Abstract
Approximately 3% of the world population is infected with hepatitis C virus (HCV), causing a serious public health burden. Like other positive-strand RNA viruses, HCV assembles replicase complexes in association with cellular membranes and produces progeny RNA genomes through negative-strand intermediates. The viral proteins required for RNA replication are nonstructural (NS) proteins NS3 to NS5B. Owing to many obstacles and limitations in structural characterization of proteins and complexes with multiple transmembrane segments, attempts to understand the assembly and action of the HCV replicase complex have been challenging. Nevertheless, great progress has been made in obtaining structural information for several replicase components, providing insights into some aspects of the viral genome replication machinery.
Introduction
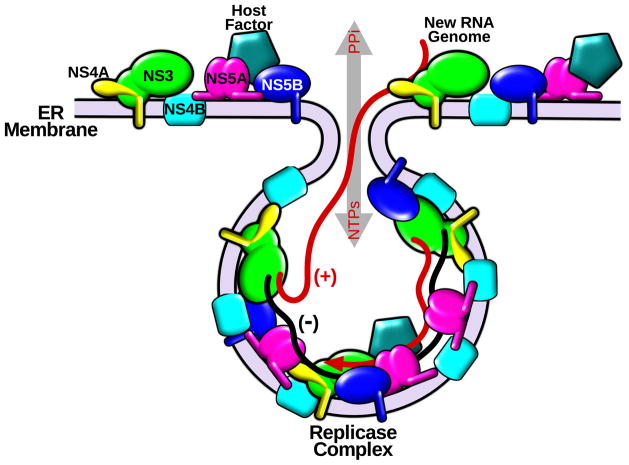
Chronic infection with hepatitis C virus (HCV) leads to liver fibrosis, cirrhosis and cancer, and is the most common cause of liver transplantation in many countries [1]. Although researchers have made progress since the discovery of HCV [2], a complete picture of the assembly and action of the HCV replicase, the central machine for synthesis of progeny RNA genomes, remains to be elucidated (Figure 1).
Figure 1. Hypothetical model of membrane-associated HCV RNA replication.
Viral NS proteins are translated and proteolytically processed on the ER membrane. A fraction of NS3 (green), NS4A (yellow), NS4B (cyan), NS5A (pink) and NS5B (blue) are associated with vesicle-like membrane structures and host factors (light blue) to assemble replicase machineries. Viral RNA is replicated through negative-strand RNA (−) colored black in the membrane wrapped environment, where replication is protected from proteases and nucleases. Newly synthesized genome RNA (+) colored red can exit and participate in other steps of the virus lifecycle.
HCV belongs to the Flaviviridae family. Its ~9.6 kb genome contains a long open reading frame flanked by 5′ and 3′ untranslated regions (UTRs). The translated polyprotein is proteolytically processed into ten proteins by host and viral proteases [3]. The nonstructural proteins NS3 to NS5B are the minimal viral protein components required for RNA replication (Figure 1) [4, 5]. Although NS2 is not a subunit of the replicase complex, its cysteine protease activity is essential for full-length genome replication as it cleaves at the NS2/NS3 junction to release a free NS3 N-terminus for functional replicase assembly [6, 7, 8]. The N-terminal third of NS3 harbors a serine protease domain, which cleaves the four downstream non-structural protein junctions, and at least three host targets [9]. The C-terminal portion of NS3 is a superfamily 2 (SF2) helicase that is essential for virus replication [10]. Its translocase activity has been well characterized with different approaches [11, 12, 13, 14]. NS4A is a transmembrane protein that acts as a cofactor for the NS3 protease, and is involved in the regulation of replicase activity [9, 15]. NS4B contains multiple transmembrane segments and is involved in remodeling the endoplasmic reticulum (ER) membrane [16], a common feature of many positive-strand RNA viruses [17]. NS5A is a multifunctional zinc-binding phosphoprotein, which has become a promising drug target [18]. NS5B is the RNA-dependent RNA polymerase (RdRp) with a C-terminal membrane-anchoring segment [19].
Analogous to other positive-strand RNA viruses, HCV assembles its replicase complexes on structurally rearranged vesicle-like host membranes often called spherules [17]. Viral RNA replication therefore occurs in a partially membrane encircled environment, which is resistant to cellular proteases and nucleases, yet allows influx of nucleotide triphosphates (NTPs) and exit of newly synthesized genome RNA (Figure 1). Despite the challenges in characterizing integral membrane components of the replicase complex, high-resolution structural information has been obtained for several replicase components, including the ectomembrane domains of the enzymatic proteins NS3 and NS5B, and the non-enzymatic NS5A domain I.
NS3 and the NS4A cofactor peptide
The polyprotein-processing NS3 serine protease adopts a chymotrypsin-like fold composed of two sub-domains consisting of mainly twisted β-sheets. The catalytic triad residues are located in a cleft between the two domains. The NS4A cofactor peptide forms a β-strand that lies between two NS3 strands, therefore completing the β-sheet and likely enhancing the NS3 protease activity (Figure 2A) [20]. The membrane localization of NS3 is determined by its α0 amphipathic helix and interaction with the transmembrane NS4A protein [21]. Structural investigations on the protease have facilitated drug development efforts, and recently two NS3-4A protease inhibitors have entered the market [22, 23].
