Abstract
microRNAs with their ability to regulate complex pathways that control cellular behavior and phenotype have been proposed as potential targets for cell engineering in the context of optimization of biopharmaceutical production cell lines, specifically of Chinese Hamster Ovary cells. However, until recently, research was limited by a lack of genomic sequence information on this industrially important cell line. With the publication of the genomic sequence and other relevant data sets for CHO cells since 2011, the doors have been opened for an improved understanding of CHO cell physiology and for the development of the necessary tools for novel engineering strategies. In the present review we discuss both knowledge on the regulatory mechanisms of microRNAs obtained from other biological models and proof of concepts already performed on CHO cells, thus providing an outlook of potential applications of microRNA engineering in production cell lines.
Keywords: MicroRNA engineering, Chinese Hamster Ovary cells, Bioprocess relevant properties
1. Introduction
1.1. MicroRNAs: from basics to applications
In the early 1990 small non-coding RNA molecules with the ability to regulate translation of target mRNAs by an antisense mechanism were discovered during developmental studies in the nematode worm (Lee et al., 1993; Wharton and Struhl, 1991). The much broader implications of these small RNAs were unacknowledged, until the identification of hundreds of similar small RNAs in a range of higher eukaryotes 7 years later. These newly discovered classes of small RNA molecules had striking similarities, such as lengths between 18 and 24 nucleotides and one or more, completely or partially complementary binding sites in 3′-UTR of mRNAs. These novel small RNAs were termed as microRNAs.
MicroRNAs were found to be expressed in a wide range of higher eukaryotes and to be highly conserved across species (Pasquinelli et al., 2000). With the availability of whole genome sequences, many more of these structurally and functionally distinct non-coding RNAs were discovered both experimentally and by bioinformatic prediction (Ambros and Lee, 2004; Ambros et al., 2003; Kozomara and Griffiths-Jones, 2011). The current version of miRBase (release 19.0) contains more than 20,000 entries from a wide variety of organisms, including viruses, plants and animals from nematodes to mammals (Griffiths-Jones, 2010). It is clear that such abundant molecules must have important functions and indeed, microRNAs have since been shown to be involved in all major cellular processes, such as cell division, death, embryonic development and timing, metabolism, host–virus interaction and tissue differentiation (Ambros, 2004; Scaria and Jadhav, 2007). Their function is based on antisense recognition of specific sequences in the target mRNA. They are able to regulate complex networks, due to the fact that a single microRNA can find targets in multiple mRNAs, while each mRNA may in turn contain binding sites for multiple microRNAs (Hobert, 2008).
The discovery and elucidation of the cellular machinery that controls this regulation enabled the development of siRNAs, a new toolbox whose discovery merited the Nobel Prize for Physiology in 2006 (Fire et al., 1998). In a recent review, the mechanism of action of microRNAs and their potential as biomarkers and novel drug targets was discussed (Bratkovic et al., 2012). In this review, we provide a detailed discussion of microRNAs with high relevance for optimizing cell lines used for biotechnological production of therapeutic proteins, with a focus on process-relevant properties such as growth, viability and apoptosis, productivity, stability and product quality. In addition to findings on microRNAs from other research areas that could be translated into cell engineering approaches, a comprehensive summary of CHO specific microRNA data will be given. The review concludes by discussing other non-coding RNAs with biological roles that might be of interest for cell culture technology.
2. CHO cell engineering — genome scale data is opening new doors
The most frequently used mammalian production cell line at industrial scales is the Chinese Hamster Ovary (CHO) cell line, isolated by Theodore Puck in the late 1950s (Puck et al., 1958). These lines were applied as recombinant hosts in 1987 with the commercial introduction of human tissue plasminogen activator (tPA) as the first recombinant therapeutic protein produced from mammalian cells (Finkle, 1988). Since then, the annual revenue of products from CHO cells has increased to more than 100 billion US dollars and continues to grow (Aggarwal, 2011). One of the major reasons for the success story of CHO cells is their adaptability and plasticity with respect to the phenotypic characteristics that are necessary for industrial production of therapeutic proteins (Jayapal et al., 2007): growth in suspension, adaptation to a number of steadily improving chemically defined media with ease, and production of proteins of high quality suitable for safe use in humans with low occurrence of immunologic reactions.
Silenced expression of specific surface proteins accounts for their low susceptibility to viral infections (Xu et al., 2011) and a high number of clones have been generated with distinct glycosylation patterns (Borisov et al., 2009; Imai-Nishiya et al., 2007; Yamane-Ohnuki et al., 2004) that enhance the therapeutic efficacy of the product (Jefferis, 2012). Despite the success story of CHO cells, the above mentioned plasticity also has drawbacks. First, a large number of clones need to be screened for each new production of cell line to identify the few that unite all the necessary phenotypic properties, such as high product quality, fast growth, high productivity and prolonged viable culture life in large scale bioprocesses. Second, once a suitable clone is identified, both phenotype and productivity can be subject to instability and phenotypic drift, resulting in a limited number of doublings over which a cell line can be used reliably.
These issues have been the focus of research over the last 25 years with only limited solutions having been found (Hacker et al., 2009). The most significant improvements to CHO culture so far have resulted from the optimization of media, feeding strategies and processes. This has resulted, at least for antibody products, in yields commonly ranging between 2 and 6 g/l (Wurm, 2004), with titers of 20 g/l being reported (Kim et al., 2012). Engineering of cell lines has resulted in improved product quality (Huang et al., 2012a; North et al., 2010; Pouilly et al., 2012; Raju et al., 2001) and some improvements in productivity (Figueroa et al., 2007); however, no successes comparable to the enhancements obtained by media optimization have been reported so far, and the most prominent problems of mammalian cell culture, such as the efficient energy utilization (Zeng et al., 1998) have not been completely resolved by metabolic engineering strategies (Kim and Lee, 2007a,b; Park et al., 2000). Most of these metabolic engineering strategies were limited to expressing single genes expected to shift metabolic pathways (Banmeyer et al., 2004; Hou and Li, 1987a,b; Jeon et al., 2011) or to making cells more resistant to apoptosis triggered by nutrient depletion or hyperosmolarity (Figueroa et al., 2007; Fussenegger et al., 2000; Lim et al., 2006; Park et al., 2000; Sauerwald et al., 2006; Sung et al., 2007; Wong et al., 2006). However, cells have redundant mechanisms to control cellular physiology, meaning that cells have different options to reach the same goal (Charaniya et al., 2009, 2010; Dinnis and James, 2005), which has been nicely demonstrated in recent years in several multiparameter -omics studies (Chong et al., 2010, 2011; Doolan et al., 2010; Meleady et al., 2012b; Wuest et al., 2012; Zhao et al., 2012). This has resulted in a paradigm shift from single gene engineering towards the control of signaling pathways, which can be achieved either by targeting regulatory hubs such as MAPK or mTOR (Dreesen and Fussenegger, 2011; Kim et al., 2011) or gene networks that control biological processes, which can be achieved by transcription factor or microRNA engineering (Barron et al., 2011b; Hackl et al., 2012a; Müller et al., 2008). microRNAs have the advantage that they do not add a translational burden onto production cells while being able to orchestrate complex gene networks in a coordinated fashion. However, as pointed out by Bratkovic et al. (2012), any research on CHO microRNA biology (or any other biology in CHO) has been severely restricted by the lack of genomic sequence information, at least until recently.
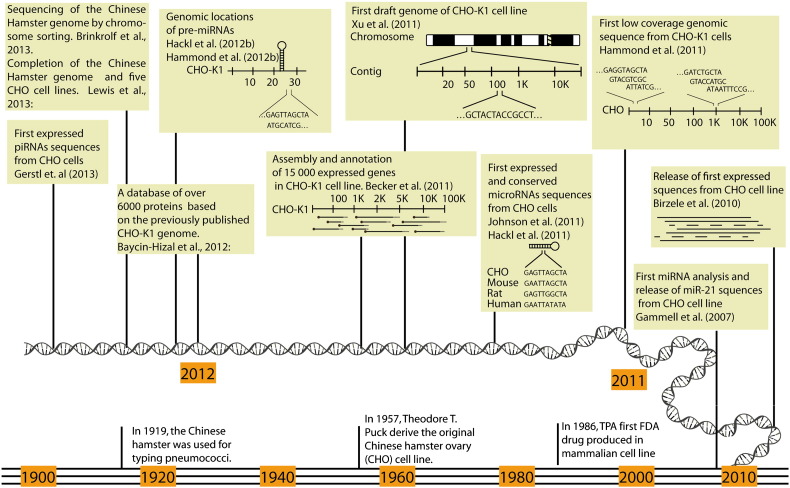
Although a consortium has been working on sequencing a CHO EST library since 2004 (Wlaschin et al., 2005), it was only during the last two years that a flood of sequencing data on CHO cells has become publically available, thus setting the stage for a new era of scientific exploration and innovation in CHO biology and engineering. The milestones of published genomic and transcriptomic sequence data are summarized in Fig. 1 (Gammell, 2007; Birzele et al., 2010; Hammond et al., 2011; Johnson et al., 2011; Hackl et al., 2011; Xu et al., 2011; Becker et al., 2011; Hackl et al., 2012b; Hammond et al., 2012b; Baycin-Hizal et al., 2012; Gerstl et al., 2013; Lewis et al., in press; Brinkrolf et al., 2013) and the respective datasets including updates are now assembled and available online at www.CHOgenome.org for easy access and reference (Hammond et al., 2012b). With this information, several essential tools can be developed that facilitate an improved understanding of CHO cell physiology and provide the impetus for groundbreaking novel engineering strategies: i) CHO-specific mRNA, cDNA or whole-genome microarrays can be designed based on CHO sequence data, and analysis of next-generation sequencing data and proteomics data will be simplified by well-annotated references; ii) sequence alignment and primer design tools will facilitate gene cloning as well as site-specific gene knock-in and knock-out approaches during cell line development; and iii) mature and stemloop microRNA sequences will enable the design of anti-sense inhibitors (termed antagomirs or anti-miRs) or mimics for overexpression (Krützfeldt et al., 2005). The importance of a sequenced host genome can be deduced from the fact that overexpression of mature microRNAs in CHO cells was higher when the autologous CHO stemloops were used rather than an artificial stemloop routinely used for expression of siRNAs (personal communication).
Fig. 1.
Timeline showing recent boom in Chinese Hamster Ovary (CHO) cell genome science with increase in publications on sequence information and annotation. These developments have significantly advanced the establishment of new and improved tools for cell engineering and bioprocess development.
Even without such tools and CHO-specific sequence data, the potential of using microRNAs to engineer the most important process-relevant properties was recognized very early, and multiple groups began to explore their use (Barron et al., 2011b; Müller et al., 2008). In the following chapter we discuss those cellular characteristics that are relevant for recombinant protein production (Fig. 2), summarizing both knowledge obtained from other biological models (Table 1) and experiments and proof of concepts already performed on CHO cells as production cell lines (Table 2), thus setting the stage for future developments of microRNA engineered cell lines.
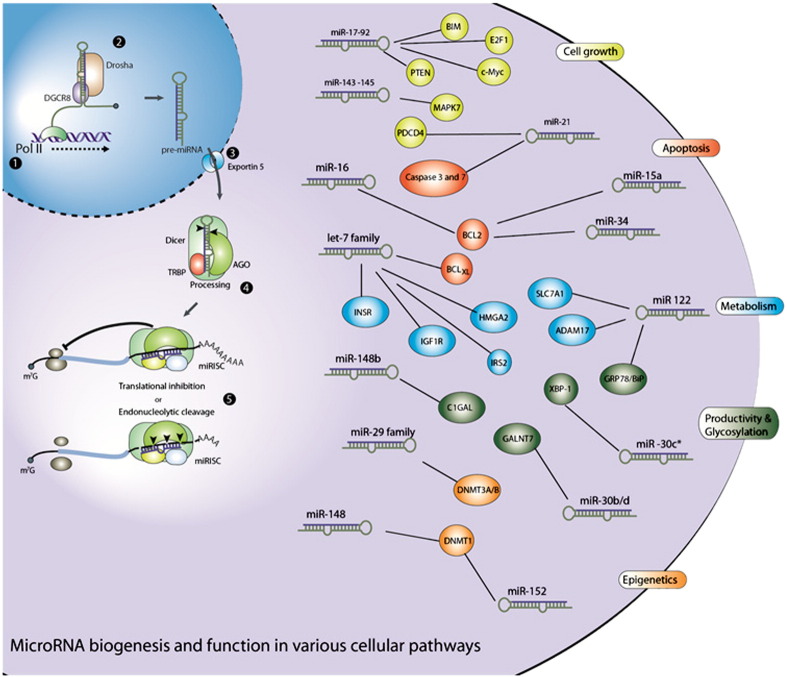
Fig. 2.
