SUMMARY
The formation of toxic aggregates composed largely of the protein α-synuclein are a hallmark of Parkinson’s disease. Evidence from both early-onset forms of the disease in humans and animal models has shown that the progression of the disease is correlated with the expression levels of α-synuclein, suggesting that cellular mechanisms that degrade excess α-synuclein are key. We and others have shown that monoubiquitinated α-synuclein can be degraded by the 26S proteasome; however, the contributions of each of the nine known individual monoubiquitination sites was unknown. Herein we determined the consequences of each of the modification sites using homogenous, semisynthetic proteins in combination with an in vitro proteasome turnover assay. The data suggest that the site-specific effects of monoubiquitination support different levels of α-synuclein degradation.
INTRODUCTION
The progressive loss of neurons in Parkinson’s disease is closely associated with the transformation of the protein α-synuclein from either α-helices closely associated with cellular membranes (Ferreon et al., 2009) or unstructured conformations in solution (Weinreb et al., 1996) to β-sheet rich, high molecular-weight aggregates termed Lewy bodies (Fink, 2006). This aggregated disease-state closely resembles the oligomers and fibers that are readily formed by isolated or recombinant α-synuclein in vitro. Additionally, when fibers of α-synuclein generated in vitro were injected into the brains of healthy mice, disease pathology spread along neuronal contacts and resulted in Parkinson’s Disease associated phenotypes, demonstrating that α-synuclein aggregates can cause disease (Luk et al., 2012). Notably, duplications and triplications of the gene encoding α-synuclein cause early-onset forms of Parkinson’s disease (Chartier-Harlin et al., 2004; Singleton et al., 2003) and several animal models were generated by α-synuclein overexpression (Magen and Chesselet, 2010), strongly suggesting that failure to maintain the appropriate cellular levels of α-synuclein can increase the rate of aggregation and progression of neurodegeneration. Therefore, a molecular understanding of the potential pathways that lead to α-synuclein degradation has a high probability to uncover targets for Parkinson’s disease therapy.
We and others have demonstrated that α-synuclein can be degraded by proteasomal, lysosomal, and autophagic pathways (Paxinou et al., 2001; Rott et al., 2008; 2011; Webb, 2003), and we have shown that the balance between these different mechanisms is controlled by the posttranslational modification of α-synuclein by monoubiquitination (Rott et al., 2011). The vast majority of ubiquitination result from the enzymatic addition of the small protein (76 residues) ubiquitin, through its C-terminus, to the side-chain amine of substrate protein lysine residues, giving an isopeptide bond (Welchman et al., 2005). α-Synuclein has been shown to be modified on 9 different lysine residues using a variety of techniques (Oueslati et al., 2010). Hasegawa and coworkers used rabbit reticulocyte lysates to demonstrate that monomeric α-synuclein can be monoubiquitinated at lysine residues K21, 23, 32, and 34 and preformed fibrils can be modified at K6, 10, and 12 (Nonaka et al., 2005). Anderson et al. then showed that α-synuclein isolated from patient Lewy bodies was monoubiquitinated at K12, 21, and 23 (Anderson et al., 2006). Despite the significant amounts of monoubiquitinated α-synuclein found in Parkinson’s disease patients, an enzyme capable of its installation was unknown until we and the Chin laboratory separately demonstrated that the E3 ubiquitin-ligases seven in absentia homolog-1 & 2 (SI-AH1/2) monoubiquitinate α-synuclein in vitro and in vivo (Lee et al., 2008; Liani et al., 2004; Rott et al., 2008). Mass spectrometry analysis indicated that α-synuclein is monoubiquitinated by SIAH at 9 lysines (K6, 10, 12, 21, 23, 32, 34, 43, and 96) as a heterogeneous mixture (Nonaka et al., 2005; Rott et al., 2008) Notably, SIAH is present in Lewy bodies and ubiquitin-modifies α-synuclein in the same lysines that were found to be monoubiquitinated in Lewy bodies, supporting its role in Parkinson’s disease (Anderson et al., 2006; Liani et al., 2004; Rott et al., 2008). Because ubiquitination is often dynamic, we used a candidate-based approach combined with co-immunoprecipitation, to identify USP9X as a physiologically relevant DUB that removes α-synuclein monoubiquitination (Rott et al., 2011). Notably, overexpression of USP9X, resulting in less α-synuclein monoubiquitination, stabilized α-synuclein protein-levels in SH-SY5Y cells, suggesting that monoubiquitination can result in α-synuclein proteasomal degradation. This finding challenged the observation that a protein requires polyubiquitination of at least four ubiquitin molecules to undergo degradation by the proteasome (Hershko and Ciechanover, 1998). However, Ciechanover and co-workers recently found that monoubiquitination at or near the N-terminus of small proteins (<~150 amino acids in length), like α-synuclein, is sufficient to promote proteasomal degradation (Shabek et al., 2012). Notably, using an expressed protein ligation strategy, they demonstrated that α-synuclein monoubiquitinated at K12 is degraded; however, they did not test the remaining eight modification sites. Furthermore, some studies have shown that SIAH overexpression has no effect on α-synuclein stability (Nagano et al., 2003).