Figure 2. Crystal structures of NS3 complexes.
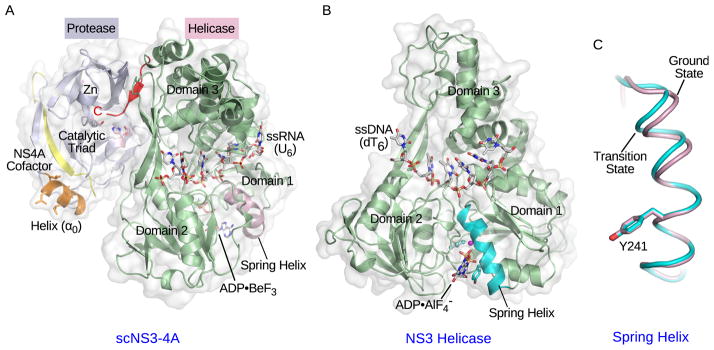
(A) Crystal structure of single-chain NS3-4A (scNS3-4A) in complex with ssRNA (U6) and ADP•BeF3 (PDB: 3O8R). The protease domain, helicase domain, NS4A cofactor peptide, and amphipathic α-helix (α0) are colored light blue, pale green, yellow and orange respectively. The catalytic triad residues in the protease domain are highlighted in light pink. The pink sphere represents the zinc ion. The C-terminal peptide bound to the protease catalytic site is highlighted in red. The ssRNA strand and ADP•BeF3 are shown as stick models. The spring helix is highlighted in light pink. (B) Crystal structure of the NS3 helicase domain in complex with ssDNA (dT6) and ADP•AlF4− (PDB: 3KQL). The helicase domain is colored pale green and the spring helix is highlighted in cyan. The ssDNA strand and ADP•AlF4− are shown as stick models. Three helicase subdomains are labeled in (A) and (B). (C) The spring helices from the ground-state (light pink, PDB: 3KQU) and transition-state (cyan, PDB: 3KQL) complexes are aligned through motif Y and the adjacent residues. The structure presentations were generated by PyMOL (www.pymol.org).
The NS3 SF2 helicase consists of two adjacent recA-like domains and a third accessory domain, similar to other SF1 and SF2 helicases [9, 10]. Nucleic-acids bind to the surface formed by the recA-like domains, which can swing toward each other upon nucleotide binding, consistent with the previously defined structural basis for an inchworm/ratchet motion (Figure 2A and 2B). The bound single-stranded (ss) DNA shows different pucker conformations in the backbone deoxyribose rings and at least one deoxyribose ring changes its pucker along the reaction pathway, explaining why ssDNA is a better substrate than ssRNA [12]. Indeed, structures of NS3 in complex with ssRNA show that the 2′ hydroxyl groups create steric hindrance that would affect association with the next ribose rings [14]. Comparison of the different conformational states captured along the reaction pathway has revealed transitions within a spring α-helix, which elongates from the ground state to the transition state of nucleotide hydrolysis (Figure 2B and 2C). This structural transition allosterically couples the catalysis of nucleotide hydrolysis to the change of nucleic-acid association on the N-terminal end of the spring helix. This newly defined structural element and the changes of protein-DNA interactions highlight the major feature distinguishing the HCV helicase from other SF2 candidates [12]. Although the kinetic steps of the helicase have been measured to be large and inconsistent in size, both crystallographic and single-molecule analyses have now concluded that one nucleotide hydrolysis is coupled to one base translocation from 3′ to 5′ [11, 12, 13, 14]. The function of the helicase in the replicase complex is unknown, although the analogous helicase domain in brome mosaic virus recruits viral RNA templates and the RdRp to ER membranes and spherule interiors [17].
Studies suggest that the covalently linked protease and helicase domain interact and regulate each other [24, 25, 26, 27]. The structure of an engineered scNS3-NS4A protein, in which the NS4A cofactor peptide is fused to the N-terminus of full-length NS3, shows that the C terminus of the helicase resides in the protease active site, supporting a cis cleavage at the NS3/NS4A junction (Figure 2A). This finding also suggests that domain rearrangement between the protease and helicase may occur so that the protease is able to perform trans cleavages at other NS protein junctions [28]. So far, no additional domain re-arrangements have been captured, presumably due to the absence of other viral or cellular factors [14].
Transmembrane proteins NS4A and NS4B
The N-terminal transmembrane α-helix of NS4A contains an unusually large number of amino acids with small side chains (Gly and Ala), a highly conserved face, and typical interface residues at both ends [21]. The central ~16 amino acids act as the aforementioned NS3 protease cofactor (Figure 1A), and interact with cellular creatine kinase B, which has been shown to enhance NS3 helicase activity in vitro and HCV replicase function in permeabilized cells [29]. The C-terminal region adopts an α-helical structure at low pH. Mutations within this region are associated with reduced NS5A hyperphosphorylation and virus RNA replication defects [15, 30].
