Abstract
Hot pepper is one of the economically important crops in Asia. A large number of gene sequences, including expressed sequence tag (EST) and genomic sequences are publicly available. However, it is still a daunting task to determine gene function due to difficulties in genetic modification of a pepper plants. Here, we show the application of the virus-induced gene silencing (VIGS) repression for the study of 459 pepper ESTs selected as non-host pathogen-induced cell death responsive genes from pepper microarray experiments in Nicotiana benthamiana. Developmental abnormalities in N. benthamiana plants are observed in the 32 (7%) pepper ESTs-silenced plants. Aberrant morphological phenotypes largely comprised of three groups: stunted, abnormal leaf, and dead. In addition, by employing the combination of VIGS and Agrobacterium-mediated transient assays, we identified novel pepper ESTs that involved in Bax or INF1-mediated cell death responses. Silencing of seven pepper ESTs homologs suppressed Bax or INF1-induced cell death, five of which suppressed both cell death responses in N. benthamiana. The genes represented by these five ESTs encode putative proteins with functions in endoplasmic reticulum (ER) stress and lipid signaling. The genes represented by the other two pepper ESTs showing only Bax-mediated cell death inhibition encode a CCCH-type zinc finger protein containing an ankyrin-repeat domain and a probable calcium-binding protein, CML30-like. Taken together, we effectively isolated novel pepper clones that are involved in hypersensitive response (HR)-like cell death using VIGS, and identified silenced clones that have different responses to Bax and INF1 exposure, indicating separate signaling pathways for Bax- and INF1-mediated cell death.
Keywords: Bax-induced cell death, hot chili pepper (Capsicum annum), hypersensitive response, INF1, Nicotiana benthamiana, virus-induced gene silencing
1. Introduction
Many plant DNA sequences, including whole genome sequences and expressed sequence tags (EST), have been released in the last decade. Developments in EST and cDNA microarray technology have allowed the responses of thousands of genes associated with intercellular and intracellular signaling pathways to be investigated. However, although this large body of information has accelerated the characterization of responses at the genomic level, the ability to functionally investigate individual genes remains limited. Some economically important crops, such as pepper, are particularly intractable for reasons such as reduced susceptibility to genetic transformation.
Generation of gain- and loss-of-function mutations is a common strategy to investigate gene function. In particular, in some plant species, knock-out mutant populations can be produced by using T-DNA insertions or site-directed mutagenesis; however, embryonic lethal mutations and redundant genes can be overlooked using this strategy. Virus-induced gene silencing (VIGS) is an alternative genetic tool that allows functional characterization by knocking-down the expression of native genes [1,2]. VIGS is particularly useful when applied to genes in which mutations are embryonic lethal and in species that are resistant to transformation. In addition, VIGS has been adapted for high-throughput functional genomics [3,4] and, by using heterologous sequences, has been successfully used for species that lack genomic sequence availability [5]. Both pepper and N. benthamiana are within the Solanaceae family and it has been approved in several publications that functional characterization of pepper genes in N. benthamiana is a useful using tobacco rattle virus (TRV)-mediated gene silencing methods [6–8].
Programmed cell death that occurs during plant-pathogen interactions is well characterized [9,10]. During incompatible plant-pathogen interactions, a rapid and localized cell death is triggered at the infection site, termed the hypersensitive response (HR); this is similar to apoptosis in animals. This HR is often associated with resistance to further pathogen multiplication and spread [9,10]. Although details of the molecular signal pathway of HR cell death remain poorly understood, production of reactive oxygen species (ROS), and gene activation have been elucidated. Plant homologs of animal apoptotic regulators such as the Bcl-2 family proteins have been identified [11,12]. Bax is a member of the Bcl-2 family of proteins that contains crucial managers of cell survival and cell death. Several reports indicate that the control and execution of programmed cell death in animal and plant cells is evolutionarily conserved. For example, transient overexpression of Bax using a viral vector induces cell death in tobacco [11], and this cell death response induced ROS production and pathogenesis-related gene expression, like HR mediated by tobacco N gene [11]. In addition, overexpression of Arabidopsis AtBI-1, a plant homolog of the mammalian Bax inhibitor, inhibits Bax-induced cell death in Arabidopsis as well as fungal elicitor-induced cell death in cultured rice cells [12,13]. INF1 is a 10 kDa extracellular protein produced by the oomycete Phytophthora infestans, the causal agent of late blight disease in potato and tomato [14]. INF1 triggers a wide range of defense responses, including cell death, in most Nicotiana species [15]. Signaling cascades following INF1 recognition, which lead to hypersensitive cell death, have been studied [16–18]. Thus far, the very limited numbers of host proteins required for Bax- or INF1-mediated cell death have been determined through transient expression study in N. benthamiana and Arabidopsis mutant analyses. To study the function of pepper ESTs that specifically regulated during pepper and non-host pathogen interaction, we employed the TRV-mediated VIGS approach. Genes were silenced in N. benthamiana plants using VIGS with 459 EST clones and the morphological phenotypes of the plants (7%) were observed. Cell death was further investigated by the transient expression of the cell death inducers Bax and INF1 in leaves of silenced N. benthamiana plants. To date, seven pepper genes, including catalytic activity-related, binding activity-coupled, and transcription factor genes, were found to be involved in Bax- and INF1-induced cell death.