Using a semisynthetic strategy, we previously demonstrated that monoubiquitination has different effects on α-synuclein aggregation depending on the site of modification (Meier et al., 2012), and we predicted that the same would be true for proteasomal degradation. To directly test this prediction, we utilized a strategy termed disulfide-directed ubiquitination that generates a disulfide-linked analog of the ubiquitin-lysine isopeptide bond (Chatterjee et al., 2010; Chen et al., 2010). This analog has been shown to be functionally equivalent to native ubiquitin and only requires a cysteine residue on the substrate protein and ubiquitin functionalized with a C-terminal thiol. Importantly, α-synuclein contains no native cysteines, making it a particularly attractive target for this technology. Here, we use this method to semisynthesize individual α-synuclein molecules bearing monoubiquitin at each of the nine different known modification sites. Subsequent incubation of these proteins or unmodified α-synuclein with purified proteasomes demonstrated that different monoubiquitination sites vary in their ability to support α-synuclein degradation. Specifically, monoubiquitination sites towards the N-terminus of α-synuclein are always degraded, while those further towards the center of the protein are typically not turned over. This supports a model where individual ubiquitin modification-sites have divergent effects on α-synuclein biology. Furthermore, it strongly suggests that in-general monoubiquitination will only result in small-protein degradation when it is located near their termini.
RESULTS
Synthesis and characterization of monoubiquitinated α-synuclein analogs
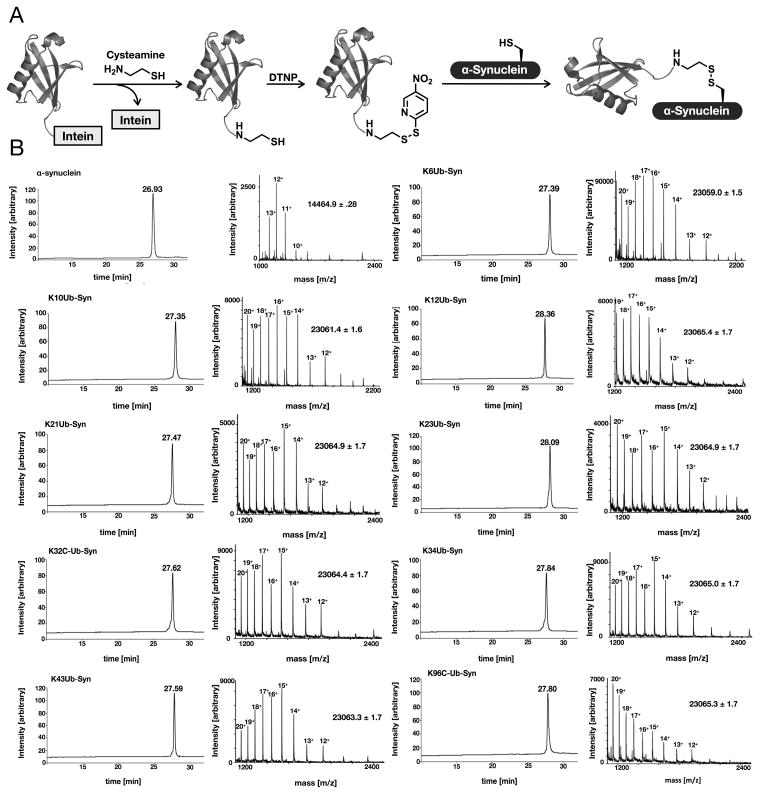
The semisynthetic route (Figure 1A) begins with recombinant expression of ubiquitin as a linear genetic fusion to the fused AvaDnaE intein (Shah et al., 2012). After purification using a HisTrap column (GE Healthcare), the ubiquitin was thiolyzed from the intein by incubation with cysteamine, yielding ubiquitin with a C-terminal aminoethanethiol linker (Figure S1A). The resulting protein was then reacted with 2,2′-dithiobis(5-nitropyridine) (DTNP) to yield the corresponding activated mixed-disulfide (Figure S1B) (Rabanal et al., 1996). At this time, wild-type and the nine lysine to cysteine mutants (K6C, K10C, K12C, K21C, K23C, K32C, K34C, K46C, and K96C) of α-synuclein were expressed heterologously in E. coli and purified by reverse-phase (RP) HPLC. To generate the monoubiquitinated analogs, the ubiquitin mixed-disulfide was incubated with each K to C mutant for 1 h, resulting in approximately complete product formation. These nine monoubiquitinated analogs were purified by RP-HPLC and characterized by mass spectrometry (Figure 1B) and SDS-PAGE analysis (Figure S2A). Because α-synuclein that is monoubiquitinated on some sites can still form protein aggregates that may affect proteasomal activity, we next characterized all ten proteins using dynamic light scattering. This analysis revealed that all proteins had Stokes radii of between 3 and 6 nm (Figure S2B), consistent with monomeric structures, with no evidence of larger oligomers or fibrils.
Figure 1. see also Figures S1 and S2. Synthesis and characterization of monoubiquitinated α-synuclein proteins.
(A) Disulfide-directed ubiquitination. Ubiquitin was expressed in E. coli as a fusion with the AvaDnaE intein and thiolyzed with cysteamine to yield ubiquitin with a C-terminal thiol. This was subsequently activated with DNTP to generate the corresponding ubiquitin mixed-disulfide. This protein was then reacted with α-synuclein cysteine point mutants to give the corresponding disulfide-directed monoubiquitinated proteins. (B) Characterization of each α-synuclein protein using RP-HPLC and mass spectrometry.
Visualization of proteasome-mediated α-synuclein degradation
To determine if the different monoubiquitinated α-synuclein proteins were degraded by the proteasome required a sensitive assay to measure both α-synuclein abundance and provide an internal loading control for signal quantitation. Therefore, we separated either purified 26S proteasome (360 ng), 26S proteasome (360 ng) with unmodified α-synuclein (150 ng), or 26S proteasome (360 ng) with monoubiquitinated α-synuclein (K96, 150 ng) using SDS-PAGE. The gel was then subjected to colloidal silver staining and imaged on a ChemiDoc XRS (Bio-Rad) (Figure 2). Importantly, both α-synuclein proteins separated completely from any proteasomal subunits that could therefore be used as an internal loading control.
Figure 2. Visualization of α-synuclein and the proteasome.
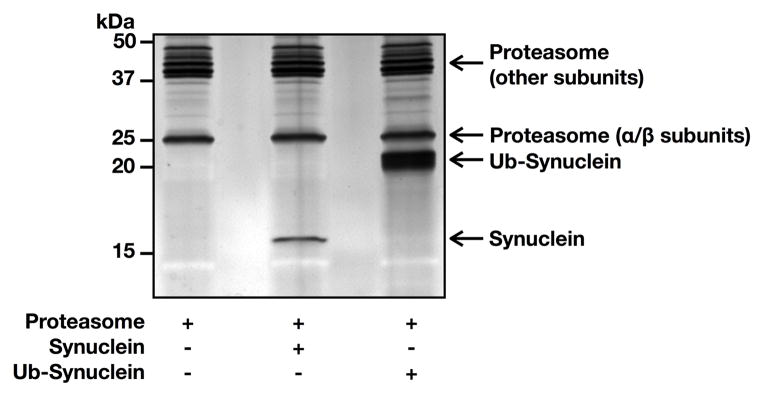
Purified proteasome (26S) alone, with unmodified α-synuclein, or with K96 monoubiquitinated α-synuclein was separated by SDS-PAGE and stained with colloidal silver. Based on molecular weight, the alpha subunits of the proteasome where chosen as a loading control.
