Significance
The inositol polyphosphate system is a major intracellular signaling modality. Inositol polyphosphate multikinase (IPMK) is essential for this system, converting inositol 1,4,5-trisphosphate (IP3) to IP4 and IP4 to IP5. IPMK also is a physiologic PI3-kinase. In a noncatalytic manner, IPMK stabilizes the mammalian target of rapamycin (mTOR) complex as well as tumor suppressor protein p53 and CREB-binding protein (CBP)/E1A binding protein p300. We now demonstrate that IPMK is a transcriptional coactivator for serum response factor (SRF) signaling, which is markedly reduced in IPMK-deleted mice and augmented by IPMK overexpression. IPMK binds directly to SRF, facilitating its interaction with serum response elements of multiple immediate early genes. Agents influencing IPMK interactions with SRF may be useful in regulating the SRF signaling system.
Abstract
Inositol polyphosphate multikinase (IPMK) is a notably pleiotropic protein. It displays both inositol phosphate kinase and phosphatidylinositol kinase catalytic activities. Noncatalytically, IPMK stabilizes the mammalian target of rapamycin complex 1 and acts as a transcriptional coactivator for CREB-binding protein/E1A binding protein p300 and tumor suppressor protein p53. Serum response factor (SRF) is a major transcription factor for a wide range of immediate early genes. We report that IPMK, in a noncatalytic role, is a transcriptional coactivator for SRF mediating the transcription of immediate early genes. Stimulation by serum of many immediate early genes is greatly reduced by IPMK deletion. IPMK stimulates expression of these genes, an influence also displayed by catalytically inactive IPMK. IPMK acts by binding directly to SRF and thereby enhancing interactions of SRF with the serum response element of diverse genes.
Inositol polyphosphate multikinase (IPMK) is one of a family of inositol phosphate kinases that generate inositol polyphosphates (1–3). IPMK appears to be a rate-limiting enzyme in the generation of inositol 1,4,5,6-tetrakisphosphate (IP4) and subsequently inositol 1,3,4,5,6-pentakisphosphate (IP5), which in turn is a requisite precursor for the energetic inositol pyrophosphates such as diphosphoinositol pentakisphosphate (PP-IP5, IP7). Additionally, IPMK is a physiologic PI3-kinase that contributes to activation of the Akt signaling pathway (4, 5). Recently, IPMK has been shown to play physiologic roles independent of its catalytic activity. Thus, IPMK stabilizes the mammalian target of rapamycin complex 1 (mTORC1), thereby regulating protein translation (6).
Since its first discovery in yeast, there has been evidence that IPMK can influence gene transcription. IPMK was identified in yeast as Arg82, a gene required for the regulation of arginine metabolism (3, 7–9). In this process, Arg82 interfaces with the yeast transcription factor MCM1, a yeast homolog of the mammalian serum response factor (SRF) (10–13). Catalytic activity of IPMK is not required for this function. More recently, evidence has been provided for IPMK acting noncatalytically as a transcriptional coactivator for p53, mediating its proapoptotic influences (14, 15). Also, IPMK binds CBP/p300, enhancing its transcriptional coactivation of CREB regulated genes involved in memory (16). In the present study, we report that IPMK is a physiologic coactivator for the transcriptional activities of SRF, thereby regulating the induction of families of immediate early genes.
Results
IPMK Is Required for Serum-Mediated Gene Activation.
The finding that IPMK is a transcriptional coactivator for p53 prompted us to consider the possibility that it regulates activation of other gene families. Because yeast IPMK regulates a homolog of SRF (10), we explored a possible role for IPMK in influencing gene transcription elicited by serum via SRF.
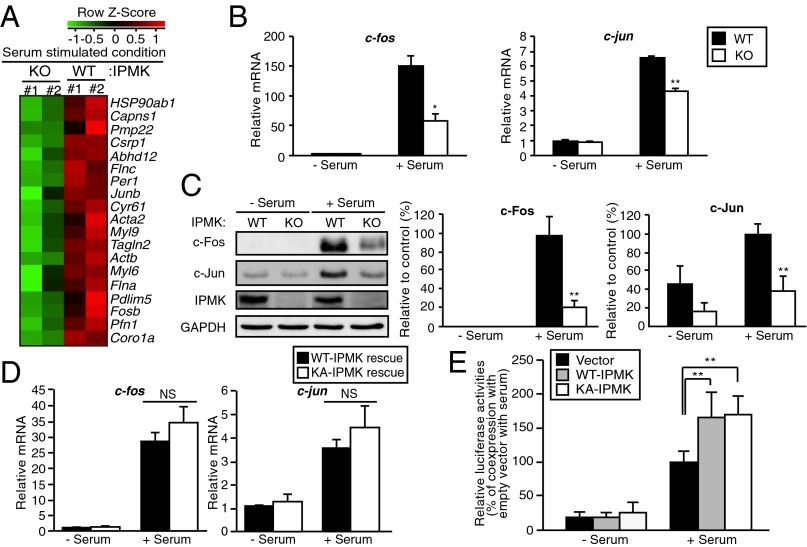
We monitored expression of RNA for a wide range of genes in a microarray analysis in wild-type and IPMK-deleted mouse embryonic fibroblasts (MEFs) in the presence of serum (Fig. 1 and Fig. S1A). Over 1,400 genes are down-regulated by IPMK deletion whereas 767 are up-regulated. Among the down-regulated genes are a substantial number of immediate early genes that contain serum response element (SRE) in their promoters and are well-known as SRF targets (Fig. 1A). We examined in greater detail the disposition of c-fos and c-jun, whose mRNA and protein levels are markedly stimulated by serum, with stimulation greatly diminished by IPMK deletion (Fig. 1 B and C). These effects do not reflect influences on SRF protein levels, which are not altered with IPMK deletion (Fig. S1B).
