translation on mRNAs involved in synaptic function and autism12,13. FMRP interacts with several proteins that associate with RNA12, one of which is Tudor domain-containing protein 3 (TDRD3)14. The FMRP-TDRD3 interaction is impaired in a disease-associated FMRP missense mutant I304N, suggesting that this interaction may contribute to the pathogenesis of FXS14. Here we show that Top3β and TDRD3 form a conserved complex that biochemically and genetically interact with FMRP. Notably, the Top3β-FMRP interaction is also disrupted by the patient-derived I304N mutation, suggesting that Top3β may contribute to pathogenesis of mental disorders. Surprisingly, we discovered that Top3β is an RNA topoisomerase that binds multiple mRNAs related to neuronal function and mental disorders, promotes expression of a schizophrenia-related gene in synapse, and is essential for normal synapse formation. Our discovery implies that RNA metabolism can create topological stress that is resolved by topoisomerases. Because DNA topoisomerases have been used as drug targets in cancer therapies, the RNA topoisomerase may also be targeted for therapeutic interventions.
Results
Top3β and TDRD3 form a complex that associates with FMRP
We purified human Top3β complex by two approaches: one, by establishing a HeLa cell line expressing 6xHis and Flag double-tagged Top3β (HF-Top3β) and immunoprecipitating the complex from the nuclear extract with a Flag antibody; and two, by immunoprecipitating the endogenous complex directly from whole-cell extracts with a Top3β antibody. Both approaches yielded two major polypeptides (Supplementary Fig. 1a; Fig. 1b), which were identified as Top3β and TDRD3 by mass spectrometry and immunoblotting (Supplementary Table 1; Fig. 1c; data not shown). The Superose 6 gel-filtration profiles of these proteins in nuclear extract were coincidental (Supplementary Fig. 1c), suggesting that they fractionate as a complex. Reciprocal immunoprecipitation (IP) with a TDRD3 antibody from nuclear extract before or after Superose 6 fractionation obtained the same two proteins as revealed by mass spectrometry and immunoblotting (Fig. 1d,e; data not shown), indicating that they are components of the same complex. The levels of these proteins were comparable by silver-staining analyses of the endogenous complexes (Fig. 1b,d), suggesting that they are present in equal molar amounts.
Mass spectrometry identified FMRP and its two homologs, FXR1 and FXR2, in the immunoprecipitates of both Top3β and TDRD3 (Supplementary Table 1; data not shown). This result was confirmed by immunoblotting (Fig. 1c,e; data not shown). These data agree with previous findings that TDRD3 interacts with FMRP and its homologs14, and further suggest that the interaction involves the entire Top3β -TDRD3 complex. Indeed, FMRP was detected in polypeptides co-immunoprecipitated with HF-Top3β (Supplementary Fig. 1b). Moreover, both endogenous and Flag-tagged FMRP co-immunoprecipitated with TDRD3 and Top3β (Fig. 1e,f). Furthermore, Top3β co-mmunoprecipitated with TDRD3 and FMRP from extracts of mouse brain (Supplementary Fig. 1c).
To rule out the possibility that the observed interaction is mediated by RNA, Top3β IP was performed using extract treated with RNase A, which degraded cellular RNA (Supplementary Fig. 1e, bottom) and eliminated the co-IP of several RNA-binding proteins (Supplementary Table 1; data not shown). Notably, the presence of TDRD3, FMRP and its homologs was retained (Fig. 1b, Supplementary Fig. 1e), indicating that their association with Top3β does not depend on RNA.
We noted that the peak of Top3β-TDRD3 by Superose 6 chromatography partially overlapped with that of FMRP (Supplementary Fig. 1c, fractions 39-41 vs. 41∼43), arguing that only a subset of Top3β-TDRD3 complexes associate with FMRP, and vice versa. In agreement with this notion, FMRP and its homologs were not major polypeptides in either TDRD3 or Top3β immunoprecipitates (Fig. 1b,d). Moreover, while both Top3β and TDRD3 were co-depleted from the input extract by Top3β IP, FMRP was not (Fig. 1e).
Several FMRP mutants have defective association with Top3β-TDRD3
A missense mutation from a FXS patient, I304N, severely impairs the association between FMRP and TDRD314. Consistent with this finding, Flag-tagged FMRP-I304N mutant transfected into HEK293 cells co-immunoprecipitated with severely reduced levels of TDRD3 and Top3β, when compared to the wildtype protein (Fig. 1f). These data suggest that the impaired association between FMRP and Top3β-TDRD3 may contribute to the pathogenesis of the FXS.
Because the Tudor domain of TDRD3 recognizes methylarginines15, and FMRP is primarily methylated on 4 arginines in its RGG motif16, we studied whether FMRP mutants with mutated methylarginines within this motif16 are defective in association with Top3β-TDRD3. Two mutants, one lacking the entire RGG motif (ΔRGG) and the other bearing substitution of all methylarginines (mR-Sub) with lysines or histidines16, co-immunoprecipitated with decreased amounts of Top3β and TDRD3 (Fig. 1f), suggesting that these methylargininines mediate interactions between FMRP and Top3β-TDRD3.
TDRD3 is a bridge connecting Top3β and FMRP
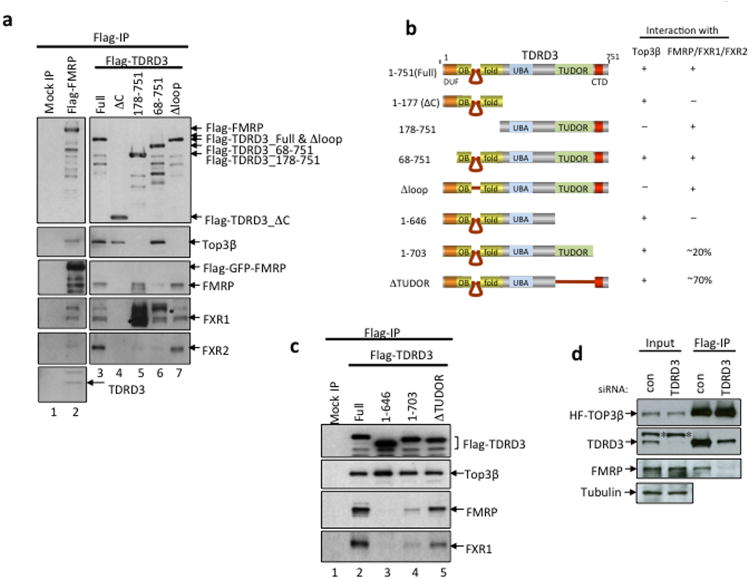
Next, we investigated whether the Tudor domain of TDRD3 is required for the association between Top3β-TDRD3 and FMRP. An earlier report showed that this domain is dispensable, whereas the region C-terminal to the Tudor domain (referred to as CTD here) is essential14. However, because this report used in vitro translated proteins that may lack sufficient methylation, we re-visited this issue by transfecting various Flag-TDRD3 deletion constructs into HEK293 cells to detect their association with endogenous FMRP. The amount of FMRP and its homolog that co-immunoprecipitated with TDRD3 was reduced by approximately 80% for the CTD-deletion mutant, 30% for the Tudor-deletion mutant (Fig. 2b,c), and was undetectable for mutants lacking both domains (Fig. 2a-c). These data argue that both CTD and Tudor domains contribute to FMRP association, with the former being more important. The data support the model that the Tudor domain of TDRD3 binds methylarginines in FMRP.
Figure 2. TDRD3 acts as a bridge connecting Top3β and FMRP.
(a-c) IP-western to assess whether various TDRD3-deletion mutants described in (b) co-immunoprecipitate with Top3β, FMRP, FXR1 and FXR2. The Flag-tagged TDRD3 variants were transfected into HEK293 cells, and co-IP was performed using a Flag antibody. The crossreactive polypeptides are indicated with asterisks. A mock IP from untransfected HEK293 cells was included as a control. (b) Schematic representation of different TDRD3-deletion mutants (left), and their ability to coimmunoprecipitate with Top3β and FMRP from HEK293 extracts (right). (d) IP-Western shows that the association between Top3β and FMRP was disrupted in HeLa cells depleted of TDRD3 by siRNA. The IP was performed using a Flag antibody from extracts of HeLa cells stably expressing HF-Top3β. Full-length pictures of the blots are in Supplementary Fig. 10. The representative images shown have been repeated at least twice, and the results are reproducible.
Having established that TDRD3 binds FMRP through its two C-terminal domains, we studied how TDRD3 interacts with Top3β. TDRD3 variants without Tudor and CTD domains had normal association with Top3β (Fig. 2a-c), hinting that Top3β may bind the N-terminus of TDRD3. This region was previously noted to have sequence similarity to RMI117: both contain DUF1767 and OB-fold domains (Fig. 1a; Supplementary Fig. 2a). The N-terminal OB-fold of RMI1 has a unique intervening region essential for interactions between RMI1 and Top3α18. We identified a similar intervening region within the TDRD3-OB-fold (Fig.1a; Supplementary Fig. 2a,b), and found that a TDRD3 mutant lacking this region failed to co-immunoprecipitate with Top3β while remaining associated with FMRP (Fig. 2a,b). Thus, both Top3 enzymes employ similar mechanisms to interact with the OB-fold domains of their respective partners.