Proteasomal turnover of site-specifically monoubiquitinated α-synuclein proteins
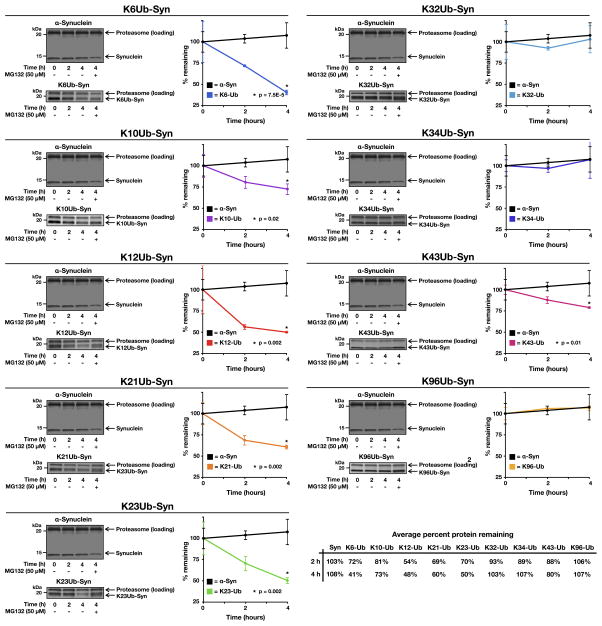
With a functional assay in hand, each of the ten different α-synuclein proteins (150 ng) were incubated in triplicate with purified 26S proteasomes (360 ng) supplemented with ATP and an ATP-regenerating system (total volume of 15 μL). Reactions were terminated after different lengths of time (0, 2, and 4 hours) by the addition of loading buffer. As a control for proteasome activity the proteasome inhibitor MG132 (100 μM) was added to a reaction that was allowed to progress for 4 hours. The proteins were separated by SDS-PAGE, stained with colloidal silver, and imaged as above (Figure 3 & S4). Amounts of α-synuclein or the monoubiquitinated derivatives were quantitated and normalized to the amounts of proteasome subunit as indicated and relative degradation rates were plotted (Figure 3). As expected, unmodified α-synuclein was not degraded by the proteasome over the course of the experiment; however, the monoubiquitinated proteins were turned over in a site-specific manner. Importantly, modification at K12 resulted in degradation of α-synuclein, consistent with results from the Ciechanover lab (Shabek et al., 2012). Other N-terminally located monoubiquitination sites (K6, K21, and K23) also resulted in high levels of degradation, between 40 and 59% of α-synuclein after 4 hours. In contrast, modifications found further into the primary sequence of α-synuclein (K32, K34, and K96) display no significant turnover when compared to unmodified α-synuclein. Notably, certain modification sites (K10 and K43) appear to be unique and only support a modest (73 to 80% of α-synuclein remaining after 4 hours) level of degradation.
Figure 3. see also Figure S3. Proteasome-dependent degradation of monoubiquitinated α-synuclein.
Unmodified or monoubiquitinated α-synuclein proteins were incubated in triplicate with 26S proteasome for the indicated lengths of time. The proteasome inhibitor MG132 (100 μM) was added as indicated. The proteins were separated by SDS-PAGE, stained with colloidal silver, and quantitated using Quantity One Analysis Software (Bio-Rad). The turnover of unmodified α-synuclein is shown in each panel for comparison. P, two-tailed t-test; error bars represent ±s.e.m.
DISCUSSION
We describe here the comprehensive analysis of the site-specific effects of monoubiquitination on the proteasomal degradation of α-synuclein. Our data demonstrate that different sites of lysine monoubiquitination have divergent consequences for α-synuclein turnover. Overall, these data support a general model where monoubiquitination events that occur towards the N-terminus of α-synuclein (first ~25 amino acids) always results in proteasome-dependent degradation, while those located further into the primary sequence, towards the middle of the protein, typically do not. Most of the monoubiquitination sites reside in amino-acid repeats in the primary sequence, including those that support degradation and those that do not. Furthermore, we have previously demonstrated that regardless of modification site monoubiquitination does not have a large impact on the secondary structure of either ubiquitin or α-synuclein (Meier et al., 2012). This argues for relative position of the monoubiquitin in relationship to the substrate protein primary-sequence as the most important determinant of proteasomal degradation. Interestingly, modification at K43 supports some α-synuclein turnover despite being located towards the middle of the protein, demonstrating that the orientation of monoubiquitin with respect to α-synuclein may also be an important factor. Importantly, we have observed similar effects on the aggregation of α-synuclein (e.g., K21 monoubiquitination blocks aggregation while K23 does not) (Meier et al., 2012).