Fig. 1.
IPMK promotes SRF-induced gene activation. (A) Microarray analysis of gene expression profiles in serum-stimulated wild-type (WT) and IPMK-deleted (KO) MEFs. Cells were deprived of serum for 12 h and stimulated with 10% FBS for 30 min. Major serum-inducible SRF target genes were clustered. Increased and decreased gene expression labeled in red and green, respectively. (B) The mRNA levels of the SRF target genes c-jun and c-fos as measured via qPCR. Cells were deprived of serum for 12 h and stimulated with 10% FBS for 30 min. (C) Immunoblot analysis of protein levels. Proteins were prepared from WT and IPMK-deleted MEFs that were stimulated with 10% FBS for 1 h. (D) Proteins were isolated from IPMK-deleted MEFs stably expressing wild-type (WT) or the catalytically inactive, mutant K129A IPMK (KA), followed by immunoblotting. (E) SRE reporter activity along with empty vector, WT-IPMK, or catalytically inactive, mutant K129A IPMK (KA) expression plasmids in transiently transfected HEK293 cells. At 36 h posttransfection, cells were incubated in serum-free DMEM overnight followed by 10% FBS treatment for 6 h. Constitutively expressing Renilla luciferase was used as an internal control for normalizing transfection efficiency. The results are presented as relative luciferase activities compared with the activity after coexpression of empty vector in serum-treated condition, which was set as 100%. Bars represent mean ± SE (B and D) or ± SD (C and E) (n = 3–5). *P < 0.05; **P < 0.01; NS, not significant.
Serum treatment leads to the activation of various transcription factors, in particular, SRF and cAMP response element binding protein (CREB) (17, 18). Both of these transcription factors have been shown to induce the expression of immediate early genes. CREB does not appear to be primarily responsible for immediate early gene induction in our experiments. Thus, cyr61, which is up-regulated by SRF (19) but down-regulated by CREB (20), is markedly stimulated by serum in an IPMK-dependent fashion (Fig. 1A and Fig. S2A). These effects are not associated with changes in cell survival as cell viability is not altered in IPMK-deleted preparations (Fig. S3A).
Mutation of lysine-129 to alanine inactivates IPMK’s catalytic activity (5). To further explore the role of catalytic activity of IPMK in the regulation of SRF/SRE targets, we overexpressed IPMK wild-type and catalytically inactive proteins in IPMK-deleted MEFs. Overexpression of either wild-type or catalytically inactive IPMK in IPMK-deleted MEFs markedly enhances levels of c-fos and c-jun mRNA upon serum treatment (Fig. 1D). The catalytically inactive IPMK is as effective as wild-type, suggesting that the gene-regulatory influences of IPMK are independent of catalytic activity. We explored whether IPMK enhances the expression of immediate early genes by regulating the serum response element (SRE) of their promoters. We examined the influence of serum on SRE reporter construct activity in HEK293 cells overexpressing IPMK (Fig. 1E). Serum markedly enhances promoter activity, and overexpression of wild-type IPMK provides further augmentation. Catalytically inactive IPMK mutant also stimulates SRE promoter activity as effectively as the wild-type enzyme, confirming that the transcription regulation of IPMK is independent of catalytic activity.
IPMK Regulates Interaction of SRF with Its Target Gene Promoters.
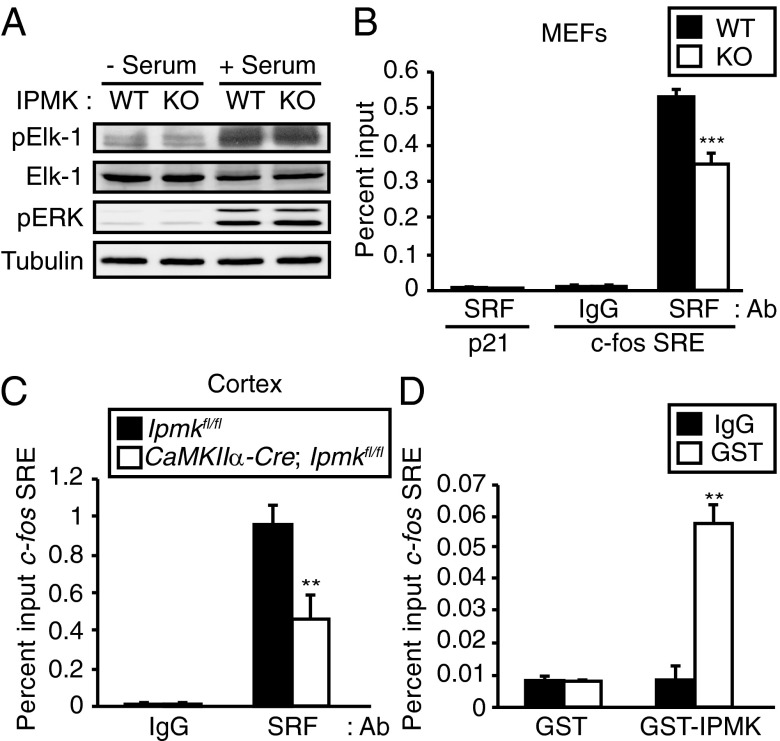
We sought mechanisms whereby IPMK regulates the influence of serum on immediate early genes. Transcriptional activation of immediate early genes in response to serum requires the presence of the Ternary Complex Factor (TCF) and SRF at the SRE (17, 18, 21–24). The Ras/MEK/ERK pathway induces phosphorylation of Elk-1, a component of TCF, leading to potentiation of transcriptional activation (25, 26). We investigated whether IPMK deletion influences the ERK-mediated phosphorylation of Elk-1 by examining levels of phospho-ERK and phospho–Elk-1 in wild-type and IPMK-deleted MEFs. Phospho-ERK and phospho–Elk-1 levels are not altered by IPMK deletion (Fig. 2A and Fig. S4).