Biogenesis and function of microRNA in controlling process relevant cellular properties.
Biogenesis: (1) canonically, microRNAs are transcribed by RNA polymerase II to generate primary transcript (pri-microRNAs), long capped and polyadenylated RNAs with hairpin structure. (2) First processing steps are mediated by the microprocessor complex, consisting of Drosha and DiGeorge syndrome critical region 8 (DGCR8) which produces a ~ 70 nt hairpin structured RNA known as a precursor-microRNA (pre-microRNA). (3) Pre-microRNA are exported from the nucleus by the Exportin-5–Ran–GTP complex. (4) In the cytosol further processing occurs by Dicer together with TRBP and Argonaut proteins 1–4 (AGO), resulting in the active microRNA-induced silencing complex (miRISC). (5) miRISC binds to its target mRNA, mediating translational inhibition or cleavage. Function of microRNA: selected examples of microRNA discussed in the text that influence process relevant cellular processing by post-transcriptionally controlling expression of genes, highlighting the complexity of the regulatory network and interactions between the different pathways.
Table 1.
miRNAs controlling cellular processes and their identified targets.
| Biological process | Manuscript section | MicroRNA identifier | Effect | Selection of confirmed targets | Annotated in CHO (miRBase v20) |
|---|---|---|---|---|---|
| Cell growth | 3.1. | let-7 | Tumor-suppressive | Ras, BCL-XL | Yes |
| miR-143–145 | Tumor-suppressive | MAPK7 | Yes | ||
| miR-17–92 | Oncogenic | c-Myc, E2F1, PTEN, Bim, HIF-1α | Yes | ||
| miR-21 | Oncogenic | PDCD4, Caspases 3 and 7 | Yes | ||
| miR-7a | Growth arrest | Stathmin | Yes | ||
| Apoptosis & cell death | 3.2. | miR-1 | Pro-apoptotic | HSP60, HSP70 | Yes |
| miR-133 | Pro-apoptotic | Caspase 9 | No | ||
| miR-144/155 | Pro-apoptotic | Caspase 3 | Yes | ||
| miR-15a-16 | Pro-apoptotic | BCL-2, BCL-XL | Yes | ||
| miR-218 | Pro-apoptotic | ECOP | Yes | ||
| miR-297–669 | Pro-apoptotic | BCL2L2, DAD1, BIRC6, STAT5a, SMO | No | ||
| miR-34 | Pro-apoptotic | BCL-2, SIRT1 Deacetylase | Yes | ||
| Hypoxia & oxidative stress | 3.2. | miR-107/210/26 | Hypoxia inducible, prevent apoptosis | Multiple pro-apoptotic genes | Yes |
| miR-31 | Supports HIF-1α induction | FIH | Yes | ||
| miR-144/451 | Oxidative stress protective | NRF2, 14-3-3ξ | Yes/no | ||
| Shear & osmotic stress | 3.2. | miR-200b/717 | Osmolarity responsive | OREBP | Yes/no |
| miR-7b | Osmolarity responsive | FOS | Yes | ||
| Energy metabolism | 3.3. | let-7 | Glucose metabolism | INSR, IGF1R, IRS2, HMGA2 | Yes |
| miR-122 | Liver metabolism | Multiple cholesterol related genes | Yes | ||
| miR-124/137/340 | Glycolysis rate | Pyruvate Kinase Isozymes (PKM1/2) | Yes | ||
| miR-23a | Glutamine metabolism | Suppressed by C-Myc, targets GLS | Yes | ||
| miR-33a/33b | Fatty acid and insulin metabolism | Multiple enzymes in cholesterol synthesis | Yes | ||
| Productivity protein expression | 3.4. | miR-122/30/181d/199a-5p | UPR | GRP78/BiP | Yes |
| miR-204 | ER-stress | SERP1/RAMP4, M6PR | Yes | ||
| miR-221/222 | Induce ER-stress | p27Kip1, MEK/ERK | Yes | ||
| miR-30c* | UPR | XBP-1 | Yes | ||
| miR-7 | Shift from growth to translation | Multiple ribosomal genes | Yes | ||
| miR-708 | ER-stress inducible | Rhodopsin | Yes | ||
| Protein quality | 3.4. | miR-148b | N-glycosylation | C1GALT1 | Yes |
| miR-30b/d | O-glycosylation | GALNT7 | Yes | ||
| Epigenetic | 3.5. | miR-29 | DNA methylation | DNMT3A/3B | Yes |
| miR-148/152 | DNA methylation | DNMT1 | Yes |
Table 2.
Summary of miRNA analysis and engineering in CHO cells.
| Experimental setting | Type of analysis | Outcome | Reference |
|---|---|---|---|
| Temperature shift | Microarray & qPCR | 26 regulated miRNAs | Gammell (2007) |
| Transcription in recombinant cell lines | qPCR | 16 miRNAs with de-regulation in recombinant DG44 cell lines | Lin et al. (2010) |
| Temperature shift | qPCR | 10 regulated miRNAs (miR-7) miR-7 reduces growth and enhances qP |
Barron et al. (2011a) |
| MicroRNA repertoire in various cell lines | NGS | 380 conserved 22 novel miRNAs |
Hackl et al. (2011) |
| MicroRNA repertoire in various cell lines | NGS | 350 conserved miRNAs | Johnson et al. (2011) |
| Batch cultivation | Microarray & qPCR | 118 miRNAs regulated during batch cultivation between lag, exponential and stationary growth phase | Bort et al. (2012) |
| Nutrient depletion & apoptosis | Microarray | 70 miRNAs with regulation upon nutrient limitation | Druz et al. (2012) |
| MicroRNA overexpression screen | Engineering | miR-17 improves growth miR-21 reduces qP |
Jadhav et al. (2012) |
| Transcription in recombinant cell lines | NGS | 190 conserved microRNAs 93 microRNA regulated in two distinct recombinant cell lines |
Hammond et al. (2012a) |
| Genomic context of microRNAs | In silico | Genomic annotation of 350 miRNAs | Hackl et al. (2012b) |
| Correlation to growth rate | Microarray | 35 miRNAs with positive correlation to growth rate 16 miRNAs with negative correlation to growth rate |
Clarke et al. (2012) |
| Specific microRNA knockdown | Engineering | miR-466h-5p knockdown improves batch performance of CHO cells | Druz et al. (2013) |
3. Process relevant properties as targets for microRNA based cell engineering
3.1. Cell growth
Since their discovery, microRNAs were thought to play critical roles in modulating cell cycle arrest, cell proliferation and cell death. A clear link came from studies of microRNA expression profiling in human cancer that lead to an understanding of the relationship between microRNA function and cancer phenotype (Blenkiron et al., 2007; Jiang et al., 2005; Porkka et al., 2007; Solomides et al., 2012; Yang et al., 2008; Yao et al., 2009). Differential expression studies revealed that the majority of microRNAs are expressed at significantly lower levels in a variety of tumors compared to normal tissues. Deregulation of microRNA expression can be both tumor suppressive or oncogenic (oncomirs), with differentially expressed microRNAs associated with pathways such as cell cycle, cell growth and cell death (Lee and Dutta, 2006).
Tumor suppressor microRNAs function by down-regulating oncogenes. The first identified tumor suppressor microRNAs shown to regulate the expression of an oncogene were let-7 family members. They regulate Ras genes, which are membrane-associated GTPases involved in signaling of cellular growth and differentiation (Johnson et al., 2005). In addition, miR-143 and miR-145 negatively regulate mitogen-activated protein kinase 7 (MAPK7) at a posttranscriptional level, thus reducing growth rate in human cell lines (Lin et al., 2009; Noguchi et al., 2011).
Similarly, oncogenic microRNAs interact with tumor-suppressor genes and have either pro-proliferative or anti-apoptotic function. Recent studies revealed that miR-17–92 cluster overexpression drives tumorigenesis under the intricate network of c-Myc and E2F, and was also shown to have bi-functional effects, acting either as oncogene or as tumor suppressor in a cell type dependent manner (Cho, 2007; Grillari et al., 2010; Mendell, 2008). For example, the miR-17-5p and miR-20a target E2F1 are transcription factors that promote cell proliferation in normal human cells but induce apoptosis in cancer cell lines (Cloonan et al., 2008; Hackl et al., 2010; Li et al., 2011c; Olive et al., 2010).
Another oncogenic microRNA – miR-21 – was described as playing a critical role in the development and progression of lung cancer by regulating multiple genes controlling several pathways including JAK/STAT, MAPK, PPAR signaling and cell cycle related pathways, based on a systematic analysis of literature and gene network studies (Frezzetti et al., 2011; Guan et al., 2012; Hatley et al., 2010). It can also promote migration and invasion of human hepatocellular carcinoma by targeting PDCD4 in a negative feedback loop (Lu et al., 2008; Si et al., 2007). Furthermore, several microRNAs were described to act similarly in support of cell proliferation in different cell types (Esquela-Kerscher and Slack, 2006).
Identification and application of oncomirs or tumor suppressor microRNAs in CHO cell engineering are interesting challenges (Müller et al., 2008). Here two different aims are of relevance: the ideal bioprocess consists of an initial extremely fast growth phase to reach full biomass quickly, followed by a non-growing, high-productivity state in which cells accumulate high yields of product. Thus microRNAs could potentially be used both for their growth-enhancing and -repressing function. In a detailed transcriptomic analysis in CHO cells aimed at understanding the microRNA–mRNA network dynamics during the course of a batch culture a set of 10 microRNAs were identified as down-regulated during stationary phase, with their target mRNA levels up-regulated (Bort et al., 2012). The functions of these mRNAs were enriched for cell cycle and programmed cell death, suggesting them as good engineering targets to control cell death and proliferation during the late stages of bioprocess. In a similar approach, Barron et al. (2011a) identified miR-7 as significantly down-regulated during a temperature shift from 37 °C to 31 °C. Interestingly, contrary to expectations, the transient overexpression of miR-7 led to growth arrest, resulting in increased recombinant protein production, generally observed during temperature shift culture conditions. In a follow-up study, the effect of miR-7 overexpression on the proteome was analyzed, revealing proteins involved in protein folding and secretion to be up-regulated, while targets that control protein translation and nucleic acid processing were down-regulated (Meleady et al., 2012a). The productivity enhancement was thus affected by an improvement in protein processing, while two mRNAs, Stathmin and catalase, were identified as potential direct targets of miR-7, which caused the growth arrest. More recent work, in which miR-7 levels were stably depleted in CHO cells using a ‘sponge’, showed a marked increase in cell proliferation and improved longevity later in batch-fed culture (personal communication).
In the microRNA sequencing experiments, Hackl et al. (2011) found a significant difference in the overall expression of microRNAs in CHO cells when comparing cells grown in serum-containing medium or adapted to protein free media. In a follow-up study on this global microRNA regulation, the relevance of Dicer, one of the key enzymes during microRNA biogenesis, for maintenance of growth in CHO cells was studied (Hackl et al., under review). Dicer mRNA and protein levels quickly decrease in response to nutrient depletion or serum removal. Conversely, Dicer expression during the exponential growth phase is 3 fold higher in fast growing, protein-free and suspension-adapted CHO cells (μ ~ 1.0 d− 1) compared to slow growing cells (μ ~ 0.5 d− 1), and siRNA mediated down-regulation of Dicer expression reduces the proliferation rate of CHO cell lines. Growth of such slow-growing cells could be increased by 20% following recombinant expression of human Dicer, suggesting a link between the overall microRNA load in CHO cells and growth behavior.