NS4B is predicted to contain at least four transmembrane segments in its central domain [16]. The S/T cluster and the GXXXG motif in the first and second transmembrane segments may be involved in protein interactions and are important for virus replication [31]. NS4B also contains a GXXXXGK P-loop for nucleotide triphosphate binding, which may be important for NS4B to mediate membrane rearrangements [32]. Recombinant NS4B has been shown to catalyze hydrolysis of ATP, GTP and GDP, releasing terminal phosphates. The protein also exhibits an adenylate kinase activity, which converts two ADP molecules into ATP and AMP [33]. However, the function of the putative P-loop remains controversial since it is not fully conserved and mutation of the lysine residue essential for P-loop function to threonine actually enhances replicon RNA replication [34]. The N-terminal portion of NS4B likely consists of two amphipathic helices. The NMR structure of amino acids 40–69 clearly demonstrates the amphipathic distribution of residues along the helix. This helix also has the potential to traverse the membrane bilayer, and is important for the assembly of the replicase complex [35]. The highly conserved C-terminal domain was also predicted to contain two α-helices [36]. The structure of amino acids 227–254 reveals two hydrophobic leucine patches and a positively charged C-terminal portion. This segment is crucial for NS4B self-interaction, which in turn is important for membranous spherule formation and functional replicase assembly [37]. Palmitoylation of two C-terminal cysteines has been reported. However, abrogation of the penultimate cysteine does not inhibit RNA replication. Mutation of the C-terminal cysteine affects polyprotein cleavage, making it difficult to determine if NS4B palmitoylation is important for virus replication [16]. Recently, NS4B has emerged as a new drug target and several small molecules have been reported to suppress HCV replication by interfering with NS4B-associated functions, such as its apparent RNA binding activity [38, 39].
Multifunctional NS5A
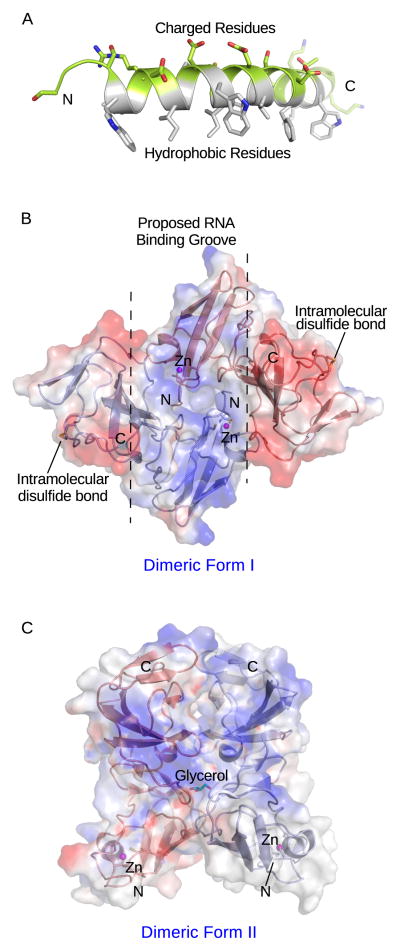
The non-enzymatic NS5A protein is required for both RNA replication and virus assembly [40, 41, 42]. It interacts with a large number of host proteins, such as cyclophilin A and phosphatidylinositol 4-Kinase IIIα, which are important for replicase assembly and function [43, 44]. Three NS5A domains have been defined. Domain I contains an N-terminal amphipathic α-helix with hydrophobic side chains clustered on one side to mediate membrane association and hydrophilic and charged residues on the opposite side, which likely participate in protein interactions (Fig. 3A) [45]. Two crystal structures of domain I without the amphipathic helix reveal a novel fold with charged surface patches [46, 47]. A zinc ion likely stabilizes NS5A domain I for its function. An intramolecular disulfide bond was found in one structure (Figure 3B), although it is not required for replicase function [46]. Interestingly, NS5A domain I can adopt different packing contacts for possible dimerization under different crystallization conditions, suggesting that this protein may be able to form alternate dimeric states that could contribute to its multifunctional roles in the virus life cycle (Fig. 3B and 3C). One type of dimer forms a positively charged groove possibly involved in RNA binding (Figure 3B) [46]. The remarkably potent RNA replication inhibitor, BMS-790052, has been suggested to target NS5A dimerization, and resistance mutations map in domain I [48]. Both domain II and III contain sequences required for binding of cyclophilin A and HCV RNA [44, 49, 50]. Domain III appears to be primarily involved in virus assembly [40]. Despite their importance in the life cycle, several structural studies have concluded that domains II and III generated in E. coli lack secondary structures [51, 52, 53]. The structure determination of these domains or full-length NS5A has therefore proven challenging.
Figure 3. Structures of NS5A domains.