2. Results and Discussion
2.1. Silencing of Pathogen-Induced Pepper ESTs in N. benthamiana Plants
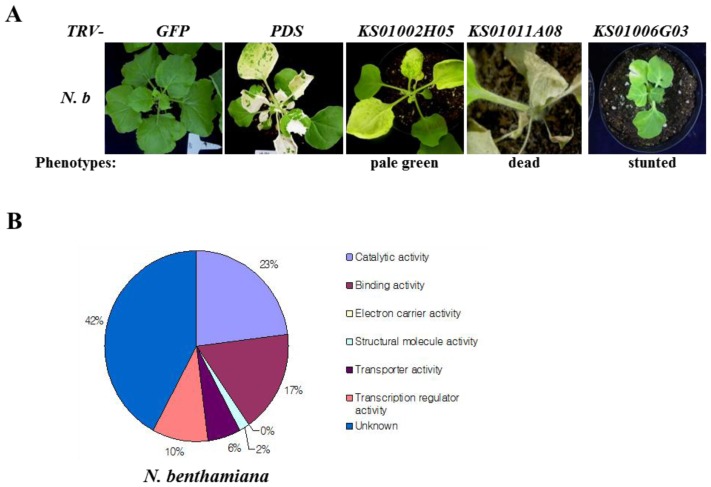
Previously, we performed hot chili pepper cDNA microarray analysis with 4685 unique EST clones, and 613 ESTs that were differentially expressed when exposed to the soybean pustule pathogen Xanthomonas axonophodis pv. glycines 8ra (Xag 8ra) were isolated [19]. Xag 8ra is not a pathogen of pepper, but does elicit an HR-like cell death in pepper leaves, as well as induce the expression of a number of pathogenesis-related (PR) genes [19]. For VIGS analysis, 459 of these ESTs were successfully ligated into the pTRV2 vector. Details of the pTRV2 derivatives are provided in Supplementary File 1. Plants infected with TRV-PDS or TRV-GFP were used as positive and negative controls for observation of the morphological change caused by VIGS experiment, respectively [20,21]. VIGS experiments were performed with two-week-old N. benthamiana plants using previously described methods [21]. At three weeks after co-infiltration of N. benthamiana plants with pTRV1 and pTRV2 constructs, morphological abnormalities such as chlorosis, leaf deformations, and stunted growth were compared with that of GFP-silenced control plants. Using data from two biologically independent VIGS experiments, plants were categorized based on their phenotypes, with most plants falling into one or more of four main groups: normal, stunted, abnormal leaf, and dead (Table 1; Figure 1A). Plants were silenced with constructs derived from 459 pepper ESTs. Of these, 32 N. benthamiana plants (7%) exhibited a different morphology from that of the corresponding GFP-silenced control plants. The low proportion of the non-host pathogen-induced genes that exhibited abnormal growth under normal growth condition in the silenced plants is consistent with their specific induction by a non-host pathogen. The pepper genes that exhibited morphological differences when silenced were categorized by molecular function (Figure 1B).
Table 1.
Pepper expressed sequence tags (ESTs) showing morphological change in VIGSed N. benthamiana plants.