The data above suggest that TDRD3 acts as a scaffold with its N-terminus interacting with Top3β, and the C-terminus with FMRP; this assembly resembles that of RMI1, which bridges Top3α and RMI2 (Fig. 1a). Consistent with this model, the amount of FMRP that co-immunoprecipitated with HF-Top3β was reduced in TDRD3-depleted HeLa cells (Fig. 2d).
Top3β associates with RNA stress granules and polyribosomes
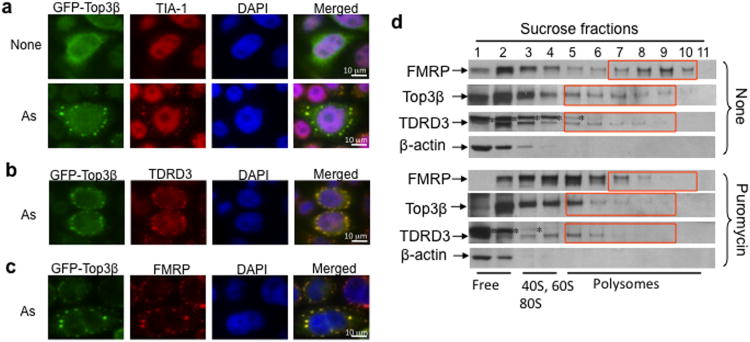
Because both partners of Top3β (FMRP and TDRD3) interact with RNA, we studied if Top3β itself has RNA association. TDRD3 and FMRP localize in RNA stress granules (SGs), which are cytoplasmic compartments containing non-translating mRNAs and form transiently in response to stress agents, such as arsenite (As)14,19. We found that GFP-tagged Top3β co-localized in SGs along with TDRD3, FMRP, and TIA-1 (a SG marker) in As-treated cells (Fig. 3a-c). In control samples, GFP alone, Top3α and its partners (RMI1 and BLM) were absent in SGs (Supplementary Fig. 3a,b; data not shown). Since most SG components are regulators of mRNA stability and translation20, our data also suggest that Top3β associates with RNA and participates in mRNA metabolism.
Figure 3. Top3β associates with TDRD3 and FMRP in SGs and polyribosomes.
(a) Immunofluorescence showing that GFP-Top3β transiently transfected into HeLa cells is redistributed into SGs after arsenite (As) treatment. TIA-1 is included as a SG marker. The nucleus is stained with DAPI. (b,c) Immunofluorescence analysis of GFP-Top3β co-localization with TDRD3 and FMRP in SGs, respectively. (d) Immunoblot analysis of the colocalization of Top3β and TDRD3 with polyribosomes. Cytoplasmic extracts were prepared from suspension HEK293 cells that had been treated with or without puromycin and centrifuged on a 15-60% w/w linear sucrose gradient. The direction of sedimentation of the gradient is left to right (heaviest fraction is 11). The positions of ribosomal subunits, and mono- and polyribosomes are detected by the absorption profile at 254 nM and indicated at the bottom. β-actin was included as a negative control. An asterisk marks a crossreactive polypeptide. Full-length pictures of the blots are in Supplementary Fig. 10. The representative images shown have been repeated at least twice, and the results are reproducible.
TDRD3 and FMRP also associate with polyribosomes19,21-23, which are clusters of ribosomes assembled on mRNA. In HEK293 cells, a fraction of Top3β associated with polyribosomes with a profile similar to that of TDRD3 (Fig. 3d). This is consistent with the notion that the two proteins bind polyribosomes as a complex. As controls, the levels of polyribosome-associated Top3β and TDRD3 (fractions 8-10) were reduced after cells were treated with either puromycin or EDTA (Fig. 3d; data not shown), which disassemble polyribosomes. Because the polyribosome is the functional unit for mRNA translation, our data suggest that Top3β-TDRD3 participates in this process.
Top3β has RNA topoisomerase activity
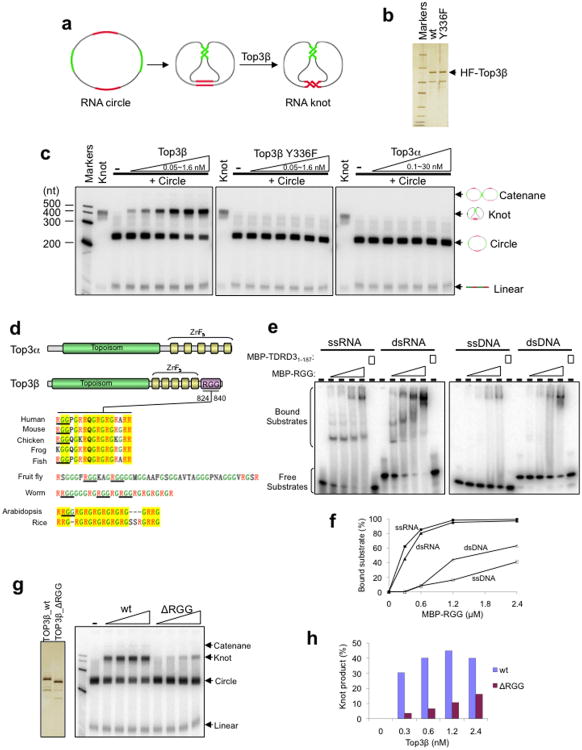
The connection between Top3β and RNA suggests that it could be an RNA topoisomerase. We investigated this possibility by employing an assay previously used to detect RNA topoisomerase activity in E. coli Top32. An RNA circular substrate was engineered containing two pairs of complementary regions intervened by single-stranded spacers (Fig. 4a). Only through a strand passage reaction can this RNA circle be converted into a knot in which the complementary regions can anneal and form double-helices.
Figure 4. Top3β has RNA topoisomerase activity that depends on a conserved RGG RNA-binding motif.
(a) Schematic representation of an RNA topoisomerase assay modified from a previous publication2. A synthetic circular RNA substrate contains two pairs of complementary regions (red and green) separated by single-stranded spacers (black). Through strand passage reactions, this substrate is converted to a knot in which the two pairs of complementary regions can form normal double helices. (b) Silver-stained SDS gel showing purified recombinant wildtype or Y336F mutant HF-Top3β proteins. This mutation is known to inactivate topoisomerase activity on DNA. (c) Autoradiographs from the RNA topoisomerase assay show that Top3β, but not its catalytic mutant or Top3α, has RNA topoisomerase activity. The reaction contains 1 nM 32P-labeled circular RNA substrate and increasing concentrations of wildtype or Y336F Top3β mutant (0.05 nM, 0.1 nM, 0.2 nM, 0.4 nM, 0.8 nM or 1.6 nM), or Top3α (0.1 nM, 0.3 nM, 1 nM, 3 nM, 10 nM or 30 nM). A ladder of RNA markers and the purified RNA knot were loaded on the left of every panel. The linear breakdown products of cyclic RNA exist in all reactions. A darker exposure of the autoradiographs shows that a small amount of catenane was also generated in the reaction (Supplementary Fig. 4a). (d) Top panel: schematic representations showing that Top3β, but not Top3α, contains a RGG motif. Bottom panel: the alignment of RGG motifs of Top3β from several higher eukaryotes. The RGG boxes are indicated by underline. Arginine and Glycine are indicated by red and green letters, respectively. Conserved regions are highlighted in yellow. (e,f) Gel-shift assay (e) and its quantification (f) show that a fusion protein containing MBP and the RGG motif of Top3β (MBP-RGG) preferentially binds RNA compared to DNA. Reaction contains 1 nM single-stranded (ss) or double-stranded (ds) RNA or DNA. MBP-TDRD31-187 was included as a negative control. (g,h) RNA topoisomerase assay (g, right panel) and its quantification (h) show that the RGG motif of Top3β is important for its RNA topoisomerase activity; (g, left panel), silver-stained SDS gel of the purified wild-type or RGG motif-deleted Top3β. The representative images shown have been repeated at least twice, and the results are reproducible.
The HF-Top3β protein (Fig. 4b) efficiently converted the RNA circle to a knot (Fig. 4c), indicating that Top3β possesses topoisomerase activity for RNA. The reaction also produced a small amount of RNA catenane (Supplementary Fig. 4a), the formation of which similarly requires a strand passage reaction. A Top3β mutant (Y336F) carrying substitution of the Tyr residue critical for DNA strand passage reactions displayed no topoisomerase activity for RNA (Fig. 4b,c), indicating that this residue is used in reactions on both DNA and RNA. The paralog of Top3β, Top3α, exhibited no RNA topoisomerase activity at protein concentrations up to 8000-fold higher than that used in Top3β assays (Fig. 4c and data not shown). As a control, both Top3 paralogs displayed comparable activity in a DNA topoisomerase assay24 (Supplementary Fig. 4b,c). Thus, the RNA topoisomerase activity of Top3β is specific.
Top3β contains a conserved RNA-binding motif
We compared the sequences of the two Top3 paralogs to determine if there exist any domains that may account for presence of RNA topoisomerase activity in one but not the other. Top3β, but not Top3α, from several species were noted to contain characteristic arginine and glycine (GR)-rich clusters that are present in several RNA-binding proteins25. We found that these clusters resemble the RGG box, a RNA-binding motif (Fig. 4d).
