Abstract
We develop a method to extract structural information from electron microscopy (EM) images of dynamic and heterogeneous molecular assemblies. To overcome the challenge of disorder in the imaged structures, we analyze each image individually, avoiding information loss through clustering or averaging. The Bayesian inference of EM (BioEM) method uses a likelihood-based probabilistic measure to quantify the consistency between each EM image and given structural models. The likelihood function accounts for uncertainties in the molecular position and orientation, variations in the relative intensities and noise in the experimental images. The BioEM formalism is physically intuitive and mathematically simple. We show that for experimental GroEL images, BioEM correctly identifies structures according to the functional state. The top-ranked structure is the corresponding X-ray crystal structure, followed by an EM structure generated previously from a superset of the EM images used here. To analyze EM images of highly flexible molecules, we propose an ensemble refinement procedure, and validate it with synthetic EM maps of the ESCRT-I-II supercomplex. Both the size of the ensemble and its structural members are identified correctly. BioEM offers an alternative to 3D-reconstruction methods, extracting accurate population distributions for highly flexible structures and their assemblies. We discuss limitations of the method, and possible applications beyond ensemble refinement, including the cross-validation and unbiased post-assessment of model structures, aand the structural characterization of systems where traditional approaches fail. Overall, our results suggest that the BioEM framework can be used to analyze EM images of both ordered and disordered molecular systems.
1. Introduction
The structural characterization of large and dynamic biomolecular assemblies is rapidly advancing, providing important insight into the function of the molecular machines and supramolecular assemblies involved in transcription and translation of genetic information, signal transduction, protein trafficking, cellular adhesion, and many other cellular processes. Electron microscopy (EM) occupies a central role in this endeavor by reporting on molecular structures with single-particle resolution, unhampered by the need to obtain crystals, and without the system size limits faced in nuclear magnetic resonance (NMR) studies (Frank, 2006). However, structural disorder in dynamic systems greatly limits the use of traditional EM methods that rely on sophisticated image pre-processing, such as class-averaging, to obtain 3D reconstructions (Saibil, 2000a; Leschziner and Nogales, 2007; Patwardhan et al., 2012). Here, we develop a method that aims to extract the maximum information by analyzing the raw EM data image-by-image within a Bayesian framework.
EM reconstructions achieve near-atomic resolution (Lerch et al., 2012; Beck et al., 2012; Ludtke et al., 2008; Zhang et al., 2013; Wang et al., 2006; Nogales et al., 1995) and reveal detailed dynamic information (Heymann et al., 2003; Ramrath et al., 2012; Cianfrocco et al., 2013). Elaborate algorithms have been developed on the modeling and simulation side to extract structural details from flexible fitting into three-dimensional (3D) electron density maps (Trabuco et al., 2008; Tama et al., 2004; Topf et al., 2008; Lindert et al., 2009; Mears et al., 2007; Schröder et al., 2007; Heymann et al., 2004; Delarue and Dumas, 2004; Loquet et al., 2012; Jaitly et al., 2010). Complementary to 3D reconstruction methods, recent integrative multi-scale protocols refine macromolecules against 2D class-averages and physico-chemical constraints. In particular, a maximum-likelihood cross-correlation metric that matches 3D models against class-averaged 2D projection images, has been used, via simulated annealing, to obtain accurate models for several multi-domain complexes (Velazquez-Muriel et al., 2012), and a Natural Moves Monte Carlo method has been successfully used to refine chaperonin (Mm-cpn) against heterogeneous projection averages (Zhang et al., 2012). Obtaining high-resolution models typically requires a large number of EM images, even for molecules exhibiting distinct features in projection that enable sophisticated clustering and reconstruction techniques. In case of highly dynamic assemblies, the traditional EM approaches face additional challenges. In particular, it is difficult to separate molecular motions from differences in the projection view if the number of relevant structural states is large (e.g., in a multidomain protein with flexible linkers, such as the ESCRT-I-II supercomplex (Boura et al., 2012)). This problem is compounded by the presence of alternative or possibly incomplete assemblies, reflecting the often weak pairwise interactions holding the assemblies together. One thus faces challenges not only in identifying the orientations of the molecules imaged, but also in assigning proper conformations and assembly states.
To classify images of heterogeneous particles, standard techniques use iterative optimization algorithms to produce the 3D density map most consistent with the 2D averaged projection views of each model (Elmlund et al., 2008; Chiu et al., 2005; Orlova and Saibil, 2010; Saibil, 2000b). Such analyses work best for images that present common features or discernible symmetries aiding in the cluster analysis (Elmlund and Elmlund, 2012, 2009). Maximum-likelihood methods that do not require the standard class-averaging techniques, have also been developed to classify conformational states (Scheres et al., 2007a), and to provide 3D density maps of macromolecules (Wang et al., 2013; Scheres et al., 2007b). Such reconstruction methods are limited by requiring large numbers of particles, making applications to dynamic systems challenging.
Here, we develop the Bayesian inference of EM (BioEM) approach, geared primarily towards the analysis of EM images of dynamic biomolecular assemblies but applicable more broadly. Importantly, we analyze the EM data image-by-image from the start, without filtering or averaging the images. As is commonly done in the refinement of protein and nucleic acid structures from X-ray crystal diffraction data or from NMR spectra (Brunger et al., 1998), we use structural models or, if needed, an entire ensemble of structures. We quantify how well any one of the models reproduces each of the observed images (in crystallography, this would correspond to calculating R factors). In the spirit of the em2D score developed in (Velazquez-Muriel et al., 2012), we determine the likelihood for each of the EM single-particle images to be created by projection of any one of the models in our ensemble. Based on earlier Bayesian inference approaches for NMR (Rieping et al., 2005), here, we use the Bayesian framework to provide a quantitative measure for comparing and analyzing structural models with respect to individual raw EM images, in contrast to earlier maximum-likelihood or Bayesian approaches for EM (Sigworth, 1998; Scheres et al., 2005; Doerschuk and Johnson, 2000; Scheres et al., 2007a; Sigworth et al., 2010; Scheres, 2012a,b; Kucukelbir et al., 2012) focused primarily on either image classification or the reconstruction of 3D maps. Our likelihood function accounts for uncertainties in the particle orientations and positions, variations in the relative image intensities, statistical noise, and the possible presence of broken particles other than the system of interest. We then feed the calculated likelihoods into a Bayesian framework to consistently and quantitatively assess how well different structures explain the data, and which structures (or structural ensembles) explain the data best.
The EM structural models can be constructed in multiple ways, for instance on the basis of our recently developed coarse-grained energy function, as implemented in the ensemble refinement of SAXS (EROS) approach (Rozycki et al., 2011; Boura et al., 2011). Our models should provide realistic, energetically meaningful structures that can account for molecular binding interactions and conformational changes. In addition, we want our mapping between structures and EM images to be computationally efficient. While our Bayesian approach is general and would work, at one end of the spectrum, with atomistic models or, at the other end, with highly coarse-grained models that treat entire domains as featureless blobs, we here concentrate on an intermediate level of residue-based coarse graining. We find that representing proteins with one site per amino acid strikes an appropriate balance between model detail, structural and energetic accuracy, flexibility and computational efficiency.
The paper is organized as follows. We first introduce the likelihood function connecting structural models and images. We then specify our Bayesian framework, including prior distributions for the model parameters. The resulting posterior provides a probabilistic measure of the degree of consistency between a structural model and an EM image. We test our method on raw experimental EM images of the unliganded chaperonin GroEL. This test demonstrates discriminatory power within a candidate pool consisting of X-ray crystal structures in different functional states, of EM structures obtained previously, and of coarse-grained models. We then test our ability to conduct an EM ensemble refinement, using synthetic images of the ESCRT-I-II supercomplex. In an ensemble of 18 model structures that jointly span the structural ensemble of ESCRT-I-II in solution (Boura et al., 2012), our probability measure correctly identifies the model from which images were generated. Moreover, we demonstrate that we can correctly identify the size of the structural ensemble. Overall, the BioEM approach should provide a useful tool to extract structural information from electron micrographs of dynamic systems, even in cases where standard EM reconstruction techniques fail.