| EST ID | BLASTX * | VIGSed phenotypes |
|---|---|---|
| Catalytic activity/ligase activity | ||
| KS01017F01 | Q8RU74_LYCES Dehydroquinate synthase (EC 4.6.1.3) | Stunted |
| KS26035G04 | Q5Z8K3_ORYSA Putative ZEITLUPE | Stunted |
|
| ||
| Catalytic activity/oxidoreductase activity | ||
| KS01002H05 | Q6S5A3_TROMA Sterol delta-7 reductase | Dead |
| KS13009F01 | Q9SVA9_ARATH Cytochrome P450-like protein | Leaf necrotic spot |
|
| ||
| Catalytic activity/transferase activity | ||
| KS13055B07 | Q5DKV0_TOBAC Adenosine kinase isoform 1T | Wrinkled leaf |
|
| ||
| Catalytic activity/hydrolase activity | ||
| KS23052B07 | chitinase/lysozyme [Nicotiana tabacum] | Pale green |
| KS08010H01 | probable pectate lyase P56-like [S. lycopersicum] | Stunted & wrinkled leaf |
| KS01052C01 | Q8H272_LYCES Metacaspase 1 (Fragment) | Wrinkled leaf |
|
| ||
| Binding/ion binding | ||
| KS12078F02 | calmodulin [Capsicum annuum] | Stunted |
| KS01006G03 | probable calcium-binding protein CML30-like [S. lycopersicum] | Stunted |
|
| ||
| Binding/protein binding | ||
| KS01047E07 | O04133_SOYBN SRC2 | Stunted |
|
| ||
| Binding/nucleic acid binding | ||
| KS01060G11 | Q93XV7_9ROSI Hydroxypyruvate reductase (EC 1.1.1.29) | Small leaf size |
| KS12047A04 | Q9XI07_ARATH F8K7.13 protein (Putative transcriptional regulatory protein) | Weak dwarf |
|
| ||
| Binding/others | ||
| KS01044F10 | BIP5_TOBAC Luminal binding protein 5 precursor (BiP 5) | Stunted |
| KS01057F02 | mannose-binding lectin [C. annuum] | Stunted |
| KS14050G06 | Q41424_SOLTU Chlorophyll a/b binding protein | Pale green |
|
| ||
| Structural molecule activity | ||
| KS10105B11 | Q9SCM3_ARATH 40S ribosomal protein S2 homolog (At3g57490) | Stunted & curling leaf |
| KS08008G04 | 60S ribosomal protein L10 [S. melongena] | Stunted & curling leaf |
|
| ||
| Transporter activity | ||
| KS24014C12 | Q93Y42_ARATH Coatomer delta subunit (Delta-coat protein) | Dead |
| KS01047B03 | Q9XF20_CAPCH Acyl carrier protein | Stunted |
| KS08017G06 | O80774_ARATH Hypothetical protein At2g34250 | Stunted & curling leaf |
|
| ||
| Transcription regulator activity | ||
| KS01043D02 | Q9XEE6_ARATH Hypothetical Cys-3-His zinc finger | Stunted |
| KS09041E04 | Q710C3_SPIOL Sigma factor (Fragment) | Pale green |
| KS09088A04 | Q7Y039_LYCES MADS-box protein 5 | Abnormal leaf |
| KS12065G09 | Q8RXK4_ARATH Hypothetical protein At4g38900 | Stunted |
|
| ||
| Unknown | ||
| KS20002B10 | later embryo abundant protein [Prunus dulcis] | Stunted |
| KS01047F02 | Q9C9T7_ARATH Hypothetical protein F25P22.17 | Stunted |
| KS01056A09 | No_hits | Complete dead |
| KS20014C01 | No_hits | Leaf local dead |
| KS07002E11 | AAD25142.1 (Hypothetical protein At3g24506) | Pale green |
| KS07012E06 | Q9FJZ7_ARATH Arabidopsis thaliana genomic DNA, chromosome 5, | Chlorosis |
| KS19065G03 | Q9C9V9_ARATH Putative golgi transport complex protein; 67058-70172 | Stunted & curling leaf |
Blast hit at http://genepool.kribb.re.kr/new/index.php-Solanaceae Gene Indices (six species) [19].
Figure 1.
Morphological phenotypes in VIGSed N benthamiana plants. (A) Representative morphological phenotypes in N. benthamiana after VIGS. GFP negative and PDS positive controls for the VIGS experiment, respectively; KS01002H05, KS01011A08, KS01006G03, characteristic aberrant phenotypes of the classes pale green, dead, and stunted, respectively. Images of the silenced plants were taken 21 days post-infiltration, respectively; and (B) Molecular characterization of pepper ESTs exhibiting morphological abnormalities. Percentages were calculated using the total numbers in N. benthamiana.