The overall trend we observed (i.e., more efficient turnover of α-synuclein monoubiquitinated at its N-terminus) is very consistent with previous experiments performed by Ciechanover and coworkers that discovered the phenomenon of monoubiquitin-dependent degradation of short proteins (Shabek et al., 2012). Specifically, several of the model systems described in this paper were generated from N-terminal genetic fusions of “lysine-less” ubiquitin to substate proteins and semisynthetic α-synuclein monoubiquitinated at K12. However, they did not test model proteins where monoubiquitination was attached in the center of the protein primary sequence. Our data begins to fill-in this experimental limitation and add to the fundamental understanding of the differences between poly- and monoubiquitin-mediated protein degradation by strongly suggesting that monoubiquitin modifications located towards the center of such a protein will not result in significant turnover. Additionally, this information is key for the complete understanding of α-synuclein clearance in Parkinson’s disease and for the potential development of therapies targeted at α-synuclein ubiquitination.
α-Synuclein has been shown to interact with cellular membranes through its N-terminal amphipathic repeat domains with lysine residues contributing key contacts (Jao et al., 2008). Therefore, monoubiquitination of specific lysines may have different effects on these interactions, with implications to degradation and aggregation. We are currently combining protein semisynthesis with cell-culture systems to elucidate the consequences of site-specific monoubiquitination on all three of these areas, with important therapeutic implications. For instance, promoting the monoubiquitination of lysine residues that both block aggregation and promote α-synuclein degradation should relieve the burden of accumulated α-synuclein and slow the progression of disease. For example, α-synuclein is known to be modified by the ubiquitin-like modifier SUMO (SUMOylation) at K96 and K102 (Dorval and Fraser, 2006; Kim et al., 2011), suggesting the possibility that SUMOylation at K96 may change the occupancy of monoubiquitin at other lysines with corresponding effects on aggregation and turnover. Thus, studies on any cross-talk between monoubiquitination and SUMOylation may shed light on molecular pathways that control α-synuclein degradation and identify enzyme targets (e.g., deSUMOylating enzymes) for Parkinson’s disease therapy.
SIGNIFICANCE
Parkinson’s disease is the second most common form of neurodegeneration, caused at least in part by the progressive accumulation and spread of toxic aggregates of the protein α-synuclein. Therefore, inhibiting or eliminating this aggregation are attractive approaches towards the development of therapies that can slow the progression of disease. Human genetics and animal models have demonstrated that higher amounts of α-synuclein increase its aggregation and accelerate the development of Parkinson’s Disease symptoms, suggesting that strategies to induce its degradation would be beneficial. To systematically investigate the consequences of monoubiquitination on the proteasome-dependent degradation of α-synuclein, a semisynthetic strategy was employed to generate homogenous protein bearing monoubiquitin at each of the nine known modification sites. Subsequent in vitro turnover experiments revealed that at least six of the monoubiquitination sites support some degradation of α-synuclein, with sites located in the N-terminus of the protein always resulting in degradation. In contrast, monoubiquitination sites located towards the middle of the protein showed less or no turnover at all. We believe that these data may explain the differences previously obtained by using heterogeneous ubiquitination of α-synuclein by SIAH1/2, they and generate a biochemical foundation for future investigations of α-synuclein degradation in cellular and animal models of Parkinson’s disease. Furthermore, they provide a working model for the ability of monoubiquitin to support the degradation of any small protein in a modification-site dependent manner. Finally, they highlight the key contributions of chemistry to revealing the consequences of protein posttranslational modifications.
EXPERIMENTAL PROCEDURES
General Methods
The thiol activating reagent 2,2′-dithiobis(5-nitropyridine) (DTNP) was purchased from Sigma Aldrich and cysteamine was purchased from Tokyo Chemical Industry CO, LTD. All other commonly used chemical reagents and solvents were obtained from commercial sources (EMD, Novagen, Invitrogen, Fluka, Sigma Aldrich) and used without any further purification. Ultrapure laboratory grade water (filtered, deionized, sterilized) was obtained from in-house MilliQ® water purification systems. All solutions were prepared in ultrapure laboratory water and filter sterilized through a 0.4 μM filter (Genesee Scientific). Media (LB broth, Miller, Novagen and S.O.C. broth, Sigma) were prepared, sterilized and stored according to the manufacturer and standard published protocols (Sambrook and Russel 2001). Media were handled in sterile conditions under an open flame. Antibiotic stock: stocks of ampicillin (Na salt, EMD) at a working concentration of 1000x (100 mg mL-1) were stored at −20 °C. Reverse phase high performance liquid chromatography (RP-HPLC) was performed using an Agilent Technologies 1200 Series HPLC with Diode Array Detector . Mass spectra were acquired on an API 3000 LC/MS-MS System (Applied Biosystems/MDS SCIEX). Light scattering data was collected with a Dynapro Titan temperature controlled microsampler (Wyatt).