2. Methods
2.1. Relating individual EM images to structures through Bayesian inference
Our first goal is to construct a quantitative measure of how well a particular set of structural models represents the observed EM single-particle images. Bayesian inference establishes such a measure by assigning a posterior probability P(model|data) to a particular model set given the image data. This probability is a product of the likelihood L of observing the data given the model set and of the prior probability of the model set and its parameters, P(model|data) ∝ L(data|model)P(model). The prior P(model) quantifies our uncertainty before the data are considered. Here, we construct the probability P(M|Ω) of a structural ensemble M with members m = 1, 2, . . . , M given a set of EM single-particle images ω = 1, 2, . . . , Ω. This posterior probability is expressed as a product of the likelihood L(Ω|M, θ) of observing the images Ω given the model ensemble M, its prior probability pM(M), and the prior p(θ)dθ of the so-called nuisance parameters θ that here account for uncertainties in the molecular orientation, statistical noise in the image, etc.,
| (1) |
For notational simplicity, we here ignore that θ may be different for each image. Anticipating a more detailed discussion below, suitable prior probabilities of the structural ensemble are the equilibrium Boltzmann distribution in configuration space, as obtained from molecular simulations, with random orientations in the EM projections. Eq. (1) can be factored in terms of contributions from individual images ω and structural models m,
| (2) |
where wm is the relative weight of structure m in the ensemble, with . The posterior probability Pmω of model m given image ω,
| (3) |
is obtained by integrating out the nuisance parameters θ in the likelihood function L(ω|m, θ). Pmω is the central quantity of the BioEM approach, with its logarithm serving as the statistical evidence for structural model m from image ω.
The key to the success of the BioEM approach lies in its ability to construct a reliable estimate of the likelihood L(ω|m, θ) of observing a particular image ω, given a model m. This likelihood function has to account not only for (1) the creation of an ideal EM projection image from a given structural model, but also for (2) contrast transfer function (CTF) effects, (3) uncertainties in the exact center of a particular molecule, (4) variations in the overall intensity, (5) background intensity offsets between different images, (6) and noise in the intensity of individual pixels. In an ideal projection image of a given structure m, with three Euler angles φ = (α, β, γ) describing the molecular orientation, the intensity at each pixel (x, y) can be approximated as (Wade, 1992)
| (4) |
where ρ(x, y, z|m, φ) is the 3D electron density contrast (positive or negative, depending on the EM technique). Here, we describe ρ simply as a collection of uniform spheres centered at the Cα atoms of the proteins, with van der Waals radii (Kim and Hummer, 2008) and number of electrons corresponding to each amino acid type. At high resolution, more refined models of the electron density, such as those used in X-ray crystallographic refinements, will become necessary. We account for inelastic scattering, CTF effects, and projection angle errors in the microscopy experiment (Penczek et al., 2006) by using a point-spread function (PSF). Specifically, we blur the ideal image with a Gaussian in the plane of the image,
| (5) |
where σ is the planar map resolution. As discussed in Suppl. Text, this Gaussian blurring is a limiting case of a general class of PSFs, which are the real-space equivalents of the product of the CTF and the envelope function in Fourier space. Whereas the Gaussian approximation captures the dominant CTF effects, more elaborate PSF models with oscillatory components, corresponding to standard CTF models (Penczek, 2010), can also be handled within our Bayesian approach (see Suppl. Text and Suppl. Figs. 7 and 8).
To account for uncertainties in the exact particle position in the plane of projection, we add a 2D translation vector d = (dx, dy),
| (6) |
Extending the em2D score (Velazquez-Muriel et al., 2012), we here also scale the intensity by a normalization parameter N and we add an offset μ to account for variations in the detailed imaging conditions. The relative intensities are not only affected by the molecule conformation and orientation but also by thermal and mechanical perturbations, variations in electron dose, ice thickness, etc. As shown in Suppl. Text, the inclusion of these parameters substantially increases the posterior, as a measure of the model quality.
We further assume that the intensity at each pixel is subject to uncorrelated Gaussian noise of zero mean and standard deviation λ, with var(I)/λ2 the signal-to-noise-ratio. The likelihood of observing an experimental image ω with intensity at pixel (x, y) for a given model m and for nuisance parameters θ = (φ, σ, d, N, μ, λ) becomes a product over all Npix pixels (x, y),
| (7) |
The optimal position and orientation of a given model are determined by the maximum of the posterior,
| (8) |
with the remaining nuisance parameters integrated out.
In constructing the prior p(θ)dθ, we assume uniform distributions of the molecular orientations, with Euler angles α and γ independently and uniformly distributed in [–π, π], and cos β uniformly distributed in [–1, 1]. However, other distributions, for example to account for preferred orientations due to the carbon grid (Rosenthal and Henderson, 2003), may also be useful. For the remaining parameters σ, d, N, θ and λ, we assume independent uniform distributions within suitably chosen intervals. The N ∈ (–∞, ∞), μ ∈ (–∞, ∞), and λ ∈ (0, ∞) integrals were carried analytically, exactly for the former two, and with a saddle-point-type approximation for the latter (see Suppl. Text). The remaining integrals over the orientation, blurring, and translation were performed numerically using grid summation.
In the calculation of the Bayesian posterior probability, for the systems studied here, we find that parameters θ = (φ, σ, d, N, μ, λ) are sufficient to account for the relevant physical observables in the microscopy experiment. However, for EM micrographs obtained under different imaging conditions, it may be necessary to include other parameters or more sophisticated functional forms accounting for CTF effects (see Suppl. Text).
A major advantage of Bayesian approaches is the explicit treatment of experimental uncertainties and statistical noise (Rieping et al., 2005). Unlike in standard measures of cross-correlation and Fourier shell correlation (Vasishtan and Topf, 2011), in our Bayesian formulation uncertainties and noise are explicitly included and their effects can be accounted for by integrating over the corresponding nuisance parameters. In our practical applications, we find that the likelihood as a function of the various parameters is sufficiently peaked, such that the choice of the prior distributions has limited influence on the final results.
For reference, we also determine the probability of generating the image from pure Gaussian noise,
| (9) |
As indicated by the angular brackets, the intensity differences ΔI are drawn from independent Gaussian distributions with standard deviation equal to that of the observed image λ, such that PNoise = (2πλ2e)–Npix/2. Here, we rescale all image intensities to zero mean and unit variances, such that λ = 1 by construction. However, this scaling is not a necessary condition for calculating the Bayesian posterior. In our analysis we will use the logarithm ln(Pmω/PNoise) as a measure of the normalized evidence for model m from image ω.
2.2. Model for damaged particles and possible contaminations
In realistic samples, we expect occasional impurities, damaged particles, or badly cropped images (Penczek et al., 2006). To account for such issues in a fully automated refinement procedure, we add a separate coarse-grained model as an alternative to the molecular models m of the system of interest. This “splotch” particle (s) is here modeled as a featureless Gaussian with variable amplitude (A), width (W ), center (c) and offset (o),
| (10) |
The likelihood of an image to be generated by a splotch particle, is calculated in analogy with Eq. (3), as the integral over the nuisance parameters θ′ = (A, W, c, o, λ) with prior p′(θ′),
| (11) |
Note that CTF effects, translation, normalization, and offset uncertainties used for the molecular models are implicitly included in W, c, A, and o, respectively. Psω can again be normalized by PNoise to assess the evidence against statistical noise.
2.3. Ensemble refinement
Overfitting is a major problem in ensemble refinement. By varying the relative weights and coordinates of multiple structures in an ensemble, one gains enormous freedom in fits to experimental data of limited resolution. The price for this freedom is the risk to fit not only signal but also noise. Here we address this problem by determining the optimal ensemble size within the Bayesian framework. Specifically, we directly compare the posteriors of model ensembles containing different numbers of structures.
According to Eq. (2), the posterior of a specific model with a single structure i is . If the model contains m ≤ M structures with relative weights wi, the posterior is
| (12) |
In our illustrative analysis here, we assume for simplicity that all ensemble weights are the same, wi = 1/m, such that
| (13) |
The logarithm Ω–1ln(P12...m/PNoise) of this posterior probability, normalized without loss of generality by the number of images Ω and the noise probability, provides us with a formal basis to select ensembles of optimal size and composition. If the respective weights of the members of the ensemble are of interest, one can instead use Eq. (12).