2.2. Screening of Pepper Genes Coupled with Bax- or INF1-Induced Cell Death Responses
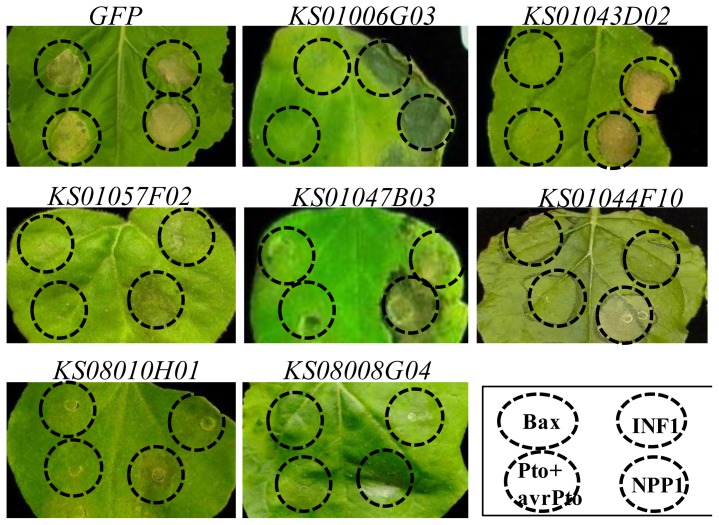
To identify genes involved in Bax- or INF1-mediated cell death responses, transient expression of Bax or INF1 was induced in silenced N. benthamiana plants, and the plants were scored for the presence of a cell death phenotype. Transient expression of Bax or INF1 in the leaves of TRV-GFP-inoculated N. benthamiana plants induced cell death within two days (Figure 2A). In an initial screen in which three replicate plants for each pepper TRV2 derivatives were exposed to Bax or INF1, 78 clones were identified in which Bax- or INF1-induced cell death was compromised. Two further independent biological replicate experiments were performed with these 78 affected clones, from which seven clones were identified that were reproducibly compromised in the cell death response. Five of these clones exhibited reduced cell death when treated with Bax or INF1, and two further clones exhibited reduced cell death only with Bax treatment (Figure 2A, Table 2). We confirmed the VIGS of the corresponding N. benthamiana genes in the silenced plants after three weeks of TRV infection by semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) using the specific primers for the each N. benthamiana homologs (Supplementary Tables S1 and S2). The expression levels of all seven N. benthamiana homologs were significantly reduced in the silenced plants compared with that of in the control plants which were silenced with the pTRV-GFP (Supplementary Figure S1). The identified genes, representing 1.5% of the set, encode proteins with putative functions in protein stability and secondary metabolism, as well as unknown functions. This is a relatively low proportion of the gene set when compared to a previous VIGS screen for HR genes [6]. This may be due to the genes used in this study, which were chosen based on their induction during infection with a non-host pathogen, Xag 8ra; accordingly, Xag 8ra-induced HR might share few genes with Bax- or INF1-induced cell death. The identification of genes involved in both INF1- and Bax-induced cell death and genes only involved in Bax-induced cell death indicates the presence of common and distinct components of these two cell death pathways.
Figure 2.
Cell death phenotypes in N. benthamiana after transient expression of Bax and INF1 in plants treated with indicated pepper TRV clones. (A) N. benthamiana plants were infected with 459 TRV-defense-associated pepper ESTs or pTRV-GFP as the control. After three weeks, the upper leaves of each silenced plant were exposed to Bax or INF1. Images of plants exhibiting suppressed cell death responses were taken three days post-inoculation; and (B) Morphological alterations of plants in which Bax and/or INF1-induced cell death-associated pepper genes were silenced. Images were taken three weeks after VIGS was initiated.
Table 2.
Pepper ESTs involved in Bax- or INF1-mediated cell death responses.
| EST ID | BLASTX * | E-value | Phenotypes | ||
|---|---|---|---|---|---|
|
| |||||
| Morphology | HR | ||||
|
| |||||
| Bax | INF | ||||
| KS01044F10 | glucose-regulated protein 78 (BiP), partial [S. lycopersicum] | 2.0 × 10−87 | stunted | X | X |
| KS01047B03 | Acyl carrier protein [C. chinense] | 3.0 × 10−58 | stunted | X | X |
| KS08008G04 | 60S ribosomal protein L10 [S. melongena] | 7.0 × 10−121 | stunted & wrinkled leaf | X | X |
| KS08010H01 | probable pectate lyase P56-like [S. lycopersicum] | 8.0 × 10−77 | stunted & wrinkled leaf | X | X |
| KS01057F02 | mannose-binding lectin [C. annuum] | 1.0 × 10−77 | stunted | X | X |
| KS01006G03 | probable calcium-binding protein CML30-like [S. lycopersicum] | 1.0 × 10−77 | stunted | X | Δ |
| KS01043D02 | Hypothetical Cys-3-His zinc finger protein [A. thaliana] | 1.0 × 10−125 | stunted | X | O |
BLAST hit at http://genepool.kribb.re.kr/new/index.php-Solanaceae Gene Indices (six species) [19]. Morphological phenotypes were observed three weeks after VIGS was performed. Suppression phenotypes varied: X: cell death was suppressed three days post-inoculation (dpi); Δ: cell death was delayed 3 dpi; O: cell death was the same as that of the GFP control plants.