A recombinant MBP fusion protein containing the RGG box of human Top3β (Supplementary Fig. 4d) indeed displayed higher binding affinity to ssRNA and dsRNA (Kd ∼300 nM) than to ssDNA and dsDNA (Kd ∼2.4 μM) (Fig. 4e,f). This binding activity was detected at the same salt concentration (200 mM) used in RNA binding studies for FMRP26 (Supplementary Fig. 4e). The activity was not derived from MBP, as MBP fused to a different protein (TDRD3 residues 1-187) had no detectable RNA-binding activity (Fig. 4e; Supplementary Fig. 4e). Importantly, a Top3β mutant lacking this RGG box (ΔRGG) had reduced strand passage activity for RNA (Fig. 4g,h), indicating that the RNA-binding activity of the RGG box is critical for the RNA topoisomerase activity. The Top3β-ΔRGG mutant also exhibited reduced DNA topoisomerase activity (Supplementary Fig. 4f), suggesting that the RGG box plays a role in reactions on DNA.
Top3β binds mRNAs that are enriched with FMRP targets
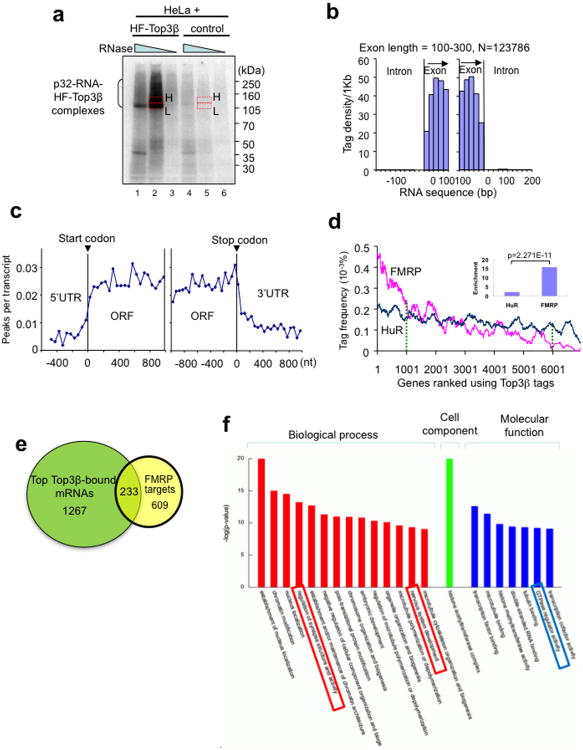
To study whether Top3β binds mRNAs in vivo and also to identify its genome-wide RNA targets, we performed HITS-CLIP (high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation)27. In this method, proteins are crosslinked with bound RNAs in vivo, thus allowing specific mapping of the sites of interaction of the protein with the RNA. The level of RNA obtained by Flag-Top3β IP from HeLa cells expressing HF-Top3β was significantly higher than the background signal from a mock IP using cells that do not express the tagged protein (Fig. 5a), indicating that Top3β binds RNA in vivo. The majority of 2.4 million sequencing reads (95.8%) had a sense orientation (Supplementary Table 2; raw data GEO accession number: GSE47502), and the density of tags within exons was 58-fold higher than within introns, suggesting that Top3β binds mRNAs (Fig. 5b; Supplementary Fig. 5a; data not shown).
Figure 5. Top3β binds coding regions of mRNAs, and its bound mRNAs are enriched with match FMRP targets.
(a) Autoradiograph of a SDS-PAGE gel from the HITS-CLIP assay shows that a significant amount of 32P-labeled RNA was crosslinked to HF-Top3β. A mock immunoprecipitation was performed using HeLa cells that do not express HF-Top3β. The RNA in gel slices marked H (high) and L (low) were extracted, reverse-transcribed, and sequenced; but only data from H are presented. (b) Histogram illustrates that Top3β binding sites, represented by sequence tags identified by HITS-CLIP, preferentially map to exons, but not introns. The number of sequence tags was counted at specific distances from exon start (left graph) and exon end (right graph) and then converted to tag density per 1 Kb. Data for longer exons are included in Supplementary Fig. 5. (c) A graph showing that Top3β binding sites on mRNAs are enriched in open-reading frames (ORFs), but not in 5′ or 3′ untranslated regions (UTRs). Peaks are locations with >15 tags. (d) A ranked plot shows that genes containing higher frequency of Top3β tags are enriched with FMRP targets (red line) than those with lower frequency of Top3β tags. The small inlet graph shows that the ratio between the number of FMRP targets in the top 1000 mRNAs with highest Top3β tag frequency vs. that in the bottom 1000 mRNAs with lowest Top3β tag frequency. The enrichment for HuR was included for comparison. The statistical analyses used Chi-square test. (e) A Venn diagram shows the number of top Top3β targets that overlap those of FMRP13. These common targets are listed in Table S2. (f) A graph shows the top gene ontology (GO) terms that are enriched in the common targets of Top3β and FMRP. The GO terms are ranked by –log (P values). The boxes highlight the categories that may be relevant to functions of Top3β and FMRP in neurons. The genes of each category are listed in Supplementary Table 4.
We compared the HITS-CLIP data of Top3β with those of FMRP13 and observed interesting similarities. First, the tag frequency of both proteins was higher in open reading frames (ORFs) than UTRs (Fig. 5c, Supplementary Fig. 5b), which is consistent with the notion that they associate with polyribosomes and participate in translation. Second, the FMRP targets were enriched by approximately 15-fold in the top 1,000 Top3β targets compared to the bottom 1,000 targets (Fig. 5d), whereas targets of a different RNA-binding protein (HuR) were enriched only 2-fold. The data indicate that Top3β and FMRP may share common mRNA targets. Indeed, about 15% of the top 1,500 Top3β targets matched those of FMRP, whereas 27% of the FMRP targets matched those of Top3β (Fig. 5e; Supplementary Table 3). Third, the common targets of the two proteins were enriched in several processes related to neurons, including regulation of synapse structure and activity (Fig. 5f; Supplementary Table 4). It should be cautioned that the HITS-CLIP data of FMRP are from mouse brain extracts13, whereas our Top3β data are from HeLa cells that do not express all the neuronal genes. Thus, our comparison may have under-estimated the overlap between the targets of the two proteins. Nevertheless, these comparisons raised a possibility that Top3β may function in neurons coordinately with FMRP.
Drosophila Top3β interacts with FMRP to promote synapse formation
We utilized Drosophila to examine functions of the Top3β and its genetic interactions with FMRP. We first identified CG13472 as the single Drosophila homologue of TDRD3 (dTDRD3) by homology searches. We then confirmed that the endogenous dTop3β biochemically interacted with dFMR1 by IP-Western using Drosophila S2 cells (Supplementary Fig. 6a,b). In S2 cells stably expressing Flag-tagged dTDRD3, all three proteins associated with each other, and the amount of dTop3β associating with dFMR1 was increased (Supplementary Fig. 6a). These data support the notion that TDRD3 connects the other two proteins.
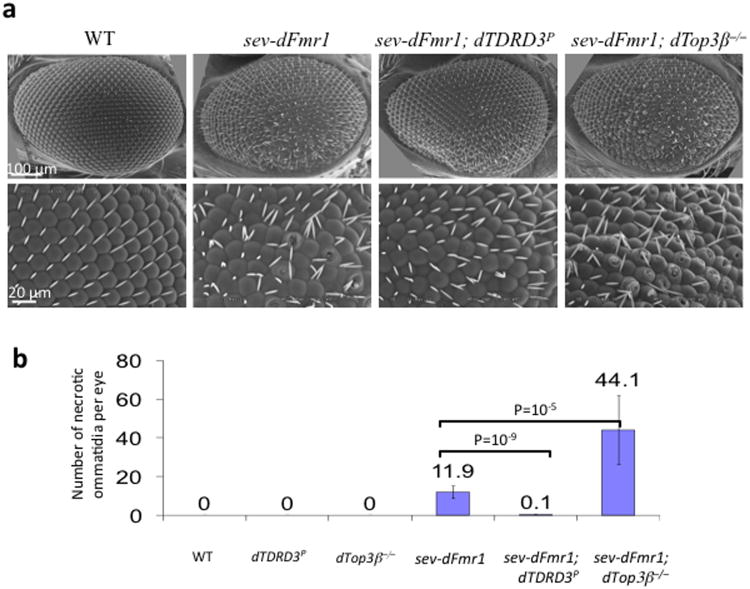
Ectopic expression of dFMR1 in Drosophila eyes causes a rough (disorganized) eye phenotype with increased necrosis of ommatidia28 (Fig. 6a,b). Modifications of this phenotype have been used to identify factors that genetically interact with dFmr129,30. Interestingly, ectopic expression of dFMR1 in a dTop3β null mutant background5 caused a 3-fold increase in necrosis and more disorganized photoreceptor cluster in each ommatidium, when compared to ectopic expression of dFMR1 in wildtype background (Fig. 6a,b; Supplementary Fig. 6c,d). The dTop3β mutant itself had normal smooth eyes, with no necrotic ommatidia detected (Fig. 6b, Supplementary Fig. 6c). These data indicate that dTop3β mutation enhances the rough eye phenotype caused by dFMR1 overexpression, and suggest that dTop3β genetically interacts with dFMR1 in an antagonistic manner.