However, since the underlying limit is likely to be found in microRNA transcription as opposed to post-transcriptional processing by Dicer, the preferential approach is to titrate the expression of specific microRNAs with high impact on specific growth to an optimal expression level that will facilitate fast proliferation. Such an approach was recently taken by Clarke et al., who performed integrated analysis of microRNA, mRNA and protein expression in a set of clones with variable growth rate (Clarke et al., 2012). In total, 35 miRNAs were identified to be up-regulated with increased growth, and 16 miRNAs that were down-regulated. By combining this information with mRNA and protein expression data, certain biological processes such as mRNA processing and protein synthesis were found to be relevant for enhanced proliferation. In silico analysis of microRNA-mediated regulation of these pathways resulted in a high-priority list of microRNAs for use as cell engineering targets or biomarkers, such as microRNA-17-92.
3.2. Apoptosis, cell viability and culture stress
Apoptosis, or programmed cell death, is a necessary physiological function which helps eliminate unhealthy cells, however, the cascade presents difficulties in maintaining high viable cell densities in mammalian bioprocess applications (Müller et al., 2008). Stress conditions in bioreactors, including nutrient limitation, byproduct accumulation, shear and oxidative stress, osmolality and hypoxia, can trigger apoptosis during CHO cell cultures. Apoptosis onset in bioreactors lowers the Integral Viable Cell Density (IVCD) and affects product yield and quality (Druz et al., 2011; Gammell, 2007; Müller et al., 2008). As a result, apoptosis prevention is one of the most widely implemented approaches in CHO cell engineering (Hacker et al., 2009; Majors et al., 2007; Zanghi et al., 1999).
The involvement of microRNAs in apoptosis regulation was initially described in peripheral blood cells of people diagnosed with chronic lymphocytic leukemia (CLL) where a deletion of miR-15a/16 cluster was reported in the majority of patients (Calin et al., 2002). Later studies revealed that the members of this cluster, miR-15a-5p and miR-16, promote apoptosis in malignant B cells by targeting Bcl-2 expression at the post-transcriptional level (Cimmino et al., 2005). Another member of miR-15a/16 cluster, miR-15a-3p, was shown to induce apoptosis by targeting Bcl-xL, thus activating Caspase-3/7 and reducing viability in several cancers (Druz et al., 2013). Another microRNA, miR-21 was found to be up-regulated in several human cancers and characterized as an oncogenic microRNA. Its silencing in glioblastoma cells led to increased apoptosis by activation of Caspases 3 and 7 (Chan et al., 2005; Meng et al., 2006; Si et al., 2007). Cheng and colleagues identified several microRNAs involved in apoptosis regulation using large-scale antisense microRNA inhibition in HeLa cells. The inhibition of miR-1d, 7, 148, 204, 210, 216 and 296 increased apoptosis by activation of Caspase 3, while the inhibition of miR-214 had the opposite effect (Cheng et al., 2005).
Other examples include miR-218 which was found to be involved in NF-kappaB response and apoptosis induction by targeting expression of ECOP gene (Gao et al., 2010). miR-34 family members function as potent mediators of the p53-induced apoptotic pathway by targeting anti-apoptotic genes including Bcl-2, and also participate in a positive feedback loop of p53 activation via increased acetylation by targeting SIRT1 deacetylase (Hermeking, 2010). miR-30 affects the levels of the Ubc9 and ITGB3 genes in breast tumor-initiating cells, thus restricting their self-renewing capacity and targeting them for apoptosis (Yu et al., 2010). miR-10a was shown to participate in the TRAIL-induced apoptosis pathway leading to Caspase 3 activation in human lung carcinomas (Ovcharenko et al., 2007). The members of let-7 microRNA family, let-7c and let-7g, target Bcl-xL directly and Mcl-1 indirectly which leads to Caspase-3/7 activation and apoptosis induction in hepatocytes (Shimizu et al., 2010).
Two of the stresses that cells are exposed to under bioreactor conditions are hypoxia and oxidative stress. Here, involvement of microRNAs has been demonstrated in several instances, however, not yet in CHO cells. miR-15a-5p, miR-16, and miR-20a are down-regulated during hypoxic conditions in human carcinomas (Hua et al., 2006), while miR-26, miR-107, and miR-210 are up-regulated in neoplastic cells. These microRNAs are likely to decrease the pro-apoptotic signaling in a hypoxic environment (Kulshreshtha et al., 2007). miR-210 is progressively up-regulated in endothelial cells in hypoxic conditions and inhibits the receptor tyrosine-kinase ligand Ephrin-A3 which is critical for vascular development (Fasanaro et al., 2008). The up-regulation of the miR-34 family, as part of the p53 network, can be implicated in stress responses to DNA damage, hyperactive cytokine signaling, and hypoxia (He et al., 2007). The miR-17–92 cluster was shown to target hypoxia-inducible factor alpha (Hif-1α), a transcriptional factor known to regulate cellular response to hypoxia and to play an important role in various biological processes such as glucose metabolism, pH regulation and angiogenesis (Taguchi et al., 2008). miR-31 was also shown to activate Hif-1α via the inhibition of factor-inhibiting hypoxia-inducible factor (FIH) (Liu et al., 2010).
With respect to oxidative stress, the bicistronic transcript miR-144/451 was shown to modulate oxidative stress in erythroid cells. miR-144 directly affects NRF2 gene expression in K562 and primary erythroid progenitor cells, which induces the expression of several antioxidant enzymes (Sangokoya et al., 2010), while miR-451 protects erythrocytes against oxidative stress and rescues erythroid cells' differentiation defect by inhibiting the intracellular regulator of cytokine signaling, 14-3-3ξ gene (Patrick et al., 2010). miR-34a and miR-93 are involved in the loss of oxidative stress defense and repress expression of genes associated with oxidative stress regulation and defense mechanism such as Sp1, Sirt1, Mgst1, and Nrf2 (Li et al., 2011b). miR-1 and miR-133 produce opposing effects on apoptosis induced by oxidative stress in rat cells (Xu et al., 2007). miR-1 has a pro-apoptotic function in response to oxidative stress by targeting heat shock proteins HSP60 and HSP70 and miR-133 seems to have an anti-apoptotic role by repressing Caspase 9 gene expression.
Some microRNAs have been associated with other stress types that might be encountered within a bioreactor such as osmotic pressure, shear stresses, and nutrient depletion/gradients. miR-200b and miR-717 are down-regulated by isotonic and hypertonic treatments in renal medullary epithelial cells. However, when up-regulated, these microRNAs inhibit the activity of a transcriptional factor called osmotic response binding protein, OREBP, a major cellular osmoregulator in kidney cells and T-lymphocytes (Huang et al., 2010). miR-7b is over-expressed in hyperosmolar conditions to down-regulate the protein levels of Fos, which reduces the activity of transcription factor activator protein 1 (AP1), a regulator of cellular processes, which is formed by dimerization of Fos and Jun proteins (Lee et al., 2006). miR-21 and miR-19a are induced by shear stress in endothelial cells (Qin et al., 2010; Weber et al., 2010).
Druz and colleagues recently showed the up-regulation of the large miR-297–669 cluster during apoptotic conditions induced by nutrient depletion in CHO cells. One member of this cluster, miR-466h-5p was shown to alter the expression of five anti-apoptotic genes from different apoptosis-initiating pathways (bcl2l2, dad1, birc6, stat5a, and smo). This microRNA was shown to be activated in response to glucose depletion (Druz et al., 2012). Antisense knockdown of miR-466h-5p delayed apoptosis onset in nutrient-depleted conditions by decreasing Caspase-3/7 activation and increasing cell viability (Druz et al., 2011). Stable inhibition of miR-466h-5p in CHO cells enhanced apoptosis resistance and increased protein production (Druz et al., 2013). One other member of miR-297–669 cluster, miR-669c-5p, has been previously associated with impairments in glutathione metabolism which activates the apoptosis cascade (Lanceta et al., 2010; Maes et al., 2008).
The utilization of microRNAs with roles in apoptosis regulation and stress response should provide researchers with an additional tool to minimize or eliminate apoptotic effects that result from different stress conditions that reduce recombinant protein production. The recent sequencing of the CHO cell genome and microRNA transcriptome revealed conserved sequences of the apoptosis and stress-regulating microRNAs such as miR-1, let-7 family, miR-7b, miR-10a, miR-15/16 cluster, miR-93, miR-107, miR-144, miR-200b, miR-210, miR-214, miR-218, and mi-R708. These microRNAs and the miR-297–669 cluster are promising targets for apoptosis pathway engineering. Due to the complexity of apoptosis and the diversity of apoptotic stimuli, it may be useful to investigate the combined effects of several microRNAs affecting genes from different stages of the apoptosis cascade as well as those that seem to be involved in global regulation of the pathway. It might be also worthwhile to consider the engineering of whole clusters of apoptosis-relevant microRNAs since clustered microRNAs are known to be transcribed together as polycistronic transcripts to regulate mRNA of genes with similar functions (Druz et al., 2011), thus enabling a more global approach to modification of cellular phenotypes. An important aspect here is the need to test CHO cell-specific microRNAs, their biological role and their effects on CHO cell-specific gene targets not only in small scale, but also in an industrial-scale bioprocess environment to finally identify the most suitable candidates and combinations.
3.3. Energy metabolism
Metabolic homeostasis is crucial for maintenance of cellular physiology for optimal growth and adaptation to culture conditions. Culture adaptations are controlled by complex regulatory networks and have to continuously evolve to monitor and respond to such changes during a bioprocess to keep it efficient. In a recent review the role of microRNAs and their integration into multilayered cellular networks functioning to maintain physiological conditions was discussed (Rottiers and Naar, 2012). The regulation of microRNA expression in response to genetic, epigenetic or environmental cues, to metabolic wastes or stress may contribute to our understanding of cellular physiology and metabolism (Lynn, 2009; Tomankova et al., 2010). The first link connecting microRNAs to metabolic control is miR-122, which is involved in lipid metabolism and regulates genes involved in cholesterol biosynthesis, such as 7-dehydrocholesterol reductase, 3-hydroxy-3-methylglutaryl-CoA synthase-1 and 3-hydroxy-3-methylglutaryl-CoA reductase (Jopling, 2012; Sacco and Adeli, 2012). Application of an LNA antagomir of miR-122 leads to 25–30% reduction of plasma cholesterol levels (Elmen et al., 2008; Esau et al., 2006). Similarly, miR-33a and miR-33b were shown to co-express from an intron of the transcription factor of sterol-regulatory element-binding proteins, a family of proteins that controls the expression of genes central to fatty acid homeostasis. They thus coordinate the regulation of fatty acid, triglyceride and cholesterol biosynthesis and uptake (Davalos et al., 2011; Rayner et al., 2010). Recent interesting findings suggested functions for let-7 in regulation of mammalian glucose metabolism by targeting IGF1R, INSR, and IRS2 components of the insulin–PI3K–mTOR pathway (Zhu et al., 2011). The ratio of Pyruvate Kinase Isozymes M1/M2 (PKM) is involved in control of glycolysis rate and it was suggested that miR-124, miR-137 and miR-340 are involved in the regulation of PKM1/2 ratios in colorectal cancer cells (Sun et al., 2012). In human leukemic Jurkat cells, it was shown that the expression of miR-23a was controlled by NF-κB and could play a critical role in glutamine metabolism (Rathore et al., 2012). Mitochondria are central to all energy, related functions of the cell, and recently it was proposed that several microRNAs are associated with them and could control their function (Carrer et al., 2012; Sripada et al., 2012a,b). Over several decades, studies have suggested that cellular and physiological responses to nutrients such as glucose, lipids, growth factors and metabolic wastes and their respective levels, induce drastic changes in gene expression patterns and altering metabolic homeostasis. The link between nutrient levels and microRNAs could be exploited by using them as metabolic sensors and/or modulators, both to fine tune and to monitor biologically controlled bioprocesses (Carrer et al., 2012; Druz et al., 2012; Kochanowski et al., 2012; Zanghi et al., 1999).
3.4. Productivity and product quality
The ability of a CHO clone to synthesize and secrete correctly modified recombinant proteins in abundance is a key attribute. Clearly, there are numerous steps, enzymes, co-factors, quality control points and internal structures involved in ensuring this process operates efficiently. All of them are subject to regulation within the cell and depend on both extracellular and intracellular stimuli, with microRNAs playing an important role in several of these processes. As mentioned above, increasing the levels of miR-7 in CHO cells has been reported to increase cellular productivity – at the expense of cell growth – which is reflected in a shift in the abundance of particular ribosomal proteins in the cell and other proteins involved in translation elongation (Meleady et al., 2012a). Lin and colleagues screened microRNAs in recombinant human IgG producing CHO cells and found miR-221 and miR-222 to be significantly down-regulated in all cell lines when compared with the parental DG44 cell line, indicating good targets for engineering high producer cell lines (Lin et al., 2010). The miR-221/222 cluster was also found down-regulated during ER stress in human hepatocellular carcinoma cells. The ectopic introduction of miR-221/222 mimics increased ER-stress and induced apoptosis which was associated with p27Kip1 and MEK/ERK-directed cell cycle regulation (Dai et al., 2010).