2.3. Silencing of the Putative Calcium-Binding Protein CML30-Like and Cys-3-His Zinc Finger Protein Suppressed Bax-Induced Cell Dath, but Not INF1-Induced Cell Death
Two pepper clones, KS01043D02 and KS01006G03, suppressed only Bax-induced cell death (Figure 2A). To investigate whether this suppression was Bax-specific, two further cell death pathways were induced in these two pepper clones, namely, those affected by Pto-avrPto [6] and NPP1 [22]. Pto is an R gene, encoding serine/threonine kinase, which confers resistance to Pseudomonas syringae. pv. tomato by recognizing and interacting with the pathogen type III effector protein AvrPto. NPP1 is necrosis-inducing Phytophthora protein 1 [22]. In both KS01043D02- and KS01006G03-silenced plants, Pto-avrPto-induced cell death was inhibited, similar to Bax-induced cell death; however, NPP1-induced cell death was not inhibited (Figure 3). In TRV-GFP control plants, cell death for all treatments was observed at two days post-inoculation.
Figure 3.
Comparison of cell death induced by Bax, Pto-avrPto, INF1, and NPP1 in plants silenced with TRV-pepper ESTs. N. benthamiana plants were inoculated with the indicated pepper TRV clones. After three weeks, upper leaves were exposed to Bax, Pto-avrPto, INF1, and NPP1, and images were taken three days later.
The putative protein encoded by KS01006G03 has high homology with a calcium-binding CML30-like protein. The biological function of this protein is unknown; however, previous research indicates that 13 calmodulin genes are differentially expressed in tobacco plants during pathogen-induced HR and wounding, and that calmodulin is one of the major signaling components involved in Tobacco Mosaic Virus-induced cell death in tobacco [23,24]. In the current study, by using VIGS, we showed that a putative calmodulin-like gene is involved in Bax- and Pto-avrPto-induced cell death, but not in INF1- or NPP1-induced cell death in N. benthamiana. We also observed that the protein encoded by this gene appears to be involved in development, as VIGS with a fragment of the gene caused growth defects (Figure 2B).
KS01043D02 encodes a hypothetical protein of unknown function that contains zinc finger and ankyrin-repeat domains. BLAST analysis indicates that putative homologs exist in tomato and Arabidopsis (LOC101258155 and At2G40140, respectively). A T-DNA insertion mutant of AtSZF1 (At2G40140) shows increased susceptibility to salt stress [25], indicating that this protein might have roles in biotic and abiotic defense mechanisms, including HR.
2.4. Bax- and INF1-Induced Cell Death Were Suppressed in N. benthamiana Plants in Which One of Five Pepper Genes Were Silenced
VIGS using five pepper ESTs, namely, KS01044F10, KS01047B03, KS01057F02, KS08010H01, and KS08008G04, resulted in a noticeable suppression of both Bax- and INF1-induced cell death (Figure 2A). This indicates that the orthologous genes in N. benthamiana of these five pepper genes might be essential for cell death induced by Bax and INF1. Interestingly, all these gene- silenced plants exhibited defected morphologies such as stunted growth and abnormal leaves, indicating that the five genes might not only be involved in cell death but, also, in plant growth and development. The five genes encode a putative luminal binding protein (BiP) (KS01044F10), an acyl carrier protein (ACP) (KS01047B03), a pectate lyase P56-like protein (KS01057F02), a mannose-binding lectin (KS08010H01), and a 60S ribosomal protein L10 (KS08008G04) (Table 2). Cell death suppression in the silenced plants was prolonged; therefore, we assumed that these genes are essential to general cell viability. However, when the five silenced plants were treated with Pto-avrPto and NPP1, they exhibited variable cell death inhibition phenotypes (Figure 3). Silencing of KS01044F10 and KS01047B03 effectively compromised Pto-avrPto-mediated HR, but did not suppress NPP1-induced cell death. KS01057F02-, KS08010H01-, and KS08008G04-silenced tobacco plants exhibited a weakened NPP1-induced cell death response as well as suppressed Pto-avrPto-induced cell death (Figure 3).
Luminal binding proteins (BiPs) are key components required for protein maturation and transport, and serve as master regulators of the unfolded protein response (UPR), the signaling pathway triggered by the accumulation of misfolded or unfolded proteins in the mammalian endoplasmic reticulum [26,27]. In plants, application of UPR inducers such as tunicamycin leads to the accumulation of transcripts corresponding to ER chaperones, such as BiP, indicating the possible conservation of UPR in plants [28]. Therefore, it is conceivable that BiP functions in Bax-, Pto-avrPto-, and INF1-mediated cell death via its role in UPR; however, NPP1-induced cell death was not affected by BiP.
ACP regulation has been the focus of several studies of lipid biosynthesis in plants [29]. Plant cells must adjust to changing lipid requirements during development and under varied environmental conditions, and so must control the expression of the many genes that are involved in the fatty acid and lipid synthesis pathways. In mammalian cells, down-regulation of mitochondrial ACP compromises protein lipolyation and the respiratory complex, and eventually results in cell death [30]. This study provides the first evidence that ACP is associated with plant cell death, in particular with Bax-, Pto-avrPto-, and INF1-coupled cell death.