Figure 6. Top3β and TDRD3 mutations modify dFMR1 function in Drosophila eyes.
(a) Scanning Electron Microcopy (SEM) of Drosophila compound eyes shows genetic interactions of dFMR1 with Top3β and TDRD3. Different genotypes are indicated. The rough eye phenotype induced by ectopic expression of dFMR1 in the eye (sev-dFMR1) is enhanced by a Top3β null mutant, but suppressed by a reduction-of-function mutant of TDRD3(dTDRD3-P) (Supplementary Fig. 6e,f). The crater-like holes are necrotic ommatidia that were quantified and shown in (b). The magnifications are: first row, 250×; second row, 800×. The tangential sections of the eyes are shown in Supplementary Fig. 6d. (b) A graph shows the number of necrotic ommatidia in the eyes of different genotypes. For flies of each genotype, 10 eyes were counted. The p values between indicated genotypes were calculated using Student t-test and are shown on the top. The error bars represent standard errors of means (s.e.m). The representative images shown have been repeated at least twice, and the results are reproducible.
In a dTDRD3 reduction-of-function mutant background (which has reduced dTDRD3 mRNA level due to a P-element insertion; Supplemental Figure 6e, f), ectopic expression of dFMR1 failed to induce significant roughness, evidenced by an almost complete absence of necrotic ommatidia and a better organized photoreceptor cluster in each ommatidia, when compared to ectopic expression of dFMR1 in the wildtype background (Fig. 6a, b, Supplemental Figure 6c and d). The dTDRD3 mutant itself had normal eye morphology. Taken together, these findings suggest that mutation of dTDRD3 suppresses rough eye phenotype caused by ectopic expression of dFMR1, and TDRD3 promotes function of FMRP. The fact that Top3β and TDRD3 have opposite effects on FMRP function (one negative and the other positive) implies that the two proteins may antagonize each other in some situations.
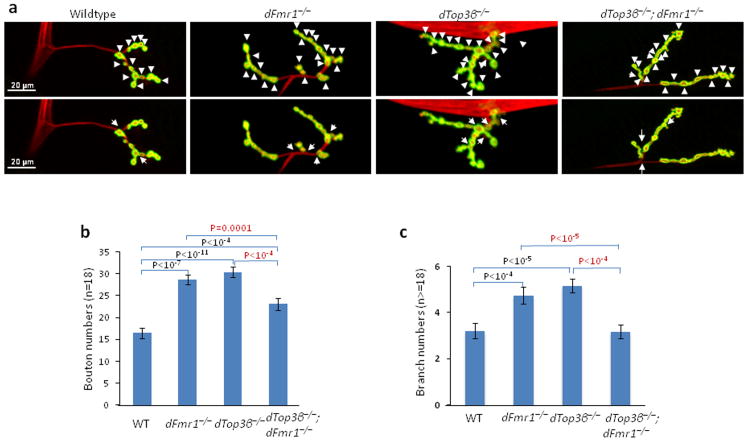
A null mutation in Drosophila dFmr1 yields neuronal and behavioral defects similar to those in Fragile X patients and Fmr1 mutant mice. One defect is abnormal neuromuscular junctions (NMJs), exemplified by over-elaboration of synaptic branches and an increased number of synaptic boutons31. dTop3β null mutant flies5 had similar NMJ abnormalities, as their branch and bouton numbers were comparable to those of dFmr1 mutants, and significantly higher than those of the wild type flies (Fig. 7a-c). Interestingly, the abnormal phenotype was partially suppressed in the dTop3β−/−;dFmr1−/− double mutant, as the numbers of branches and boutons were fewer in the double mutant than the single mutants. These results are reminiscent of genetic interactions between dAdar and dFmr1: whereas either single mutant exhibits abnormal NMJ phenotypes, reduction of dAdar dosage in dFmr1 mutant background corrected the abnormality32. Our data suggest that dTop3β and dFMR1 functionally antagonize each other in a common pathway to promote formation of normal NMJs.
Figure 7. Drosophila Top3β and dFmr1 work in a common pathway to promote formation of NMJs.
(a) Representative immunofluorescence images of neuromuscular junctions at muscle 4 (NMJ4) of third instar wandering Drosophila larvae of different genotypes as indicated. The NMJ4 was co-labeled with a presynaptic marker (anti-HRP, red) and a postsynaptic marker (anti-DJG, green). The same set of images is shown in both top and bottom, but the arrowheads in the top images mark synaptic boutons, and the arrows in bottom mark synaptic branches. (b) Quantification of synaptic boutons from NMJ4 segments 3 to 5 (n>=18) of different genotypes as indicated. (c) Quantification of synaptic branches at NMJ4 from segments 3, 4, and 5 of both sides of wandering third instar larvae (n>=18). The graphs show the means of the branches, and error bars represent standard errors of mean (s.e.m.). The p-values shown above each bar were calculated between wildtype (WT) and a mutant using Student' t-test. The representative images shown have been repeated at least twice, and the results are reproducible.
Top3β works with FMRP to promote expression of a schizophrenia gene
The accompanying manuscript reported that Top3β is a risk factor for schizophrenia and intellectual disability33. In addition, an earlier study has identified a de novo single nucleotide variant (SNV; R472Q) and a copy number variant (CNV) of Top3β in a different schizophrenia population34. Notably, a de novo SNV (C666R) and several CNVs of Top3β have been found in individuals with autism spectrum disorders (ASDs)35 (CNV references at http://autismkb.cbi.pku.edu.cn). R472Q and C666R substituted conserved residues in the catalytic and Zn-finger domains of Top3β, respectively; and are likely to impair function of the protein. Because schizophrenia and autism share common etiology factors36,37, Top3β could be a risk factor for both disorders. In support of this notion, Top3β-bound mRNAs identified by HITS-CLIP matched multiple schizophrenia and/or autism related genes (Supplementary Table 5), suggesting that Top3β may affect disease pathogenesis by modulating expression of these mRNAs.
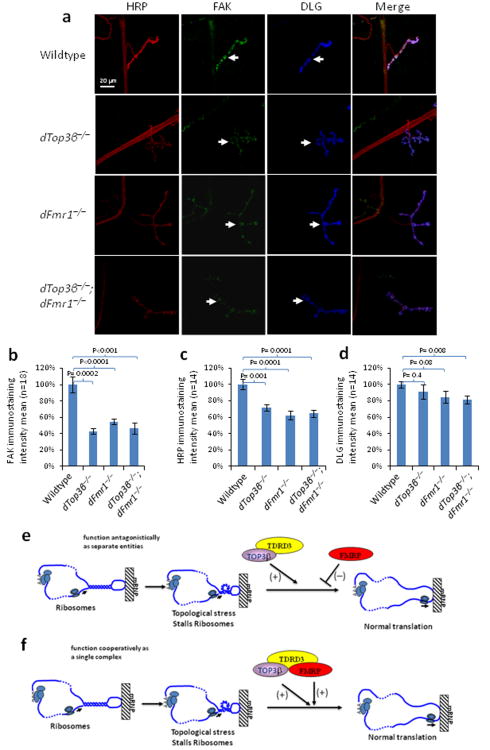
We selected ptk2/FAK (protein tyrosine kinase 2/focal adhesion kinase), a common target of Top3β and FMRP (Supplementary Table 5) that is also a schizophrenia related-gene38, for further study. Ptk2/FAK is crucial for axonal growth and synapse formation39,40, and its null mutant has increased number of synaptic boutons and branches in Drosophila41, similar to those observed in the dTop3β or dFmr1 mutants. Immunostaining revealed that ptk2/FAK expression was reduced in NMJs of dTop3β and dFmr1 mutant flies (Fig. 8a,b). Moreover, the level of ptk2/FAK in the double mutant was comparable to those of the single mutants, suggesting that the two proteins work in the same pathway to promote synaptic expression of ptk2/FAK.
Figure 8. Drosophila Top3β and dFmr1 work in the same pathway to promote expression of ptk2/FAK.
(a) Representative immunofluorescence images, and (b) their quantification, show that the level of ptk2/FAK protein (green) is reduced in NMJs of the 3rd instar larvae of Top3β and dFmr1 single and double mutant flies. Presynapses of NMJs and axons were labeled with anti-HRP antibody (Red), whereas postsynapses were labeled with anti-DLG antibody (Blue). The FAK staining of the entire NMJ area, which was marked by DLG staining [arrows in (a)], was quantified by using Imaris imaging software and shown in (b). Eighteen sets of the NMJs from wild-type or top3β mutants were stained and quantified. The p-values in the graph were calculated using Student' t-test. Error bars represent standard errors of means (s.e.m.). (c and d) Quantification of immunofluorescence signals of HRP and DLG to serve as comparisons. The representative images shown have been repeated at least twice, and the results are reproducible. (e) A model to illustrate how Top3β -TDRD3 complex may work antagonistically with FMRP to regulate translation of an mRNA. An mRNA may become topologically constrained by circularization through protein-mediated interactions between its 5′-UTR and 3′-poly A tail. The mRNA may contain local duplex regions or hairpin structures. When ribosomes or RNA helicases unwind such structures, it may create topological stress that enhances ribosomal stalling which is facilitated by FMRP binding. Top3β may reduce the topological stress and thus antagonize FMRP-mediated ribosomal stalling. Our biochemistry experiments showed that a large fraction of FMRP in cells does not associate with Top3β-TDRD3. We hypothesize that this fraction of FMRP antagonizes Top3β action. (f) A model that illustrates how Top3β -TDRD3 may cooperate with FMRP to enhance mRNA translation. Our biochemistry experiments show that a fraction of Top3β-TDRD3 complex associates with FMRP and vise versa. We hypothesize that this fraction of FMRP may enhance Top3β-TDRD3 to bind its target mRNA, reduce topological stress, and stimulate mRNA translation.