3. Results
3.1. Analysis of experimental EM images of GroEL
To validate our method against experiment, we test its ability to distinguish different structures of the chaperonin GroEL in individual experimental EM images. The challenge is to identify the correct structure, as determined by the functional state of the protein, within a pool of 8 experimental candidate structures of GroEL in a variety of conformational states (Suppl. Table 1). These structures differ by Cα-backbone root-mean-square distances of 0-12 Å at the monomer level, and 2-12 Å over the entire protein (Suppl. Tables 2-4). GroEL is a multisubunit chaperonin with a molecular weight of ~800 kDa, formed by two symmetrically stacked rings of seven identical subunits. Its structure has been studied extensively, in particular by X-ray crystallography (Braig et al., 1994; Bartolucci et al., 2005; Xu et al., 1997; Wang and Boisvert, 2003; Chaudhry et al., 2004; Cabo-Bilbao et al., 2006) and cryo-EM (Ranson et al., 2001; Ludtke et al., 2008; Falke et al., 2005). For our validation, we used the Bsoft program (Heymann and Belnap, 2007; Heymann et al., 2008) to collect 1,283 native unliganded GroEL particles from cryo-EM micrographs that are distributed by the National Center for Macromolecular Imaging (Ludtke et al., 2008). The image size was 170×170 pixels of 1.05 Å length. Each image was normalized to intensities of zero mean and unit variance.
Finding proper orientations of individual GroEL EM images
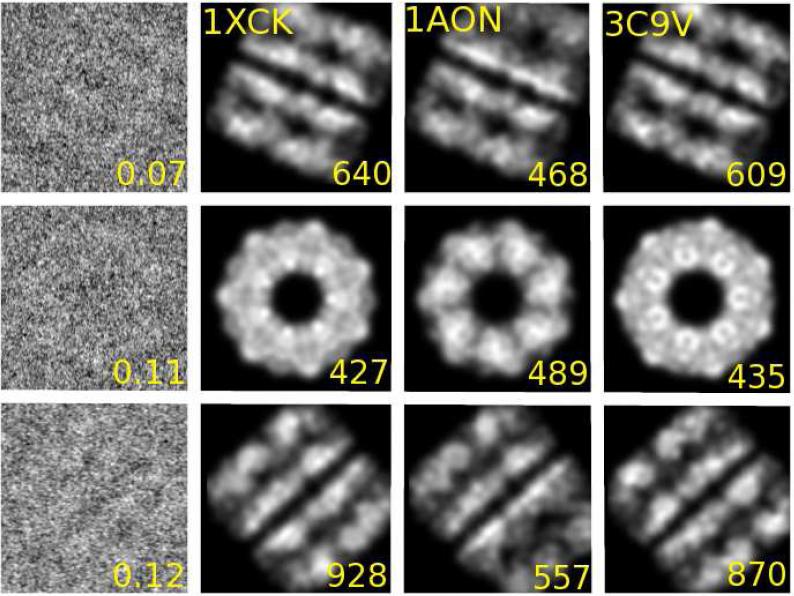
Addressing the first challenge, Fig. 1 illustrates that our fully automated Bayesian approach identifies the positions and orientations of the structural models consistent with a visual inspection of individual EM images. Representative EM images in different orientations are shown together with calculated intensities for several different GroEL models in their optimal position and orientation, as determined by Eq. (8). Results are shown for native GroEL solved by X-ray crystallography (PDB code 1XCK) (Bartolucci et al., 2005) in the state corresponding to the EM experiments, the GroEL part (chains A-N) of the crystallographic complex GroEL-GroES-(ADP)7 (1AON) (Xu et al., 1997), and GroEL with C7 symmetry as determined directly from the cryo-EM images (PDB code 3C9V) (Ludtke et al., 2008). We find that visually the different structural models are consistently and correctly orientated to match the individual EM images. We also find that the log-posterior identifies the 7-fold symmetry of GroEL (see Suppl. Fig. 2) when calculated as a function of the planar rotational angle for the top-view GroEL image shown in Fig. 1 (second row).
Figure 1.
Representative experimental EM images of GroEL (Ludtke et al., 2008) (left column; numbers indicate estimated signal-to-noise ratio) together with calculated projection images for structures 1XCK, 1AON, and 3C9V in optimal position and orientation (columns 2 to 4, respectively), as determined by the posterior probability, Eq. (8). The number in the bottom right of the calculated projection images indicates the logarithm of the posterior probability ln(Pmω/PNoise), with high values indicating strong agreement between the model and the EM image.
Identifying the proper GroEL structure from individual EM images
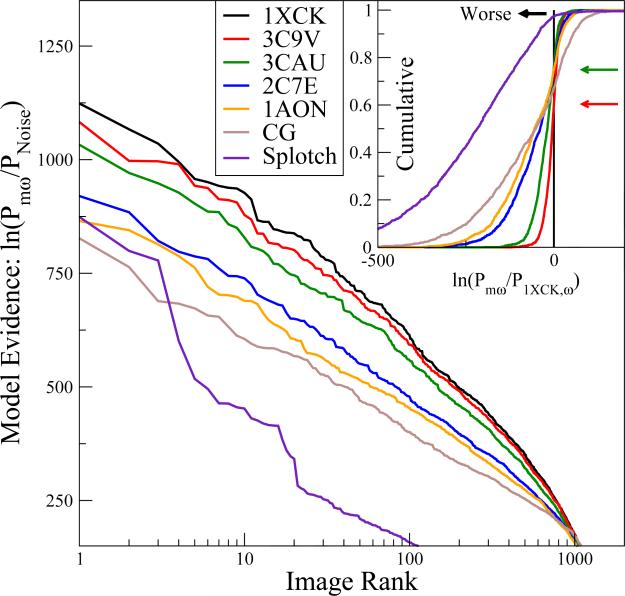
Addressing the major challenge, Fig. 2 demonstrates that we can consistently and quantitatively identify the correct structure within a pool of different candidates. We calculated the log model evidence defined as the natural logarithm of the posterior probability relative to statistical noise, ln(Pmω/PNoise), for each of the models m and each of the images ω. For each model, we then rank-ordered the different EM images according to Pmω. Fig. 2 shows the resulting rank-ordered posteriors for each of the images and models. In addition to structures 1XCK, 3C9V, and 1AON, we also include results for the cryo-EM GroEL structure 3CAU with D7 symmetry (Ludtke et al., 2008), and structure 2C7E of GroEL bound to ATP (Ranson et al., 2001), with all nucleotides removed in our calculations. Results for three additional GroEL PDB structures are shown in Suppl. Fig. 1. In calculating Pmω, we assumed independent uniform prior distributions for the parameters θ, within intervals of 1 < σ < 6 Å for the planar map resolution, and –16 < dx, dy < 16 Å for the translation vectors. Uniform priors in (–∞, ∞) were assumed for the normalization (N) and offset (μ), and in (0, ∞) for the standard deviation of the noise (λ). Euler angles α and γ were integrated uniformly within [–π, π], and cos β uniformly within [–1, 1]. All structural models were considered equiprobable with prior probabilities pM(m) = 1. We note that the log-posteriors are for the entire image.
Figure 2.
Statistical evidence for different GroEL structural models from individual experimental EM images, with the top curve corresponding to the structure most consistent with the EM images. The logarithm ln(Pmω/PNoise) of the posterior probability relative to statistical noise is plotted for different models m for individual EM images ω. The images are rank-ordered according to their respective posterior for PDB structures 1XCK (black), 3C9V (red), 3CAU (green), 2C7E (blue), and 1AON (orange), and for the splotch (purple) and coarse-grained (magenta) models (i.e., the rank order is different for each model; for a direct one-on-one comparison see Fig. 4; for a global comparison see Fig. 3; see also Suppl. Text and Suppl. Fig. 1 for other models). The inset shows the cumulative distribution of the logarithm of the posterior ln(Pmω/P1XCK,ω) relative to the X-ray crystal structure 1XCK, as calculated over all EM images. The cumulative probability at the point of crossing of the vertical black line indicates the fraction of EM images ω for which the model performs worse than 1XCK according to the posterior. Red and green arrows indicate these points for structures 3C9V and 3CAU, respectively.