A previous VIGS Pto-AvrPto-induced HR screen identified diverse ribosomal proteins as cell death components [6], and ribosomal proteins are differentially expressed upon elicitation of a defense response in tomato [31]. The HR requires transcriptional up-regulation of various defense-related genes [32], and it may require an increase in ribosomal proteins for the concomitant increase in translational activity.
The KS08010H01, mannose-binding lectin (CaMBL1), has been shown previously to play a role in plant cell death and defense responses [33]. Transient overexpression of the CaMBL1 induced cell death on pepper leaves and transgenic Arabidopsis plants expressing the CaMBL1 increased defense responses to bacterial pathogen invasion [33]. Although, they have not studied a suppression of the HR in the CaMBL1-silenced pepper plants, it is tempting to speculate that the mannose-binding lectin we observed here could be involved in the general cell death responses or in the defense responses to fungal pathogen.
The pectate lyase P56-like protein (KS01057F02) has been previously shown to be involved in extension of pollen tube. The role of pectate lyase neither in plant innate immunity nor in cell death has been previously investigated. In this study, the observed loss-of-function phenotype suggests that pectate lyase plays an important role as a positive regulator of basal defense such as HR, and also has a role in plant development.
3. Experimental Section
3.1. Plant Materials and VIGS
Nicotiana benthamiana seeds were germinated in soil, and plants were grown in a glasshouse at 23 °C. DNA from 459 pepper EST clones [19], was double-digested with EcoRI/XhoI and, in a separate reaction, with BamHI/KpnI. The resulting fragments were blunted by T4 DNA polymerase and then ligated into the pTRV2 silencing vector. pTRV RNA1 vector (pTRV1) or pTRV2 derivatives (contained pepper ESTs) were mobilized into Agrobacterium tumefaciens strain GV2260 by the freeze-thaw method of transformation [20,21]. A. tumefaciens cells carrying pTRV1 and pTRV2 derivatives were initially cultured in 5 mL of liquid Luria-Bertani (LB) media containing 50 μg/mL kanamycin and 100 μg/mL rifampicin and grown overnight at 28 °C. A. tumefaciens cells were harvested and re-suspended in infiltration media (10 mM MgCl2, 10 mM MES buffer, pH 5.6, 200 μM acetosyringone), adjusted to an absorbance (OD600) of 0.8–1.0, and incubated at room temperature for 3 h. Subsequently, the mixtures of A. tumefaciens containing pTRV1 and pTRV2 derivative plasmids were mixed together in a ratio of 1:1 and infiltrated into the fully expanded leaf of ~three-week-old N. benthamiana plants using a 1 mL needleless syringe. VIGS experiment was conducted two times with three biological replications for each construct. VIGS of chili pepper was performed as previously described [21].
3.2. Agrobacterium-Mediated Transient Expression
Overnight cultures of A. tumefaciens strain GV3101 containing the following plasmids were harvested by centrifugation: 35S-promoter driven Bax [11], Pto and avrPto [6], and pGR106 INF1 and NPP1 [15,22] cells were re-suspended in infiltration media (10 mM MgCl2, 10 mM MES buffer, pH 5.6, 200 μM acetosyringone) to an OD600 of 0.5–0.1 and incubated for 2 h at room temperature. A. tumefaciens were infiltrated into the silenced tobacco leaves two weeks after infection with pTRV plasmids. Experiment was conducted three times with six replications for each experiment.
3.3. Non-Host Pathogen-Induced Pepper ESTs
Hot chili pepper EST candidate genes were categorized into functional groups. Gene sequences were identified from the pepper consensus sequence [http://genepool.kribb.re.kr/new/index.php-Solanaceae Gene Indices (six species)] and these sequences were used to perform BLASTN and BLASTX analysis using the GO (http://www.geneontology.org/) and TAIR (http://www.Arabidopsis.org) databases.
3.4. RT-PCR Analysis
Total RNA was extracted from leaf tissues of the silenced plants using the TRI reagent according to the manufacturer’s instructions (MRC, Cincinnati, OH, USA). Total extracted RNA was treated with 1 unit of RNase-free DNase (Promega, Madison, WI, USA) for 10 min at 37 °C, and purified using the TRI reagent. The first-strand cDNA was synthesized using 1 μg of DNase-treated RNA, oligo d(T) primer, and Moloney murine leukemia virus reverse transcriptase(M-MLV RT, Invitrogen, Rockville, MD, USA) following standard protocols and Semi-quantitative PCR was performed in a total volume of 20 μL in AccuPower PCR PreMix (Bioneer, Daejeon, Korea). To ensure that only host genes and not the viral RNA transcripts were amplified, the RT transcriptase reactions were performed using oligo d(T) primers. The forward primer F1 and reverse primer R2 were used to detect endogenous gene transcripts. The primers used in RT-PCR analysis were shown in the Supplementary Table S2.