(A) NMR structure of the N-terminal amphipathic helix (PDB: 1R7G). The hydrophobic residues are colored white, while charged residues are highlighted in green. (B) and (C) Two crystal structures of NS5A domain I without the amphipathic helix (PDB: 1ZH1 and 3FQM). Blue represents positive surface charge, while red represents negative charge. The N- and C- the termini are labeled. The putative RNA binding groove (blue) is denoted by dashed lines.
NS5A exists in hypophosphorylated (~56 kDa) and hyperphosphorylated (~58 kDa) forms [54]. Mutagenesis studies and in vitro phosphorylation of recombinant proteins suggest a number of phosphorylation sites, and mass spectrometry analysis of NS5A has identified at least two serine acceptors [55, 56]. Replication analysis of phosphomimetic mutations of serine 222 in the linker between domain I and II suggest that hyperphosphorylation negatively affects RNA replication [55]. It is thought that hypophosphorylated NS5A interacts with human vesicle-associated membrane protein-associated protein subtype A, which in turn may facilitate the interaction between NS5A and NS5B for replicase assembly and function [54, 55].
Tail-anchored NS5B
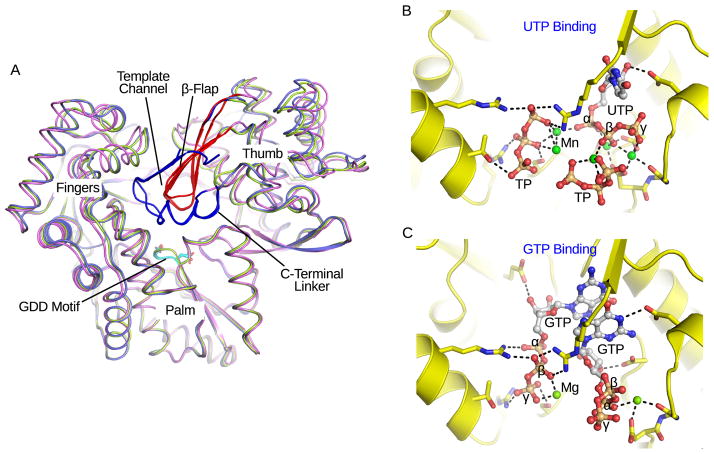
The NS5B RdRp, which is thought to mediate de novo RNA synthesis in vivo, is the catalytic core of the HCV replication machinery. The structure of NS5B without its C-terminal transmembrane tail is homologous to other viral RdRp enzymes [57]. It consists of fingers, palm and thumb subdomains (Figure 4A). The palm domain contains a four-stranded β-sheet flanked by three α-helices. Five of the eight highly conserved viral RdRp motifs are found in the palm domain, including the GDD motif VI that coordinates two catalytic magnesium ions. HCV NS5B possesses a large thumb domain that forms extensive contacts with the fingers domain and encircles the active site, a structural feature that distinguishes Flaviviridae RdRps from other viral polymerases. A β-flap motif in the thumb domain protrudes toward the active site, allowing only ssRNA to enter the active site during de novo initiation (Figure 4A). The HCV RdRp was crystallized in different conformations, consistent with the hypothesis that the enzyme is flexible and can adopt a closed conformation for de novo initiation and an opened template-primer form for replication elongation (Figure 4A) [57, 58]. A high concentration of GTP facilitates de novo RNA synthesis. Nucleotides, such as GTP and UTP soaked into preformed crystals, bind to the catalytic pocket. A number of residues from all three domains are involved in the coordination of nucleotides. The nucleotides, however, adopt different orientations, and in some cases only the triphosphate moieties can be visualized (Figure 4B) [59, 60]. Mutagenesis has shown that several GTP coordinating residues are important for de novo initiation [61]. A GTP binding site on the NS5B (HCV BK strain) surface has also been found [59]. This site has been suggested to play a role in the transition to elongation, although at least one coordinating residue is not conserved. A short ssRNA strand soaked into NS5B crystals associates with the fingers domain in the putative template channel. It likely represents a template strand being directed to the catalytic site [62]. In most structures, a C-terminal linker before the transmembrane segment occludes the catalytic cleft (Figure 4A). A recent study has suggested that it is directly involved in the very first steps of de novo initiation [60]. NS5B is an important target for drug development. Nucleotide inhibitors bind to the catalytic pocket, while non-nucleotide inhibitors target variable allosteric pockets leading to conformational changes of the polymerase active site [63].
Figure 4. Comparison of NS5B structures.
(A) Three NS5B crystal structures are aligned, and conformational differences can be seen in the fingers and thumb domains (PDB: 1YUY, 1YV2 and 3I5K). The GDD motif, β-flap, and C-terminal linker are colored cyan, red and blue respectively. The template channel is noted. (B) and (C) Structural comparison of nucleotides coordinated in catalytic pockets (PDB: 2XI3 and 1GX6 for GTP and UTP soaked complexes respectively). The proteins are shown as ribbon and stick models, and colored yellow. The nucleotides are shown as sticks and spheres, and colored by elements. Metal ions are green spheres. TP stands for triphosphate. The α-, β-, and γ-phosphate groups are labeled for the complete nucleotides. The dashed lines represent possible atomic interactions.