To identify the GroEL structural model that best explains the entire set of experimental EM images, we sum the logarithms of the posterior probabilities of individual images,
| (14) |
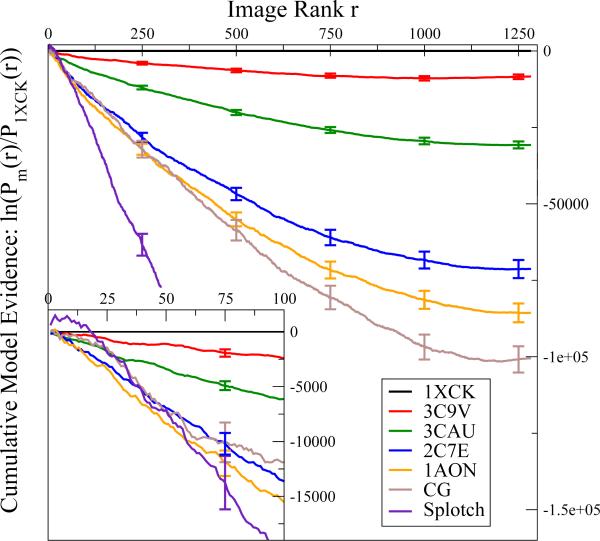
over images rank ordered for each model m such that Pm1 ≥ Pm2 ≥ . . . ≥ Pmr ≥ . . . ≥ PmΩ. In Fig. 3, we plot the cumulative log evidence for each model m relative to the top-ranked structure 1XCK, ln[Pm(rank r)/P1XCK(rank r)], as a function of the image rank r determined individually for each model. We find that these curves decrease more or less linearly from zero up to ~1000 images. The log evidence in favor of the X-ray structure 1XCK thus increases at a more or less constant rate per image for 80-90 % of the images. The remaining 10-20 % of images individually provide little evidence either way, as indicated by a plateau in the running sum of Eq. (14) (Fig. 3). Overall, the cumulative evidence in favor of 1XCK is >8,000 natural-log units, even over the structure 3C9V obtained by refinement against the EM data used also here. Over, say, the GroEL structure 1AON in a different ligation and activation state, the evidence in favor of 1XCK is even stronger, ~80,000 log units. Moreover, by plotting the overall cumulative evidence as a function of the Cα RMSDs from structure 1XCK (see Suppl. Fig. 5), we find that the two measures anti-correlate, indicating that the more a model deviates (in RMSD) from 1XCK, the less likely it is to have generated the images.
Figure 3.
Cumulative evidence, ln[Pm(rank r)/P1XCK(rank r)], for structural models relative to the overall best model 1XCK as a function of the image rank for each model. Negative values indicate that 1XCK has a higher posterior probability and is thus favored. The image rank r was determined individually for each model. The inset shows a zoom-in on the cumulative evidence for each model from the respective 100 top-ranked images. Error bars were calculated using the bootstrap technique.
Whereas the clear distinction between different structures in the global comparisons of Figs. 2 and 3 is encouraging, for the ultimate goal of ensemble refinement of EM data it is important that we can also assign the correct structure to individual EM images. In the inset of Fig. 2, we plot the cumulative distribution of the evidence of different models over the top-ranked crystal structure 1XCK. In this plot we find that for a small but significant number of images, the evidence favors models other than 1XCK. For instance, for about 40 % of the images, the EM structure 3C9V performs better than 1XCK, as indicated by the red arrow in Fig. 2 inset. However, in the majority of these images the difference in evidence is only small, such that the cumulative distributions shown in the inset of Fig. 2 rise sharply to one after having crossed the vertical zero line indicating a toss-up. By contrast, the cumulative distributions show broad tails on the left, indicating that 1XCK typically outperforms the other models by more significant amounts. As a result, the overall evidence points clearly toward 1XCK, against all other models considered.
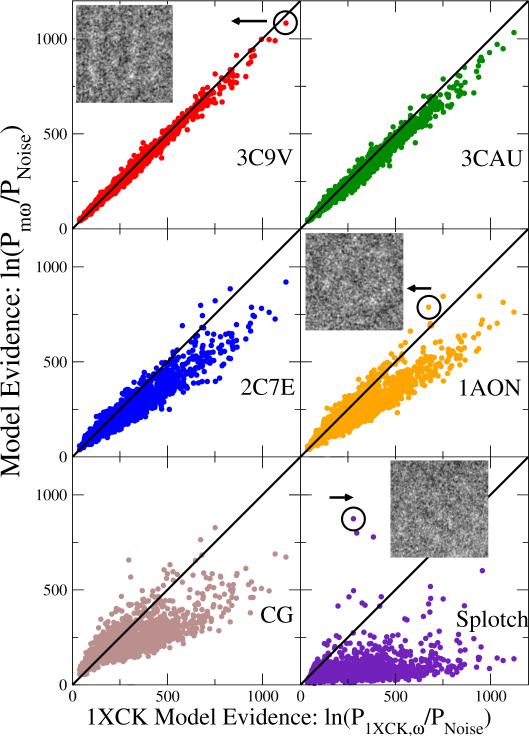
To strengthen this important point, Fig. 4 shows a scatter plot of the posteriors against the top-ranked structural model 1XCK. Points above the diagonal line indicate images for which the statistical evidence is in favor of a particular model over 1XCK; for points below the diagonal, 1XCK performs better than the competing model. Consistent with the preceding discussion, we find that for 3C9V, 60 % of the points are below the diagonal, i.e., most images favor 1XCK; and images that favor 3C9V do so by a small margin, i.e., the points are close to the diagonal. The models in the wrong GroEL activation state, 2C7E and 1AON, are favored by very few images, and those images typically have only small absolute posteriors, i.e., they are in the bottom left quadrant of the scatter plots.
Figure 4.
Image-by-image comparison of calculated log-posteriors for different GroEL models relative to that of 1XCK. Each point corresponds to the posterior probabilities of one of the structural models (vertical axis) and of the X-ray crystal structure 1XCK (horizontal axis) for an individual EM image. Points above the diagonal indicate images for which the respective model outperforms 1XCK. For 3C9V, 1AON, and the splotch model, representative EM images are shown, with corresponding points indicated by circles.
For each of the five PDB models, we also classified the fractions of GroEL particles assigned by the model with the highest score (see Suppl. Fig. 4). We found that for images in which at least one model received intermediate and high probabilities (log-posterior > 250 of the top-scoring model relative to noise), model 1XCK had the highest score for > 60 % of the particles, and 3C9V claimed another ~20 % of the particles. This winner-take-all scoring further demonstrates that one can use individual EM images to identify the correct structures from a pool with some confidence. Remarkably, we found that only a small number of particles is needed to establish with statistical significance 1XCK as the better structural model than 3C9V, two structures that differ by less than 5 Å in RMSD. Specifically, we picked random samples of n images out of the 1283 images total, and calculated for each n the fraction of samples in which 3C9V overall ranks higher than 1XCK. This fraction corresponds to the p-value. In this way, we found that n ≈ 28 images suffice to achieve a p-value of 0.05, and n ≈ 52 images to achieve a p-value of 0.01, respectively.
The images and data in the center row of Fig. 1 and the center right panel of Fig. 4 help rationalize the few cases where a likely incorrect structure has a high statistical evidence. In both cases illustrated, 1AON has a higher posterior than 1XCK. These images are top views of GroEL. In this projection along the rotation axis, the two models cannot be distinguished easily on the basis of a single image, despite a relatively clear EM intensity signal (also see Suppl. Fig. 2). The problem that certain models cannot be distinguished easily in some projections is also reflected in the branching of the scatter plots for 2C7E and 1AON in Fig. 4, with the upper branch staying close to the diagonal. Such ambiguous projections could cause difficulty for class-averaging techniques in cryo-EM. Moreover, poor class-averages analyzed with low-pass filtering techniques can cause over-fitting and cryo-EM reconstructions may be subject to errors (van Heel and Schatz, 2005; Scheres and Chen, 2012). Here we avoid such problems, with challenging projection views producing little relative gains in evidence for one model over another.
Excluding artifacts through a splotch model
As another serious challenge in the analysis of EM images, one has to exclude artifacts that arise, for instance, from damaged sample molecules, the formation of aggregates, the presence of impurities, or imaging issues. Since here we, ultimately, aim to analyze each image individually as part of an ensemble refinement, it is critical that we minimize the resulting unfavorable effects. We used a simple splotch model to identify artifacts that can then be excluded from further analysis (see Methods).