Fig. 2.
IPMK regulates interaction of SRF to its target gene promoters. (A) IPMK deletion does not affect Elk-1 phosphorylation. Wild-type and IPMK-deleted MEFs were deprived of serum for 12 h and stimulated with 10% FBS for 1 h. Cell lysates were prepared and analyzed for Elk-1 and ERK phosphorylation by immunoblotting. (B) The ChIP assay was used to assess SRF recruitment to the c-fos promoter SRE. Deletion of IPMK in MEFs decreased SRF recruitment to the c-fos promoter. (C) CaMKIIα-Cre; Ipmkfl/fl mice exhibited ∼60% less SRF recruitment to the c-fos promoter compared with littermate Ipmkfl/fl control. The cortical tissue was analyzed for the ChIP assay. (D) IPMK is recruited to the SRE in the c-fos promoter. GST or GST-human IPMK (GST-IPMK) was overexpressed in HEK293 cells. The ChIP assay was performed by using GST antibody. Bars represent mean ± SE (n = 3). **P < 0.01; ***P < 0.001.
We explored the possibility that IPMK acts as a regulator of SRF by examining the binding of SRF to SRE in IPMK-deleted preparations. Using ChIP assay, we monitored the binding of SRF to c-fos SRE in wild-type and IPMK-deleted MEFs (Fig. 2B). Binding in IPMK-deleted MEFs is substantially less than in wild-type preparations. To ensure the specificity of the ChIP procedure, we examined the p21 promoter DNA sequence as a non-SRE DNA control and observed no background signal (Fig. 2B). Specificity is further endured by the failure of IPMK deletion to affect SP1 function (Fig. S5).
To ascertain whether the actions of SRF are dependent upon IPMK in intact organisms, we constructed IPMK knockout mice in which the IPMK gene is selectively deleted in excitatory neurons of the forebrain using a CaM kinase IIα-Cre construct (Fig. S6A). We used this selective deletion of IPMK because total deletion of IPMK is embryonic lethal (27). In the mutant mice, IPMK is undetectable in forebrain regions such as the hippocampus and cerebral cortex, but not in cerebellum, which is derived from the hindbrain (Fig. S6B). In cerebral cortical tissues, we monitored recruitment of SRF to the c-fos SRE (Fig. 2C). Interactions of SRF with SRE are reduced more than 50% in brains of the IPMK mutants.
If IPMK acts as a transcriptional coactivator with SRF, then one would anticipate recruitment of IPMK to SRE of immediate early genes. Thus, ChIP analysis using the SRE of the c-fos gene reveals robust binding of overexpressed IPMK to SRE (Fig. 2D).