Despite the many reported NS5B structures, all lack the C-terminal transmembrane α-helix, an element that is essential for the assembly of functional replicase complexes [64, 65, 66]. In addition, the enzymatic activity of NS5B by itself does not efficiently complete the RNA replication cycle in vitro [67]. It will likely be necessary to obtain full-length NS5B in complex with other replicase subunits and host factors to better understand its activity in the multicomponent replicase. Furthermore, the model of HCV RNA-dependent RNA polymerization is still based on a set of co-complex structures of the bacteriophage φ6 polymerase [57]. Analogous complexes are, however, difficult to capture for the Flaviviridae RdRps, as no candidate has been crystallized with an open active site that can accommodate double-stranded RNA. Further structural investigation on HCV NS5B is expected to reveal the mechanism by which conformational changes allow the active site to coordinate both RNA substrates and incoming NTPs for catalysis.
Conclusion
Since the discovery of HCV, high-resolution structures of several viral replicase NS proteins have been successfully obtained. The structures have helped us understand the functions and mechanisms of the important viral factors, and stimulated drug development efforts. Despite great progress, structures for the NS4A and NS4B integral membrane proteins/domains and unstructured regions of NS5A are missing, yet these components are essential for replicase function and have emerged as drug targets. To facilitate structural characterization, appropriate expression approaches are required to generate recombinant NS4A and NS4B proteins in large quantity, and obtain well-folded NS5A with possible posttranslational modifications and/or in association with key binding partner(s). It is also worth noting that studies aimed at probing the roles of host factors required for viral replicase assembly in vivo should facilitate a complete structural and biochemical understanding of the HCV replicase complex [68, 69]. One important goal is to obtain a high-resolution structure of the RNA replication machine, which should help demonstrate how replicase subunits interact with each other to remodel cellular membranes and accomplish genome RNA replication through negative-strand intermediates. Future research in this field would reveal common principles in positive-strand RNA virus replication and has the potential to open up new avenues for broad-spectrum virus control.
Highlights.
Membrane-associated positive-strand RNA virus replication
HCV NS proteins involved in replicase assembly and function
Structures, mechanisms and functional implications of HCV NS proteins
HCV antivirals targeting the replicase NS proteins
Acknowledgments
M.G. was supported in part by a Marie-Josée and Henry R. Kravis Fellowship at The Rockefeller University. Additional financial support came from an NIH grant (CA057973), and generous gifts from the Greenberg Medical Research Institute, the Starr Foundation, the Richard Salomon Family Foundation, the Ronald A. Shellow, M.D. Memorial Fund, Paul Nash and the MGM Mirage Voice Foundation, Gregory F. Lloyd Memorial contributions, and anonymous donors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.WHO. URL: http://www.who.int/csr/disease/hepatitis/whocdscsrlyo2003/en/index2.html.
- 2•.Murray CL, Rice CM. Turning Hepatitis C into a Real Virus. Annual Review of Microbiology. 2011;65:307–327. doi: 10.1146/annurev-micro-090110-102954. This review discusses the development of model systems for laboratory investigation of HCV replication. [DOI] [PubMed] [Google Scholar]
- 3•.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nature Reviews Microbiology. 2007;5:453–463. doi: 10.1038/nrmicro1645. This review describes the replication cycle of HCV. [DOI] [PubMed] [Google Scholar]
- 4.Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 5.Lohmann V, Korner F, Koch JO, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 6.Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 7.Welbourn S, Green R, Gamache I, Dandache S, Lohmann V, Bartenschlager R, Meerovitch K, Pause A. Hepatitis C virus NS2/3 processing is required for NS3 stability and viral RNA replication. J Biol Chem. 2005;280:29604–29611. doi: 10.1074/jbc.M505019200. [DOI] [PubMed] [Google Scholar]
- 8.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morikawa K, Lange CM, Gouttenoire J, Meylan E, Brass V, Penin F, Moradpour D. Nonstructural protein 3–4A. the Swiss army knife of hepatitis C virus. J Viral Hepat. 2011;18:305–315. doi: 10.1111/j.1365-2893.2011.01451.x. [DOI] [PubMed] [Google Scholar]