For the vast majority of images, splotch is a very poor model, as shown in Fig. 2, performing worse than any of the GroEL structural models in almost all cases. However, the thin tail to the right in the cumulative distribution in Fig. 2 and the scatter plot in the bottom right of Fig. 4 show that for a few images, the splotch model performs substantially better than even the overall best GroEL model, 1XCK. Remarkably, whereas for none of the images any of the alternative GroEL structures outperforms 1XCK by more than ~50 natural-log units, the splotch model outperforms 1XCK for a handful of images by up to ~500 log units. In these rare cases, we are thus nearly certain that the object in the EM image is not an intact GroEL molecule. Indeed, the cumulative log evidence for the splotch model relative to 1XCK, as shown in the inset of Fig. 3, is initially positive, before turning strongly negative. By including a splotch model with a flexible parameter range, we can identify such images and exclude them from further analysis, even at low signal-to-noise ratios.
We also find that for some images, the coarse-grained model (see Suppl. Text) performs significantly better than 1XCK (tail to the right in the cumulative distribution in Fig. 2 and Fig. 4 bottom left). These images all have small posteriors for 1XCK, as was found also for the splotch model. Viewing the coarse-grained model as an extension of the splotch model, images in which this low-resolution description scores best could thus also be marked for exclusion in a final analysis.
3.2. Minimal ensemble method applied to ESCRT I-II supercomplex
To test the applicability of our method in EM studies of dynamic systems with diverse structures instead of a pure state, we performed an ensemble refinement study for the ESCRT I-II supercomplex (Boura et al., 2012). Above, we validated the EM single-image analysis method against experimental data, showing that we could identify the correct structural model with high confidence and consistency in a system where we expect only a single dominant structure. To test whether we could identify the correct ensemble of structures from a set of EM images containing a mixture not only of orientations but also of molecular conformations, we used synthetic data. With full control over the composition of the ensemble, we can perform a quantitative assessment of the ensemble refinement method.
As a test system we used 18 models of the 244-kDa ESCRT I-II supercomplex that were previously obtained from a combination of X-ray crystallography, small-angle X-ray scattering, single-molecule fluorescence resonance energy transfer (smFRET), and double electron-electron resonance (DEER) spin-label distance measurements (Boura et al., 2012). ESCRT-I-II is a dynamic supercomplex that is critical for the function of the ESCRT machinery involved in membrane protein trafficking, the budding of enveloped viruses including HIV, and other normal and pathological cell processes (Hurley and Hanson, 2010). Protein domains are connected by long disordered linkers and through transient binding interactions, making ESCRT-I-II an intrinsically flexible complex with structures ranging from compact to highly extended conformations about 400 Å in length. Due to its structural variability, the ESCRT I-II complex thus offers an ideal benchmark to test our method on a system with heterogeneous conformations.
For four of these models (with extensions DMax = 360, 313, 267, and 227 Å, respectively (Boura et al., 2012)), we generated 200 synthetic EM maps each, using projections with random orientations, center and blurring. Images had 150x150 pixels, with 2 Å per pixel. We used planar map resolutions of 8 < σ < 22 Å and translation vectors –30 < dx, dy < 30 Å. The normalization (N) and noise standard deviation (λ) were selected to create images with signal-to-noise ratios between 0.005 and 0.15. The maps obtained after adding uncorrelated Gaussian noise were normalized to have zero mean and unit standard deviation.
Images from a single model
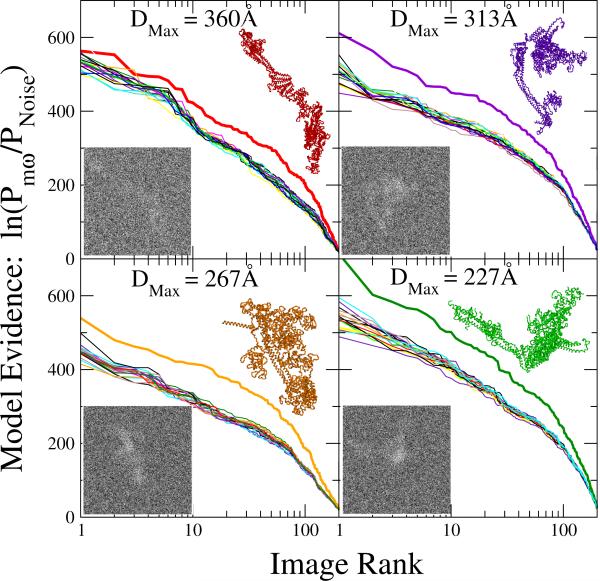
In Fig. 5 we plot the logarithm ln(Pmω/PNoise) of the posterior probabilities relative to statistical noise for each of the 18 models m and each of the 4 × 200 images ω. For each model, we rank-ordered the different EM images according to the posteriors, which jointly serve as statistical evidence for the particular model. We find that the curve corresponding to the model used to generate the images is consistently at the top. Fig. 5 thus provides strong evidence that our method systematically identifies the correct model among the set of 18 candidate structures. The most challenging structure is that of the model with DMax = 360 Å. The needle-like character without distinct features makes it relatively difficult to distinguish this model from the 17 others, in particular in projections along the main axis. For the three other, more feature-rich models, the gap separating the curves for the correct and the incorrect structures is significantly larger.
Figure 5.
Statistical evidence for different ESCRT-I-II structural models from individual EM images, with the top curve indicating the model of highest consistency. 200 synthetic EM maps each were created by projection of models with extensions DMax = 360 (top left), 313 (top right), 267 (bottom left), and 227 Å (bottom right). The logarithm ln(Pmω/PNoise) of the posterior probability relative to statistical noise is plotted for 18 ESCRT-I-II structural models m for individual EM images ω. The images are rank-ordered for each model m according to the respective posteriors. The thick line indicates the posteriors for the structural model used to create the EM image. Insets show representative synthetic EM maps generated from the respective ESCRT-I-II models.
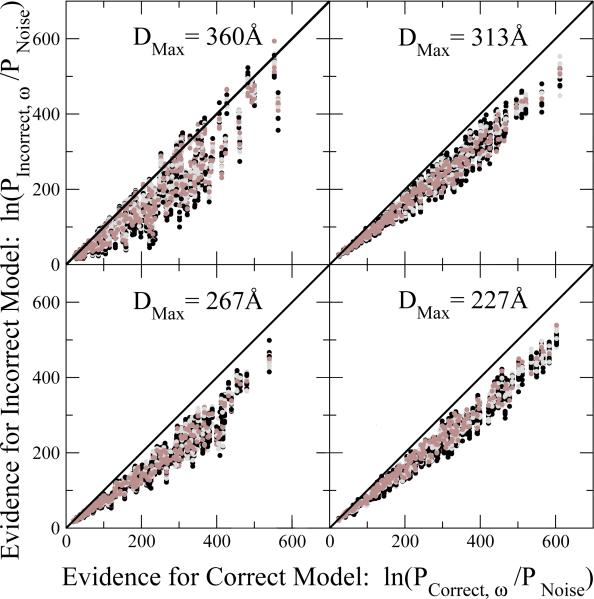
In a more challenging test, we compare the calculated evidence for the correct model against the evidence for each of the 17 incorrect models. Fig. 6 shows the corresponding scatter plots, with almost all points falling below the diagonal that separates correct assignments (below) from incorrect ones (above). We hence find that our posterior distinguishes with high reliability between the correct and incorrect structures for individual images. Consistent with Fig. 5, the relatively featureless model with DMax = 360 Å is the most challenging structure, with incorrect models achieving comparable posteriors for a few images.
Figure 6.
Image-by-image comparison of calculated log-posteriors for “incorrect” ESCRTI-II supercomplex models (vertical axis) relative to the correct structure (horizontal axis). Each point corresponds to the posterior probabilities of one of the 17 “incorrect” structural models (different colors; vertical axis) and of the correct model used to obtain the EM projection image (horizontal axis). Results are shown for EM projections of ESCRT-I-II models with extensions DMax = 360 (top left), 313 (top right), 267 (bottom left), and 227 Å (bottom right).
