Abstract
Central and peripheral nervous systems are lipid rich tissues. Lipids, in the context of lipid-protein complexes, surround neurons and provide electrical insulation for transmission of signals allowing neurons to remain embedded within a conducting environment. Lipids play a key role in vesicle formation and fusion in synapses. They provide means of rapid signaling, cell motility and migration for astrocytes and other cell types that surround and play supporting roles neurons. Unlike many other signaling molecules, lipids are capable of multiple signaling events based on the different fragments generated from a single precursor during each event. Lipidomics, until recently suffered from two major disadvantages: (1) level of expertise required an overwhelming amount of chemical detail to correctly identify a vast number of different lipids which could be close in their chemical reactivity; and (2) high amount of purified compounds needed by analytical techniques to determine their structures. Advances in mass spectrometry have enabled overcoming these two limitations. Mass spectrometry offers a great degree of simplicity in identification and quantification of lipids directly extracted from complex biological mixtures. Mass spectrometers can be regarded to as mass analyzers. There are those that separate and analyze the product ion fragments in space (spatial) and those which separate product ions in time in the same space (temporal). Databases and standardized instrument parameters have further aided the capabilities of the spatial instruments while recent advances in bioinformatics have made the identification and quantification possible using temporal instruments.
Keywords: Mass spectrometry, Lipidomics, Phospholipids, Serial signaling, Neuroscience
Core tip: Mass spectrometry offers a degree of simplicity and sophistication to the biological sciences. In this review we are focusing on its application towards the analysis of lipids in neuroscience. Lipids have a variety of functions, they surround neurons, provide insulation for transmission of signals, an environment for facilitating motility and migration of astrocytes and other cell types, among many other functions. Recent advances in mass spectrometry have enabled quantification of lipids directly extracted from complex biological mixtures in the neuronal system with the help of databases, standardized instrument parameters and bioinformatics. In this review, we intend to highlight all recent efforts with an emphasis on its application to neuroscience.
INTRODUCTION
The central nervous system (CNS) is lipid rich. A significant part of the dry weight of the human brain, close to 50% is contributed to by lipids[1]. Processing of behavior by organisms depends on the brain, and lipids play an active biological role towards those complex functions. Lipids provide much needed electrical insulation to the neurons enabling signal transduction through them and serving as a guiding influence in membrane fission and fusion processes. Structural properties are important for membrane protrusion and fusion. Depending on the head and tail of the individual lipids, especially phospholipids, they may form either a cone or an inverse cone[1]. It is likely that membrane fission and fusion events in neurons involve critical roles of lipids. In the fusion sites between two mating cells of protozoan Tetrahymena thermophila, high-resolution imaging of lipid composition using time-of-flight secondary-ion mass spectrometry has revealed that small membrane regions containing the largest number of fusion pores are highly enriched in the cone-shaped form (2-aminoethylphosphonolipid) suggesting that the localized changes in lipid geometry are likely to play critical roles in the fusion process[2]. Many lipid intermediates are potent intracellular signal transduction molecules themselves. Lipids also are central to vesicle formation, fusion and fission, all processes that are central to synaptic transmission of nerve impulses[1].
Lipids in cellular systems offer different features such as permutability of constituent fragments to generate diverse species, the capability of generating multiple types of signaling, some which can be rapid and on demand; others being localized due to their membrane bound nature. The maintenance of lipid heterogeneity across the plasma and organelle membranes facilitate lipid mediated localized signaling[1,3].
Rearrangement of structural units of individual lipids within the mammalian lipidome provides the potential for generating a vast diversity of lipid species. The capability of generating a vast number of entities provides the potential for many different types of signaling to originate from lipids. Lipid rearrangements can occur confined to a space within tissue or cells, or within the same space but at different times during the life cycle of a given cell or tissue. Lipids are capable of serial signaling, that is, application of a single biochemical route for multiple signaling events[1,3]. An example is the conversion of adenosine triphosphate into cyclic adenosine monophosphate (cAMP), which is catalyzed by adenylyl cyclase. Adenylyl cyclase is activated by binding of several neurotransmitters with neuronal surface receptors. However, the hydrolysis of cAMP stops this signaling process. Such a signaling process is a “one pathway-one signal” model of transmembrane signaling[4,5].
On the other hand, an example of serial signaling in neurons by the lipids is the 1,2-Diacyl-sn-glycerols (1,2 DAG) cascade. In this cascade, phospholipases cleave membrane bound phosphatidylinositol (PI) 4,5 bisphosphate to generate 1,2-DAG and Inositol (Ins) 1,4,5 trisphosphate. While Ins 1,4,5 trisphosphate is hydrolyzed and deactivated after signaling just as it happens to cAMP, 1,2-DAG, on the other hand often serves as the starting material for a series of competing conversions generating various intermediates. 1,2-DAG is phosphorylated into Phosphatidic acid (PA) by DAG-kinases[6]. The PA can be converted into (1) Lysophosphatidic acid (LPA) by losing a fatty-acid residue catalyzed by phospholipase A2; (2) into cytidyldiphosphate-DAG; or (3) can be dephosphorylated into 1,2-DAG by a phosphatase. 1,2-DAG can be hydrolyzed by DAG-lipases to generate Monoacylglycerol (MAG)[7,8], which itself is subsequently hydrolyzed into fatty acid and glycerol by MAG lipase[9]. 1,2-DAG could also be hydrolyzed to generate 2-arachidonoylglycerol (2-AG) and arachidonic acid, which in turn acts as substrates for lipid oxygenases[10] converting them into oxidized metabolites which are termed as eicosanoids[11]. The metabolic intermediates of 1,2-DAG, for example PA, activates raf protein kinase and PI(4)monophosphate-5-kinase[12,13]; LPA and 2-AG are high-affinity agonists for LPA and cannabinoid receptors respectively. 1,2-DAG also activates various members of the protein kinase C family[14].