- 10.Raney KD, Sharma SD, Moustafa IM, Cameron CE. Hepatitis C virus non-structural protein 3 (HCV NS3): a multifunctional antiviral target. J Biol Chem. 2010;285:22725–22731. doi: 10.1074/jbc.R110.125294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng W, Arunajadai SG, Moffitt JR, Tinoco R, Bustamante C. Single-Base Pair Unwinding and Asynchronous RNA Release by the Hepatitis C Virus NS3 Helicase. Science. 2011;333:1746–1749. doi: 10.1126/science.1206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci USA. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Appleby TC, Anderson R, Fedorova O, Pyle AM, Wang R, Liu X, Brendza KM, Somoza JR. Visualizing ATP-dependent RNA Translocation by the NS3 Helicase from HCV. J Mol Biol. 2011;405:1139–1153. doi: 10.1016/j.jmb.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Phan T, Kohlway A, Dimberu P, Pyle AM, Lindenbach BD. The acidic domain of hepatitis C virus NS4A contributes to RNA replication and virus particle assembly. J Virol. 2011;85:1193–1204. doi: 10.1128/JVI.01889-10. The authors report mutagenesis analyses that probe the functions of NS4A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouttenoire J, Penin F, Moradpour D. Hepatitis C virus nonstructural protein 4B: a journey into unexplored territory. Rev Med Virol. 2010;20:117–129. doi: 10.1002/rmv.640. [DOI] [PubMed] [Google Scholar]
- 17••.den Boon JA, Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. This review describes how different positive-strand RNA viruses utilize host membranes to accomplish a common principle of genome replication. [DOI] [PubMed] [Google Scholar]
- 18.Schmitz U, Tan SL. NS5A--from obscurity to new target for HCV therapy. Recent Pat Antiinfect Drug Discov. 2008;3:77–92. doi: 10.2174/157489108784746597. [DOI] [PubMed] [Google Scholar]
- 19.Behrens SE, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JL, Morgenstern KA, Lin C, Fox T, Dwyer MD, Landro JA, Chambers SP, Markland W, Lepre CA, O’Malley ET, et al. Crystal Structure of the Hepatitis C Virus NS3 Protease Domain Complexed with a Synthetic NS4A Cofactor Peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 21.Brass V, Berke JM, Montserret R, Blum HE, Penin F, Moradpour D. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3–4A complex. Proc Natl Acad Sci USA. 2008;105:14545–14550. doi: 10.1073/pnas.0807298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romano KP, Ali A, Aydin C, Soumana D, Ozen A, Deveau LM, Silver C, Cao H, Newton A, Petropoulos CJ, et al. The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors. PLoS Pathog. 2012;8:e1002832. doi: 10.1371/journal.ppat.1002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett F, Huang Y, Hendrata S, Lovey R, Bogen SL, Pan W, Guo Z, Prongay A, Chen KX, Arasappan A, et al. The introduction of P4 substituted 1-methylcyclohexyl groups into Boceprevir: a change in direction in the search for a second generation HCV NS3 protease inhibitor. Bioorg Med Chem Lett. 2010;20:2617–2621. doi: 10.1016/j.bmcl.2010.02.063. [DOI] [PubMed] [Google Scholar]
- 24.Zhang C, Cai Z, Kim YC, Kumar R, Yuan F, Shi PY, Kao C, Luo G. Stimulation of hepatitis C virus (HCV) nonstructural protein 3 (NS3) helicase activity by the NS3 protease domain and by HCV RNA-dependent RNA polymerase. J Virol. 2005;79:8687–8697. doi: 10.1128/JVI.79.14.8687-8697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beran RK, Pyle AM. Hepatitis C viral NS3–4A protease activity is enhanced by the NS3 helicase. J Biol Chem. 2008;283:29929–29937. doi: 10.1074/jbc.M804065200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beran RK, Serebrov V, Pyle AM. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J Biol Chem. 2007;282:34913–34920. doi: 10.1074/jbc.M707165200. [DOI] [PubMed] [Google Scholar]
- 27.Frick DN, Rypma RS, Lam AM, Gu B. The nonstructural protein 3 protease/helicase requires an intact protease domain to unwind duplex RNA efficiently. J Biol Chem. 2004;279:1269–1280. doi: 10.1074/jbc.M310630200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao N, Reichert P, Taremi SS, Prosise WW, Weber PC. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure. 1999;7:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 29.Hara H, Aizaki H, Matsuda M, Shinkai-Ouchi F, Inoue Y, Murakami K, Shoji I, Kawakami H, Matsuura Y, Lai MM, et al. Involvement of creatine kinase B in hepatitis C virus genome replication through interaction with the viral NS4A protein. J Virol. 2009;83:5137–5147. doi: 10.1128/JVI.02179-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindenbach BD, Prágai BM, Montserret R, Beran RK, Pyle AM, Penin F, Rice CM. The C terminus of hepatitis C virus NS4A encodes an electrostatic switch that regulates NS5A hyperphosphorylation and viral replication. J Virol. 2007;81:8905–8918. doi: 10.1128/JVI.00937-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han Q, Aligo J, Manna D, Belton K, Chintapalli SV, Hong Y, Patterson RL, van Rossum DB, Konan KV. Conserved GXXXG- and S/T-Like Motifs in the Transmembrane Domains of NS4B Protein Are Required for Hepatitis C Virus Replication. J Virol. 2011;85:6464–6479. doi: 10.1128/JVI.02298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Einav S, Elazar M, Danieli T, Glenn JS. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J Virol. 2004;78:11288–11295. doi: 10.1128/JVI.78.20.11288-11295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson AA, Zou A, Yan J, Duggal R, Hao W, Molina D, Cronin CN, Wells PA. Biochemical characterization of recombinant hepatitis C virus nonstructural protein 4B. evidence for ATP/GTP hydrolysis and adenylate kinase activity. Biochemistry. 2009;48:906–916. doi: 10.1021/bi801747p. [DOI] [PubMed] [Google Scholar]