In other model systems, microRNAs have been implicated in regulating proteins involved in the unfolded protein response — a key cellular stress response that impacts on recombinant protein production. miR-30c-2* can down-regulate the expression of XBP-1, a critical mediator of cellular adaptation to increased protein processing load (Byrd et al., 2012). This gene has been successfully engineered in CHO cells to improve productivity in the past (Tigges and Fussenegger, 2006).
Several microRNAs including miR-122 (Yang et al., 2011), miR-30, 181d and 199a-5p have been shown to suppress GRP78/BiP, another cellular chaperone involved in UPR and one that has been given some attention in the context of recombinant protein production in CHO cells (Morris et al., 1997; Van Dyk et al., 2003). miR-708 was shown to be induced during ER stress by the transcription factor CCAAT enhancer-binding homologous protein (CHOP) and may facilitate the enhancement of ER protein-folding capacity under the stress of accelerated protein synthesis (Behrman et al., 2011). miR-204 supported ER and oxidative stress induction in human trabecular meshwork cells by inhibition of two genes involved in the elimination of damaged and misfolded proteins (SERP1/RAMP4 and M6PR), thus enhancing the expression of carbonylated proteins (Li et al., 2011a). microRNAs are also known to be key regulators of pancreatic beta cell function. In particular, miR-375 can influence glucose-induced insulin secretion by modulating the expression of myotrophin, a protein potentially involved in cytoskeleton dynamics (Poy et al., 2004). In addition, a number of microRNAs are involved in various aspects of exocytosis that have implications in other human diseases (Lovis et al., 2008; Sullivan et al., 2012; Zhang et al., 2011). Finally, mTOR overexpression has recently been shown to confer benefit to both CHO cell productivity and growth (Dreesen and Fussenegger, 2011) and mTOR has also been identified as a protein whose expression can be regulated by microRNA binding (Liu and Wilson, 2012). These observations demonstrate the potential that manipulation of particular microRNAs may have in engineering aspects of the secretory and high productivity function of CHO cells. This is also supported by consistent patterns of microRNA expression observed between different host cell lines and their recombinant, high producing subclones (Hackl et al., 2011; Lin et al., 2010).
A major aspect of recombinant protein production is product quality, specifically the pattern of glycosylation on the therapeutic product. Several instances indicate that microRNAs may also play a role in the control of this important property. It was recently found that a specific microRNA, miR-148b, modulates the expression of β1,3-galactosyltransferase-1 (C1GALT1), an important enzyme in the synthesis of O-glycosylation (Coppo and Amore, 2004; Novak et al., 2001).
In another study focused on the regulation of glycosylation and its impact on cancer metastasis, it was shown that the up-regulation of microRNA clusters suppresses N-acetylgalactosamine transferases (GALNTs) which initiate O-linked glycosylation (Gaziel-Sovran and Hernando, 2012). Specifically, miR-30b/30d expression was shown to silence GALNT7, resulting in defective glycosylation and changes in protein exocytosis. To this end, no study has looked in detail on microRNA target sites in enzymes mediating the formation of N- and O-glycosylation in animal cells. However, the above examples provide evidence that microRNAs are involved in these processes. Therefore, this justifies a systematic analysis of microRNA target site enrichment in CHO glycosylation genes, which will unveil the potential value of microRNAs as diagnostic and engineering tools to control the precise pattern of glycosylation required for production of biosimilars.
3.5. Clonal stability and epigenetics
At an early stage of the biopharmaceutical product development pipeline, a major R&D challenge is to accelerate progress from the point of having cloned an appropriate gene for a biopharmaceutical product into a CHO cell, to having established the best CHO cell clone – in terms of growth rate, productivity, product quality and stability – to place in the bioreactor for manufacture of clinical trial batches (and with a reasonable level of confidence that the same clone can be used for subsequent large-scale production, in order to avoid expensive and time-consuming new cell line development and re-validation of the process for regulatory approval).
Clone to clone variation is hugely important with respect to many bioprocess-relevant cellular phenotypes, including productivity (O'Callaghan et al., 2010; Pichler et al., 2011; Pilbrough et al., 2009; Porter et al., 2010; Prieto et al., 2011; Sigal et al., 2006; Sunley et al., 2008). Currently, clone selection is done on a trial-and-error basis, and many initially promising clones prove, at a later stage in the process, to be unstable, thus making the process unpredictable. Methylation of the viral promoters commonly used in mammalian expression vectors is believed to play a role (especially in slow loss of productivity over time) (Osterlehner et al., 2011; Yang et al., 2010). Loss of transgene amplification when certain selection systems (e.g., DHFR) have been used and DNA rearrangements in and around the transgene can occur; these mechanisms are probably most important in rapid loss of productivity (Kim et al., 2011).
A considerable literature exists in relation to stability/instability of cells in culture (e.g., Bailey et al., 2012). Substantial recent research has been done on the effects of including features in the expression vector that discourage epigenetic silencing. These include insulators such as the chicken β-globin HS4 element; (S) MARs — ((Scaffold) Matrix Attachment Factors); STARs (stabilizing anti-repression elements); and UCOEs (ubiquitous chromosome opening elements) (Allen and Antoniou, 2007; Galbete et al., 2009; Harraghy et al., 2012). While the proximate biochemical mechanisms of silencing (e.g. DNA methylation, histone deacetylation, reduction of copy number, and DNA rearrangement) and the identity of corresponding enzymes are reasonably well understood, there is no in-depth understanding of the molecular regulatory events that lead to these modifications in some clones, but not in their sister clones, which are being cultured at the same time under the same conditions. Of course, the impact of epigenetic changes in CHO cells goes beyond transgene stability; it may also influence the stability of other characteristics including growth and product quality.
Since microRNAs are believed to regulate expression of over 50% of proteins, it is likely that they have a role in regulation of expression of the enzymes involved in DNA and chromatin modification (Lorio et al., 2010). However, limited information exists in this regard, and no information at all is available for CHO. One of the few microRNAs with a recognized role in epigenetic modifications in cancer is the miR-29 family, which was shown to target DNA Methyltransferases (DNMT) 3A and 3B (Fabbri et al., 2007). In doing so, these microRNAs prevent inappropriate methylation at the promoters of tumor-suppressor genes and their expression has been shown to be down-regulated in tumor cells. On the other hand there are several reports of epigenetic changes impacting on microRNA expression.
A recent publication by Druz et al. (2012) demonstrated how the biogenesis of microRNAs can be influenced by epigenetic events. In this case, glucose depletion in the culture medium led to histone deacetylase inhibition, increased promoter acetylation and subsequently increased transcription of miR-466h-5p. Although this work was performed in mouse cells, this microRNA had previously been shown by the same group to increase resistance to apoptosis in CHO cells (Druz et al., 2011). As with most genetic regulatory networks, there is evidence of feedback loops between microRNAs and their target genes or proteins. A good example of this feedback is the link between miR-148 and miR-152 and DNMT1. The promoters of these microRNAs are silenced by DNMT1-dependent methylation, and DNMT1 itself is, in turn, a target for repression by these miRs (Xu et al., 2012). It is becoming more apparent, as an increasing number of reports appear, that there is a strong inter-relationship between microRNA activity and the epigenetic status of cells, and it will be interesting to see how this relationship might be exploited in the bioprocessing area.
3.6. Tools for probing and engineering microRNA function in CHO cells
Sections 3.1 to 3.5. give clear evidence of microRNA-mediated regulation of gene expression in a range of biological processes that are of high relevance for a robust and excellent performing CHO cell line (Table 1). But should one directly translate these valuable insights into experimental engineering approaches in CHO? The answer unfortunately has to be no, since microRNA function strongly depends on the cellular mRNA transcriptome, and can therefore be extremely diverse between different cell types or even different states of the same cell type (Shu et al., 2012) (which likely includes different CHO host cell lines as well). Hence, two roads can be taken in order to prioritize microRNAs for stable engineering: i) promising microRNA candidates from expanded literature searches can be probed for their applicability as engineering targets in CHO cells using classical reverse genetic approaches (Jadhav et al., 2012; Müller et al., 2008) and ii) transcriptomic experiments can be helpful in reducing the list of engineering candidates to a few microRNAs that can be directly taken to transient and stable functional analyses, as in the case of miR-7 (Barron et al., 2011a) and miR-466h-5p (Druz et al., 2013). In the following an overview of currently available methods for transient and stable overexpression and knockdown of microRNA is given.
3.6.1. miRNA loss of function by antagomirs
Antagomirs are antisense miRNA oligonucleotides, which are currently the most widely used molecules for targeted miRNA inhibition (Kaur et al., 2007; Krützfeldt et al., 2005). They have been applied successfully to test miRNA function in cell culture systems and as well as animal models (Pasquinelli, 2012). Some antagomirs contain chemical modifications to increase their binding to a target miRNA and/or serve as protection from nucleases. One of the most common chemical modifications are 2-O-methyl or 2-O-methoxyethyl and locked nucleic acid (LNA) (Fabani and Gait, 2008). A prominent example for using antagomirs is the blocking of miR-122 in the liver in order to reduce replication of hepatitis-C virus, which is dependent on high miR-122 levels in the liver (Gottwein, 2013). Besides microRNA sequestration through antagomirs, targeted microRNA cleavage was established by introducing a catalytic domain from DNAzymes to the antagomir. These molecules are called “antagomirzymes” and function by binding and cleaving complementary microRNA sequences (Jadhav et al., 2009).
Antagomirs are well suited as transient tools for testing microRNA function in animal cell models if they can be efficiently delivered into the cytoplasm of a cell. For long-term effect (i.e. during a fed-batch or continuous bioprocess), antagomirs would need to be repeatedly delivered to the cell via a media feed, thus, requiring highly effective and cheap delivery methods (Stein et al., 2010) for suspension cells. While there have been reports of transfection reagent free uptake of small RNAs into cells (“naked” or “gymniotic” uptake) (Lingor et al., 2004; Moschos et al., 2011), these are challenging protocols and still would require large amounts of synthetic RNA to be delivered to the culture media. Therefore, methods have been developed that employ synthetic antagomir molecules produced by transcription from simple expression vectors and are termed “decoys” or “sponges” (Yang et al., 2012).
3.6.2. miRNA loss-of-function by miRNA sponges
miRNA sponges are synthetic RNA molecules that act as pseudo target by presenting a dominant amount of miRNA binding sites to a cell, thus acting as scavenger for miRNA function. Fittingly, these synthetic RNA molecules are termed “sponges” or “decoys”. Ebert and colleagues described for the first time the application of miRNA sponges for knockdown of miRNA function. They designed and engineered tandem repeats of specific miRNA binding sites into the 3′-UTR of green fluorescent protein reporter genes and demonstrated more effective inhibition of miRNA function compared to other methods such as antagomirs. Furthermore sponges can be designed in such a way, that an entire family of miRNAs can be inhibited. Thereafter, several studies have successfully applied miRNA sponges in both in vitro cell culture systems and in vivo for inhibiting miRNA function (Brown and Naldini, 2009). Druz and colleagues have already applied this tool to generate engineered CHO cells (Druz et al., 2013), which overexpress miR-466h-5p sponge and thereby exhibit a modulated growth behavior. The future directions in CHO cell engineering include the design and development of microRNA-sponge expression systems that are based on an inducible system for culture-stage specific fine tuning of microRNA activity.
3.6.3. miRNA gain-of-function by miRNA mimics and vector based expression
Modulating miRNA function by overexpression is a prominent alternative for cell engineering and commonly termed “miRNA-targeting” or “miRNA-gain-of-function” strategy. Two main approaches have been developed for ectopic expression, which depend on delivery of a synthetic microRNA (termed “microRNA mimic”) (Wang, 2011) or vector-based transcription of microRNA precursors, which are further processed to yield the mature microRNA of interest.