It is pertinent to mention that lipids are players in neuronal retrograde signaling as well. Retrograde signaling mechanisms altering the strength of incoming synaptic inputs have been shown to be modulated by endocannabinoid lipids 2-AG or its derivatives in pyramidal neurons during their depolarization. Similar signaling is initiated by the activation of postsynaptic glutamate metabotropic receptors and acetylcholine muscarinic receptors[10,15,16].
Another important aspect of lipid based signaling is their rapid nature. Lipid signaling that can be achieved in a relatively short period of time because the signaling molecules can be generated catalytically from the existing precursor at a very accelerated pace, that which cannot be performed for larger macromolecules such as proteins. Lipids also often enable localized signaling due to their heterogeneous adherence to a specific subcellular site. Several lipids that are intermediates of the 1,2-DAG pathway serve critical functions in neuronal development, synaptic plasticity and behavior[15,17]. For example, arachidonic acid modulates the activities of neuronal ion channels[18,19]; and the eicosanoids modulate G protein-coupled signaling that underlie a myriads of neuronal functions[18,20].
The two properties, (1) dynamic changes in membrane lipids that underlie several cellular property changes; and (2) ability to generate several signaling molecules from a single precursor, simultaneously leading to multiple signaling events warrant the studying of the lipidome rather than the biochemical analyses of single lipids or only a handful of lipids. Capturing lipidome changes is therefore critical to understanding the behavior of neurons and to gather a greater insight into the functioning of the CNS. Until recently the analyses of lipids posed two challenges, (1) the inability to identify lipids without a vast arsenal of chemical reagents and a deep knowledge of chemistry; and (2) the necessity of a large amount of purified lipids for structure determination. However, the advent of mass spectrometry has largely removed these two barriers. We present here an overview of mass spectrometric techniques enabling the identification and relative quantitation of cellular lipidome (categorized into class specific and pan lipidome analyses) and briefly discuss their utility for neuroscience related research.
CLASS SPECIFIC ANALYSES OF LIPIDS ON MASS ANALYZER WHERE FRAGMENTS ARE RESOLVED IN SPACE
Electrospray ionization mass spectrometry enables the identification and quantification of the cellular or tissue lipidomes directly from crude extracts of cell, tissue or organ samples[21-23]. The mass spectrometers that resolve fragments of entrant precursor ions of biomolecules in space domains enable identification and quantification in a class specific manner, referred to as automated shotgun lipid profiling[24]. Mass spectrometers where fragmentation and analysis occur in separated chambers (different chamber than that of the entrant precursor ions) enable precise control of collision energy and hence enable class specific scans for lipids with one class scan at a time. Ion mode and collision energy are the two key parameters which remain somewhat standardized for class specific identification of lipids among different instrumenPt platforms. The knowledge of class specific parameters has been derived from control experiments with purified lipids from many lipid chemistry groups over several decades of research in synthetic organic chemistry. The most common of space resolving instruments are the triple quadrupole mass spectrometers, Figure 1 shows a typical spectra. The instrument parameters vary on instruments manufactured by different vendors (referred to as platforms). The settings of other parameters for class specific identification need fine tuning from one platform to another and enable quantification of lipids on moderate and higher resolution triple quadruple mass spectrometers with only little optimization. The moderate resolution space domain resolving instruments (approximately 1amu resolution) enable lipid profiling from extracts of crude biological samples. The specific lipid entities of interest may then be confirmed for identities using a combination of high resolution instruments and chemical derivatization for identification of correct isomers.
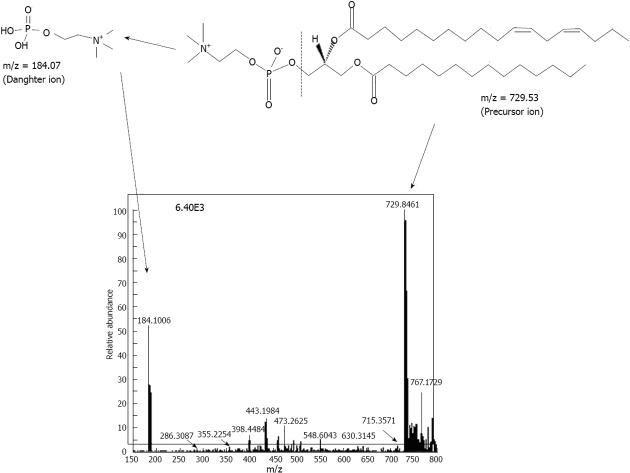
Figure 1.
Electrospray ionization mass spectrometer of 1-tetradecanoyl-2-(11Z,14Z-octadecadienoyl)-sn-glycero-3-phosphocholine (m/z 729 for singly charged species). The scan was performed in the third quadrupole (Q3) of a Triple stage quadrupole Quantum Access Max instrument with collision energy of 35 V in Q2. The precursor (or parent) ion and daughter ion have been shown in the spectrum. The corresponding molecular structures of daughter ion (184.07) and precursor ion (729) have been shown above the spectrum and corresponding m/z in the spectrum has been identified by dashed arrows. The total ion intensity was 6.4E3 as indicated. The arrow head and thick dashed arrows in the precursor ion structure indicate the fragmentation point leading to the generation of the daughter ion.