- 34.Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol. 2003;77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gouttenoire J, Castet V, Montserret R, Arora N, Raussens V, Ruysschaert JM, Diesis E, Blum HE, Penin F, Moradpour D. Identification of a novel determinant for membrane association in hepatitis C virus nonstructural protein 4B. J Virol. 2009;83:6257–6268. doi: 10.1128/JVI.02663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gouttenoire J, Montserret R, Kennel A, Penin F, Moradpour D. An amphipathic alpha-helix at the C terminus of hepatitis C virus nonstructural protein 4B mediates membrane association. J Virol. 2009;83:11378–11384. doi: 10.1128/JVI.01122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Paul D, Romero-Brey I, Gouttenoire J, Stoitsova S, Krijnse-Locker J, Moradpour D, Bartenschlager R. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J Virol. 2011;85:6963–6976. doi: 10.1128/JVI.00502-11. The authors report the contribution of the highly conserved C-terminal domain in NS4B oligomerization and membranous vesicle induction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rai R, Deval J. New opportunities in anti-hepatitis C virus drug discovery: targeting NS4B. Antiviral Res. 2011;90:93–101. doi: 10.1016/j.antiviral.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Einav S, Gerber D, Bryson PD, Sklan EH, Elazar M, Maerkl SJ, Glenn JS, Quake SR. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat Biotechnol. 2008;26:1019–1027. doi: 10.1038/nbt.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathogens. 2008;4:e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benga WJ, Krieger SE, Dimitrova M, Zeisel MB, Parnot M, Lupberger J, Hildt E, Luo G, McLauchlan J, Baumert TF, et al. Apolipoprotein E interacts with hepatitis C virus nonstructural protein 5A and determines assembly of infectious particles. Hepatology. 2010;51:43–53. doi: 10.1002/hep.23278. [DOI] [PubMed] [Google Scholar]
- 42.Shimakami T, Hijikata M, Luo H, Ma YY, Kaneko S, Shimotohno K, Murakami S. Effect of Interaction between Hepatitis C Virus NS5A and NS5B on Hepatitis C Virus RNA Replication with the Hepatitis C Virus Replicon. J Virol. 2004;78:2738–2748. doi: 10.1128/JVI.78.6.2738-2748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim YS, Hwang SB. Hepatitis C Virus NS5A Protein Interacts with Phosphatidylinositol 4-Kinase Type IIIα and Regulates Viral Propagation. J Biol Chem. 2011;286:11290–11298. doi: 10.1074/jbc.M110.194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foster TL, Gallay P, Stonehouse NJ, Harris M. Cyclophilin A interacts with domain II of hepatitis C virus NS5A and stimulates RNA binding in an isomerase-dependent manner. J Virol. 2011;85:7460–7464. doi: 10.1128/JVI.00393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Penin F, Brass V, Appel N, Ramboarina S, Montserret R, Ficheux D, Blum HE, Bartenschlager R, Moradpour D. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J Biol Chem. 2004;279:40835–40843. doi: 10.1074/jbc.M404761200. [DOI] [PubMed] [Google Scholar]
- 46.Tellinghuisen TL, Marcotrigiano J, Rice CM. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature. 2005;435:374–379. doi: 10.1038/nature03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love RA, Brodsky O, Hickey MJ, Wells PA, Cronin CN. Crystal Structure of a Novel Dimeric Form of NS5A Domain I Protein from Hepatitis C. J Virol. 2009;83:4395–4403. doi: 10.1128/JVI.02352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48••.Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun JH, O’Boyle DR, 2nd, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465:96–100. doi: 10.1038/nature08960. The authors report the discovery of a small molecule that inhibits HCV RNA replication at picomolar EC(50) concentrations. This symmetric inhibitor has been hypothesized to target dimeric NS5A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verdegem D, Badillo A, Wieruszeski JM, Landrieu I, Leroy A, Bartenschlager R, Penin F, Lippens G, Hanoulle X. Domain 3 of NS5A protein from the hepatitis C virus has intrinsic α-helical propensity and is a substrate of cyclophilin A. J Biol Chem. 2011;286:20441–20454. doi: 10.1074/jbc.M110.182436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foster TL, Belyaeva T, Stonehouse NJ, Pearson AR, Harris M. All three domains of the hepatitis C virus nonstructural NS5A protein contribute to RNA binding. J Virol. 2010;84:9267–9277. doi: 10.1128/JVI.00616-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanoulle X, Verdegem D, Badillo A, Wieruszeski JM, Penin F, Lippens G. Domain 3 of non-structural protein 5A from hepatitis C virus is natively unfolded. Biochem Biophys Res Commun. 2009;381:634–638. doi: 10.1016/j.bbrc.2009.02.108. [DOI] [PubMed] [Google Scholar]