The miRNA mimic technology results in silenced target gene translation by introducing synthetic double-stranded RNA molecules with sequences equivalent to an endogenous miRNA, thus reinforcing the biological effect. Compared to vector-based screening of microRNA function, mimics have the advantage of being readily available without the need for cloning. Hence, high-throughput screenings of microRNA function usually employ large synthetic libraries that cover the entire miRNome available for a specific organism. The shortcomings of this strategy are i) the limitation to transient overexpression with short-term effects, especially in cell lines exhibiting rapid proliferation and therefore dilution of synthetic RNA sequences, ii) the synthetic nature of these molecules and their chemical modifications, which could have cytotoxic effects, and iii) the usually high initial increase in microRNA copies per cell (usually several hundred-folds) might result in off-target effects. Therefore, the alternative strategy uses the endogenous miRNA maturation pathway of a cell to ectopically produce mature miRNAs from plasmids containing a primary microRNA transcript. Such constructs can be used for both transient and stable expression of miRNAs to study long term effect in gain-of-function. Furthermore, the choice of specific promoters allows to time microRNA expression with the entry of specific culture stages and provides a further opportunity to control miRNA function. Jadhav and colleagues have developed vector based expression system for screening microRNA function in CHO cells. The reported protocol uses in silico designed stem-loop sequences that are synthesized and readily cloned into a commercial expression vector. Thus, albeit the need for cloning, a medium-throughput protocol for gain-of-function screen of selected microRNAs could be established, which identified positive effects for miR-17 on CHO growth performance (Jadhav et al., 2012).
3.7. The need for better computational tools to predict microRNA function
Theoretically, the labor- and time-intensive phase of functional screening could be replaced by computational tools that reliably predict the biological function of a microRNA in a certain transcriptomic environment. However, the improvement of tools for the identification of interactions between microRNAs and their respective targets remains a key challenge of microRNA research. When microRNAs were first identified, sequence analysis tools were developed for the prediction of such interactions, exploiting conserved seeds and sequence complementarity (Lewis et al., 2005). Limitations of early seed-motif matching approaches led to the integration of thermodynamic models for binding strength. The challenges of reliable de novo prediction are, however, reflected in the lack of agreement between different tools (Rajewsky, 2006; Sethupathy et al., 2006). While focusing on predictions common to several tools has been a popular strategy to try and reduce false positives, this comes at a considerable cost in terms of false negatives. High false positive/negative rates therefore motivate more elaborate attempts at integration of multiple tools (Zhang and Verbeek, 2010).
In a complementary trend, individual tools now consider and combine additional data sources. Algorithms may (1) incorporate other relevant computational predictions, like target site accessibility (Kertesz et al., 2007), (2) take advantage of multi-track measurements, such as matched microRNA and mRNA expression profiles (Bonnet et al., 2010), or (3) use expression profiles to refine sequence based predictions (Stingo et al., 2010; Wang and Li, 2009). Increased enrichment of targets in known pathways suggests that the incorporation of additional information as filters for sequence based predictions may be a promising strategy for reducing false positive rates (Muniategui et al., 2012).
Lack of agreement across methods has nevertheless remained an issue and concordance with experimentally validated or refuted interactions was found to be poor in an evaluation of a large independent data set (Shridhar and Kreil, in press). Prediction performance as well as true interactions may be specific to certain experimental settings. Indeed, some approaches have been tailored for particular experimental designs like time-course data (Jayaswal et al., 2009) or try to exploit data from different tissues or heterogeneous cell mixtures to identify additive interactions between microRNAs and multiple targets (Wang and Li, 2009). The unavailability of an experimentally validated comprehensive ‘gold standard’ list of interactions and absence of interactions, on the other hand, has been a major limitation for the development of improved analysis tools. Recent collection of data from high-throughput techniques like HITS-CLIP and TAP-tar for probing physical interactions (Yang et al., 2011) may over time, fill this need. Collections of evidence covering a variety of experimental scenarios will provide more powerful data to train and validate new analysis methods.
4. Conclusions and future perspectives
With respect to microRNA engineering of CHO cells for enhanced phenotypes, an important aspect is that many of the phenotypes discussed in the previous sections overlap (Fig. 2). Therefore, it is likely that a cell line with optimal properties needs to express a variety of microRNAs in a coordinated and balanced way. Apart from new engineering strategies, a better understanding of the microRNAs' roles in determining phenotypes could also lead to the development of novel screening tools that allow better prediction of cell behavior at industrial scale, by analyzing the precise expression pattern of those microRNAs that were previously identified to control these properties. With improvements in analysis methods, these could be assessed during an early stage in cell line development, from small scale cultures, as a novel screening tool for the identification of suitable clones and as tools for monitoring cell behavior and state in production processes. Emerging technologies, such as flow cytometry assisted RNA quantification (Chapin et al., 2011) that could be directly applied to a cell homogenate without prior RNA isolation, might become valuable tools for integration of biomarkers in the process of cell line development.
Finally, even with the research on microRNAs and their application for CHO cell engineering in full swing, we would like to point out that microRNAs are by no means the only interesting non-coding RNAs that could be used for cell line optimization, specifically in the field of genomic and phenotypic stability. A recent manuscript identifies piRNAs expressed in CHO cells (Fig. 1) (Gerstl et al., 2013), another group of small non-coding RNAs that have been linked to epigenetic and post-transcriptional gene silencing, specifically of retrotransposons, by their interaction with PIWI proteins (Sienski et al., 2012). In addition, long non-coding RNAs are considered to be involved in the regulation of gene transcription, both by controlling the basal transcriptional machinery and by gene-specific regulation via recruitment of epigenetic modifying factors to genomic loci, and modulation of mRNA splicing, as well as translation (Huang et al., 2012b). Both of these classes of RNA promise to enable exciting new approaches to cell line optimization for industrial purposes in the near future.
Acknowledgments
VJ and SS were funded by Biotop, a PhD program funded by the Austrian Science Fund, W1224. MH is the recipient of a BOKU DOC grant. AD and YS acknowledge support by the intramural research program of the NIDDK/NIH. NBarron is funded by the Science Foundation Ireland. DPK acknowledges funding by the Vienna Science and Technology Fund (WWTF), the Baxter AG, the Austrian Research Centre Seibersdorf and the Austrian Centre of Biopharmaceutical Technology. NBorth acknowledges support by the Austrian Center of Industrial Biotechnology, ACIB.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Aggarwal S. What's fueling the biotech engine—2010 to 2011. Nat Biotechnol. 2011;29:1083–1089. doi: 10.1038/nbt.2060. [DOI] [PubMed] [Google Scholar]
- Allen M.L., Antoniou M. Correlation of DNA methylation with histone modifications across the HNRPA2B1-CBX3 ubiquitously-acting chromatin open element (UCOE) Epigenetics-Us. 2007;2:227–236. doi: 10.4161/epi.2.4.5231. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ambros V., Lee R.C. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey L.A., Hatton D., Field R., Dickson A.J. Determination of Chinese hamster ovary cell line stability and recombinant antibody expression during long-term culture. Biotechnol Bioeng. 2012;109(8):2093-03. doi: 10.1002/bit.24485. [DOI] [PubMed] [Google Scholar]
- Banmeyer I., Marchand C., Verhaeghe C., Vucic B., Rees J.F., Knoops B. Overexpression of human peroxiredoxin 5 in subcellular compartments of Chinese Hamster Ovary cells: effects on cytotoxicity and DNA damage caused by peroxides. Free Radic Biol Med. 2004;36:65–77. doi: 10.1016/j.freeradbiomed.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Barron N., Kumar N., Sanchez N., Doolan P., Clarke C., Meleady P. Engineering CHO cell growth and recombinant protein productivity by overexpression of miR-7. J Biotechnol. 2011;151:204–211. doi: 10.1016/j.jbiotec.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Barron N., Sanchez N., Kelly P., Clynes M. MicroRNAs: tiny targets for engineering CHO cell phenotypes? Biotechnol Lett. 2011;33:11–21. doi: 10.1007/s10529-010-0415-5. [DOI] [PubMed] [Google Scholar]
- Baycin-Hizal D., Tabb D.L., Chaerkady R., Chen L., Lewis N.E., Nagarajan H. Proteomic analysis of Chinese Hamster Ovary cells. J Proteome Res. 2012;11:5265–5276. doi: 10.1021/pr300476w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J., Hackl M., Rupp O., Jakobi T., Schneider J., Szczepanowski R. Unraveling the Chinese Hamster Ovary cell line transcriptome by next-generation sequencing. J Biotechnol. 2011;156(3):227–235. doi: 10.1016/j.jbiotec.2011.09.014. [Dec 10] [DOI] [PubMed] [Google Scholar]
- Behrman S., Acosta-Alvear D., Walter P. A CHOP-regulated microRNA controls rhodopsin expression. J Cell Biol. 2011;192:919–927. doi: 10.1083/jcb.201010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birzele F., Schaub J., Rust W., Clemens C., Baum P., Kaufmann H. Into the unknown: expression profiling without genome sequence information in CHO by next generation sequencing. Nucleic Acids Res. 2010;38:3999–4010. doi: 10.1093/nar/gkq116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron C., Goldstein L.D., Thorne N.P., Spiteri I., Chin S.F., Dunning M.J. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E., Michoel T., YVd Peer. Prediction of a gene regulatory network linked to prostate cancer from gene expression, microRNA and clinical data. Bioinformatics. 2010;26:i638–i644. doi: 10.1093/bioinformatics/btq395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov O.V., Field M., Ling V.T., Harris R.J. Characterization of oligosaccharides in recombinant tissue plasminogen activator produced in Chinese Hamster Ovary cells: two decades of analytical technology development. Anal Chem. 2009;81:9744–9754. doi: 10.1021/ac901498k. [DOI] [PubMed] [Google Scholar]
- Bort J.A.H., Hackl M., Hoflmayer H., Jadhav V., Harreither E., Kumar N. Dynamic mRNA and microRNA profiling of CHO-K1 suspension cell cultures. Biotechnol J. 2012;7:500–515. doi: 10.1002/biot.201100143. [DOI] [PubMed] [Google Scholar]
- Bratkovic T., Glavan G., Strukelj B., Zivin M., Rogelj B. Exploiting microRNAs for cell engineering and therapy. Biotechnol Adv. 2012;30:753–765. doi: 10.1016/j.biotechadv.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Brinkrolf K., Rupp O., Laux H., Kollin F., Ernst W., Linke B. Chinese hamster genome sequenced from sorted chromosomes. Nat Biotechnol. 2013;31:694–695. doi: 10.1038/nbt.2645. [DOI] [PubMed] [Google Scholar]