A comparison of different mass spectrometers used for lipidomics has been provided in Table 1. With the advent of knowledge of lipid chemistry, further advancement has been made to identify lipid side chains using mass spectrometry and also to incorporate a second quantification step. The parameters for these determinations have been presented in Tables 2 and 3. Three chief parameters contribute towards class specific identification of the lipids: (1) ion mode of operation (positive or negative); (2) collision energy (CE usually in volts); and (3) scan type, that is, precursor-ion (often also referred to as parent-ion) or neutral loss scan for the daughter ions that is generated due to the collision induced dissociation of the parent ion. The class specific identification and quantification is based on the scan type due to generation of daughter ion of specific mass under a given collision energy[24]. All mass spectrometers usually are equipped with software for data acquisition. The optimal parameters for a specific instrument platform are not universal but need to be tuned for another platform. A lipid species may belong to more than one class due to presence of common parts encompassing more than a class, thus enabling their analysis using more than one class setting. The identification of lipids in these instruments necessitates comparison of experimental spectra to a reference spectrum or comparison of m/z ratios to databases. These comparisons are achieved through the use of analytical softwares.
Table 1.
Comparison of leading mass spectrometers for lipidomics
| Resolution | Mass range(m/z) | Mass accuracy(ppm) | |
| ToF | 2-5 typical | ||
| Waters LCT premier | 10000 | 18000 | NA |
| Agilent LC/ToF | > 22000 | 50-20000 | < 1 |
| Bruker microToF | 10000 | 3000 | NA |
| QToF | Same as ToF | ||
| Agilent 6540 | > 40000 | 50-10000 | < 1 |
| Waters QToF ultima | 17500 | 32000 | 1 |
| Waters QToF micro | 5000 | 20000 | 1 |
| Bruker max is 4G | > 60000 | 10000 | < 1 |
| LTQ velos | 3000 | 4000 | 0.1 Da |
| LTQ-orbitrap velos | > 100000 | 4000 | < 2 |
| TSQ quantum access max | 0.4 (FWHM) | 3000 | 0.1 Da |
| TSQ quantiva | 0.2 (FWHM) | 1850 | 0.1 Da |
| Q-exactive | 140000 | 6000 | < 1 |
| Orbitrap fusion tribrid | 450000 | 4000 | < 1 |
| Synapt G2-HDMS | 40000 | 32000 (1000001) | < 1 |
| Synapt G2S-HDMS | 20000 | 32000 (1000001) | Approx- imately 2 |
When fitted with a quadrupole. m/z: Mass to charge ratio, ppm: Parts per million; LCT: Liquid chromatograph; LTQ: Linear trap quadropole; TSQ: Triple stage quadrupole; HDMS: High definition mass spectrometry; FWHM: Full width at half maximum; ToF: Time of flight; NA: Not available.
Table 2.
Select performance parameters of some commercially available mass spectrometers
| Parameter | TSQ quantum access max | TSQ quantiva | LTQ-orbitrap velos | Qq-ToF | Synapt G2-S HDMS | Q-exactive | Agilent 6540 | Orbitrap fusion tribrid |
| Acquisition rate | 5000 amu/s | 15000 amu/s | 1/s@60000 RP | 3/s@30000 RP | 30/s MS or MSe mode | 13/s@17500 | 20 MS/s | 15/sec@15000 |
| 3/s@15000 RP | 2000/s IMS mode | 1.5/s@140000 | 10 MS2/s | |||||
| Linear dynamic range | 5 orders | 6 orders | 5-6 orders | 2-3 orders | > 5 orders In-spectrum1 | > 5 orders | 5 orders | > 5 orders |
| Mass accuracy | 0.1 Da | 0.1 Da | < 3 ppm MS | < 5 ppm Full scan w/Lock Mass | < 1 ppm MS MSe or MS/MS | < 1 ppm | < 1 ppm MS | < 1 ppm |
| < 3 ppm MSn | > 100 ppm MS/MS | < 2 ppm MS2 | < 1 ppm MSn | |||||
| (no lock mass) | ||||||||
| Resolution | 0.4 Da FWHM | 0.2 Da FWHM | > 100000 RP | Up to 30000 RP | Up to 40000RP | 140000 | > 15000 RP@ 1522 | 450000 |
| Sensitivity | Sub-femto-mol | Atto-mol | Sub atto-mol | Sub femto-mol | Approximately 10 charges at the detector | Atto-mol | Femto-mol | Sub atto-mol |
At full resolution and maximum acquisition rate. ToF: Time of flight; LTQ: Linear trap quadropole; TSQ: Triple stage quadrupole; HDMS: High definition mass spectrometry; MS: Mass spectrometry; IMS: Ion mobility spectometry.
Table 3.