In summary, IPMK appears to regulate SRF actions by influencing its binding to the SRE of target genes. These actions of IPMK are independent of its catalytic activity, reminiscent of the catalytically independent regulation of p53 by IPMK (14).
Binding of IPMK to SRF Mediates Immediate Early Gene Regulation.
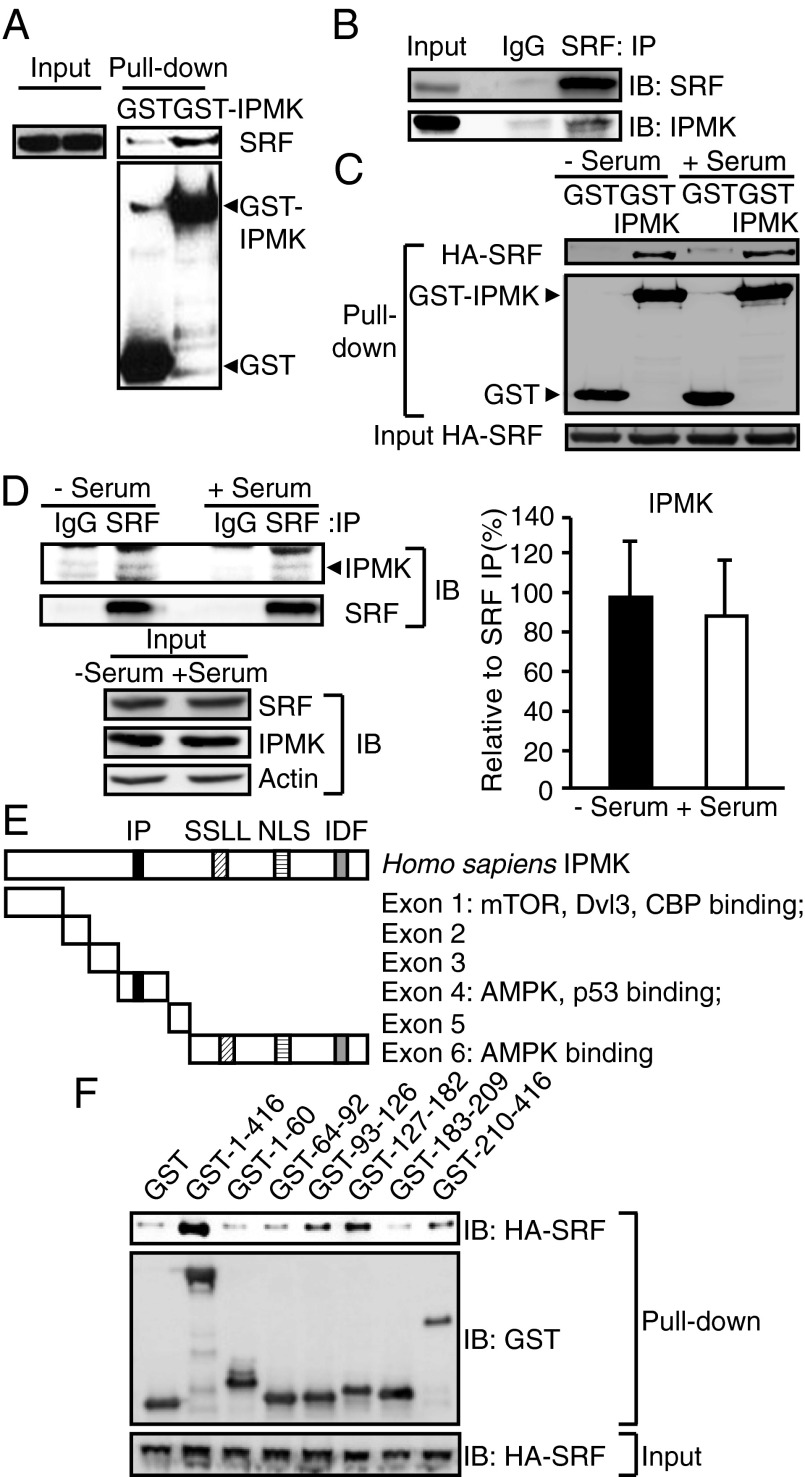
We explored mechanisms whereby IPMK modulates the SRF binding to SRE of immediate early genes. Recruitment of IPMK to the SRE of the c-fos gene despite its lack of a DNA binding domain raised the possibility that IPMK binds to SRF and is thereby recruited to the regulatory elements of SRF target genes. GST-labeled IPMK binds robustly to endogenous SRF (Fig. 3A). The physiologic relevance of this binding is evident in the avid binding of endogenous SRF and IPMK (Fig. 3B). Binding between IPMK and SRF does not appear to be regulated by the levels of serum in culture media (Fig. 3 C and D).
Fig. 3.
IPMK physically interacts with SRF. (A) Identification of SRF using GST-IPMK pull-down. GST or GST-IPMK was transfected in HEK293T cells followed by pull-down and immunoblot analysis for endogenous SRF. (B) Immunoprecipitation of IPMK and SRF proteins. MEF lysates were used for immunoprecipitaion (IP) against control IgG or SRF antibody to determine the binding between endogenous IPMK and SRF. (C) HA-SRF and GST-IPMK were cotransfected in HEK293T cells that were deprived of serum for 12 h and stimulated with 10% FBS for 30 min. GST-IPMK pull-down and immunoblotting were conducted. (D) SRF immunoprecipitates were prepared from MEFs that were deprived of serum for 12 h and stimulated with 10% FBS for 30 min. IPMK and SRF levels were analyzed by immunoblotting. Quantification of IPMK level normalized to coimmunoprecipitated SRF is presented as a graph. Bars represent mean ± SD (n = 4). (E) A schematic diagram of IPMK exon fragments with the numbers of amino acid sequences used for binding studies. IPMK-binding proteins were indicated. Key domains for inositol binding (IP), kinase activity (SSLL and IDF), and nuclear localization signal (NLS) are shown. (F) Mapping of binding region of IPMK responsible for SRF interaction. GST, GST-IPMK, or GST-IPMK exon fragments (exon 1, 1–63; exon 2, 64–92; exon 3, 93–124; exon 4, 125–182; exon 5, 183–208; and exon 6, 209–416) were pulled-down from HEK293T cells cotransfected with HA-SRF. SRF levels in IPMK pull-down were determined by immunoblotting.