We noticed that the immunostaining signal of the HRP antibody, which was used as a marker for Drosophila neurons, was reduced in the NMJs in the dTop3β and dFmr1 single as well as double-mutants (Fig. 8a,c), suggesting that the two proteins work together to promote expression of one or more proteins that carrying the HRP-epitope. In comparison, expression of a postsynapse marker, DLG, was not significantly altered in the dTop3β mutant (Fig. 8a, d), suggesting that Top3β is not required for expression of all mRNAs at the synapse.
Top3β is required for normal synaptogenesis in mice
Both Fmr1 mutant flies and mice display abnormal synaptogenesis31,42. We therefore investigated if Top3β-knockout (KO) mice exhibit abnormal synapse formation similar to the Drosophila mutant.
Immunostaining of cultured primary cortical neurons revealed that the density of presynaptic vesicles labeled by synaptophysin was reduced in dendrites from Top3β-KO mice compared to its wildtype and heterozygous littermates (Supplementary Figure 7a). In addition, the density of synapses, labeled by co-staining of synpatophysin and PSD95, a postsynaptic marker, was also reduced (Supplementary Figure 7b). The data suggest that Top3β is required for normal synapse formation in not only Drosophila, but also mice.
Discussion
It is generally assumed that RNA should not have topological problems, because it is single-stranded linear and thus not topologically constrained. Our discovery of Top3β as an RNA topoisomerase necessitates reconsideration of this assumption. We envision at least three scenarios where topological complexity may arise in RNA. First, A linear mRNA can be circularized with both ends interacting with a common protein bridge43, or by direct base-pairing interactions between 5′ and 3′ UTR44. A circularized mRNA is topologically constrained, so that if two mRNA circles form a catenane, their decatenation may need a topoisomerase (Supplementary Fig. 8a,b). An RNA topoisomerase can catalyze catenation or decatenation of circularized mRNAs, thus affecting their translation, transport or segregation. Second, an mRNA can fold into higher-order structures with localized duplex regions, such as hairpins and pseudoknots45,46. Unwinding of these structures by the translation machinery and RNA helicases during transport may create topological stress whose resolution may be facilitated by a topoisomerase (Supplementary Fig. 8c). Third, circular RNAs (cirRNAs) have recently been identified in higher eukaryotes, which function as miRNA sponges to regulate mRNA expression47,48. Top3β may interconvert these RNAs between forms of circles, knots and catenanes, thus affecting their activities (Supplemental Figure 8d).
The RNA topoisomerase activity is specific for Top3β and absent in Top3α. Because E. coli Top3 can catalyze both DNA and RNA topoisomerase reactions2, this single bacterial enzyme may have evolved into two homologous complexes in eukaryotes: Top3α-RMI1 for DNA and Top3β-TDRD3 for RNA (Supplementary Fig. 9). One role of RMI1 is to connect Top3α at its N-terminus to other proteins at its C-terminus. This feature is shared by TDRD3, which binds Top3β with its N-terminus and FMRP through its C-terminus. The TDRD3-Tudor domain is capable of interacting with other RNA-binding proteins that carry methylarginines19. We propose that TDRD3 could be a multiprotein adaptor linking Top3β to different RNA binding molecules, thus enabling the topoisomerase to regulate different mRNA metabolic reactions. Notably, TDRD3 depletion reduced FMRP-Top3β association, a feature similar to that of the FMRP-I304N mutation, hinting that TDRD3 may also be a risk factor for mental disorders. Indeed, TDRD3 CNVs have been detected in autism patients (references at http://autismkb.cbi.pku.edu.cn).
Top3β interacts with FMRP in vitro and modifies FMRP function in vivo. The accompanying manuscript and other studies indicate that Top3β itself is a risk factor for schizophrenia, intellectual disability, and autism33,34,35. As both Top3β and FMRP are contributing factors for mental disorders, they may work together to promote normal neurodevelopment and prevent mental disorders. Several lines of evidence support this notion. First, FMRP-Top3β interaction is disrupted by the patient-derived I304N mutation. Second, Top3β-bound mRNAs are enriched with FMRP targets, which include multiple genes related to schizophrenia and autism. Some of these targets play crucial roles in neurodevelopment, which is often compromised in mental disorders36,38. Third, one common target of Top3β and FMRP, ptk2/FAK, which is related to schizophrenia and plays a crucial role in neurodevelopment, has reduced expression in synapse of dTop3β and dFmr1 mutant flies. Fourth, synapse formation is abnormal in both dTop3β and dFmr1 mutant flies, as well as in their mutant mice42. Thus, Top3β and FMRP may work coordinately to regulate expression of mRNAs in synapse that are crucial for neurodevelopment. Mutation of either gene or disruption of their interactions compromises synaptic gene expression and neurodevelopment, contributing to mental disorders.
Our genetic evidence suggests that Top3β and FMRP interact either antagonistically, as during Drosophila eye development and NMJ formation; or cooperatively, as in promoting synaptic expression of a specific gene, ptk2/FAK. The two opposite modes of interactions may be due to the ability of FMRP to either repress12 or enhance translation49,50, depending on the specific mRNAs involved. Thus, if the role of Top3β is to resolve mRNA topological stress to stimulate translation, FMRP may antagonize Top3β action on some mRNAs, but facilitate it on others. Because only a fraction of Top3β and FMRP associate with each other, we speculate that the FMRP fraction without the association may antagonize Top3β action, whereas those with the association may facilitate (Figure 8e,f). In the first scenario, the topological stress in mRNA may increase FMRP-mediated ribosomal stalling, which can be counteracted by Top3β because it can relieve the topological stress (Fig. 8e). Perhaps, the balanced actions of the two proteins can fine-tune expression of certain mRNAs within their optimal range to allow formation of normal synapses: these mRNA levels may become too high in dFmr1−/− mutant flies, too low in dTop3β−/− mutant, but near normal in dFmr1−/−/dTop3β−/− double mutant. In the second scenario, FMRP may assist Top3β to bind certain mRNAs to relieve topological stress and stimulate translation (Fig. 8f). Disruption of FMRP-Top3β association may lead to reduced translation of its target mRNAs, such as ptk2/FAK, contributing to abnormal neurodevelopment and mental disorders.
Methods
Cell culture and transfection
HeLa and HEK293 cells were cultured in DMEM supplemented with 10% fetal calf serum at 37 °C, 5% CO2. Transfections of plasmids and siRNAs were carried out with Lipofectamine 2000 and Lipofectamine RNAiMAX (Invitrogen), respectively, according to manufacturer' protocols. Top3β and TDRD3 expression vectors were made by linking their amino terminus in-frame with a double-tag containing 6xHistidine and a Flag51; the products obtained were then subcloned into pIRESpuro3 vector (Clonetech). The expression vectors for wildtype and mutant FMRP, which are all double-tagged with Flag and EGFP, were kindly provided by Dr. S. Ceman16. TDRD3 Smart-Pool siRNA oligos were purchased from Dharmacon. FMR1 siRNA oligos were purchased from Santa Cruz Biotech. Top3β−/− MEF cells were derived and cultured as previously described7.
DT40 cells were cultured at 39.5 °C, 5% CO2 in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% chicken serum, 10 mM Hepes and 1% penicillin-streptomycin mixture. Transfection was carried out by electroporation using the Amaxa Nucleofector2 in Solution T. For selection of stable clones, growth medium containing G418 (2 mg/ml), puromycin (0.5μg/ml) or Zeocin (0.5mg/ml) was used.