Class specific scanning parameters of different lipid classes using spatial resolution triple quadrupole mass spectrometer1
| Lipid class |
Class-specific pre screen |
Second-step quantification |
|||||
| Ion mode | Daughter ion mass (m/z) | Collision energy (V) | Scan type | Daughter ion mass (m/z) | Collision energy (V) | Scan type | |
| PC | + | 184 | 35 | PIS | 189, 183.1, 59 | 35, 35, 24 | NLS |
| lysoPC | + | 59 | 22 | NLS | 205, 59 | 34, 22 | NLS |
| PE, lysoPE | - | 196 | 50 | PIS | 222.2 | 30 | NLS |
| PI, LysoPI | - | 241.1 | 45 | PIS | 241.1 | 45 | PIS |
| PS, lysoPS | - | 87.1 | 24 | NLS | 87.1 | 24 | NLS |
| PG, lysoPG | - | 153.1 | 35 | PIS | 153.1 | 35 | PIS |
| PA, lysoPA | - | 153.1 | 35 | PIS | 153.1 | 35 | PIS |
| CL, monolysoCL | - | Full MS at high resolution | |||||
| Triglycerides | + | ||||||
| Spingomyelin | + | 213.2 | 50 | NLS | 213.2 | 50 | NLS |
| Ceramide | - | 256.2 | 32 | NLS | NLS | ||
| Sphingoid base | + | 48 | 18 | NLS | 48 | 18 | NLS |
| Psychosine | + | 180 | 24 | NLS | 180 | 24 | NLS |
| Acyl -carnitine | + | 85.1 | 30 | PIS | 85.1 | 30 | PIS |
| Acyl-CoA | - | 134 | 30 | PIS | 134 | 30 | PIS |
1The parameters have been adopted from a review of vast literature, which has been reviewed in Yang et al[24]. Some lipid extraction for analyses in positive ion mode performed in presence of Sodium or Lithium ion will change the mass to charge ratio (m/z) of ions. Similarly, extraction performed for analyses in negative ion mode in presence of ammonium acetate may change the m/z of ions. MS: Mass spectrometry; PIS: Precursor ion scan; NLS: Neutral loss scan; PC: Phosphocholine; lysoPC: Lysophosphocholine; PE: Phosphoethanolamine; PI: Phosphoinositol; PS: Phosphoserine; PG: Glycerophosphoglycerols; PA: Phosphatidic acid; CL: Cardiolipin.
In mass spectrometric lipidomics, the quantification of lipids is ratiometric and performed with a synthetic or purified standard, often termed as internal standard usually in a two-step process[23-25]. The most abundant lipid species are quantified using direct comparison of the peak intensities to those of the added internal standard for the lipid class in the mass spectrum from the first step. This is done after correcting for the isotopic differences of 13C, which are often present in low abundance in the internal standard molecules[25]. The basic premise of this quantification is that the lipid species ions linearly correlate with its concentration in the low-concentration region of the mass spectrometer (MS) spectrum[25,26]. In the second step, the ratiometrically determined concentrations of the abundant lipids using internal standard are used for additional refinement of quantification. Different lipids of the same class may undergo different fragmentation kinetics[26], thus multiple standards belonging to a single class and representing different physical properties (side chain and degrees of unsaturation) enable better quantification[27-29]. Ratiometric quantification encompasses the entire linear dynamic range of ion peak intensities offered by the spatial resolving instruments and a two-step approach enables quantification of low abundance region of lipid species from organic extracts. However, one of the limitations of class specific identification using spatial instruments is the ready identification of entities that are present in the databases. The structures of lipid entities that are absent in databases can also be deduced particularly with different collision energies or utilizing high resolution mass spectrometers with chemical derivatization.
ROLE OF DATABASES
Databases play an important role in the identification of lipids in the class based lipid identification approach. Databases are usually built on either single instruments or very closely related instrument platforms, with most parameters being normalized, except for variation in parameters that enable determination of the given lipid class (Tables 2 and 3). The parameters that could be normalized are sheath gas flow rate and spray voltage, while the collision energy and mode (negative or positive ion) are parameters which have to remain largely unaltered for class based lipid identification. The limitation of identification in this approach thus depends on the presence of a given entry in the database. To a certain extent, platform dependency is not a limiting factor. Most often, normalization of analytical parameters across instrument platforms is achievable and verifiable with known standards that enable validation of class specific identification on different instrument platforms, thus enabling class specific lipid identification.
BIOINFORMATICS APPROACHES AND SOFTWARE
There exists a few databases for lipids such as the yeast metabolomics and human metabolomics databases (www.ymdb.ca and www.hmdb.ca respectively). The most complete and easily downloadable database is lipidmaps database (www.lipidmaps.org), which enables downloading specific lipid class entries in *.csv and a few other formats that can then be locally used for searching using different softwares. A few different softwares are available. Multidimensional mass spectrometry-based shotgun lipidomics (MDMS-SL) and MZmine (recent version 2.10.0) are freely downloadable while Simlipid 3.0 is a commercially available software. Ratiometic quantification in these softwares is achieved as a two-step process. In the first step a specific class based known standard that does not naturally exist in mammalian systems is first used for quantification of the most abundant ion species within the lipid class in the previously identified entities. In the second step the standard and abundant lipids are used for quantification of low abundance species in the experimentally obtained ion spectrum.
TOTAL LIPID ANALYSES AND DE NOVO LIPID IDENTIFICATION AND QUANTIFICATION USING MASS ANALYZERS THAT RESOLVE FRAGMENT IONS IN TIME ON THE SAME SPACE
Many instruments harboring ion-trap type of mass analyzers enable generation of fragment ions from precursors in the same space but over different time spans and are termed time resolving instruments. Time resolving mass spectrometers enable the capture of ions with minimal loss as ions are not lost in a vast space that they would have to travel otherwise. The ability to acquire and align multiple related high resolution spectra enables de novo analyses of new species that may not be present in the database. The acquisition and alignment of related MS/MS spectra reduces the false positive assignments and greatly improves the ion statistics. Time domain resolving instruments capture all precursors and their fragments in parallel and in a single scan. However, they utilize a single collision energy that fragments different lipids with different efficiencies. The analyses of the data involves relating the fragments to their precursors, a task that poses a great challenge. The analyses of such data is handled by bioinformatics. Another approach is specific chemical derivatization that selectively reacts to specific or class specific lipids eliminating them from the total spectra upon chemical derivatization. Thus, mass spectra with and without chemical derivatization enables distinguishing specific lipids from scans performed in time domain resolving instruments[30-32].