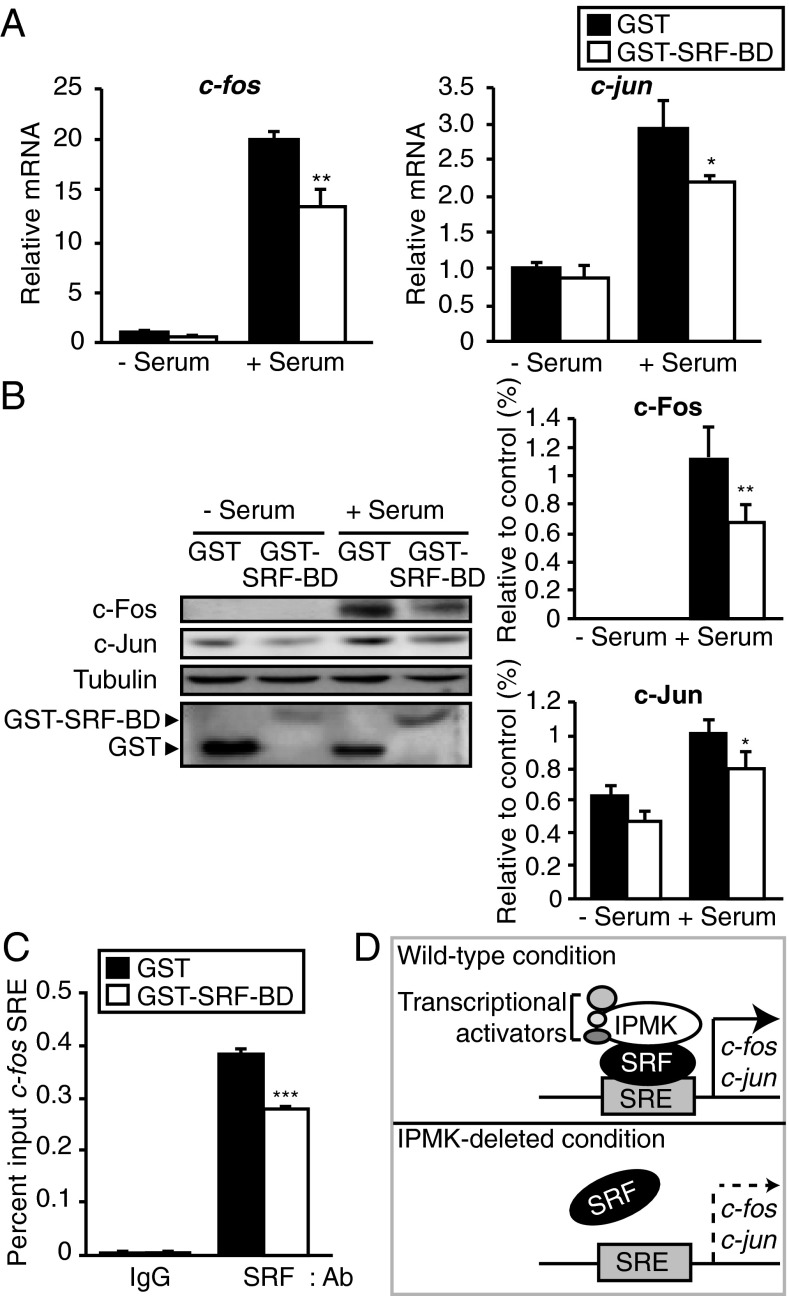
The striking influences of IPMK upon serum activation of immediate early genes and the interaction of IPMK with SRF imply that regulation of these genes is dependent upon IPMK–SRF associations. However, immediate early genes are also regulated by influences of various other factors on SRF. To investigate the importance of IPMK–SRF binding for gene regulation, we mapped SRF binding sites on IPMK and developed a dominant-negative construct to prevent such binding. The mapping analysis reveals that IPMK–SRF binding is largely determined by exons 3, 4, and 6 of IPMK (Fig. 3F). We constructed a dominant-negative structure comprising amino acids 93–182 of IPMK, which we designate the SRF binding domain (SRF-BD). In HEK293 cells, overexpression of SRF-BD significantly diminishes serum-elicited induction of c-fos, c-jun, and cyr61 RNA (Fig. 4A and Fig. S2B) and protein (Fig. 4B). This action of SRF-BD is specific as overexpression of exon1 of IPMK fails to alter serum stimulation of c-fos, c-jun, and cyr61 mRNA (Fig. S7A) or protein levels of c-Fos and c-Jun (Fig. S7B). Moreover, SRF-BD does not alter cell viability (Fig. S3B). We mapped SRF for areas mediating binding to IPMK (Fig. S8). The DNA binding domain of SRF (the MADS domain) appears to be a principal determinant of IPMK binding (Fig. S8). Addition of a partial MADS sequence (amino acids 180–205) to the C-terminal portion of SRF enhances its binding to IPMK (Fig. S8).
Fig. 4.
Dominant-negative IPMK peptide interferes with immediate early gene expression. (A) The mRNA levels of the SRF target genes c-jun and c-fos as measured via qPCR. GST or GST-SRF-BD was transfected in HEK293 cells. Cells were deprived of serum for 12 h and stimulated with 10% FBS for 30 min. For immunoblot analysis in B, cells were stimulated with 10% FBS for 1 h. (C) The ChIP assay was used to assess SRF recruitment to the c-fos promoter SRE by using HEK293 cells overexpressing GST or GST-SRF-BD. Bars represent mean ± SE (A and C) or ± SD (B) (n = 3–5). *P < 0.05; **P < 0.01; ***P < 0.001. (D) A model depicting regulation of immediate early gene expression by IPMK. IPMK binds to SRF, enhancing its stable interaction with DNA. In the absence of IPMK, SRF dissociates from its DNA targets, at least partially. By the loss of signals from SRF and its transcriptional regulators, immediate early genes cannot be fully transcribed.
Our experiments have indicated that IPMK and SRF bind together. If this binding is physiologically relevant, then displacement of endogenous IPMK from SRF should disrupt signaling to SRE. Accordingly, we monitored influences of SRF-BD upon SRF interactions with c-fos SRE in ChIP assays (Fig. 4C). SRF-BD significantly diminishes SRF–SRE interactions, indicating that endogenous IPMK participates with SRF in binding SRE.
Discussion
In the present study, we demonstrate that IPMK is a transcriptional coactivator for SRF and thereby regulates a wide range of immediate early genes whose expression is drastically reduced in IPMK-deleted preparations. IPMK acts by binding to SRF and enhancing its interactions with SRE. The regulation by IPMK of SRF targets is determined by IPMK-SRF binding, as dominant-negative constructs of IPMK block IPMK–SRF binding and prevent immediate early gene induction. The in vivo importance of these findings is highlighted by the diminution of immediate early gene activation in the brains of IPMK-deleted mice.