Ubiquitin-thiol Expression and Purification
E. coli BL21 (DE3) cells transformed with pTXB1-Ub-AvaDnaE plasmid were grown in 1 L of LB medium containing ampicillin (100 μg/mL) at 37 °C until OD600 = 0.6. Then, expression was induced by addition of 0.5 mM IPTG and incubation for 16 hours at 18 °C. Cells were harvested by centrifugation (5,000 × g, 30 min, 4 °C). The cell pellets were resuspended by adding 10 mL of lysis buffer (50 mM phosphate, 300 mM NaCl, 5 mM imidazole, pH 8.0) supplemented with Complete protein inhibitor cocktail. Cells were lysed by sonication (35% amplitude, 6× 30 second pulses separated by 30 seconds on ice). The soluble fraction was recovered by centrifugation (13,000 × g, 30 min, 4 °C). The soluble fraction was then loaded on to a 1mL HisTrap column (GE Healthcare). After discarding the flow-through, the column was washed with 5 column volumes (CV) of lysis buffer, 10 CV of buffer A (lysis buffer with 50 mM imidazole. The protein was eluted with buffer B (lysis buffer with 250 mM imidazole) in 4.5 CV elution fractions. The elution fractions were analyzed by SDS-PAGE and the pure fractions containing Ub-AvaDnaEIntein-His fusion were pooled and dialyzed against buffer C (100 mM sodium phosphate monobasic, 150 mM NaCl, 1 mM EDTA, 1 mM TCEP, pH 7.75). Ubiquitin was cleaved from AvaDnaE-His fusion by incubating with 100 mM cysteamine and 50 mM TCEP (in buffer C, pH7.75) for 12 h at 25 °C. Ubiquitin with C-terminal aminoethane thiol linker was purified by C4 semi-preparative RP-HPLC with a 25–60 %B gradient over 60 min (buffer A: 0.1% TFA in H2O, buffer B: 0.1% TFA, 90% ACN in H2O). Yield of 10.0 mg of Ub-SH/L of culture was obtained.
Synthesis of disulfide-linked monoubiquitinated α-synuclein
In a typical reaction 20.0 equivalents of DTNP (7.20 mg, 23.2 μmol) dissolved in 2.0 mL of a 3:1 (v/v) acetic acid:water mixture was added to Ub-SH (2.50 mg, 1.2 μmol). The reaction was allowed to proceed for 72 h at 25 °C with stirring. Disulfide activated ubiquitin was purified by C4 semi-preparative RP-HPLC with a 30–60%B gradient over 60 min (buffer A: 0.1% TFA in H2O, buffer B: 0.1% TFA, 90% ACN in H2O). This yielded 6.00 mg (59%) of the pure Ub-S-nitro-2-pyridinesulfenyl disulfide adduct (Ub-DTNP).Then 1.0 equivalent of α-synuclein cysteine mutant (1.50 mg, 0.065 μmol) and 2.0 equivalents of Ub-DTNP (1.20 mg, 0.136 μmol) were dissolved in 500 μL of reaction buffer (1M HEPES, 6 M Gn-HCl, pH 6.93). The reaction was allowed to proceed for 1 h at 25 °C with continuous shaking. Ubiquitin conjugated α-synuclein was purified by C4 semi-preparative RP-HPLC with a 30–60%B gradient over 60 min (buffer A: 0.1% TFA in H2O, buffer B: 0.1% TFA, 90% ACN in H2O) to yield typically ~1.00 mg of α-synuclein K#-Ub.