The time domain instruments produce a comprehensive dataset of MS precursor ions and the MS/MS spectra comprising all fragment ions derived from all lipid precursor ions[33]. In these instruments a survey or MS spectrum is acquired to determine m/z and abundances of precursor ions, which follows acquisition of MS/MS fragment spectra from automatically selected precursors. The acquisition of MS and MS/MS spectra is repeated. Each acquisition comprises a large number of MS and MS/MS spectra from selected precursors. The MS and MS/MS spectra share common attributes: (1) mass accuracy (ppm, Da or amu); (2) mass resolution (FWHM); and (3) occupancy of peaks. Mass accuracy and mass resolution are properties of individual mass spectrometers and applies equally to all peaks. The peak occupancy is dependent on: (1) instrument performance; and (2) intrinsic characteristics of the sample. Repetitive acquisitions do not often fully compensate for low abundant precursors, which are often affected by poor signal-to-noise ratio. The low abundant precursors are often not fragmented in all acquisitions and often occur with non-equal efficiency. The peak occupancy attribute is the frequency with which a particular peak is encountered in individual acquisitions within the full series of acquisitions[34]. Normalizing for peak occupancy is often used for enhancing coverage and reproducibility of peak detection.
BIOINFORMATIC APPROACHES AND INSTRUMENT INDEPENDENT IDENTIFICATION OF LIPIDS
As stated above, many freeware (for example, MZmine 2.10) as well as commercial software (Simlipid 3.0) exist for analyses of mass spectrometric lipid identification. A number of programs have been developed for analyses of MS and MS/MS datasets from high resolution and time domain mass spectrometers. LipidXplorer is a recently developed program that has been built from the experience of several previous programs[34]. The LipidXplorer software is based on three axioms: (1) that the software should utilize spectra acquired on any tandem mass spectrometer; (2) should identify and quantify species from any lipid class; and (3) should handle large datasets composed of highly redundant MS and MS/MS spectra, with several technical and biological replicates acquired from each analyzed sample. LipidXplorer therefore enables de novo or database-independent identification of lipids. Spectra interpretation rules are flexible and not encoded into the software engine. In LipidXplorer users can define new rules or modify the existing rules and apply any number of interpretation rules in parallel. LipidXplorer employs a two-step process for lipid identification employing a two-step analyses in a fully database-independent manner[34,35]. In step 1, a full pool of acquired MS and MS/MS spectra is organized into a single flat-file database termed as MasterScan. In step 2, the MasterScan is interpreted using molecular fragmentation query language (MFQL). The findings of MFQL are exported in a user-defined format. These two steps eliminate the reliance of comparison of experimental and reference spectra for lipid identification.
CHEMICAL MODIFICATION APPROACHES FOR COMPLEMENTARY CONFIRMATION
Chemical derivatization using reagents that are relatively specific in reacting with one group versus others enables selective elimination of peaks in a given spectra and thus allow identification of those species. Derivatization is used for conversion of nonpolar or less polar lipids into polar lipids in electrospray ionization mass spectrometry[36,37]. The conversion into polar lipids necessitates imparting an inherent charge. Addition of sulfate group to cholesterol[36] or oxosteroids into their oximes[38] are examples of rendering these entities polar. Derivatization enhances detection of specific lipids or members of lipid classes on time domain resolved instruments. Derivatization also often assists localization of lipids on cell and organelle membranes or even identification of lipid bound proteins in the membranes[31,32,39].
UTILITY IN NEUROSCIENCE
Mass spectrometric lipidomic analyses is poised to identify specific lipid players at different locations of neuronal cells of various types that serve diverse specialized functions. One of the key feature of neurons is (1) insulation, a process so central to neuronal function in neurotransmission. The insulation in neurons is achieved by the association of lipids with proteins[40,41]. For example, Myelin basic protein’s organization and proper functioning are optimized at a particular combination of both the amounts of and ratio between the charged lipids and Myelin basic protein[42]; (2) another important biological aspect of neurons is transport across rather long region of the body of cells termed soma. Neuronal cells employ a wide variety of molecular motors[43] and the hindrance to axonal transport is part of the pathology in several neurological diseases. In this context, the role of specific lipids remains very important to analyze. Mass spectrometric detection and quantification of lipids associated with various transport components will provide a rare insight into the biological chemistry of neuronal transport; and (3) is signaling. In neuronal systems signaling occurs broadly in three different ways: (1) within the cells of similar types, the connected neurons are an example of this; (2) with the cells of dissimilar types, for example amacrine cells signaling to neurons[44,45]; and (3) with environmental factors serving as environmental cues (for example, trophic factors[46]) to neurons. These signaling events are important for neuronal patterning in development, their maintenance in adulthood, the health and day to day function of the organism responding to the opportunities and threats in the environment and in health and disease, when their aberrations results in progressive neurological deficits. For example, a form of retrograde signaling is initiated by a voltage-dependent influx of Ca2+ into hippocampal neurons that leads to the inhibition of glutamate-mediated or γ-aminobutyric acid (GABA)-mediated inputs[16,47,48]. This retrograde signaling mechanism appears to be widespread in the CNS, and is mediated by a diffusible endocannabinoid lipid[49,50]. The endocannabinoid 2-AG has been directly implicated in mGluR-induced retrograde signaling in the hippocampus, cerebellum and other regions of the brain[51,52]. 2-AG is likely also produced in dendritic spines via a phospholipase C/DAG lipase (DGL) pathway. The fidelity of the 2-AG mediated signaling sequence depends on the precise localization of DGL-α. The latter is the major biosynthetic enzyme for 2-AG in neurons[7,53]. Some lipid messengers use stable signaling junctions. In the brain, most lipid signals such as 2-AG travels short distances from their sites of production and engage G-protein-coupled receptors on neighboring neurons and/or glial cells. The lysophosphatidic acid[54,55], platelet-activating factor[56,57] and anandamide[15,58], a neurotrophic signal that play a role in neuropathic pain, a retrograde messenger in hippocampal long-term potentiation, and an endocannabinoid ligand have already been identified as lipid messengers for signaling. Neurosteroids, oleoylethanolamide and its derivates are some examples of lipid messengers that do not require G-protein-coupled receptors to exert their function. Neurosteroids interact with membrane GABA-gated receptor channels to enhance neuronal inhibition[59]. The oleoylethanolamide and its analogue palmitoylethanolamide regulate peroxisome proliferator-activated receptors-α in the cell cytosol and nucleus that contribute to feeding[60] and pain sensations[61].