CREB is another well-characterized transcription factor that augments the induction of immediate early genes (28–33). However, there are distinctions between influences of IPMK upon CREB and SRF. For instance, cyr61, which is down-regulated by CREB, is induced by serum via SRF in an IPMK-dependent fashion. CREB and SRF can also be discriminated in terms of signaling systems that influence their disposition. For instance, CREB is classically activated by cAMP as well as cGMP (34–40) whereas SRF is inhibited by cGMP (41, 42).
Besides impacting SRF, IPMK displays fairly direct regulatory influences on at least two other transcription regulators, and, as with SRF, these actions do not require IPMK catalytic activity. Thus, IPMK binds directly to p53, serving as a transcriptional coactivator (14). In this instance, IPMK noncatalytically enhances the transcription of genes that mediate the p53 cell death program. IPMK also binds CBP/p300, a transcriptional coactivator of CREB, and noncatalytically augments the readout of CREB-regulated genes (16). Different conformations of IPMK might mediate these diverse binding events. Thus, mapping studies indicate that the middle portion of IPMK interacts with SRF (amino acids 93–182) and p53 (amino acids 125–184) (14). By contrast, the N-terminal 75 amino acids of IPMK are responsible for binding to CBP (16). Factors determining the dynamics of IPMK binding differentially to these three proteins are unclear. Conceivably, modifications of IPMK, such as phosphorylation, influence binding to one or the other of them.
SRF is generally regarded as a relatively weak transcription factor. However, its actions are enhanced substantially by a number of coactivators besides IPMK (21, 43–47). Most of these are recruited to regulatory elements in genes adjacent to binding sites for SRF (43–46, 48–50) whereas some of these cofactors, such as GATA-4 and p65, potentiate transcription of SRF targets by SRF binding only and, like IPMK, do not require direct interaction with the regulatory elements of target genes (47, 51–53). However, except for IPMK, all known cofactors of SRF have DNA binding ability and also participate in transcriptional activation of their target genes independent of SRF. Thus, IPMK is a cofactor of SRF that influences the ability of SRF to activate the expression of immediate early genes but lacks any intrinsic DNA binding ability.
Two families of SRF coactivators have been particularly well-characterized. One is the ETS-domain-containing ternary complex factor (TCF), which includes Elk-1, SAP-1, and SAP-2/Net (18, 21, 22, 24, 26, 46). Growth-factor stimulation and mitogen-activated protein kinase signaling lead to the activation of TCFs, resulting in the activation of growth-related genes (26, 54, 55). IPMK deletion does not influence Elk-1 activity. However, it is conceivable that SRF–Elk-1 multimerization is impacted by IPMK. Such interactions are thought to play a role in SRF-associated transactivation. The myocardin family, another group of SRF coactivators, predominantly influences gene expression in the heart (44, 45, 56–58). Indeed, cardiac function is one of the best-studied targets for SRF as embryonic lethality in SRF-deleted mice primarily reflects defects in heart function and vascular circulation (59). Our findings that IPMK regulates SRF in brain tissue fit well with studies of Ginty and co-workers demonstrating major defects in long-term depression upon SRF inhibition (60) and impaired axonal growth and synaptic plasticity of SRF gene knockout mice (61, 62). Synaptic plasticity is also determined by CREB. However, SRF deletion selectively influences synaptic plasticity without affecting neuronal viability whereas CREB deletion leads to major neuronal degeneration (62).
The transcriptional influences of IPMK add to a surprisingly broad range of other functions, much more than are encountered with most proteins. Thus, IPMK displays two distinct catalytic activities. It is a major inositol phosphate kinase, the key physiologic generator of IP4 and IP5, which are the requisite precursors for the inositol pyrophosphates such as IP7 (1, 2). Additionally, IPMK is a lipid kinase with physiologic PI3-kinase activity, acting in conjunction with the p85/p110 PI3-kinase to activate the Akt signaling cascade (4, 5). These two kinase activities of IPMK appear to be reciprocal, suggesting that posttranslational modifications of IPMK mediate transitions of the enzyme between these two functions (4). The present study describes a noncatalytic role of IPMK as a transcriptional coactivator for SRF, analogous to such influences upon p53 (14) and CBP (16). Similar transcriptional regulatory activities occur in yeast, where IPMK regulates genes involved in arginine and phosphate disposition, actions that also appear to be independent of catalytic activity (63). However, another noncatalytic action of IPMK is evident in its binding to mTOR to stabilize the mTORC1 complex and enhance protein translation (6).
Materials and Methods
Mice.
To generate forebrain-specific IPMK knockout mice, Ipmkfl/fl mice (6) were mated with CaMKIIα-Cre mice (line 93) (64) obtained from EUCOMM. Male mice were used for experiments at 10–12 wk of age. All mice were housed with a 12 h light–dark schedule and received food and water ad libitum. Animal protocols were performed in accordance with guidelines approved by the Korea Advanced Institute of Science and Technology-Animal Care and Use Committee.
Chromatin Immunoprecipitaton Assay.