- Brown B.D., Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- Byrd A.E., Aragon I.V., Brewer J.W. MicroRNA-30c-2* limits expression of proadaptive factor XBP1 in the unfolded protein response. J Cell Biol. 2012;196:689–698. doi: 10.1083/jcb.201201077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer M., Liu N., Grueter C.E., Williams A.H., Frisard M.I., Hulver M.W. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci U S A. 2012;109:15330–15335. doi: 10.1073/pnas.1207605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.A., Krichevsky A.M., Kosik K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chapin S.C., Appleyard D.C., Pregibon D.C., Doyle P.S. Rapid microRNA profiling on encoded gel microparticles. Angew Chem. 2011;50:2289–2293. doi: 10.1002/anie.201006523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charaniya S., Karypis G., Hu W.S. Mining transcriptome data for function–trait relationship of hyper productivity of recombinant antibody. Biotechnol Bioeng. 2009;102:1654–1669. doi: 10.1002/bit.22210. [DOI] [PubMed] [Google Scholar]
- Charaniya S., Le H.O., Rangwala H., Mills K., Johnson K., Karypis G. Mining manufacturing data for discovery of high productivity process characteristics. J Biotechnol. 2010;147:186–197. doi: 10.1016/j.jbiotec.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Cheng A.M., Byrom M.W., Shelton J., Ford L.P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.C. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong W.P.K., Reddy S.G., Yusufi F.N.K., Lee D.Y., Wong N.S.C., Heng C.K. Metabolomics-driven approach for the improvement of Chinese Hamster Ovary cell growth: overexpression of malate dehydrogenase II. J Biotechnol. 2010;147:116–121. doi: 10.1016/j.jbiotec.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Chong W.P.K., Yusufi F.N.K., Lee D.Y., Reddy S.G., Wong N.S.C., Heng C.K. Metabolomics-based identification of apoptosis-inducing metabolites in recombinant fed-batch CHO culture media. J Biotechnol. 2011;151:218–224. doi: 10.1016/j.jbiotec.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Cimmino A., Calin G.A., Fabbri M., Iorio M.V., Ferracin M., Shimizu M. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C., Henry M., Doolan P., Kelly S., Aherne S., Sanchez N. Integrated miRNA, mRNA and protein expression analysis reveals the role of post-transcriptional regulation in controlling CHO cell growth rate. BMC Genomics. 2012;13:656. doi: 10.1186/1471-2164-13-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N., Brown M.K., Steptoe A.L., Wani S., Chan W.L., Forrest A.R. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008;9:R127. doi: 10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppo R., Amore A. Aberrant glycosylation in IgA nephropathy (IgAN) Kidney Int. 2004;65:1544–1547. doi: 10.1111/j.1523-1755.2004.05407.x. [DOI] [PubMed] [Google Scholar]
- Dai R., Li J., Liu Y., Yan D., Chen S., Duan C. miR-221/222 suppression protects against endoplasmic reticulum stress-induced apoptosis via p27(Kip1)- and MEK/ERK-mediated cell cycle regulation. Biol Chem. 2010;391:791–801. doi: 10.1515/BC.2010.072. [DOI] [PubMed] [Google Scholar]
- Davalos A., Goedeke L., Smibert P., Ramirez C.M., Warrier N.P., Andreo U. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnis D.M., James D.C. Engineering mammalian cell factories for improved recombinant monoclonal antibody production: lessons from nature? Biotechnol Bioeng. 2005;91:180–189. doi: 10.1002/bit.20499. [DOI] [PubMed] [Google Scholar]
- Doolan P., Meleady P., Barron N., Henry M., Gallagher R., Gammell P. Microarray and proteomics expression profiling identifies several candidates, including the Valosin-Containing Protein (VCP), involved in regulating high cellular growth rate in production CHO cell lines. Biotechnol Bioeng. 2010;106:42–56. doi: 10.1002/bit.22670. [DOI] [PubMed] [Google Scholar]
- Dreesen I.A.J., Fussenegger M. Ectopic expression of human mTOR increases viability, robustness, cell size, proliferation, and antibody production of Chinese Hamster Ovary cells. Biotechnol Bioeng. 2011;108:853–866. doi: 10.1002/bit.22990. [DOI] [PubMed] [Google Scholar]
- Druz A., Chu C., Majors B., Santuary R., Betenbaugh M., Shiloach J. A novel microRNA mmu-miR-466h affects apoptosis regulation in mammalian cells. Biotechnol Bioeng. 2011;108:1651–1661. doi: 10.1002/bit.23092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druz A., Betenbaugh M., Shiloach J. Glucose depletion activates mmu-miR-466h-5p expression through oxidative stress and inhibition of histone deacetylation. Nucleic Acids Res. 2012;40:7291–7302. doi: 10.1093/nar/gks452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druz A., Son Y.J., Betenbaugh M., Shiloach J. Stable inhibition of mmu-miR-466h-5p improves apoptosis resistance and protein production in CHO cells. Metab Eng. 2013;16:87–94. doi: 10.1016/j.ymben.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J., Lindow M., Schutz S., Lawrence M., Petri A., Obad S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Esau C., Davis S., Murray S.F., Yu X.X., Pandey S.K., Pear M. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Slack F.J. Oncomirs — microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Fabani M.M., Gait M.J. miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14:336–346. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanaro P., D'Alessandra Y., Di Stefano V., Melchionna R., Romani S., Pompilio G. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa B., Jr., Ailor E., Osborne D., Hardwick J.M., Reff M., Betenbaugh M.J. Enhanced cell culture performance using inducible anti-apoptotic genes E1B-19K and Aven in the production of a monoclonal antibody with Chinese Hamster Ovary cells. Biotechnol Bioeng. 2007;97:877–892. doi: 10.1002/bit.21222. [DOI] [PubMed] [Google Scholar]
- Finkle B. New medicines from industry. J Chem Technol Biotechnol. 1988;43:313–327. [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Frezzetti D., De Menna M., Zoppoli P., Guerra C., Ferraro A., Bello A.M. Upregulation of miR-21 by Ras in vivo and its role in tumor growth. Oncogene. 2011;30:275–286. doi: 10.1038/onc.2010.416. [DOI] [PubMed] [Google Scholar]
- Fussenegger M., Fassnacht D., Schwartz R., Zanghi J.A., Graf M., Bailey J.E. Regulated overexpression of the survival factor bcl-2 in CHO cells increases viable cell density in batch culture and decreases DNA release in extended fixed-bed cultivation. Cytotechnology. 2000;32:45–61. doi: 10.1023/A:1008168522385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbete J.L., Bucetaz M., Mermod N. MAR elements regulate the probability of epigenetic switching between active and inactive gene expression. Mol Biosyst. 2009;5:143–150. doi: 10.1039/b813657b. [DOI] [PubMed] [Google Scholar]
- Gammell P. MicroRNAs: recently discovered key regulators of proliferation and apoptosis in animal cells: identification of miRNAs regulating growth and survival. Cytotechnology. 2007;53:55–63. doi: 10.1007/s10616-007-9049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Zhang Z., Liu W., Xiao S., Gu W., Lu H. Reduced microRNA-218 expression is associated with high nuclear factor kappa B activation in gastric cancer. Cancer. 2010;116:41–49. doi: 10.1002/cncr.24743. [DOI] [PubMed] [Google Scholar]
- Gaziel-Sovran A., Hernando E. miRNA-mediated GALNT modulation of invasion and immune suppression: a sweet deal for metastatic cells. Oncoimmunology. 2012;1:746–748. doi: 10.4161/onci.19535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstl M.P., Hackl M., Graf A.B.., Borth N., Grillari J. Prediction of transcribed PIWI-interacting RNAs from CHO RNAseq data. J Biotechnol. 2013;166:51–57. doi: 10.1016/j.jbiotec.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Gottwein E. Roles of microRNAs in the life cycles of mammalian viruses. Curr Top Microbiol Immunol. 2013;371:201–227. doi: 10.1007/978-3-642-37765-5_8. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: microRNA sequences and annotation. In: Baxevanis Andreas D., editor. Current protocols in bioinformatics. 2010. [Chapter 12:Unit 12 9 1-0] [DOI] [PubMed] [Google Scholar]
- Grillari J., Hackl M., Grillari-Voglauer R. miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology. 2010;11:501–506. doi: 10.1007/s10522-010-9272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P., Yin Z.H., Li X.L., Wu W., Zhou B.S. Meta-analysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. J Exp Clin Cancer Res. 2012;31 doi: 10.1186/1756-9966-31-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker D.L., De Jesus M., Wurm F.M. 25 years of recombinant proteins from reactor-grown cells — where do we go from here? Biotechnol Adv. 2009;27:1023–1027. doi: 10.1016/j.biotechadv.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Hackl M., Brunner S., Fortschegger K., Schreiner C., Micutkova L., Muck C. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell. 2010;9:291–296. doi: 10.1111/j.1474-9726.2010.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl M., Jakobi T., Blom J., Doppmeier D., Brinkrolf K., Szczepanowski R. Next-generation sequencing of the Chinese Hamster Ovary microRNA transcriptome: identification, annotation and profiling of microRNAs as targets for cellular engineering. J Biotechnol. 2011;153:62–75. doi: 10.1016/j.jbiotec.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl M., Jadhav V., Jakobi T., Rupp O., Brinkrolf K., Goesmann A. Computational identification of microRNA gene loci and precursor microRNA sequences in CHO cell lines. J Biotechnol. 2012;158:151–155. doi: 10.1016/j.jbiotec.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl M., Borth N., Grillari J. miRNAs — pathway engineering of CHO cell factories that avoids translational burdening. Trends Biotechnol. 2012;30:405–406. doi: 10.1016/j.tibtech.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Hackl M., Jadhav V., Klanert G., Grillari J., Borth N. Analysis of microRNA transcription and post-transcriptional processing by Dicer in the context of CHO cell proliferation. Biotechnol Bioeng. 2013 doi: 10.1016/j.jbiotec.2013.12.018. [under review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S., Swanberg J.C., Kaplarevic M., Lee K.H. Genomic sequencing and analysis of a Chinese Hamster Ovary cell line using Illumina sequencing technology. BMC Genomics. 2011;12 doi: 10.1186/1471-2164-12-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S., Swanberg J.C., Polson S.W., Lee K.H. Profiling conserved microRNA expression in recombinant CHO cell lines using Illumina sequencing. Biotechnol Bioeng. 2012;109:1371–1375. doi: 10.1002/bit.24415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S., Kaplarevic M., Borth N., Betenbaugh M.J., Lee K.H. Chinese hamster genome database: an online resource for the CHO community at www.CHOgenome.org. Biotechnol Bioeng. 2012;109:1353–1356. doi: 10.1002/bit.24374. [DOI] [PubMed] [Google Scholar]
- Harraghy N., Buceta M., Regamey A., Girod P.A., Mermod N. Using matrix attachment regions to improve recombinant protein production. Methods Mol Biol. 2012;801:93–110. doi: 10.1007/978-1-61779-352-3_7. [DOI] [PubMed] [Google Scholar]
- Hatley M.E., Patrick D.M., Garcia M.R., Richardson J.A., Bassel-Duby R., van Rooij E. Modulation of K-ras-dependent lung tumorigenesis by microRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- Hou E.W., Li S.S.L. Cyclic Amp-induced expression of the mouse lactate dehydrogenase—a promoter-cat fusion gene in Chinese-Hamster Ovary wild-type cells, but not in cAMP-dependent protein-kinase mutant-cells. Biochem Biophys Res Commun. 1987;147:501–505. doi: 10.1016/s0006-291x(87)80149-3. [DOI] [PubMed] [Google Scholar]
- Hou E.W., Li S.S.L. Functional expression of the cDNA-encoding for human lactate dehydrogenase—a in Chinese-Hamster Ovary cells. Cell Biol Int Rep. 1987;11:729–733. doi: 10.1016/0309-1651(87)90132-9. [DOI] [PubMed] [Google Scholar]
- Hua Z., Lv Q., Ye W., Wong C.K., Cai G., Gu D. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Liu H., Wang T., Zhang T., Kuang J., Luo Y. Tonicity-responsive microRNAs contribute to the maximal induction of osmoregulatory transcription factor OREBP in response to high-NaCl hypertonicity. Nucleic Acids Res. 2010;39:475–485. doi: 10.1093/nar/gkq818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Giddens J., Fan S.Q., Toonstra C., Wang L.X. Chemoenzymatic glycoengineering of intact IgG antibodies for gain of functions. J Am Chem Soc. 2012;134:12308–12318. doi: 10.1021/ja3051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Liu N., Wang J.P., Wang Y.Q., Yu X.L., Bin Wang Z. Regulatory long non-coding RNA and its functions. J Physiol Biochem. 2012;68:611–618. doi: 10.1007/s13105-012-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai-Nishiya H., Mori K., Inoue M., Wakitani M., Iida S., Shitara K. Double knockdown of alpha1,6-fucosyltransferase (FUT8) and GDP-mannose 4,6-dehydratase (GMD) in antibody-producing cells: a new strategy for generating fully non-fucosylated therapeutic antibodies with enhanced ADCC. BMC Biotechnol. 2007;7:84. doi: 10.1186/1472-6750-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M.V., Piovan C., Croce C.M. Interplay between microRNAs and the epigenetic machinery: an intricate network. Biochim Biophys Acta. 2010;1799:694–701. doi: 10.1016/j.bbagrm.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Jadhav V.M., Scaria V., Maiti S. Antagomirzymes: oligonucleotide enzymes that specifically silence microRNA function. Angew Chem. 2009;48:2557–2560. doi: 10.1002/anie.200805521. [DOI] [PubMed] [Google Scholar]