CONCLUSION
Lipids are involved in almost all steps of critical function of nervous systems. Several lipid changes occur simultaneously that regulate the membrane structure and function which underlie the function of neurons at the cellular level. Lipids also are involved in serial signaling resulting in multiple signaling with single precursor molecules. It is important therefore to understand changes in several lipids simultaneously. Advances in mass spectrometry have made such investigations simpler by allowing to profile and quantitate lipids simultaneously from complex biological mixtures of cells, tissues or organ extracts. Both, class specific identification of known lipids based on database as well as de novo identification of new lipids are possible by judiciously utilization of different types of mass spectrometers.
Footnotes
P- Reviewers: Bian ZX, Masocha W, Osaka H S- Editor: Gou SX L- Editor: A E- Editor: Wu HL
References
- 1.Piomelli D, Astarita G, Rapaka R. A neuroscientist’s guide to lipidomics. Nat Rev Neurosci. 2007;8:743–754. doi: 10.1038/nrn2233. [DOI] [PubMed] [Google Scholar]
- 2.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Mass spectrometric imaging of highly curved membranes during Tetrahymena mating. Science. 2004;305:71–73. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piomelli D. The challenge of brain lipidomics. Prostaglandins Other Lipid Mediat. 2005;77:23–34. doi: 10.1016/j.prostaglandins.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Greengard P. Possible role for cyclic nucleotides and phosphorylated membrane proteins in postsynaptic actions of neurotransmitters. Nature. 1976;260:101–108. doi: 10.1038/260101a0. [DOI] [PubMed] [Google Scholar]
- 5.Francis SH, Corbin JD, Bischoff E. Cyclic GMP-hydrolyzing phosphodiesterases. Handb Exp Pharmacol. 2009;(191):367–408. doi: 10.1007/978-3-540-68964-5_16. [DOI] [PubMed] [Google Scholar]
- 6.Mérida I, Avila-Flores A, Merino E. Diacylglycerol kinases: at the hub of cell signalling. Biochem J. 2008;409:1–18. doi: 10.1042/BJ20071040. [DOI] [PubMed] [Google Scholar]
- 7.Bisogno T, Howell F, Williams G, Minassi A, Cascio MG, Ligresti A, Matias I, Schiano-Moriello A, Paul P, Williams EJ, et al. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J Cell Biol. 2003;163:463–468. doi: 10.1083/jcb.200305129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinh TP, Carpenter D, Leslie FM, Freund TF, Katona I, Sensi SL, Kathuria S, Piomelli D. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc Natl Acad Sci USA. 2002;99:10819–10824. doi: 10.1073/pnas.152334899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsson M, Contreras JA, Hellman U, Tornqvist H, Holm C. cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J Biol Chem. 1997;272:27218–27223. doi: 10.1074/jbc.272.43.27218. [DOI] [PubMed] [Google Scholar]
- 10.Katona I. Endocannabinoid receptors: CNS localization of the CB1 cannabinoid receptor. Curr Top Behav Neurosci. 2009;1:65–86. doi: 10.1007/978-3-540-88955-7_3. [DOI] [PubMed] [Google Scholar]
- 11.Guindon J, Hohmann AG. A physiological role for endocannabinoid-derived products of cyclooxygenase-2-mediated oxidative metabolism. Br J Pharmacol. 2008;153:1341–1343. doi: 10.1038/bjp.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDermott M, Wakelam MJ, Morris AJ. Phospholipase D. Biochem Cell Biol. 2004;82:225–253. doi: 10.1139/o03-079. [DOI] [PubMed] [Google Scholar]
- 13.Sigal YJ, McDermott MI, Morris AJ. Integral membrane lipid phosphatases/phosphotransferases: common structure and diverse functions. Biochem J. 2005;387:281–293. doi: 10.1042/BJ20041771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irvine RF. 20 years of Ins(1,4,5)P3, and 40 years before. Nat Rev Mol Cell Biol. 2003;4:586–590. doi: 10.1038/nrm1152. [DOI] [PubMed] [Google Scholar]
- 15.Piomelli D. The molecular logic of endocannabinoid signalling. Nat Rev Neurosci. 2003;4:873–884. doi: 10.1038/nrn1247. [DOI] [PubMed] [Google Scholar]
- 16.Alger BE. Retrograde signaling in the regulation of synaptic transmission: focus on endocannabinoids. Prog Neurobiol. 2002;68:247–286. doi: 10.1016/s0301-0082(02)00080-1. [DOI] [PubMed] [Google Scholar]
- 17.Ye X, Fukushima N, Kingsbury MA, Chun J. Lysophosphatidic acid in neural signaling. Neuroreport. 2002;13:2169–2175. doi: 10.1097/00001756-200212030-00002. [DOI] [PubMed] [Google Scholar]
- 18.Rao JS, Kellom M, Kim HW, Rapoport SI, Reese EA. Neuroinflammation and synaptic loss. Neurochem Res. 2012;37:903–910. doi: 10.1007/s11064-012-0708-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilius B, Honoré E. Sensing pressure with ion channels. Trends Neurosci. 2012;35:477–486. doi: 10.1016/j.tins.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Piomelli D, Volterra A, Dale N, Siegelbaum SA, Kandel ER, Schwartz JH, Belardetti F. Lipoxygenase metabolites of arachidonic acid as second messengers for presynaptic inhibition of Aplysia sensory cells. Nature. 1987;328:38–43. doi: 10.1038/328038a0. [DOI] [PubMed] [Google Scholar]