Silver staining
SDS-PAGE was performed using 18% Criterion™ Tris-HCl polyacrylamide pre-cast gel. Silver staining of the gels was performed according to the instructions provided in the Bio-Rad Silver Stain Plus Kit. Briefly, the resolved proteins were fixed by incubating the gel in 200 mL of fixative enhancer solution (50% methanol, 20% acetic acid, 10% fixative enhancer concentrate, 30% deionized distilled water) for 20 min at 25 °C. Following that the gel was rinsed twice with 100 mL of deionized distilled water for 10 min. Gel was then stained by incubating in 100 mL of staining solution (35 mL of deionized distilled water, 5 mL of silver complex solution, 5 mL of reduction moderator solution, 5 mL of image development reagent, 50 mL of development accelerator solution) until protein bands became visible (approximately 10–15 min). Then the staining reaction was stopped by incubating the gel in 100 mL of 5% (v/v) acetic acid for 15 min. Stained gel was then imaged using ChemiDoc XRS (Bio-Rad).
Proteasome Turnover Assay
Ubiquitin conjugated α-synuclein K#-Ub or wild type protein (150 ng) was incubated at 37 °C for indicated times with Bovine 26S proteasome (360 ng, Ubiquitin Proteasome Biotechnologies), 40 mM Tris-HCl (pH 7.6), 5 mM MgCl2, 2 mM ATP, 10 mM creatine phosphate, .1 mg/mL creatine phosphokinase at a final volume of 15 μL. When indicated, the proteasome inhibitor MG132 was added to a final concentration of 100 μM. Reactions were terminated by the addition of 4-fold concentrated sample buffer. Samples were then boiled, resolved via SDS-PAGE, stained with colloidal silver, and imaged on ChemiDoc XRS (Bio-Rad).
Supplementary Material
HIGHLIGHTS.
Nine monoubiquitinated α-synuclein proteins were prepared semisynthetically
Monoubiquitination towards the N-terminus resulted in degradation by the proteasome
Three sites of modification did not result in α-synuclein degradation
There are site-specific differences in monoubiuqitin-mediated protein degradation
Acknowledgments
This research was supported by the University of Southern California to M.R.P and by the Isreal Academy of Sciences, the B. Rappaport Foundation, Dears Foundation, Elsa Fendler and Sternheim Research Funds for Parkinson’s and Alzheimer’s Research, and the Technion Research Funds to S.E., and in part by the National Cancer Institute (P30CA014089). M.R.P acknowledges additional support from the Damon Runyon Cancer Research Foundation and the Concern Foundation. We thank Prof. J. Camarero for instrumentation and N. Marotta for assistance with the graphical abstract. Dynamic light scattering data were generated at the USC NanoBiophysics Core Facility.
Footnotes
Supplemental Data include 3 figures and can be found with this article online at
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson JP, Walker DE, Goldstein JM, de Laat R, Banducci K, Caccavello RJ, Barbour R, Huang J, Kling K, Lee M, et al. Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J Biol Chem. 2006;281:29739–29752. doi: 10.1074/jbc.M600933200. [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet Neurol. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, Mcginty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- Chen J, Ai Y, Wang J, Haracska L, Zhuang Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat Chem Biol. 2010;6:270–272. doi: 10.1038/nchembio.316. [DOI] [PubMed] [Google Scholar]
- Dorval V, Fraser PE. Small ubiquitin-like modifier (SUMO) modification of natively unfolded proteins tau and α-synuclein. J Biol Chem. 2006;281:9919–9924. doi: 10.1074/jbc.M510127200. [DOI] [PubMed] [Google Scholar]
- Ferreon ACM, Gambin Y, Lemke EA, Deniz AA. Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc Natl Acad Sci USA. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AL. The Aggregation and Fibrillation of α-Synuclein. Acc Chem Res. 2006;39:628–634. doi: 10.1021/ar050073t. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Jao CC, Hegde BG, Chen J, Haworth IS, Langen R. Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc Natl Acad Sci USA. 2008;105:19666–19671. doi: 10.1073/pnas.0807826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Jang WH, Quezado MM, Oh Y, Chung KC, Junn E, Mouradian MM. Proteasome inhibition induces α-synuclein SUMOylation and aggregate formation. Journal of the Neurological Sciences. 2011;307:157–161. doi: 10.1016/j.jns.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Wheeler T, Li L, Chin L. Ubiquitination of {alpha}-synuclein by Siah-1 promotes {alpha}-synuclein aggregation and apoptotic cell death. Hum Mol Genet. 2008;17:906. doi: 10.1093/hmg/ddm363. [DOI] [PubMed] [Google Scholar]