- Jadhav V., Hackl M., Bort J.A., Wieser M., Harreither E., Kunert R. A screening method to assess biological effects of microRNA overexpression in Chinese Hamster Ovary cells. Biotechnol Bioeng. 2012;109:1376–1385. doi: 10.1002/bit.24490. [DOI] [PubMed] [Google Scholar]
- Jayapal K.R., Wlaschin K.F., Hu W.S., Yap M.G.S. Recombinant protein therapeutics from CHO cells — 20 years and counting. Chem Eng Prog. 2007;103:40–47. [Google Scholar]
- Jayaswal V., Lutherborrow M., Ma D.D., Hwa Yang Y. Identification of microRNAs with regulatory potential using a matched microRNA-mRNA time-course data. Nucleic Acids Res. 2009;37(8):e60. doi: 10.1093/nar/gkp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis R. Isotype and glycoform selection for antibody therapeutics. Arch Biochem Biophys. 2012;526:159–166. doi: 10.1016/j.abb.2012.03.021. [DOI] [PubMed] [Google Scholar]
- Jeon M.K., Yu D.Y., Lee G.M. Combinatorial engineering of ldh-a and bcl-2 for reducing lactate production and improving cell growth in dihydrofolate reductase-deficient Chinese Hamster Ovary cells. Appl Microbiol Biotechnol. 2011;92:779–790. doi: 10.1007/s00253-011-3475-0. [DOI] [PubMed] [Google Scholar]
- Jiang J., Lee E.J., Gusev Y., Schmittgen T.D. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnson K.C., Jacob N.M., Nissom P.M., Hackl M., Lee L.H., Yap M. Conserved microRNAs in Chinese Hamster Ovary cell lines. Biotechnol Bioeng. 2011;108:475–480. doi: 10.1002/bit.22940. [DOI] [PubMed] [Google Scholar]
- Jopling C.L. Liver-specific microRNA-122 biogenesis and function. RNA Biol. 2012;9 doi: 10.4161/rna.18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H., Babu B.R., Maiti S. Perspectives on chemistry and therapeutic applications of Locked Nucleic Acid (LNA) Chem Rev. 2007;107:4672–4697. doi: 10.1021/cr050266u. [DOI] [PubMed] [Google Scholar]
- Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Lee G.M. Down-regulation of lactate dehydrogenase-A by siRNAs for reduced lactic acid formation of Chinese Hamster Ovary cells producing thrombopoietin. Appl Microbiol Biotechnol. 2007;74:152–159. doi: 10.1007/s00253-006-0654-5. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Lee G.M. Functional expression of human pyruvate carboxylase for reduced lactic acid formation of Chinese Hamster Ovary cells (DG44) Appl Microbiol Biotechnol. 2007;76:659–665. doi: 10.1007/s00253-007-1041-6. [DOI] [PubMed] [Google Scholar]
- Kim Y.G., Han Y.K., Kim J.Y., Lee E.G., Lee H.W., Lee G.M. Effect of constitutively active ras overexpression on cell growth in recombinant Chinese Hamster Ovary cells. Biotechnol Prog. 2011;27:577–580. doi: 10.1002/btpr.567. [DOI] [PubMed] [Google Scholar]
- Kim M., O'Callaghan P.M., Droms K.A., James D.C. A mechanistic understanding of production instability in CHO cell lines expressing recombinant monoclonal antibodies. Biotechnol Bioeng. 2011;108(10):2434–2446. doi: 10.1002/bit.23189. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Kim Y.G., Lee G.M. CHO cells in biotechnology for production of recombinant proteins: current state and further potential. Appl Microbiol Biotechnol. 2012;93:917–930. doi: 10.1007/s00253-011-3758-5. [DOI] [PubMed] [Google Scholar]
- Kochanowski K., Volkmer B., Gerosa L., RHvR B., Schmidt A., Heinemann M. Functioning of a metabolic flux sensor in Escherichia coli. Proc Natl Acad Sci U S A. 2012;110:1130–1135. doi: 10.1073/pnas.1202582110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R., Ferracin M., Wojcik S.E., Garzon R., Alder H., Agosto-Perez F.J. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanceta J., Prough R.A., Liang R., Wang E. MicroRNA group disorganization in aging. Exp Gerontol. 2010;45:269–278. doi: 10.1016/j.exger.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Dutta A. MicroRNAs: small but potent oncogenes or tumor suppressors. Curr Opin Investig Drugs. 2006;7:560–564. [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Palkovits M., Young W.S., III miR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation. Proc Natl Acad Sci U S A. 2006;103:15669–15674. doi: 10.1073/pnas.0605781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis N.E., Liu X., Li Y., Nagarajan H., Yerganian G., O'Brien E., Bordbar A., Roth A.M., Rosenbloom J., Bian C., Xie M., Chen W., Li N., Baycin-Hizal D., Latif H., Forster J., Betenbaugh M.J., Famili I., Xu X., Wang J., Palsson B.O. Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat Biotechnol. 2013 doi: 10.1038/nbt.2624. [Epub ahead of print, in press] [DOI] [PubMed] [Google Scholar]
- Li G., Luna C., Qiu J., Epstein D.L., Gonzalez P. Role of miR-204 in the regulation of apoptosis, endoplasmic reticulum stress response, and inflammation in human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2011;52:2999–3007. doi: 10.1167/iovs.10-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Muthusamy S., Liang R., Sarojini H., Wang E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech Ageing Dev. 2011;132:75–85. doi: 10.1016/j.mad.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Li Y., Li Y., Zhang H., Chen Y. MicroRNA-mediated positive feedback loop and optimized bistable switch in a cancer network Involving miR-17–92. PLoS One. 2011;6:e26302. doi: 10.1371/journal.pone.0026302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.F., Chuan K.H., Liu S., Loh S.O.H., Chung B.Y.F., Ong C.C. RNAi suppression of Bax and Bak enhances viability in fed-batch cultures of CHO cells. Metab Eng. 2006;8:509–522. doi: 10.1016/j.ymben.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Lin T., Dong W., Huang J., Pan Q., Fan X., Zhang C. MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol. 2009;181:1372–1380. doi: 10.1016/j.juro.2008.10.149. [DOI] [PubMed] [Google Scholar]
- Lin N., Davis A., Bahr S., Borgschulte T., Achtien K., Kayser K. Profiling highly conserved microRNA expression in recombinant IgG-producing and parental Chinese Hamster Ovary cells. Biotechnol Prog. 2010;27(4):1163–1171. doi: 10.1002/btpr.556. [DOI] [PubMed] [Google Scholar]
- Lingor P., Michel U., Scholl U., Bahr M., Kugler S. Transfection of “naked” siRNA results in endosomal uptake and metabolic impairment in cultured neurons. Biochem Biophys Res Commun. 2004;315:1126–1133. doi: 10.1016/j.bbrc.2004.01.170. [DOI] [PubMed] [Google Scholar]
- Liu P., Wilson M.J. miR-520c and miR-373 upregulate MMP9 expression by targeting mTOR and SIRT1, and activate the Ras/Raf/MEK/Erk signaling pathway and NF-kappaB factor in human fibrosarcoma cells. J Cell Physiol. 2012;227:867–876. doi: 10.1002/jcp.22993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.J., Tsai M.M., Hung P.S., Kao S.Y., Liu T.Y., Wu K.J. miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010;70:1635–1644. doi: 10.1158/0008-5472.CAN-09-2291. [DOI] [PubMed] [Google Scholar]
- Lovis P., Gattesco S., Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem. 2008;389:305–312. doi: 10.1515/BC.2008.026. [DOI] [PubMed] [Google Scholar]
- Lu Z., Liu M., Stribinskis V., Klinge C.M., Ramos K.S., Colburn N.H. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn F.C. Meta-regulation: microRNA regulation of glucose and lipid metabolism. Trends Endocrinol Metab. 2009;20:452–459. doi: 10.1016/j.tem.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Maes O.C., An J., Sarojini H., Wang E. Murine microRNAs implicated in liver functions and aging process. Mech Ageing Dev. 2008;129:534–541. doi: 10.1016/j.mad.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Majors B.S., Betenbaugh M.J., Chiang G.G. Links between metabolism and apoptosis in mammalian cells: applications for anti-apoptosis engineering. Metab Eng. 2007;9:317–326. doi: 10.1016/j.ymben.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Meleady P., Gallagher M., Clarke C., Henry M., Sanchez N., Barron N. Impact of miR-7 over-expression on the proteome of Chinese Hamster Ovary cells. J Biotechnol. 2012;160:251–262. doi: 10.1016/j.jbiotec.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Meleady P., Hoffrogge R., Henry M., Rupp O., Bort J.H., Clarke C. Utilization and evaluation of CHO-specific sequence databases for mass spectrometry based proteomics. Biotechnol Bioeng. 2012;109:1386–1394. doi: 10.1002/bit.24476. [DOI] [PubMed] [Google Scholar]
- Mendell J.T. miRiad roles for the miR-17–92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Henson R., Lang M., Wehbe H., Maheshwari S., Mendell J.T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- Morris J.A., Dorner A.J., Edwards C.A., Hendershot L.M., Kaufman R.J. Immunoglobulin binding protein (BiP) function is required to protect cells from endoplasmic reticulum stress but is not required for the secretion of selective proteins. J Biol Chem. 1997;272:4327–4334. doi: 10.1074/jbc.272.7.4327. [DOI] [PubMed] [Google Scholar]
- Moschos S.A., Frick M., Taylor B., Turnpenny P., Graves H., Spink K.G. Uptake, efficacy, and systemic distribution of naked, inhaled short interfering RNA (siRNA) and locked nucleic acid (LNA) antisense. Mol Ther. 2011;19:2163–2168. doi: 10.1038/mt.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D., Katinger H., Grillari J. MicroRNAs as targets for engineering of CHO cell factories. Trends Biotechnol. 2008;26:359–365. doi: 10.1016/j.tibtech.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Muniategui A., Nogales-Cadenas R., Vázquez M., X L. Aranguren, Agirre X., Luttun A. Quantification of miRNA–mRNA interactions. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi S., Mori T., Hoshino Y., Maruo K., Yamada N., Kitade Y. MicroRNA-143 functions as a tumor suppressor in human bladder cancer T24 cells. Cancer Lett. 2011;307:211–220. doi: 10.1016/j.canlet.2011.04.005. [DOI] [PubMed] [Google Scholar]
- North S.J., Huang H.H., Sundaram S., Jang-Lee J., Etienne A.T., Trollope A. Glycomics profiling of Chinese Hamster Ovary cell glycosylation mutants reveals N-glycans of a novel size and complexity. J Biol Chem. 2010;285:5759–5775. doi: 10.1074/jbc.M109.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak J., Julian B.A., Tomana M., Mesteck J. Progress in molecular and genetic studies of IgA nephropathy. J Clin Immunol. 2001;21:310–327. doi: 10.1023/a:1012284402054. [DOI] [PubMed] [Google Scholar]
- O'Callaghan P.M., McLeod J., Pybus L.P., Lovelady C.S., Wilkinson S.J., Racher A.J. Cell line-specific control of recombinant monoclonal antibody production by CHO cells. Biotechnol Bioeng. 2010;106:938–951. doi: 10.1002/bit.22769. [DOI] [PubMed] [Google Scholar]
- Olive V., Jiang I., He L. miR-17–92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlehner A., Simmeth S., Gopfert U. Promoter methylation and transgene copy numbers predict unstable protein production in recombinant Chinese Hamster Ovary cell lines. Biotechnol Bioeng. 2011;108:2670–2681. doi: 10.1002/bit.23216. [DOI] [PubMed] [Google Scholar]
- Ovcharenko D., Kelnar K., Johnson C., Leng N., Brown D. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 2007;67:10782–10788. doi: 10.1158/0008-5472.CAN-07-1484. [DOI] [PubMed] [Google Scholar]
- Park H.S., Kim I.H., Kim I.Y., Kim K.H., Kim H.J. Expression of carbamoyl phosphate synthetase I and ornithine transcarbamoylase genes in Chinese Hamster Ovary dhfr-cells decreases accumulation of ammonium ion in culture media. J Biotechnol. 2000;81:129–140. doi: 10.1016/s0168-1656(00)00282-0. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A.E. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A.E., Reinhart B.J., Slack F., Martindale M.Q., Kuroda M.I., Maller B. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Patrick D.M., Zhang C.C., Tao Y., Yao H., Qi X., Schwartz R.J. Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3zeta. Genes Dev. 2010;24:1614–1619. doi: 10.1101/gad.1942810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler J., Galosy S., Mott J., Borth N. Selection of CHO host cell subclones with increased specific antibody production rates by repeated cycles of transient transfection and cell sorting. Biotechnol Bioeng. 2011;108:386–394. doi: 10.1002/bit.22946. [DOI] [PubMed] [Google Scholar]