Antibodies and immunoblotting
Rabbit TDRD3 and Top3β polyclonal antibodies were made against MBP-fused proteins (New England Biolabs) containing the regions of TDRD3 (residues 620-751) and TOP3β (residues 61-148), respectively. These antibodies were affinity-purified using the corresponding immunogens as the affinity matrix and were subsequently used for immunoprecipitation and Western assays. A Top3β antibody was purchased from Sigma (WH0008940M1) and used to immunoprecipitate the endogenous Top3β complex from whole cell lysates. A FMRP mouse antibody was purchased from Millipore (MAB2160). A TIA-1 antibody was from Santa Cruz Biotech (SC-1751). An anti-Flag M2 monoclonal antibody was from Sigma, and anti-β-actin polyclonal antibodies were from Bethyl (A300-491). A synaptophysin antibody was from Cell Signaling Technology (#5461). A PSD95 antibody was from Thermol Scientific (MA1-046). A MAP2 antibody was from Sigmal-Aldrich (M9942). FXR1 and FXR2 antibodies were kindly provided by Dr. A. Hoogeveen. The Drosophila Top3β antibody was provided by Dr. T. Hiseh. A mouse monoclonal antibody to Drosophila FMRP was provided by Dr. D. Dreyfuss. The Drosophila FAK(FAK56D) antibody was from Dr. R. Hynes. We typically dilute primary antibody 1:1000 for immunoblotting with the exceptions of the following. The actin antibody was diluted 1:2000. The Drosophila Top3β and FMRP antibodies was diluted 1:3000. The representative images (including Western blots and immunohistochemistry staining) shown in the paper have been repeated at least twice, and the results are reproducible.
Fractionation, gel-filtration chromatography and immunoprecipitation
Preparation of nuclear and cytoplasmic extracts, fractionation of the extract by Superose 6 gelfiltration chromatography, and immunoprecipitation were performed as described previously52. Preparation of whole cell extracts and subsequent immunoprecipitation were performed as described 53. Mass spectrometry analysis was done by the Taplin Biological Mass Spectrometry Facility of Harvard Medical School, and the data are available upon request. The original purification of the Top3β-TDRD3 complex was performed using nuclear extract (Supplemental Figure 1a and data not shown), based on the assumption that Top3β is a DNA topoisomerase like Top3α. However, we later found out that Top3β-TDRD3 is an RNA toposiomerase complex, and is present in both nuclear and cytoplasmic extracts. We were able to purify the entire complex from unfractionated whole cell extract using a Top3β antibody (see Fig. 1b).
In control experiments to exclude the effect of RNA in mediating protein-protein interactions, RNase A was added to the whole cell extracts at final concentration of 0.1 mg/ml prior to immunoprecipitation, whereas RNase-out (0.4 unit/ml) was added to the control extract to inhibit RNA degradation by the endogenous RNases. The efficiency of RNase A digestion was verified by ethidium bromide staining of total RNA and by RT-PCR (data not shown).
Polyribosome Analyses
We performed the polyribosome assay as described 54.
HITS-CLIP assay
We performed HITS-CLIP assay as described27. Briefly, we cultured a HeLa cells stably-expressing HF-Top3β and irradiated the cells with UV to crosslink RNA and proteins in vivo. The level of HF-Top3β was about 2.5-fold of that of the endogenous protein. Following cell lysis, RNA was partially digested using various concentrations of RNase A. The RNA crosslinked to HF-Top3β was then immunoprecipitated with anti-Flag M2 argarose beads (Sigma). As a control, we also performed a mock immunoprecipitation using HeLa cells that do not express HF-Top3β. The 3′ ends of the purified RNAs were ligated to a p32-labeled RNA adapter, fractionated by SDS-PAGE, and transferred to a nitrocellulose membrane. The p32-labeled RNA-Top3β complexes (marked “H” and “L” in Figure 4A) were detected by autoradiography and recovered from the membrane. The complexes were treated with Proteinase K to remove proteins. The RNA was ligated at their 5′end to another RNA adaptor, and reverse-transcribed to cDNA. The cDNA obtained was fractionated a denaturing 6% TBE-Urea gel (InVitrogen), amplified by PCR, and subjected to high-throughput sequencing by Genome Analyzer II (Illumina). Sequenced tags were mapped to the human genome (hg18, NCBI/NIH) and to human RefSeq genes using ELAND program (Illumina)55. Only the tags from the “H” sample were shown in this manuscript. To minimize the potential off-target effects of the over-expressed HF- Top3β, we chose study the top 1500 mRNA targets of Top3β which contain at least 10 peaks of sequencing tags. These mRNAs represent less than 20% of total Top3β-bound mRNAs.
The top Top3β -bound mRNAs were compared to the published FMRP targets by Drs Darnell and Tuschl' groups13,56. In both cases, enrichment of FMRP targets in Top3β-bound mRNAs was observed (Figure 5; and data not shown). The top Top3β-bound mRNAs were also compared to a combined list of schizophrenia-related genes10,57,58, one of which was downloaded from the website: http://bioinfo.mc.vanderbilt.edu/SZGR/. The Top3β-bound mRNAs were also compared to autism spectrum disorder-related genes (https://gene.sfari.org/autdb/HG_Home.do).
Single-stranded DNA decatenation assay
Single-stranded DNA decatenation assays were performed as described24. Briefly, 400 fmol substrate was incubated with the indicated proteins in 15 μl of reaction buffer containing 50 mM Tris-HCl (pH 7.5), 40 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 0.1 mg/ml BSA at 37°C for 30 minutes. Reactions were stopped by the addition of 1% SDS and 20 mM EDTA. Samples were deproteinized with 1 mg/ml Proteinase K at 50°C for 1 hour before being subjected to 12% polyacrylamide + 8 M urea gel electrophoresis in TBE buffer at 175 V for 75 minutes. Gels were fixed in 10% MeOH/5% acetic acid for 30 minutes, and equilibrated in 1% glycerol/10% acetic acid for another 30 minutes prior to drying. Gels were then exposed to a storage phosphor screen and subsequently analyzed with a Typhoon scanner (GE Pharmacia) and ImageJ software.
RNA topoisomerase assay
The RNA substrates for topoisomerase assay were constructed and distinguished as described2, with several modifications. Briefly, a single-strand RNA (GGGAGAUUUUUUUUUUUUUUUUUUUUGUCAGACGGAUCUUUUUUUUUUUUUUUUUUUUUCUCCCGACUGGUUUUUUUUUUUUUUUUUUUUGAUCCGUCUGACUUUUUUUUUUUUUUUUUUUUCCAGUC) was transcribed by MEGAshortscript™ T7 Kit (Ambion) from an annealed DNA template consisting of synthetic oligos K128f and K128r (Supplementary Table 6). The transcription reaction was treated with 2 units of TURBO DNase (Ambion) at 37°C for 15 min to remove DNA. The RNA product was purified by electrophoresis on a 6% TBE-Urea gel (Ambion) for 40 min at 180 V and then labeled with [γ-32] ATP using KinaseMax Kit (Ambion).
For generation of the RNA knot, the above 32P-labeled RNA (1 μM) was denatured at 90°C for 5 min and cooled down slowly to 16°C to allow formation of intramolecular double strand helices. Ligation of RNA was performed in 50 μl ligation buffer using 0.1 unit/μl T4 RNA ligase (Ambion) at 16°C overnight. The products were purified by electrophoresis on a 15% TBE-Urea gel (Invitrogen) for 4-5 hour at 150 V.
For generation of the RNA circle, the above 32P-labeled RNA (2 μM) was annealed with 4 μM DNA oligo K128link (Supplementary Table 6). Ligation was performed in 50 μl ligation buffer using 0.1 unit/μl T4 RNA ligase (Ambion) at 37°C for 3 hours. The reaction was treated with 2 units of TURBO DNase (Ambion) at 37°C for 15 min to remove DNA. The RNA products were purified on a 15% TBE-Urea gel as above.
The RNA topoisomerase assay followed a previously published protocol2. Briefly, the 32P -labled purified RNA circle substrate (1 nM) was incubated with the indicated topoisomerase in 10 μl of reaction buffer containing 20 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 2 mm DTT, 0.1 mg/ml BSA, 5% glycerol and 10% PEG400 at 37 °C for 90 min. Reactions were terminated by adding 10 μl stop buffer (0.5% SDS, 20 mm EDTA and 0.2 mg/ml proteinase K) and incubated at 50 °C for 30 min. After phenol/chloroform extraction and ethanol precipitation, the mixtures were fractionated on 15% TBE-Urea gels and analyzed by autoradiography or PhosphoImager. The linear RNA was identified because its relative position to known RNA markers is unchanged in different percentage of PAGE gels. In contrast, the relative position of a knot versus its circle is altered: the knot has slower mobility than the circle on high percentage gels, but has equal or faster mobility on low percentage gels. In addition, conversion from a circle to a knot can be blocked when the self-annealing of the two complementary regions of the circle is inhibited by addition of a competing oligo that is also complementary to one of the regions (data not shown).
Gel-shift assays
We used a gel-shift assay to detect RNA and DNA binding activity of a MBP-fusion protein containing the RGG domain of Top3β (MBP-RGG). As a negative control, we included a fusion protein containing MBP and the N-terminal region of TDRD3 (residues 1-187) in the assay. The RNA and DNA oligos used in the assays are listed in Table S5. For ssRNA or ssDNA substrates: poly(rA)40, poly (rU)40 or poly (rC)40, RNA oligo rH1, and DNA oligo H1 (Supplementary Table 6) were end-labeled with Lsqb;γ-32P] ATP using T4 polynucleotide kinase (Roche) and used in the gel-shift assays. The dsRNA and dsDNA substrates were made by annealing oligo rH1 and rH5, oligo H1 and H5, respectively (Supplementary Table 6). The indicated amounts of proteins and 1 nM 32P-labelled RNA or DNA subtracts were incubated at 25 °C in 10 μl reaction buffer (25 mM Tris–HCl at pH 7.5, 4 mM MgCl2, 50 mM KCl, 1 mM DTT, 0.1 mg/ml BSA, and 5% glycerol) for 15 min. For competition assay, the indicated amounts of unlabeled subtracts were added for another 30 min. The reaction mixture was analyzed on a 4–12% TBE gel (Invitrogen).