4. Conclusions
In this study, we transiently overexpressed cell death induction components in a population of N. benthamiana plants, in which various candidate HR genes from pepper were silenced using VIGS. Using this strategy, we identified seven pepper ESTs that are involved in INF1- and/or Bax-induced cell death responses. Further studies of these seven candidate genes are required to determine whether they are also required for other defense signaling pathways, and at which stages the proteins act in the signal transduction cascades leading to HR.
Supplementary Information
Acknowledgments
We thank Dinesh-Kumar for providing pTRV1 and pTRV2 and Sophien Kamoun for providing pGR106 INF1 and NPP1. This work was supported by a Korea Research Foundation Grant funded by the Korean Government (MOEHRD) (KRF-2006-532-F00004) awarded to Lee, J.H. and by the Cabbage Genomics assisted breeding supporting Center funded by Ministry for Food, Agriculture, and Forestry of the Korean Government (to Park, J.M.) and partly by the KRIBB initiative program.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Baulcombe D.C. Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 1999;2:109–113. doi: 10.1016/S1369-5266(99)80022-3. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz M.T., Voinnet O., Baulcombe D.C. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terauchi R., Bin Nasir K.H., Ito A., Saitoh H., Berberich T., Takahashi Y. High-throughput functional screening of plant and pathogen genes in planta. Plant Biotechnol. 2005;22:455–459. [Google Scholar]
- 4.Anand A., Vaghchhipawala Z., Ryu C.M., Kang L., Wang K., Del-Pozo O., Martin G.B., Mysore K.S. Identification and characterization of plant genes involved in Agrobacterium-mediated plant transformation by virus-induced silencing. Mol. Plant Microbe Interact. 2007;20:41–52. doi: 10.1094/MPMI-20-0041. [DOI] [PubMed] [Google Scholar]
- 5.Senthil-Kumar M., Udayakumar M. High-throughput virus-induced gene-silencing approach to assess the functional relevance of a moisture stress-induced cDNA homologous to lea4. J. Exp. Bot. 2006;57:2291–2302. doi: 10.1093/jxb/erj200. [DOI] [PubMed] [Google Scholar]
- 6.Lu R., Malcuit I., Moffett P., Ruiz M.T., Peart J., Wu A.J., Rathjen J.P., Bendahmane A., Day L., Baulcombe D.C. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 2003;22:5690–5699. doi: 10.1093/emboj/cdg546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bae C., Kim S.M., Lee D.J., Choi D. Multiple classes of immune-related proteases associated with the cell death response in pepper plants. PLoS One. 2013;8:e63533. doi: 10.1371/journal.pone.0063533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senthil-Kumar M., Hema R., Anand A., Kang L., Udayakumar M., Mysore K.S. A systematic study to determine the extent of gene silencing in Nicotiana benthamiana and other Solanaceae species when heterologous gene sequences are used for virus-induced gene silencing. New Phytol. 2007;176:782–791. doi: 10.1111/j.1469-8137.2007.02225.x. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg J.T., Yao N. The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 2004;6:201–211. doi: 10.1111/j.1462-5822.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- 10.Lam E. Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol. 2004;5:305–315. doi: 10.1038/nrm1358. [DOI] [PubMed] [Google Scholar]
- 11.Lacomme C., Santa Cruz S. Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. USA. 1999;96:7956–7961. doi: 10.1073/pnas.96.14.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai-Yamada M., Jin L., Yoshinaga K., Hirata A., Uchimiya H. Mammalian Bax-induced plant cell death can be down-regulated by overexpression of Arabidopsis Bax Inhibitor-1 (AtBI-1) Proc. Natl. Acad. Sci. USA. 2001;98:12295–12300. doi: 10.1073/pnas.211423998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumura H., Nirasawa S., Kiba A., Urasaki N., Saitoh H., Ito M., Kawai-Yamada M., Uchimiya H., Terauchi R. Overexpression of Bax inhibitor suppresses the fungal elicitor-induced cell death in rice (Oryza sativa L.) cells. Plant J. 2003;33:425–434. doi: 10.1046/j.1365-313x.2003.01639.x. [DOI] [PubMed] [Google Scholar]
- 14.Kamoun S., van West P., Vleeshouwers V.G., de Groot K.E., Govers F. Resistance of nicotiana benthamiana to phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell. 1998;10:1413–1426. doi: 10.1105/tpc.10.9.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamoun S., Young M., Glascok C., Tyler B.M. Extracellular protein elicitors from Phytophthora: Host-specificity and induction of resistance to fungal and bacterial phytopathogens. Mol. Plant Microbe Interact. 1993;10:13–20. [Google Scholar]