Ensemble refinement: prevention of overfitting
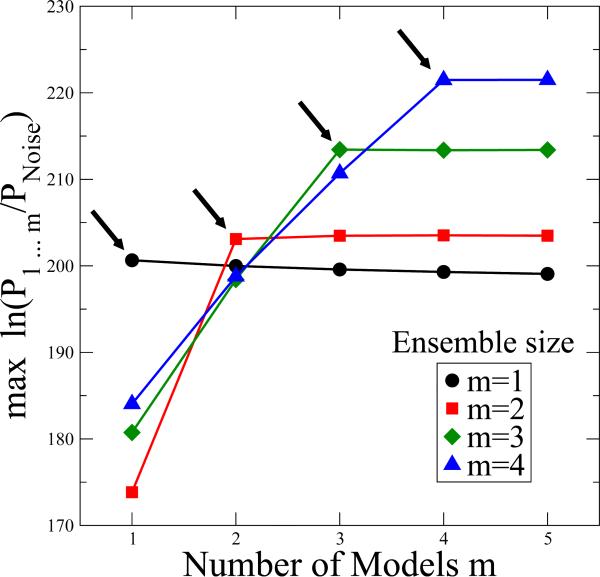
We also determined whether the ensemble size can be estimated reliably using the Bayesian formalism described in the Methods section. The specific aim is to identify both the size of the ensemble and its members for different sets of ESCRT-I-II EM images, containing 200 projections each of 1, 2, 3, or 4 structures. In Fig. 7, for each image set, we plot the maximum over all one-member ensembles, maxm ln Pm, all two-member ensembles, maxm<n ln Pmn, all three-member ensembles, maxl<m<n ln Plmn, and all four-member ensembles, maxk<l<m<n ln Pklmn. Note that in the plot we divided all probabilities by PNoise, which does not change the results. We find that for ensembles with only one model, the posterior probability slightly decreases when the model ensemble size is increased beyond one. This decrease results from the factor m–Ω in the posterior, Eq. (13), which penalizes large ensemble sizes. For EM ensembles of two structures we see a sharp increase in the posterior when increasing the number of models in the fit ensemble from one to two, but again no significant gains occur beyond. This pattern is repeated for the larger EM ensembles, with the posterior increasing with the number of models only until it reaches the correct number. We also confirmed that the m models in the optimal ensemble were consistent with those used to generate the EM images. The results in Fig. 7 thus show that the BioEM probabilistic approach can produce a meaningful bound on the ensemble size.
Figure 7.
ESCRT-I-II EM ensemble refinement, with minimal ensemble size deduced from the logarithm of the posterior probability. Synthetic EM images were created by projection of ESCRT-I-II supercomplexes with ensembles of M = 1 (circle), M = 2 (square), M = 3 (diamond), and M = 4 (triangle) different structural models. Shown is maxm models Ω–1 ln(P12...m/PNoise), the maximum of the logarithm of the posterior per image, calculated for different sets of m = 1 to 5 models included in the refinement ensemble. The posterior increases sharply until the number of models reaches the actual ensemble size, as indicated by arrows.
4. Discussion
We developed a Bayesian framework to extract structural information from EM images of dynamic biomolecular assemblies, where traditional approaches face major challenges (Patwardhan et al., 2012). The central idea is to analyze each EM particle individually from the start, without filtering, clustering or averaging the images. By analyzing single raw particles, we avoid information loss when lumping different structures into the same class (see Suppl. Fig. 6). The key quantity in the BioEM approach is the likelihood that a particular EM image corresponds to a given molecular structure. This likelihood function accounts for uncertainties in the molecular position and orientation, and for noise in the experimental EM image, similarly as in (Velazquez-Muriel et al., 2012), and additionally accounts for CTF effects and overall variations in the relative intensities that further boost the performance (see Suppl. Text). Weighted with suitable prior distributions of the parameters describing these uncertainties, the resulting Pmω defined in Eq. (3) provides a probabilistic measure of the consistency of a given molecular structure with a particular EM image. The formalism to calculate Pmω is mathematically simple, based on the physics of EM imaging, and can thus easily be extended to include complex imaging effects if needed (see Suppl. Text).
We showed that for experimental GroEL EM images, our methodology correctly identified the most probable structures within a diverse pool of plausible conformations. Our method correctly ranked models according to their conformational state and the level of detail. In particular, the top-ranked structure was the X-ray crystal structure of GroEL in a state corresponding to that of the EM images, followed closely by the EM structure generated from a super-set of EM images that included the maps used here. Structures corresponding to GroEL in different states with minor or major conformational changes ranked significantly lower. Interestingly, we found that the splotch model helped in discriminating images of intact GroEL from damaged particles or other objects.
We also showed that we can determine the optimal size and identify the correct members of a structural ensemble by using synthetic EM maps of the ESCRT-I-II supercomplex. The dynamic structures of ESCRT-I-II were correctly identified even in images with low resolution and low signal-to-noise ratio. These results suggest that the BioEM framework indeed provides us with the tools for a quantitative ensemble refinement of structures against individual EM images.
The BioEM single-image analysis can be readily combined with the traditional class-averaging approaches. As we showed here for GroEL, BioEM post-analysis may provide additional insight into the quality of different structural models, independent of the 3D density maps obtained from the traditional approach. Ludtke et al. (2008) generated reconstructions using C7 and D7 symmetry, respectively, from experimental apo-GroEL EM data with resolution 4.7 and 4.2 Å. Interestingly, for the EM images used here, but collected as part of the original study (Ludtke et al., 2008), we found that the two models differ significantly in their likelihood, with the C7 model 3C9V of lower enforced symmetry being more probable than the D7 model 3CAU of higher symmetry. More generally, to address the need for validation methods (Patwardhan et al., 2012; Henderson et al., 2012), the BioEM framework could become a tool to cross-validate models generated from 3D EM reconstructions, when over-fitting is a concern (van Heel and Schatz, 2005; Scheres and Chen, 2012). In this way, the traditional reconstruction and BioEM approaches should complement each other even in cases where a single structure is sufficient to describe the images.
This complementarity of the current approaches and BioEM can possibly be extended to the optimization of atomic-level structures obtained from fits of (x-ray crystal) structures into EM maps. Again, while class-averaging is not a pre-requisite for all methods, the potential advantage of BioEM is that structures are fitted to essentially raw image data without the unavoidable loss of structural information because of image processing and averaging. Indeed, we have found that filtering, clustering and averaging the images leads to a decrease in the discriminatory power of the Bayesian scoring function (see Suppl. Text). Our method could thus be used in the last steps of 3D reconstruction to extract additional structural information.
Our approach can also be extended to electron tomography (Bohm et al., 2000; Medalia et al., 2002; Bartesaghi et al., 2012). Specifically, the posterior for a given particle and model is then the product of the posteriors calculated for the different projection views, making full use of the additional tomographic information.
A possible weak point of the BioEM approach is that it appears to depend heavily on the availability and quality of the initial structural models. In many practical cases, unlike the GroEL and ESCRT-I-II systems studied here, we may not have any good starting structures. To address this problem, we again envision a multi-resolution approach. At the coarsest level, the model could simply be the 3D density map in Eq. (4), suitably represented in terms of grids, spherical harmonics, etc., similar to what is done in traditional approaches. Again highlighting the complementarity of the current approaches and BioEM, one could thus start from a 3D map obtained from a reconstruction, and then test and, if needed, refine it using the BioEM probability measure. Alternatively, or in addition, models at progressively finer levels of resolution can be used, from simple geometric shapes to residue and possibly atom resolution.
The BioEM provides a quantitative framework to compare different structural models against each other, and to compare models against noise. Importantly, the log-posterior does not provide an absolute measure of the quality of a model per se, due to its dependence on various model parameters and properties of the input data (number of pixels, observed image variance, etc.) and also the highly peaked local maxima in the underlying likelihood function. Thus, the main focus here is on calculating differences in log-posteriors for different models.
Several limitations can be encountered with the ensemble refinement methodology: first, the necessity of a good model ensemble is essential; second, even though the minimal number of models is well determined in the ideal case, the error and resolution in the general case of imperfect models and uncertain ensemble sizes are not known. For experiments with significantly less detailed data, such as solution scattering, maximum entropy and minimal ensemble methods (Rozycki et al., 2011; Boura et al., 2011) have proved useful in preventing overfitting in ensemble refinement.