Intact cells were treated with 1.5 mM dithiobis [succinimidylpropionate] (Sigma) for 30 min at room temperature to cross-link protein complexes, followed by 1% formaldehyde for 10 min at room temperature to covalently link proteins to DNA. Quenching reaction was done by 50 mM glycine for 15 min. For in vivo experiments, 100 mg of cortical tissue lysates were used per chromatin immunoprecipitaton (ChIP) assay. Samples were chopped into small pieces, fixed in 1% (vol/vol) formaldehyde, quenched with 0.125 M glycine, and homogenized by using a glass homogenizer in cold PBS with protease inhibitor mixtures. Homogenized tissues were centrifuged at 100 × g for 10 min. Steps for immunoprecipitation, elution, and reverse cross-linking were performed by using a ChIP assay kit according to the manufacturer’s protocol (Millipore). ChIP primers were as follows: mouse c-fos promoter SRE, 5′-CGTCAATCCCTCCCTCCTTT-3′, 5′-CCGTCTTGGCATACATCTTT-3′; mouse c-fos promoter SP1 binding, 5′-GTTGAAAGCCTGGGGCGTAG-3′, 5′-GGAGTAGTAGGCGCCTCAGC-3′; human c-fos promoter SRE, 5′-GGATGTCCATATTAGGACATCT-3′, 5′-AGATGTCCTAATATGGACATCC-3; mouse p21 promoter, 5′-GTGGCTCTGATTGGCTTTCTG-3′, 5′-CTGAAAACAGGCAGCCCAAG-3′.
Microarray.
Microarray experiments were carried out by Macrogen. Total RNA was extracted from serum-stimulated MEFs by using Tri reagent according to the manufacturer’s protocol. After processing with DNase digestion and clean-up procedures, RNA samples were quantified, aliquot and stored at −80 °C until use. For quality control, RNA purity and integrity were evaluated by denaturing gel electrophoresis, OD 260/280 ratio, and analyzed on an Agilent 2100 Bioanalyzer (Agilent Technologies). Total RNA was amplified and purified using the Ambion Illumina RNA amplification kit (Ambion) to yield biotinylated cRNA according to the manufacturer’s instructions. Briefly, 550 ng of total RNA was reverse-transcribed to cDNA using a T7 oligo (dT) primer. Second-strand cDNA was synthesized, in vitro transcribed, and labeled with biotin-NTP. After purification, the cRNA was quantified using the ND-1000 Spectrophotometer (NanoDrop). Seven hundred fifty nanograms of labeled cRNA samples were hybridized to each mouse-6 expression bead array for 16–18 h at 58 °C, according to the manufacturer's instructions (MouseWG-6 v2.0 Expression BeadChip; Illumina). Detection of array signal was carried out using Amersham fluorolink streptavidin-Cy3 (GE Healthcare Bio-Sciences) following the bead array manual. Arrays were scanned with an Illumina bead array Reader confocal scanner according to the manufacturer's instructions. The quality of hybridization and overall chip performance were monitored by visual inspection of both internal quality-control checks and the raw scanned data. Raw data were extracted using the software provided by the manufacturer [Illumina GenomeStudio v2011.1 (Gene Expression Module v1.9.0)]. The statistical significance of the expression data was determined using Fold change and LPE (Local Pooled Error) test in which the null hypothesis was that no difference exists between two groups. The false discovery rate was controlled by adjusting P values using the Benjamini–Hochberg algorithm. All data analysis and visualization of differentially expressed genes were conducted using R 2.15.1 (www.r-project.org).
Image Quantification and Statistical Analysis.
Immunoblot results were quantified by using ImageJ software. Data are presented as means ± SE (qPCR) or ± SD (immunoblot) from at least three independent experiments. Data were analyzed with Student t test.
Supplementary Material
Acknowledgments
We thank R. Barrow and other members of the S.K. and S.H.S. laboratory for their helpful discussions. This work was supported by National Research Foundation of Korea Grants NRF-2012R1A1A1040873, NRF-2012M3A9C7050093, and NRF-2013M3C7A1056102 (to S.K.); the Korea Advanced Institute of Science and Technology High Risk High Return Project (S.K.); a POSCO TJ Park Science Fellowship (to S.K.); a Korean Government Global PhD Fellowship (to E.K.); and National Institutes of Health Grant MH-18501 (to S.H.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320171110/-/DCSupplemental.
References
- 1.Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9(22):1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 2.Saiardi A, et al. Mammalian inositol polyphosphate multikinase synthesizes inositol 1,4,5-trisphosphate and an inositol pyrophosphate. Proc Natl Acad Sci USA. 2001;98(5):2306–2311. doi: 10.1073/pnas.041614598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287(5460):2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 4.Maag D, et al. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc Natl Acad Sci USA. 2011;108(4):1391–1396. doi: 10.1073/pnas.1017831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Resnick AC, et al. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc Natl Acad Sci USA. 2005;102(36):12783–12788. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S, et al. Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell Metab. 2011;13(2):215–221. doi: 10.1016/j.cmet.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bercy J, Dubois E, Messenguy F. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene. 1987;55(2-3):277–285. doi: 10.1016/0378-1119(87)90287-3. [DOI] [PubMed] [Google Scholar]
- 8.Dubois E, Bercy J, Messenguy F. Characterization of two genes, ARGRI and ARGRIII required for specific regulation of arginine metabolism in yeast. Mol Gen Genet. 1987;207(1):142–148. doi: 10.1007/BF00331501. [DOI] [PubMed] [Google Scholar]
- 9.Bechet J, Greenson M, Wiame JM. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12(1):31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 10.Messenguy F, Dubois E. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13(4):2586–2592. doi: 10.1128/mcb.13.4.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Bakkoury M, Dubois E, Messenguy F. Recruitment of the yeast MADS-box proteins, ArgRI and Mcm1 by the pleiotropic factor ArgRIII is required for their stability. Mol Microbiol. 2000;35(1):15–31. doi: 10.1046/j.1365-2958.2000.01665.x. [DOI] [PubMed] [Google Scholar]