- Pilbrough W., Munro T.P., Gray P. Intraclonal protein expression heterogeneity in recombinant CHO cells. PLoS One. 2009;4:e8432. doi: 10.1371/journal.pone.0008432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porkka K.P., Pfeiffer M.J., Waltering K.K., Vessella R.L., Tammela T.L., Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- Porter A.J., Racher A.J., Preziosi R., Dickson A.J. Strategies for selecting recombinant CHO cell lines for cGMP manufacturing: improving the efficiency of cell line generation. Biotechnol Prog. 2010;26:1455–1464. doi: 10.1002/btpr.443. [DOI] [PubMed] [Google Scholar]
- Pouilly S., Piller V., Piller F. Metabolic glycoengineering through the mammalian GalNAc salvage pathway. FEBS J. 2012;279:586–598. doi: 10.1111/j.1742-4658.2011.08448.x. [DOI] [PubMed] [Google Scholar]
- Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X.S., MacDonald P.E. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Prieto C., Fontana D., Etcheverrigaray M., Kratje R. A strategy to obtain recombinant cell lines with high expression levels. Lentiviral vector-mediated transgenesis. BMC Proc. 2011;5(Suppl. 8):7. doi: 10.1186/1753-6561-5-S8-P7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puck T.T., Cieciura S.J., Robinson A. Genetics of somatic mammalian cells. III. Long-term cultivation of euploid cells from human and animal subjects. J Exp Med. 1958;108:945–956. doi: 10.1084/jem.108.6.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Wang X., Wang Y., Tang Z., Cui Q., Xi J. MicroRNA-19a mediates the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells. Proc Natl Acad Sci U S A. 2010;107:3240–3244. doi: 10.1073/pnas.0914882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Raju T.S., Briggs J.B., Chamow S.M., Winkler M.E., Jones A.J.S. Glycoengineering of therapeutic glycoproteins: in vitro galactosylation and sialylation of glycoproteins with terminal N-acetylglucosamine and galactose residues. Biochemistry-Us. 2001;40:8868–8876. doi: 10.1021/bi010475i. [DOI] [PubMed] [Google Scholar]
- Rathore M.G., Saumet A., Rossi J.F., de Bettignies C., Tempe D., Lecellier C.H. The NF-kappaB member p65 controls glutamine metabolism through miR-23a. Int J Biochem Cell Biol. 2012;44:1448–1456. doi: 10.1016/j.biocel.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Rayner K.J., Suarez Y., Davalos A., Parathath S., Fitzgerald M.L., Tamehiro N. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V., Naar A.M. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco J., Adeli K. MicroRNAs: emerging roles in lipid and lipoprotein metabolism. Curr Opin Lipidol. 2012;23:220–225. doi: 10.1097/MOL.0b013e3283534c9f. [DOI] [PubMed] [Google Scholar]
- Sangokoya C., Telen M.J., Chi J.T. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116:4338–4348. doi: 10.1182/blood-2009-04-214817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerwald T.M., Figueroa B., Jr., Hardwick J.M., Oyler G.A., Betenbaugh M.J. Combining caspase and mitochondrial dysfunction inhibitors of apoptosis to limit cell death in mammalian cell cultures. Biotechnol Bioeng. 2006;94:362–372. doi: 10.1002/bit.20874. [DOI] [PubMed] [Google Scholar]
- Scaria V., Jadhav V. microRNAs in viral oncogenesis. Retrovirology. 2007;4:82. doi: 10.1186/1742-4690-4-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P., Megraw M., Hatzigeorgiou A.G. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Shimizu S., Takehara T., Hikita H., Kodama T., Miyagi T., Hosui A. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52:698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- Shridhar S., Kreil D.P. Sample size considerations and the efficiency of extracting regulatory connections from a combined miRNA and gene expression data set. Syst Biomed. 2013;1(2) [in press] [Google Scholar]
- Shu J., Xia Z., Li L., Liang E.T., Slipek N., Shen D. Dose-dependent differential mRNA target selection and regulation by let-7a–7f and miR-17–92 cluster microRNAs. RNA Biol. 2012;9:1275–1287. doi: 10.4161/rna.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si M.L., Zhu S., Wu H., Lu Z., Wu F., Mo Y.Y. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Sienski G., Donertas D., Brennecke J. Transcriptional silencing of transposons by Piwi and Maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal A., Milo R., Cohen A., Geva-Zatorsky N., Klein Y., Liron Y. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- Solomides C.C., Evans B.J., Navenot J.M., Vadigepalli R., Peiper S.C., Wang Z.X. MicroRNA profiling in lung cancer reveals new molecular markers for diagnosis. Acta Cytol. 2012;56:645–654. doi: 10.1159/000343473. [DOI] [PubMed] [Google Scholar]
- Sripada L., Tomar D., Prajapati P., Singh R., Singh A.K., Singh R. Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS One. 2012;7:e44873. doi: 10.1371/journal.pone.0044873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripada L., Tomar D., Singh R. Mitochondria: one of the destinations of miRNAs. Mitochondrion. 2012;12:593–599. doi: 10.1016/j.mito.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Stein C.A., Hansen J.B., Lai J., Wu S., Voskresenskiy A., Hog A. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010;38:e3. doi: 10.1093/nar/gkp841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingo F.C., Chen Y.A., Vannucci M., Barrier M., Mirkes P.E. A Bayesian graphical modeling approach to microRNA regulatory network inference. Ann Appl Stat. 2010;4:2024–2048. doi: 10.1214/10-AOAS360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan R.P., Leong J.W., Schneider S.E., Keppel C.R., Germino E., French A.R. MicroRNA-deficient NK cells exhibit decreased survival but enhanced function. J Immunol. 2012;188:3019–3030. doi: 10.4049/jimmunol.1102294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zhao X., Zhou Y., Hu Y. miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol Rep. 2012;28:1346–1352. doi: 10.3892/or.2012.1958. [DOI] [PubMed] [Google Scholar]
- Sung Y.H., Lee J.S., Park S.H., Koo J., Lee G.M. Influence of co-down-regulation of caspase-3 and caspase-7 by siRNAs on sodium butyrate-induced apoptotic cell death of Chinese Hamster Ovary cells producing thrombopoietin. Metab Eng. 2007;9:452–464. doi: 10.1016/j.ymben.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Sunley K., Tharmalingam T., Butler M. CHO cells adapted to hypothermic growth produce high yields of recombinant beta-interferon. Biotechnol Prog. 2008;24:898–906. doi: 10.1002/btpr.9. [DOI] [PubMed] [Google Scholar]
- Taguchi A., Yanagisawa K., Tanaka M., Cao K., Matsuyama Y., Goto H. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17–92 microRNA cluster. Cancer Res. 2008;68:5540–5545. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- Tigges M., Fussenegger M. Xbp1-based engineering of secretory capacity enhances the productivity of Chinese Hamster Ovary cells. Metab Eng. 2006;8(3):264–272. doi: 10.1016/j.ymben.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Tomankova T., Petrek M., Kriegova E. Involvement of microRNAs in physiological and pathological processes in the lung. Respir Res. 2010;11 doi: 10.1186/1465-9921-11-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyk D.D., Misztal D.R., Wilkins M.R., Mackintosh J.A., Poljak A., Varnail J.C. Identification of cellular changes associated with increased production of human growth hormone in a recombinant Chinese Hamster Ovary cell line. Proteomics. 2003;3:147–156. doi: 10.1002/pmic.200390023. [DOI] [PubMed] [Google Scholar]
- Wang Z. The guideline of the design and validation of MiRNA mimics. Methods Mol Biol. 2011;676:211–223. doi: 10.1007/978-1-60761-863-8_15. [DOI] [PubMed] [Google Scholar]
- Wang H., Li W.-H. Increasing microRNA target prediction confidence by the relative R2 method. J Theor Biol. 2009;259:793–798. doi: 10.1016/j.jtbi.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M., Baker M.B., Moore J.P., Searles C.D. MiR-21 is induced in endothelial cells by shear stress and modulates apoptosis and eNOS activity. Biochem Biophys Res Commun. 2010;393:643–648. doi: 10.1016/j.bbrc.2010.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton R.P., Struhl G. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell. 1991;67:955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- Wlaschin K.F., Nissom P.M., Gatti M.D., Ong P.F., Arleen S., Tan K.S. EST sequencing for gene discovery in Chinese Hamster Ovary cells. Biotechnol Bioeng. 2005;91:592–606. doi: 10.1002/bit.20511. [DOI] [PubMed] [Google Scholar]
- Wong D.C., Wong K.T., Nissom P.M., Heng C.K., Yap M.G. Targeting early apoptotic genes in batch and fed-batch CHO cell cultures. Biotechnol Bioeng. 2006;95:350–361. doi: 10.1002/bit.20871. [DOI] [PubMed] [Google Scholar]
- Wuest D.M., Harcum S.W., Lee K.H. Genomics in mammalian cell culture bioprocessing. Biotechnol Adv. 2012;30:629–638. doi: 10.1016/j.biotechadv.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm F.M. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004;22:1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- Xu C., Lu Y., Pan Z., Chu W., Luo X., Lin H. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- Xu X., Nagarajan H., Lewis N.E., Pan S., Cai Z., Liu X. The genomic sequence of the Chinese Hamster Ovary (CHO)-K1 cell line. Nat Biotechnol. 2011;29:735–741. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Jiang Y., Yin Y., Li Q., He J., Jing Y. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 2012;5(1):3–13. doi: 10.1093/jmcb/mjs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane-Ohnuki N., Kinoshita S., Inoue-Urakubo M., Kusunoki M., Iida S., Nakano R. Establishment of FUT8 knockout Chinese Hamster Ovary cells: an ideal host cell line for producing completely defucosylated antibodies with enhanced antibody dependent cellular cytotoxicity. Biotechnol Bioeng. 2004;87:614–622. doi: 10.1002/bit.20151. [DOI] [PubMed] [Google Scholar]
- Yang H., Kong W., He L., Zhao J.J., O'Donnell J.D., Wang J. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- Yang Y., Mariati Chusainow J., Yap M.G. DNA methylation contributes to loss in productivity of monoclonal antibody-producing CHO cell lines. J Biotechnol. 2010;147:180–185. doi: 10.1016/j.jbiotec.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Yang F., Zhang L., Wang F., Wang Y., Huo X.S., Yin Y.X. Modulation of the unfolded protein response is the core of microRNA-122-involved sensitivity to chemotherapy in hepatocellular carcinoma. Neoplasia. 2011;13:590–600. doi: 10.1593/neo.11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Rutnam Z.J., Jiao C., Wei D., Xie Y., Du J. An anti-let-7 sponge decoys and decays endogenous let-7 functions. Cell Cycle. 2012;11:3097–3108. doi: 10.4161/cc.21503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Suo A.L., Li Z.F., Liu L.Y., Tian T., Ni L. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2:963–970. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
- Yu F., Deng H., Yao H., Liu Q., Su F., Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29:4194–4204. doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
- Zanghi J.A., Fussenegger M., Bailey J.E. Serum protects protein-free competent Chinese Hamster Ovary cells against apoptosis induced by nutrient deprivation in batch culture. Biotechnol Bioeng. 1999;64:108–119. doi: 10.1002/(sici)1097-0290(19990705)64:1<108::aid-bit12>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Zeng A.P., Deckwer W.D., Hu W.S. Determinants and rate laws of growth and death of hybridoma cells in continuous culture. Biotechnol Bioeng. 1998;57:642–654. [PubMed] [Google Scholar]
- Zhang Y., Verbeek F.J. Comparison and integration of target prediction algorithms for microRNA studies. J Integr Bioinform. 2010;7 doi: 10.2390/biecoll-jib-2010-127. [DOI] [PubMed] [Google Scholar]
- Zhang X.M., Zhao H.W., Gao S., Wang W.C., Katiyar-Agarwal S., Huang H.D. Arabidopsis argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell. 2011;42:356–366. doi: 10.1016/j.molcel.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Yang B., Datta P., Gasmili L., Zhang F.M., Linhardt R.J. Cell-based microscale isolation of glycoaminoglycans for glycomics study. J Carbohydr Chem. 2012;31:420–435. doi: 10.1080/07328303.2012.658126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Shyh-Chang N., Segre A.V., Shinoda G., Shah S.P., Einhorn W.S. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]