- 52.Hanoulle X, Badillo A, Verdegem D, Penin F, Lippens G. The domain 2 of the HCV NS5A protein is intrinsically unstructured. Protein Pept Lett. 2010;17:1012–1018. doi: 10.2174/092986610791498920. [DOI] [PubMed] [Google Scholar]
- 53.Liang Y, Ye H, Kang CB, Yoon HS. Domain 2 of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfolded. Biochemistry. 2007;46:11550–11558. doi: 10.1021/bi700776e. [DOI] [PubMed] [Google Scholar]
- 54.Huang Y, Staschke K, De Francesco R, Tan SL. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology. 2007;364:1–9. doi: 10.1016/j.virol.2007.01.042. [DOI] [PubMed] [Google Scholar]
- 55.Lemay KL, Treadaway J, Angulo-Herrera I, Tellinghuisen TL. A Hepatitis C Virus NS5A Phosphorylation Site that Regulates RNA Replication. J Virol. 2012 doi: 10.1128/JVI.02154-12. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nordle Gilliver A, Griffin S, Harris M. Identification of a novel phosphorylation site in hepatitis C virus NS5A. J Gen Virol. 2010;91:2428–2432. doi: 10.1099/vir.0.023614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Choi KH, Rossmann MG. RNA-dependent RNA polymerases from Flaviviridae. Curr Opin Struct Biol. 2009;19:746–751. doi: 10.1016/j.sbi.2009.10.015. This review covers current knowledge of Flaviviridae RdRps and implications regarding the mechanism of de novo RNA synthesis. [DOI] [PubMed] [Google Scholar]
- 58.Biswal BK, Cherney MM, Wang M, Chan L, Yannopoulos CG, Bilimoria D, Nicolas O, Bedard J, James MN. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J Biol Chem. 2005;280:18202–18210. doi: 10.1074/jbc.M413410200. [DOI] [PubMed] [Google Scholar]
- 59.Bressanelli S, Tomei L, Rey FA, De Francesco R. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J Virol. 2002;76:3482–3492. doi: 10.1128/JVI.76.7.3482-3492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harrus D, Ahmed-El-Sayed N, Simister PC, Miller S, Triconnet M, Hagedorn CH, Mahias K, Rey FA, Astier-Gin T, Bressanelli S. Further insights into the roles of GTP and the C terminus of the hepatitis C virus polymerase in the initiation of RNA synthesis. J Biol Chem. 2010;285:32906–32918. doi: 10.1074/jbc.M110.151316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ranjith-Kumar CT, Sarisky RT, Gutshall L, Thomson M, Kao CC. De novo initiation pocket mutations have multiple effects on hepatitis C virus RNA-dependent RNA polymerase activities. J Virol. 2004;78:12207–12217. doi: 10.1128/JVI.78.22.12207-12217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Farrell D, Trowbridge R, Rowlands D, Jäger J. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): Structural evidence for nucleotide import and de-novo initiation. J Mol Biol. 2003;326:1025–1035. doi: 10.1016/s0022-2836(02)01439-0. [DOI] [PubMed] [Google Scholar]
- 63.Mayhoub AS. Hepatitis C RNA-dependent RNA polymerase inhibitors: a review of structure-activity and resistance relationships; different scaffolds and mutations. Bioorg Med Chem. 2012;20:3150–3161. doi: 10.1016/j.bmc.2012.03.049. [DOI] [PubMed] [Google Scholar]
- 64.Moradpour D, Brass V, Bieck E, Friebe P, Gosert R, Blum HE, Bartenschlager R, Penin F, Lohmann V. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J Virol. 2004;78:13278–13284. doi: 10.1128/JVI.78.23.13278-13284.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee KJ, Choi J, Ou JH, Lai MM. The C-terminal transmembrane domain of hepatitis C virus (HCV) RNA polymerase is essential for HCV replication in vivo. J Virol. 2004;78:3797–3802. doi: 10.1128/JVI.78.7.3797-3802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66•.Brass V, Gouttenoire J, Wahl A, Pal Z, Blum HE, Penin F, Moradpour D. Hepatitis C virus RNA replication requires a conserved structural motif within the transmembrane domain of the NS5B RNA-dependent RNA polymerase. J Virol. 2010;84:11580–11584. doi: 10.1128/JVI.01519-10. This research highlights the importance of the C-terminal transmembrane segment of NS5B, which is required for the assembly of a functional replication complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lohmann V, Körner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moriishi K, Matsuura Y. Host factors involved in the replication of hepatitis C virus. Reviews in Medical Virology. 2007;17:343–354. doi: 10.1002/rmv.542. [DOI] [PubMed] [Google Scholar]
- 69.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci USA. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]