- 21.Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 22.Han X, Yang J, Cheng H, Ye H, Gross RW. Toward fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal Biochem. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharya SK. Recent advances in shotgun lipidomics and their implication for vision research and ophthalmology. Curr Eye Res. 2013;38:417–427. doi: 10.3109/02713683.2012.760742. [DOI] [PubMed] [Google Scholar]
- 24.Yang K, Cheng H, Gross RW, Han X. Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal Chem. 2009;81:4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han X, Gross RW. Shotgun lipidomics: multidimensional MS analysis of cellular lipidomes. Expert Rev Proteomics. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 26.Lehmann WD, Koester M, Erben G, Keppler D. Characterization and quantification of rat bile phosphatidylcholine by electrospray-tandem mass spectrometry. Anal Biochem. 1997;246:102–110. doi: 10.1006/abio.1996.9941. [DOI] [PubMed] [Google Scholar]
- 27.Brügger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci USA. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ekroos K, Chernushevich IV, Simons K, Shevchenko A. Quantitative profiling of phospholipids by multiple precursor ion scanning on a hybrid quadrupole time-of-flight mass spectrometer. Anal Chem. 2002;74:941–949. doi: 10.1021/ac015655c. [DOI] [PubMed] [Google Scholar]
- 29.Liebisch G, Lieser B, Rathenberg J, Drobnik W, Schmitz G. High-throughput quantification of phosphatidylcholine and sphingomyelin by electrospray ionization tandem mass spectrometry coupled with isotope correction algorithm. Biochim Biophys Acta. 2004;1686:108–117. doi: 10.1016/j.bbalip.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Gołębiowski M, Boguś MI, Paszkiewicz M, Stepnowski P. Cuticular lipids of insects as potential biofungicides: methods of lipid composition analysis. Anal Bioanal Chem. 2011;399:3177–3191. doi: 10.1007/s00216-010-4439-4. [DOI] [PubMed] [Google Scholar]
- 31.Ibrahim H, Caudron E, Kasselouri A, Prognon P. Interest of fluorescence derivatization and fluorescence probe assisted post-column detection of phospholipids: a short review. Molecules. 2010;15:352–373. doi: 10.3390/molecules15010352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arneson KO, Roberts LJ. Measurement of products of docosahexaenoic acid peroxidation, neuroprostanes, and neurofurans. Methods Enzymol. 2007;433:127–143. doi: 10.1016/S0076-6879(07)33007-3. [DOI] [PubMed] [Google Scholar]
- 33.Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal Chem. 2006;78:585–595. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- 34.Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;12:R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herzog R, Schuhmann K, Schwudke D, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A. LipidXplorer: a software for consensual cross-platform lipidomics. PLoS One. 2012;7:e29851. doi: 10.1371/journal.pone.0029851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandhoff R, Brügger B, Jeckel D, Lehmann WD, Wieland FT. Determination of cholesterol at the low picomole level by nano-electrospray ionization tandem mass spectrometry. J Lipid Res. 1999;40:126–132. [PubMed] [Google Scholar]
- 37.Griffiths WJ, Liu S, Alvelius G, Sjövall J. Derivatisation for the characterisation of neutral oxosteroids by electrospray and matrix-assisted laser desorption/ionisation tandem mass spectrometry: the Girard P derivative. Rapid Commun Mass Spectrom. 2003;17:924–935. doi: 10.1002/rcm.1002. [DOI] [PubMed] [Google Scholar]
- 38.Liu S, Sjövall J, Griffiths WJ. Analysis of oxosteroids by nano-electrospray mass spectrometry of their oximes. Rapid Commun Mass Spectrom. 2000;14:390–400. doi: 10.1002/(SICI)1097-0231(20000331)14:6<390::AID-RCM882>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 39.Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Persaud R, Boggs JM, Wood DD, Moscarello MA. Interaction of glycosylated human myelin basic protein with lipid bilayers. Biochemistry. 1989;28:4209–4216. doi: 10.1021/bi00436a013. [DOI] [PubMed] [Google Scholar]