Protein purification
Flag-tagged TOP3β and its mutants were expressed and purified from HEK293 cells. Briefly, suspension HEK293 cells were transfected with pcDNA-Flag-Top3β or its mutants with polyethylenimine (PEI). Cells were harvested after 3 days, washed with cold PBS twice, and lysed in 3.5 volume of lysis buffer (20 mM Tris-HCl pH 7.5, 500 mM NaCl, 10% glycerol, 0.5% NP40, 1 mM PMSF, 10 mM NaF and protease inhibitor cocktail) on ice. Cell Lysates was added with 2 volumes of cold Tris buffer (20 mM Tris-HCl pH 7.5) and ultracentrifuged at 90,000 rpm for 15 min in a TLA110 rotor (Beckman). The supernatant was then incubated with the anti-Flag M2-agarose (Sigma) at 4 °C for 3 hours. The agarose beads were washed three times with high salt washing buffer (50 mM Tris-HCl pH 7.5, 500 mM NaCl, 0.5% NP40, 1 mM EDTA, 10% glycerol and 1 mM PMSF) and once with elution buffer (25mM Tris-HCl pH7.5, 100 mM NaCl and 10% glycerol) at room temperature for 8 min each. Proteins were eluted from the beads using elution buffer containing 400 μg/ml 3×Flag peptide (Sigma).
Recombinant Top3α was purified as described previously24. MBP-RGG and MBP-TDRD31-187 fusion proteins were expressed and purified as MBP-Rif1C described previously 52.
Fly culture and strains
Flies were cultured on standard cornmeal food at 25±1°C and 60±5% humidity and under a 12h/12hr light/dark cycle. Canton S and w1118 lines served as wild type controls. w; cn/PES-dFmr1,CyO (sev-dFmr1) was kindly provided by Dr. D. Zarnescu; top3β mutants, top3β26 and top3β16, were gifts from Dr. T.S. Hsieh 5. All the other strains, including CG13472EY06846, Canton S, W; elav-gal4/Cyo, w1118; dfmr1Δ50M/TM6B, Tb1, y1w 67c23; P{EPgy2CG13472 EY06848 were obtained from the Bloomington stock center (Bloomington, IN, USA). The transgenic flies are made in w1118 background. We used both males and females in the experiments, and did not notice any significant differences. For studies of the Drosophila rough eye phenotype, we used age-matched wildtype and mutant adult flies between 2-3 weeks.
Immunoprecipitation and Western blot analyses of Drosophila cells
The entire coding region of dTDRD3 was amplified from a full length CG13472 cDNA clone (RE01471, Drosophila Genomics Resource Center, Indiana University). A Flag tag was added to the N-terminus of dTDRD3 coding sequence, which was then inserted into the expression vector pMT/V5-His (Cat. # K5120-01, Invitrogen) at NotI and XbaI sites. The dTDRD3 expression plasmid was transfected into Drosophila S2 cells, and the expression of Flag-dTDRD3 fusion protein was induced by incubating cells in 500 μM copper sulfate for 24 hours before harvesting. The cell lysates were incubated with anti-Flag M2-agarose (Cat. # A2220, Sigma-Aldrich) or protein G beads with specific antibodies to immunoprecipitate dTDRD3 and its interacting proteins. Samples were processed according to the manufacturer' instructions. The eluates from the antibody beads were loaded on 8-16% Tris-Glycine Gel (Invitrogen) for Western blot analysis.
Immunohistochemistry analysis of flies
Drosophila 3rd instar wandering larvae were dissected in cold Schneider's Drosophila Medium (Invitrogen) and fixed in 1xPBS with 4% paraformaldehyde (pH 7.4) for 25 minutes at room temperature. Samples were rinsed three times with 1xPBT (0.2% Triton X-100 in 1xPBS) and blocked with 1% normal goat serum (NGS) for 1 hour at room temperature. Primary and secondary antibodies were diluted with 1xPBT with 1% NGS and incubated overnight at 4°C and 2 hours at room temperature, respectively. Primary antibodies used in this study include: anti-Discs large (DLG) [4F3; 1:500 dilution] from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa city, IA) and DyLight 549 anti-horseradish peroxidase (1:500 dilution) from Jackson Immuno Research. Stained preparations were imaged with the Zeiss confocal microscopy system LSM 710 (Zeiss).
Morphological analyses of flies
For neuromuscular junction (NMJ) analyses, the NMJs at the muscle 4 of in the abdominal segments 3, 4, and 5 were imaged and used for branch quantification. Values were determined for both left and right hemisegments. A total of more than 6 larvae from each genotype were analyzed for branch numbers. An axonal project with at least two synaptic boutons is counted as a branch. Quantification of the number of boutons was performed at the NMJ of the muscle 4 in segments 3 to 5. At least a total of eighteen larvae were selected for bouton quantification. The experiment was performed blindly with regard to the genotype.
For eye morphological analyses, the compound eyes were imaged by the scanning electron microscope performed by Electron Microscopy Laboratory, NCI at Frederick. Eye section and staining were performed as described by Gaengel and Mlodzik (Drosophila Methods in Molecular Biology, 2008, Volume 420, 277-287).
Immunostaining analysis of the ptk2/FAK protein in Drosophila NMJs and Axons
Muscle 4 NMJs from 3rd instar larvae were co-stained with HRP (1:500 dilution), DLG (1:500 dilution) and ptk2/FAK (Fak56D; 1:1000 dilution) antibodies. Presynapses of NMJs and axons were labeled with an anti-HRP antibody (Red), whereas postsynapses were labeled with anti-DLG antibody (Blue). The FAK staining of the entire NMJ area, which was marked by DLG staining, was quantified using Imaris imaging software. After staining, eighteen sets of the NMJs from wild-type or top3β mutants were quantified.
Mouse primary neuronal culture
The Top3β knockout mice are in C57/Bl6 background and are kindly provided by Dr. J.C. Wang59. All animals are housed in compliance with National Institutes of Health (NIH) guidelines. Animal procedures are approved by the National Institute on Aging Animal Care and Use Committee. We followed a published protocol for cortical neuronal culture60. Briefly, embryonic day 17.5 mouse embryos were obtained from surgically sacrificed pregnant mouse and separated cerebral cortex under surgical stereomicroscope. Separated tissues were trypsinized (0.125%) for 10 min in 37°C. Finally, dissociated neurons were cultured on PDL/laminin coated coverslips. The number of total plated cell was adjusted to approximately 1× 106 cells/well. Culture media was Neurobasal medium plus B-27 supplement (2%). Culture media was changed once every 3 days through the entire in vitro period. At each time of media change, only half of the media was removed and replaced with the same amount of fresh media. The cells were cultured at 37°C and 5% CO2 for 10 days before fixation.
Immunostaining of synaptic makers in cultured primary neurons
For immunofluorescent labeling experiments, the cultured primary neurons were fixed with 4% paraformaldehyde for 20 min at 4°C and permeabilized for 15 min in a buffer containing 0.2% Triton X-100 in phosphate-buffered saline (PBS) on ice. Unspecific binding was blocked with 2.5% BSA in PBS for 1 h at room temperature. Incubation with the primary antibodies directed against the presynaptic synaptophysin (rabbit, Cell Signaling Technology, 1:400), the postsynaptic PSD95 (mouse, Thermol Scientific, 1:400) and dentritic MAP2 (mouse, Sigmal-Aldrich, 1:500) proteins was performed overnight at 4°C. The Alexa-488- and/or Alexa-568-coupled secondary antibodies (Invitrogen, 1:1000) were applied for 1 h. Counterstaining was performed with 4,6-diamidino-2-phenylindole (DAPI). Subsequently, images of cells were acquired and analyzed by using a fluorescence microscope (Delta Vision RT microscopy system). The stained synaptic markers were counted for at least 6 areas and 24 dendrites of each mouse. The experiment was performed blindly with regard to the genotype.
Statistics
Student' t-tests were performed to determine statistical significance of the differences in means between samples in experiments reported in Fig 6-8 and Supplementary Fig. 7. P<0.05 was considered statistically significant. Error bars represent standard errors (s.e.m.). We designed our experiments to have the sample size large enough so that the difference observed between the tested sample and the control sample is statistically significant (p value<0.05). No randomization was used. Data distribution was assumed normal but this was not formally tested. The data obtained (Fig. 6, 7, 8, Supplementary Fig. 7) are statistically different (p<0.001). Chi-square test was performed for data in Fig. 5d. Hypergeometric distribution test was performed for data in Supplementary Table 4.