- 16.Kanzaki H., Saitoh H., Takahashi Y., Berberich T., Ito A., Kamoun S., Terauchi R. NbLRK1, a lectin-like receptor kinase protein of Nicotiana benthamiana, interacts with Phytophthora infestans INF1 elicitin and mediates INF1-induced cell death. Planta. 2008;228:977–987. doi: 10.1007/s00425-008-0797-y. [DOI] [PubMed] [Google Scholar]
- 17.Yoshioka H., Numata N., Nakajima K., Katou S., Kawakita K., Rowland O., Jones J.D.G., Doke N. Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell. 2003;15:706–718. doi: 10.1105/tpc.008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanzaki H., Saitoh H., Ito A., Fujisawa S., Kamoun S., Katou S., Yoshioka H., Terauchi R. Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to pseudomonas cichorii in Nicotiana benthamiana. Mol. Plant Pathol. 2003;4:383–391. doi: 10.1046/j.1364-3703.2003.00186.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee S., Kim S.Y., Chung E., Joung Y.H., Pai H.S., Hur C.G., Choi D. EST and microarray analyses of pathogen-responsive genes in hot pepper (Capsicum annuum L.) non-host resistance against soybean pustule pathogen (Xanthomonas axonopodis pv. glycines) Funct. Integr. Genomics. 2004;4:196–205. doi: 10.1007/s10142-003-0099-1. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y., Schiff M., Dinesh-Kumar S.P. Virus-induced gene silencing in tomato. Plant J. 2002;31:777–786. doi: 10.1046/j.1365-313x.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 21.Chung E., Seong E., Kim Y.C., Chung E.J., Oh S.K., Lee S., Park J.M., Joung Y.H., Choi D. A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. cv Bukang) Mol. Cells. 2004;17:377–380. [PubMed] [Google Scholar]
- 22.Fellbrich G., Romanski A., Varet A., Blume B., Brunner F., Engelhardt S., Felix G., Kemmerling B., Krzymowska M., Nürnberger T. NPP1 a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J. 2002;32:375–390. doi: 10.1046/j.1365-313x.2002.01454.x. [DOI] [PubMed] [Google Scholar]
- 23.Yamakawa H., Mitsuhara I., Ito N., Seo S., Kamada H., Ohashi Y. Transcriptionally and post-transcriptionally regulated response of 13 calmodulin genes to tobacco mosaic virus-induced cell death and wounding in tobacco plant. Eur. J. Biochem. 2001;268:3916–3929. doi: 10.1046/j.1432-1327.2001.02301.x. [DOI] [PubMed] [Google Scholar]
- 24.Takabatake R., Karita E., Seo S., Mitsuhara I., Kuchitsu K., Ohashi Y. Pathogen-induced calmodulin isoforms in basal resistance against bacterial and fungal pathogens in tobacco. Plant Cell Physiol. 2006;48:414–423. doi: 10.1093/pcp/pcm011. [DOI] [PubMed] [Google Scholar]
- 25.Sun J., Jiang H., Xu Y., Li H., Wu X., Xie Q., Li C. The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 2007;48:1148–1158. doi: 10.1093/pcp/pcm088. [DOI] [PubMed] [Google Scholar]
- 26.Haas I.G. BiP (GRP78) an essential hsp70 resident protein in the endoplasmic reticulum. Experientia. 1994;50:1012–1020. doi: 10.1007/BF01923455. [DOI] [PubMed] [Google Scholar]
- 27.Rutkowski D.T., Kaufman R.J. A trip to the ER: Coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Koizumi N., Ujino T., Sano H., Chrispeels M.J. Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol. 1999;121:353–361. doi: 10.1104/pp.121.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonaventure G., Ohlrogge J.B. Differential regulation of mRNA levels of acyl carrier protein isoforms in Arabidopsis. Plant Physiol. 2002;128:223–235. [PMC free article] [PubMed] [Google Scholar]
- 30.Feng D., Witkowski A., Smith S. Down-regulation of mitochondrial acyl carrier protein in mammalian cells compromises protein lipoylation and respiratory complex I and results in cell death. J. Biol. Chem. 2009;284:11436–11445. doi: 10.1074/jbc.M806991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakravarthy S., Tuori R.P., D’Ascenzo M.D., Fobert P.R., Despres C., Martin G.B. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell. 2003;15:3033–3050. doi: 10.1105/tpc.017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zago E., Morsa S., Dat J.F., Alard P., Ferrarini A., Inze D., Delledonne M., van Breusegem F. Nitric oxide- and hydrogen peroxide-responsive gene regulation during cell death induction in tobacco. Plant Physiol. 2006;141:404–411. doi: 10.1104/pp.106.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang I.S., Hwang B.K. The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol. 2011;155:447–463. doi: 10.1104/pp.110.164848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.