- 41.Chattopadhyay A, Paila YD. Lipid-protein interactions, regulation and dysfunction of brain cholesterol. Biochem Biophys Res Commun. 2007;354:627–633. doi: 10.1016/j.bbrc.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 42.Min Y, Kristiansen K, Boggs JM, Husted C, Zasadzinski JA, Israelachvili J. Interaction forces and adhesion of supported myelin lipid bilayers modulated by myelin basic protein. Proc Natl Acad Sci USA. 2009;106:3154–3159. doi: 10.1073/pnas.0813110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cantuti Castelvetri L, Givogri MI, Hebert A, Smith B, Song Y, Kaminska A, Lopez-Rosas A, Morfini G, Pigino G, Sands M, et al. The sphingolipid psychosine inhibits fast axonal transport in Krabbe disease by activation of GSK3β and deregulation of molecular motors. J Neurosci. 2013;33:10048–10056. doi: 10.1523/JNEUROSCI.0217-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Rosello T, Anderson CT, Schopfer FJ, Zhao Y, Gilad D, Salvatore SR, Freeman BA, Hershfinkel M, Aizenman E, Tzounopoulos T. Synaptic Zn2+ inhibits neurotransmitter release by promoting endocannabinoid synthesis. J Neurosci. 2013;33:9259–9272. doi: 10.1523/JNEUROSCI.0237-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agudo-Barriuso M, Lahoz A, Nadal-Nicolás FM, Sobrado-Calvo P, Piquer-Gil M, Díaz-Llopis M, Vidal-Sanz M, Mullor JL. Metabolomic changes in the rat retina after optic nerve crush. Invest Ophthalmol Vis Sci. 2013;54:4249–4259. doi: 10.1167/iovs.12-11451. [DOI] [PubMed] [Google Scholar]
- 46.Schael S, Nüchel J, Müller S, Petermann P, Kormann J, Pérez-Otaño I, Martínez SM, Paulsson M, Plomann M. Casein kinase 2 phosphorylation of protein kinase C and casein kinase 2 substrate in neurons (PACSIN) 1 protein regulates neuronal spine formation. J Biol Chem. 2013;288:9303–9312. doi: 10.1074/jbc.M113.461293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chevaleyre V, Takahashi KA, Castillo PE. Endocannabinoid-mediated synaptic plasticity in the CNS. Annu Rev Neurosci. 2006;29:37–76. doi: 10.1146/annurev.neuro.29.051605.112834. [DOI] [PubMed] [Google Scholar]
- 48.Mackie K. Signaling via CNS cannabinoid receptors. Mol Cell Endocrinol. 2008;286:S60–S65. doi: 10.1016/j.mce.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson RI, Nicoll RA. Endogenous cannabinoids mediate retrograde signalling at hippocampal synapses. Nature. 2001;410:588–592. doi: 10.1038/35069076. [DOI] [PubMed] [Google Scholar]
- 50.Ohno-Shosaku T, Maejima T, Kano M. Endogenous cannabinoids mediate retrograde signals from depolarized postsynaptic neurons to presynaptic terminals. Neuron. 2001;29:729–738. doi: 10.1016/s0896-6273(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 51.Makara JK, Mor M, Fegley D, Szabó SI, Kathuria S, Astarita G, Duranti A, Tontini A, Tarzia G, Rivara S, et al. Selective inhibition of 2-AG hydrolysis enhances endocannabinoid signaling in hippocampus. Nat Neurosci. 2005;8:1139–1141. doi: 10.1038/nn1521. [DOI] [PubMed] [Google Scholar]
- 52.Uchigashima M, Narushima M, Fukaya M, Katona I, Kano M, Watanabe M. Subcellular arrangement of molecules for 2-arachidonoyl-glycerol-mediated retrograde signaling and its physiological contribution to synaptic modulation in the striatum. J Neurosci. 2007;27:3663–3676. doi: 10.1523/JNEUROSCI.0448-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung KM, Astarita G, Zhu C, Wallace M, Mackie K, Piomelli D. A key role for diacylglycerol lipase-alpha in metabotropic glutamate receptor-dependent endocannabinoid mobilization. Mol Pharmacol. 2007;72:612–621. doi: 10.1124/mol.107.037796. [DOI] [PubMed] [Google Scholar]
- 54.Herr DR, Chun J. Effects of LPA and S1P on the nervous system and implications for their involvement in disease. Curr Drug Targets. 2007;8:155–167. doi: 10.2174/138945007779315669. [DOI] [PubMed] [Google Scholar]
- 55.Inoue M, Rashid MH, Fujita R, Contos JJ, Chun J, Ueda H. Initiation of neuropathic pain requires lysophosphatidic acid receptor signaling. Nat Med. 2004;10:712–718. doi: 10.1038/nm1060. [DOI] [PubMed] [Google Scholar]
- 56.Kato K, Clark GD, Bazan NG, Zorumski CF. Platelet-activating factor as a potential retrograde messenger in CA1 hippocampal long-term potentiation. Nature. 1994;367:175–179. doi: 10.1038/367175a0. [DOI] [PubMed] [Google Scholar]
- 57.Chen C, Bazan NG. Lipid signaling: sleep, synaptic plasticity, and neuroprotection. Prostaglandins Other Lipid Mediat. 2005;77:65–76. doi: 10.1016/j.prostaglandins.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 58.Di Marzo V, Fontana A, Cadas H, Schinelli S, Cimino G, Schwartz JC, Piomelli D. Formation and inactivation of endogenous cannabinoid anandamide in central neurons. Nature. 1994;372:686–691. doi: 10.1038/372686a0. [DOI] [PubMed] [Google Scholar]
- 59.Belelli D, Herd MB, Mitchell EA, Peden DR, Vardy AW, Gentet L, Lambert JJ. Neuroactive steroids and inhibitory neurotransmission: mechanisms of action and physiological relevance. Neuroscience. 2006;138:821–829. doi: 10.1016/j.neuroscience.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 60.Fu J, Gaetani S, Oveisi F, Lo Verme J, Serrano A, Rodríguez De Fonseca F, Rosengarth A, Luecke H, Di Giacomo B, Tarzia G, et al. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature. 2003;425:90–93. doi: 10.1038/nature01921. [DOI] [PubMed] [Google Scholar]
- 61.Calignano A, La Rana G, Loubet-Lescoulié P, Piomelli D. A role for the endogenous cannabinoid system in the peripheral control of pain initiation. Prog Brain Res. 2000;129:471–482. doi: 10.1016/s0079-6123(00)29034-1. [DOI] [PubMed] [Google Scholar]