Supplementary Material
Acknowledgments
We thank Drs. T. Hsieh for Drosophila Top3β reagents; J.C. Wang for Top3β knockout mice; D. Zarnescu, T. Jongens and G. Dreyfuss for dfmr1 fly strains and antibodies; S. Warren, S. Ceman and Y. Feng for vectors of FMR1 variants, U. Fischer for vectors of TDRD3 and FMR1 orthologs; A. Hoogeveen for FXR1 and FXR2 antibodies; R. Hynes and R. Palmer for FAK reagents; T. Enomoto for Top3β−/− DT40 cells, R. Hanai for Top3β vector. We thank Drs. E. Chen, DJ Pan, and Y. Feng for advice and assistance, S.K. Lee for assistance, and Dr. D. Schlessinger for support and critical reading of the manuscript. This work is supported in part by the Intramural Research Program of the National Institute on Aging (Z01 AG000657-08), National Institutes of Health, the Johns Hopkins Center for Neuroscience Research (NS050274), the Canadian Institutes of Health Research Grant MOP-79368 (to G. W. B.), the National Basic Research Program of China (2013CB911002) and National Natural Science Foundation of China (31271435). This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Footnotes
Database accession numbers: The raw HITS-CLIP data of Top3β have been deposited in GEO (accession number: GSE47502).
Author Contributions: DX, WS, RG, YX, WP, JS, JY, SS, JY, DF, JLM, YP, JM, SRJ, and SM conducted experiments; ACW, TEL, GWB, MSHK, MG, SZ and WW supervised the project; AS, YQ, DX, WS, RG, YX, WP, JS, JY, SS, ACS, TEL, GWB, MSHK, MG, SZ and WW analyzed the data; DX, WS, MG, SZ and WW wrote the manuscript with input from the other authors.
Competing financial interests: The authors declare that they have no competing interest in publishing this work.
References
- 1.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Wang H, Di Gate RJ, Seeman NC. An RNA topoisomerase. Proc Natl Acad Sci U S A. 1996;93:9477–9482. doi: 10.1073/pnas.93.18.9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, Wang JC. Mammalian DNA topoisomerase IIIalpha is essential in early embryogenesis. Proc Natl Acad Sci U S A. 1998;95:1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plank JL, Chu SH, Pohlhaus JR, Wilson-Sali T, Hsieh TS. Drosophila melanogaster topoisomerase IIIalpha preferentially relaxes a positively or negatively supercoiled bubble substrate and is essential during development. J Biol Chem. 2005;280:3564–3573. doi: 10.1074/jbc.M411337200. [DOI] [PubMed] [Google Scholar]
- 5.Wu J, Hou JH, Hsieh TS. A new Drosophila gene wh (wuho) with WD40 repeats is essential for spermatogenesis and has maximal expression in hub cells. Dev Biol. 2006;296:219–230. doi: 10.1016/j.ydbio.2006.04.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwan KY, et al. Development of autoimmunity in mice lacking DNA topoisomerase 3beta. Proc Natl Acad Sci U S A. 2007;104:9242–9247. doi: 10.1073/pnas.0703587104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohanty S, et al. Defective p53 engagement after the induction of DNA damage in cells deficient in topoisomerase 3beta. Proc Natl Acad Sci U S A. 2008;105:5063–5068. doi: 10.1073/pnas.0801235105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seki M, et al. Bloom helicase and DNA topoisomerase IIIalpha are involved in the dissolution of sister chromatids. Mol Cell Biol. 2006;26:6299–6307. doi: 10.1128/MCB.00702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu D, et al. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22:2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schizophrenia Psychiatric Genome-Wide Association Study C. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verkerk AJ, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 12.Bhakar AL, Dolen G, Bear MF. The Pathophysiology of Fragile X (and What It Teaches Us about Synapses) Annu Rev Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linder B, et al. Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum Mol Genet. 2008;17:3236–3246. doi: 10.1093/hmg/ddn219. [DOI] [PubMed] [Google Scholar]
- 15.Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 16.Stetler A, et al. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum Mol Genet. 2006;15:87–96. doi: 10.1093/hmg/ddi429. [DOI] [PubMed] [Google Scholar]
- 17.Yin J, et al. BLAP75, an essential component of Bloom's syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, et al. Crystal structures of RMI1 and RMI2, two OB-fold regulatory subunits of the BLM complex. Structure. 2010;18:1159–1170. doi: 10.1016/j.str.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goulet I, Boisvenue S, Mokas S, Mazroui R, Cote J. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum Mol Genet. 2008;17:3055–3074. doi: 10.1093/hmg/ddn203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khandjian EW, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- 22.Tamanini F, et al. FMRP is associated to the ribosomes via RNA. Hum Mol Genet. 1996;5:809–813. doi: 10.1093/hmg/5.6.809. [DOI] [PubMed] [Google Scholar]
- 23.Feng Y, et al. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Bachrati CZ, Ou J, Hickson ID, Brown GW. Human topoisomerase IIIalpha is a single-stranded DNA decatenase that is stimulated by BLM and RMI1. J Biol Chem. 2010;285:21426–21436. doi: 10.1074/jbc.M110.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson TM, Chen AD, Hsieh T. Cloning and characterization of Drosophila topoisomerase IIIbeta. Relaxation of hypernegatively supercoiled DNA. J Biol Chem. 2000;275:1533–1540. doi: 10.1074/jbc.275.3.1533. [DOI] [PubMed] [Google Scholar]
- 26.Siomi H, Choi M, Siomi MC, Nussbaum RL, Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 27.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan L, Dockendorff TC, Jongens TA, Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol Cell Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarnescu DC, et al. Fragile X protein functions with lgl and the par complex in flies and mice. Dev Cell. 2005;8:43–52. doi: 10.1016/j.devcel.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 30.Cziko AM, et al. Genetic modifiers of dFMR1 encode RNA granule components in Drosophila. Genetics. 2009;182:1051–1060. doi: 10.1534/genetics.109.103234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang YQ, et al. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- 32.Bhogal B, et al. Modulation of dADAR-dependent RNA editing by the Drosophila fragile X mental retardation protein. Nat Neurosci. 2011;14:1517–1524. doi: 10.1038/nn.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoll G, et al. Deletion of Top3b, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nature Neuroscience. 2013;16:1228–37. doi: 10.1038/nn.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu B, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilman SR, et al. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci. 2012;15:1723–1728. doi: 10.1038/nn.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan PF, et al. Family history of schizophrenia and bipolar disorder as risk factors for autism. Arch Gen Psychiatry. 2012;69:1099–1103. doi: 10.1001/archgenpsychiatry.2012.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 39.Rico B, et al. Control of axonal branching and synapse formation by focal adhesion kinase. Nat Neurosci. 2004;7:1059–1069. doi: 10.1038/nn1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu G, et al. Netrin requires focal adhesion kinase and Src family kinases for axon outgrowth and attraction. Nat Neurosci. 2004;7:1222–1232. doi: 10.1038/nn1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai PI, et al. Fak56 functions downstream of integrin alphaPS3betanu and suppresses MAPK activation in neuromuscular junction growth. Neural development. 2008;3:26. doi: 10.1186/1749-8104-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeiffer BE, Huber KM. Fragile X mental retardation protein induces synapse loss through acute postsynaptic translational regulation. J Neurosci. 2007;27:3120–3130. doi: 10.1523/JNEUROSCI.0054-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 44.Chen J, Kastan MB. 5′-3′-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev. 2010;24:2146–2156. doi: 10.1101/gad.1968910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svoboda P, Di Cara A. Hairpin RNA: a secondary structure of primary importance. Cell Mol Life Sci. 2006;63:901–908. doi: 10.1007/s00018-005-5558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brierley I, Gilbert RJ, Pennell S. RNA pseudoknots and the regulation of protein synthesis. Biochem Soc Trans. 2008;36:684–689. doi: 10.1042/BST0360684. [DOI] [PubMed] [Google Scholar]
- 47.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 48.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 49.Todd PK, Mack KJ, Malter JS. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci U S A. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwan KY, et al. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell. 2012;149:899–911. doi: 10.1016/j.cell.2012.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ling C, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26:2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu D, et al. Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J. 2010;29:3140–3155. doi: 10.1038/emboj.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Islam MN, Fox D, 3rd, Guo R, Enomoto T, Wang W. RecQL5 promotes genome stabilization through two parallel mechanisms--interacting with RNA polymerase II and acting as a helicase. Mol Cell Biol. 2010;30:2460–2472. doi: 10.1128/MCB.01583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee EK, et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat Struct Mol Biol. 17:732–739. doi: 10.1038/nsmb.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Ascano M, Jr, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun J, et al. A multi-dimensional evidence-based candidate gene prioritization approach for complex diseases-schizophrenia as a case. Bioinformatics. 2009;25:2595–6602. doi: 10.1093/bioinformatics/btp428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chandrasekaran S, B DG. A network view on Schizophrenia related genes. Network Biology. 2012;2:16–25. [Google Scholar]
- 59.Kwan KY, Wang JC. Mice lacking DNA topoisomerase IIIbeta develop to maturity but show a reduced mean lifespan. Proc Natl Acad Sci U S A. 2001;98:5717–5721. doi: 10.1073/pnas.101132498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim YG, Lee YI. Differential Expressions of Synaptogenic Markers between Primary Cultured Cortical and Hippocampal Neurons. Experimental neurobiology. 2012;21:61–67. doi: 10.5607/en.2012.21.2.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.