Computational requirements are another concern. The computational cost scales roughly as the product of the number of images, models, and pixels per image, ΩMNpix, for a compact PSF in the convolution Eq. (5), or as ΩMNpix ln Npix, if fast-Fourier transforms are used for non-compact PSFs. Significant computational challenges are posed by a typically large number Ω of images, and by the orientational averaging in the calculation of Pmω. Currently our code runs approximately at ~ 10–7s per integration grid point per pixel per processor. To compare one GroEL structure to one map of 170×170 pixels with one million integration grid points takes ~1 min on 64 cores. However, a number of avenues are possible to speed up the calculations, from the use of importance sampling strategies in the evaluation of the integrals, over reducing the resolution both of the images and of the model detail in an initial search, to exploiting the power of GPU/multicore computing for both the geometric transformation and the matrix operations required for the posterior calculation, as in other EM algorithms (Tagare et al., 2010; Li et al., 2010). Moreover, a hierarchical approach should cut down on computational cost by working with a smaller number of down-sampled images in the initial phase, when the number of possible models is large, and then gradually increasing the image number and resolution as the space of possible models is thinned out.
Despite these challenges, the BioEM approach offers an alternative to 3D-reconstruction methods by its ability to extract structural information and accurate population distributions (1) of asymmetric systems that are difficult to class-average because of an absence of easily identifiable features for proper grouping and orientation; (2) of dynamic or heterogeneous systems for which it is not possible to collect sufficient numbers of different projections for each conformation; and (3) of intermediate size complexes that are too large to study with solution NMR but smaller (yet identifiable) than those commonly reconstructed through EM.
Supplementary Material
Acknowledgments
To obtain the source code please contact the corresponding author. The authors thank Dr. J. Bernard Heymann for help with the Bsoft program, and Drs. James Hurley and Alasdair Steven for stimulating discussions. We also thank Dr. Jürgen Köfinger for useful discussions concerning the Bayesian methodology. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, and by the Max Planck Society, and used the biowulf computing resource at the National Institutes of Health, and the high-performance computers at the Rechenzentrum Garching of the Max Planck Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartesaghi A, Lecumberry F, Sapiro G, Subramaniam S. Protein secondary structure determination by constrained single-particle cryo-electron tomography. Structure. 2012;20:2003–2013. doi: 10.1016/j.str.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolucci C, Lamba D, Grazulis S, Manakova E, Heumann H. Crystal structure of wild-type chaperonin GroEL. J. Mol. Biol. 2005;354:940–951. doi: 10.1016/j.jmb.2005.09.096. [DOI] [PubMed] [Google Scholar]
- Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, Forster F. Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. USA. 2012;109:14870–5. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm J, Frangakis AS, Hegerl R, Nickell S, Typke D, Baumeister W. Toward detecting and identifying macromolecules in a cellular context: Template matching applied to electron tomograms. Proc. Natl. Acad. Sci. USA. 2000;97:14245–14250. doi: 10.1073/pnas.230282097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E, Rozycki B, Chung HS, Herrick DZ, Canagarajah B, Cafiso DS, Eaton WA, Hummer G, Hurley JH. Solution Structure of the ESCRT-I and -II Supercomplex: Implications for Membrane Budding and Scission. Structure. 2012;20:874–886. doi: 10.1016/j.str.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E, Rozycki B, Herrick DZ, Chung HS, Vecer J, Eaton WA, Cafiso DS, Hummer G, Hurley JH. Solution structure of the ESCRT-I complex by small-angle X-ray scattering, EPR, and FRET spectroscopy. Proc. Natl. Acad. Sci. USA. 2011;108:9437–9442. doi: 10.1073/pnas.1101763108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig K, Otwinowski Z, Hegde R, Boisvert D, Joachimiak A, Horwich A, Sigler P. The crystal-structure of the bacterial chaperonin GroEL at 2.8-Ångstrom. Nature. 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system. A new software suite for macromolecular structure determination. Acta Cryst. Sect. D Biol. Cryst. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cabo-Bilbao A, Spinelli S, Sot B, Agirre J, Mechaly AE, Muga A, Guerin DMA. Crystal structure of the temperature-sensitive and allosteric-defective chaperonin GroEL(E461K). J. Struct. Biol. 2006;155:482–492. doi: 10.1016/j.jsb.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Chaudhry C, Horwich A, Brunger A, Adams P. Exploring the structural dynamics of the E-coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states. J. Mol. Biol. 2004;342:229–245. doi: 10.1016/j.jmb.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Chiu W, Baker M, Jiang W, Dougherty M, Schmid M. Electron cryomicroscopy of biological machines at subnanometer resolution. Structure. 2005;13:363–372. doi: 10.1016/j.str.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, Nogales E. Human TFIID binds to core promoter dna in a reorganized structural state. Cell. 2013;152:120–31. doi: 10.1016/j.cell.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Dumas P. On the use of low-frequency normal modes to enforce collective movements in refining macromolecular structural models. Proc. Natl. Acad. Sci. USA. 2004;101:6957–6962. doi: 10.1073/pnas.0400301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerschuk P, Johnson J. Ab initio reconstruction and experimental design for cryo electron microscopy. IEEE Trans. Inf. Theory. 2000;46:1714–1729. [Google Scholar]
- Elad N, Clare DK, Salbil HR, Orlova EV. Detection and separation of heterogeneity in molecular complexes by statistical analysis of their two-dimensional projections. J. Struct. Biol. 2008;162:108–120. doi: 10.1016/j.jsb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Elmlund D, Elmlund H. High-resolution single-particle orientation refinement based on spectrally self-adapting common lines. J. Struct. Biol. 2009;167:83–94. doi: 10.1016/j.jsb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Elmlund D, Elmlund H. SIMPLE: Software for ab initio reconstruction of heterogeneous single-particles. J. Struct. Biol. 2012;180:420–427. doi: 10.1016/j.jsb.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Elmlund H, Lundqvist J, Al-Karadaghi S, Hansson M, Hebert H, Lindahl M. A new cryo-EM single-particle ab initio reconstruction method visualizes secondary structure elements in an ATP-fueled AAA+ motor. J. Mol. Biol. 2008;375:934–947. doi: 10.1016/j.jmb.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Falke S, Tama F, Brooks C, Gogol E, Fisher M. The 13 angstrom structure of a chaperonin GroEL-protein substrate complex by cryo-electron microscopy. J. Mol. Biol. 2005;348:219–230. doi: 10.1016/j.jmb.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. Oxford Univ. Press; New York: 2006. [Google Scholar]
- Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng Z, Frank J, Grigorieff N, Jiang W, Ludtke SJ, Medalia O, Penczek PA, Rosenthal PB, Rossmann MG, Schmid MF, Schroeder GF, Steven AC, Stokes DL, Westbrook JD, Wriggers W, Yang H, Young J, Berman H, Chiu W, Kleywegt GJ, Lawson CL. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann JM, Hoenger A, Mastronarde DN. Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J. Struct. Biol. 2011;175:288–299. doi: 10.1016/j.jsb.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann J, Cheng N, Newcomb W, Trus B, Brown J, Steven A. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nature Struct. Biol. 2003;10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- Heymann J, Conway J, Steven A. Molecular dynamics of protein complexes from four-dimensional cryo-electron microscopy. J. Struct. Biol. 2004;147:291–301. doi: 10.1016/j.jsb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J. Struct. Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Cardone G, Winkler DC, Steven AC. Computational resources for cryo-electron tomography. Bsoft. J. Struct. Biol; 4th International Conference on Electron Tomography; San Diego, CA. NOV 05-08, 2006; 2008. pp. 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery. It's all in the neck. Nature Rev. Mol. Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitly N, Brubaker MA, Rubinstein JL, Lilien RH. A Bayesian method for 3D macromolecular structure inference using class average images from single particle electron microscopy. Bioinformatics. 2010;26:2406–2415. doi: 10.1093/bioinformatics/btq456. [DOI] [PubMed] [Google Scholar]