- Liani E, Eyal A, Avraham E, Shemer R, Szargel R, Berg D, Bornemann A, Riess O, Ross CA, Rott R, et al. Ubiquitylation of synphilin-1 and alpha-synuclein by SIAH and its presence in cellular inclusions and Lewy bodies imply a role in Parkinson’s disease. Proc Natl Acad Sci USA. 2004;101:5500–5505. doi: 10.1073/pnas.0401081101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Kehm V, Carroll J, Zhang B, O’Brien P, Trojanowski JQ, Lee VMY. Pathological -Synuclein Transmission Initiates Parkinson-like Neurodegeneration in Nontransgenic Mice. Science. 2012;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magen I, Chesselet MF. Genetic mouse models of Parkinson’s disease The state of the art. Prog Brain Res. 2010;184:53–87. doi: 10.1016/S0079-6123(10)84004-X. [DOI] [PubMed] [Google Scholar]
- Meier F, Abeywardana T, Dhall A, Marotta NP, Varkey J, Langen R, Chatterjee C, Pratt MR. Semisynthetic, Site-Specific Ubiquitin Modification of α-Synuclein Reveals Differential Effects on Aggregation. J Am Chem Soc. 2012;134:5468–5471. doi: 10.1021/ja300094r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano Y, Yamashita H, Takahashi T, Kishida S, Nakamura T, Iseki E, Hattori N, Mizuno Y, Kikuchi A, Matsumoto M. Siah-1 facilitates ubiquitination and degradation of synphilin-1. J Biol Chem. 2003;278:51504–51514. doi: 10.1074/jbc.M306347200. [DOI] [PubMed] [Google Scholar]
- Nonaka T, Iwatsubo T, Hasegawa M. Ubiquitination of alpha-synuclein. Biochemistry. 2005;44:361–368. doi: 10.1021/bi0485528. [DOI] [PubMed] [Google Scholar]
- Oueslati A, Fournier M, Lashuel HA. Role of post-translational modifications in modulating the structure, function and toxicity of alpha-synuclein Implications for Parkinson’s disease pathogenesis and therapies. Prog Brain Res. 2010;183C:115–145. doi: 10.1016/S0079-6123(10)83007-9. [DOI] [PubMed] [Google Scholar]
- Paxinou E, Chen Q, Weisse M, Giasson BI, Norris EH, Rueter SM, Trojanowski JQ, Lee VM, Ischiropoulos H. Induction of alpha-synuclein aggregation by intracellular nitrative insult. Journal of Neuroscience. 2001;21:8053–8061. doi: 10.1523/JNEUROSCI.21-20-08053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabanal F, DeGrado WF, Dutton PL. Use of 2, 2′-dithiobis (5-nitropyridine) for the heterodimerization of cysteine containing peptides. Introduction of the 5-nitro-2-pyridinesulfenyl group. Tetrahedron Letters. 1996;37:1347–1350. [Google Scholar]
- Rott R, Szargel R, Haskin J, Shani V, Shainskaya A, Manov I, Liani E, Avraham E, Engelender S. Monoubiquitylation of α-Synuclein by Seven in Absentia Homolog (SI-AH) Promotes Its Aggregation in Dopaminergic Cells. Journal of Biological Chemistry. 2008;283:3316–3328. doi: 10.1074/jbc.M704809200. [DOI] [PubMed] [Google Scholar]
- Rott R, Szargel R, Haskin J, Bandopadhyay R, Lees AJ, Shani V, Engelender S. α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc Natl Acad Sci USA. 2011;108:18666–18671. doi: 10.1073/pnas.1105725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N, Herman-Bachinsky Y, Buchsbaum S, Lewinson O, Haj-Yahya M, Hejjaoui M, Lashuel HA, Sommer T, Brik A, Ciechanover A. The Size of the Proteasomal Substrate Determines Whether Its Degradation Will Be Mediated by Mono- or Polyubiquitylation. Mol Cell. 2012;48:87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- Shah NH, Dann GP, Vila-Perelló M, Liu Z, Muir TW. Ultrafast Protein Splicing is Common among Cyanobacterial Split Inteins: Implications for Protein Engineering. J Am Chem Soc. 2012;134:11338–11341. doi: 10.1021/ja303226x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Webb JL. alpha-Synuclein Is Degraded by Both Autophagy and the Proteasome. Journal of Biological Chemistry. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT. NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- Welchman R, Gordon C, Mayer R. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.