- Kim YC, Hummer G. Coarse-grained models for simulations of multiprotein complexes: application to ubiquitin binding. J. Mol. Biol. 2008;375:1416–1433. doi: 10.1016/j.jmb.2007.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. A Bayesian adaptive basis algorithm for single particle reconstruction. J. Struct. Biol. 2012;179:56–67. doi: 10.1016/j.jsb.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch TF, O'Donnell JK, Meyer NL, Xie Q, Taylor KA, Stagg SM, Chapman MS. Structure of AAV-DJ, a retargeted gene therapy vector: cryo-electron microscopy at 4.5 Ångstrom resolution. Structure. 2012;20:1310–1320. doi: 10.1016/j.str.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leschziner AE, Nogales E. Visualizing flexibility at molecular resolution: Analysis of heterogeneity in single-particle electron microscopy reconstructions. Annu. Rev. Biophys. Biomol. Struct. 2007;36:43–62. doi: 10.1146/annurev.biophys.36.040306.132742. [DOI] [PubMed] [Google Scholar]
- Li X, Grigorieff N, Cheng Y. GPU-enabled FREALIGN: Accelerating single particle 3D reconstruction and refinement in Fourier space on graphics processors. J. Struct. Biol. 2010;172:407–412. doi: 10.1016/j.jsb.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindert S, Staritzbichler R, Woetzel N, Karakas M, Stewart PL, Meiler J. EM-Fold: De Novo Folding of alpha-Helical Proteins Guided by Intermediate-Resolution Electron Microscopy Density Maps. Structure. 2009;17:990–1003. doi: 10.1016/j.str.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, Griesinger C, Kolbe M, Baker D, Becker S, Lange A. Atomic model of the type III secretion system needle. Nature. 2012;486:276–279. doi: 10.1038/nature11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baker ML, Chen D-H, Song J-L, Chuang DT, Chiu W. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Mears JA, Ray P, Hinshaw JE. A corkscrew model for dynamin constriction. Structure. 2007;15:1190–1202. doi: 10.1016/j.str.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medalia O, Weber I, Frangakis AS, Nicastro D, Gerisch G, Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf S, Khan I, Luduena R, Downing K. Structure of tubulin at 6.5 Angstrom and location of the taxol-binding site. Nature. 1995;375:424–427. doi: 10.1038/375424a0. [DOI] [PubMed] [Google Scholar]
- Orlova EV, Saibil HR. Jensen GJ, editor. Methods for three-dimensional reconstruction of heterogeneous assemblies. Methods in Enzymology, Vol. 482: Cryo-EM, PART B: 3-D Reconstruction. Vol. 482 of Methods in Enzymology. 2010:321–341. doi: 10.1016/S0076-6879(10)82013-0. [DOI] [PubMed] [Google Scholar]
- Patwardhan A, Carazo J-M, Carragher B, Henderson R, Heymann JB, Hill E, Jensen GJ, Lagerstedt I, Lawson CL, Ludtke SJ, Mastronarde D, Moore WJ, Roseman A, Rosenthal P, Sorzano C-OS, Sanz-Garcia E, Scheres SHW, Subramaniam S, Westbrook J, Winn M, Swedlow JR, Kleywegt GJ. Data management challenges in three-dimensional EM. Nature Struct. Mol. Biol. 2012;19:1203–1207. doi: 10.1038/nsmb.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek P, Frank J, Spahn C. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J. Struct. Biol. 2006;154:184–194. doi: 10.1016/j.jsb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Penczek PA. Image restoration in cryo-electron microscopy. Methods in Enzymology. 2010;482:35–72. doi: 10.1016/S0076-6879(10)82002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrath DJF, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, Loerke J, Scheerer P, Ivanov P, Teraoka Y, Shpanchenko O, Nierhaus KH, Spahn CMT. The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Nature. 2012;485:526–U140. doi: 10.1038/nature11006. [DOI] [PubMed] [Google Scholar]
- Ranson N, Farr G, Roseman A, Gowen B, Fenton W, Horwich A, Saibil H. ATP-bound states of GroEL captured by cryo-electron microscopy. Cell. 2001;107:869–879. doi: 10.1016/s0092-8674(01)00617-1. [DOI] [PubMed] [Google Scholar]
- Rieping W, Habeck M, Nilges M. Inferential structure determination. Science. 2005;309:303–306. doi: 10.1126/science.1110428. [DOI] [PubMed] [Google Scholar]
- Rosenthal P, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Rozycki B, Kim YC, Hummer G. SAXS ensemble refinement of ESCRT-III CHMP3 conformational transitions. Structure. 2011;19:109–116. doi: 10.1016/j.str.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibil H. Conformational changes studied by cryo-electron microscopy. Nature Struct. Biol. 2000a;7:711–714. doi: 10.1038/78923. [DOI] [PubMed] [Google Scholar]
- Saibil H. Macromolecular structure determination by cryo-electron microscopy. Acta Cryst. Sect. D Biol. Cryst. 2000b;56:1215–1222. doi: 10.1107/s0907444900010787. [DOI] [PubMed] [Google Scholar]
- Scheres S, Valle M, Nunez R, Sorzano C, Marabini R, Herman G, Carazo J. Maximum-likelihood multi-reference refinement for electron microscopy images. J. Mol. Biol. 2005;348:139–149. doi: 10.1016/j.jmb.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Scheres SHW. A Bayesian view on cryo-em structure determination. J. Mol. Biol. 2012a;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012b;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature Meth. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW, Gao H, Valle M, Herman GT, Eggermont PPB, Frank J, Carazo J-M. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nature Meth. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- Scheres SHW, Núñez-Ramírez R, Gómez-Llorente Y, San Martín C, Eggermont PPB, Carazo JM. Modeling Experimental Image Formation for Likelihood-Based Classification of Electron Microscopy Data. Structure. 2007;15:1167–1177. doi: 10.1016/j.str.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder GF, Brunger AT, Levitt M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigworth F. A maximum-likelihood approach to single-particle image refinement. J. Struct. Biol. 1998;122:328–339. doi: 10.1006/jsbi.1998.4014. [DOI] [PubMed] [Google Scholar]
- Sigworth FJ, Doerschuk PC, Carazo J-M, Scheres SHW. Jensen GJ, editor. An introduction to maximum-likelihood methods in cryo-EM. Methods in Enzymology. Vol. 482 of Methods in Enzymology. 2010:263–294. doi: 10.1016/S0076-6879(10)82011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagare HD, Barthel A, Sigworth FJ. An adaptive expectation-maximization algorithm with GPU implementation for electron cryomicroscopy. J. Struct. Biol. 2010;171:256–265. doi: 10.1016/j.jsb.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tama F, Miyashita O, Brooks C. Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryo-EM. J. Struct. Biol. 2004;147:315–326. doi: 10.1016/j.jsb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M, Patwardhan A. Single-particle electron cryo-microscopy towards atomic resolution. Quart. Rev. Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- van Heel M, Schatz M. Fourier shell correlation threshold criteria. J. Struct. Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Vasishtan D, Topf M. Scoring functions for cryoEM density fitting. J. Struct. Biol. 2011;174:333–343. doi: 10.1016/j.jsb.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Velazquez-Muriel J, Lasker K, Russel D, Phillips J, Webb BM, Schneidman-Duhovny D, Sali A. Assembly of macromolecular complexes by satisfaction of spatial restraints from electron microscopy images. Proc. Natl. Acad. Sci. USA. 2012;109:18821–18826. doi: 10.1073/pnas.1216549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade R. A brief look at imaging and contrast transfer. Ultramicroscopy. 1992;46:145–156. [Google Scholar]
- Wang J, Boisvert D. Structural basis for GroEL-assisted protein folding from the crystal structure of (GroEL-KMgATP)14 at 2.0 Ångstrom resolution. J. Mol. Biol. 2003;327:843–855. doi: 10.1016/s0022-2836(03)00184-0. [DOI] [PubMed] [Google Scholar]
- Wang Q, Matsui T, Domitrovic T, Zheng Y, Doerschuk PC, Johnson JE. Dynamics in cryo EM reconstructions visualized with maximum-likelihood derived variance maps. J. Struct. Biol. 2013;181:195–206. doi: 10.1016/j.jsb.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YA, Yu X, Overman S, Tsuboi M, Thomas GJ, Jr., Egelman EH. The structure of a filamentous bacteriophage. J. Mol. Biol. 2006;361:209–215. doi: 10.1016/j.jmb.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Xu Z, Horwich A, Sigler P. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- Zhang J, Minary P, Levitt M. Multiscale natural moves refine macromolecules using single-particle electron microscopy projection images. Proc. Natl. Acad. Sci. USA. 2012;109:9845–9850. doi: 10.1073/pnas.1205945109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Kirnmel M, Spahn CMT, Penczek PA. Heterogeneity of Large Macromolecular Complexes Revealed by 3D Cryo-EM Variance Analysis. Structure. 2008;16:1770–1776. doi: 10.1016/j.str.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH. Cryo-EM structure of the mature dengue virus at 3.5-Ångstrom resolution. Nature Struct. Mol. Biol. 2013;20:105–U133. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.