- 12.Dubois E, Messenguy F. In vitro studies of the binding of the ARGR proteins to the ARG5,6 promoter. Mol Cell Biol. 1991;11(4):2162–2168. doi: 10.1128/mcb.11.4.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christ C, Tye BK. Functional domains of the yeast transcription/replication factor MCM1. Genes Dev. 1991;5(5):751–763. doi: 10.1101/gad.5.5.751. [DOI] [PubMed] [Google Scholar]
- 14.Xu R, et al. Inositol polyphosphate multikinase is a coactivator of p53-mediated transcription and cell death. Sci Signal. 2013;6(269):ra22. doi: 10.1126/scisignal.2003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu R, Snyder SH. Gene transcription by p53 requires inositol polyphosphate multikinase as a co-activator. Cell Cycle. 2013;12(12):1819–1820. doi: 10.4161/cc.25119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu R, et al. Inositol polyphosphate multikinase is a transcriptional coactivator required for immediate early gene induction. Proc Natl Acad Sci USA. 2013;110(40):16181–16186. doi: 10.1073/pnas.1315551110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46(4):567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- 18.Shaw PE, Schröter H, Nordheim A. The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c-fos promoter. Cell. 1989;56(4):563–572. doi: 10.1016/0092-8674(89)90579-5. [DOI] [PubMed] [Google Scholar]
- 19.Kim KH, et al. Expression of angiogenic factor Cyr61 during neuronal cell death via the activation of c-Jun N-terminal kinase and serum response factor. J Biol Chem. 2003;278(16):13847–13854. doi: 10.1074/jbc.M210128200. [DOI] [PubMed] [Google Scholar]
- 20.Dobroff AS, et al. Silencing cAMP-response element-binding protein (CREB) identifies CYR61 as a tumor suppressor gene in melanoma. J Biol Chem. 2009;284(38):26194–26206. doi: 10.1074/jbc.M109.019836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janknecht R, Nordheim A. Elk-1 protein domains required for direct and SRF-assisted DNA-binding. Nucleic Acids Res. 1992;20(13):3317–3324. doi: 10.1093/nar/20.13.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latinkić BV, Zeremski M, Lau LF. Elk-1 can recruit SRF to form a ternary complex upon the serum response element. Nucleic Acids Res. 1996;24(7):1345–1351. doi: 10.1093/nar/24.7.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treisman R. The serum response element. Trends Biochem Sci. 1992;17(10):423–426. doi: 10.1016/0968-0004(92)90013-y. [DOI] [PubMed] [Google Scholar]
- 24.Treisman R, Marais R, Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 1992;11(12):4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gille H, et al. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14(5):951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73(2):381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 27.Frederick JP, et al. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc Natl Acad Sci USA. 2005;102(24):8454–8459. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4(4):477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 29.Sheng M, McFadden G, Greenberg ME. Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron. 1990;4(4):571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- 30.Greenberg ME, Thompson MA, Sheng M. Calcium regulation of immediate early gene transcription. J Physiol Paris. 1992;86(1-3):99–108. doi: 10.1016/s0928-4257(05)80013-0. [DOI] [PubMed] [Google Scholar]
- 31.Shaywitz AJ, Greenberg ME. CREB: A stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 32.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35(4):605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 33.Bébien M, et al. Immediate-early gene induction by the stresses anisomycin and arsenite in human osteosarcoma cells involves MAPK cascade signaling to Elk-1, CREB and SRF. Oncogene. 2003;22(12):1836–1847. doi: 10.1038/sj.onc.1206334. [DOI] [PubMed] [Google Scholar]
- 34.Nichols M, et al. Phosphorylation of CREB affects its binding to high and low affinity sites: Implications for cAMP induced gene transcription. EMBO J. 1992;11(9):3337–3346. doi: 10.1002/j.1460-2075.1992.tb05412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dash PK, Karl KA, Colicos MA, Prywes R, Kandel ER. cAMP response element-binding protein is activated by Ca2+/calmodulin- as well as cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1991;88(11):5061–5065. doi: 10.1073/pnas.88.11.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto KK, Gonzalez GA, Biggs WH, 3rd, Montminy MR. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988;334(6182):494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59(4):675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 38.Ciani E, Guidi S, Bartesaghi R, Contestabile A. Nitric oxide regulates cGMP-dependent cAMP-responsive element binding protein phosphorylation and Bcl-2 expression in cerebellar neurons: Implication for a survival role of nitric oxide. J Neurochem. 2002;82(5):1282–1289. doi: 10.1046/j.1471-4159.2002.01080.x. [DOI] [PubMed] [Google Scholar]
- 39.Lu YF, Kandel ER, Hawkins RD. Nitric oxide signaling contributes to late-phase LTP and CREB phosphorylation in the hippocampus. J Neurosci. 1999;19(23):10250–10261. doi: 10.1523/JNEUROSCI.19-23-10250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gudi T, Casteel DE, Vinson C, Boss GR, Pilz RB. NO activation of fos promoter elements requires nuclear translocation of G-kinase I and CREB phosphorylation but is independent of MAP kinase activation. Oncogene. 2000;19(54):6324–6333. doi: 10.1038/sj.onc.1204007. [DOI] [PubMed] [Google Scholar]
- 41.Pilz RB, Casteel DE. Regulation of gene expression by cyclic GMP. Circ Res. 2003;93(11):1034–1046. doi: 10.1161/01.RES.0000103311.52853.48. [DOI] [PubMed] [Google Scholar]
- 42.Gudi T, et al. cGMP-dependent protein kinase inhibits serum-response element-dependent transcription by inhibiting rho activation and functions. J Biol Chem. 2002;277(40):37382–37393. doi: 10.1074/jbc.M204491200. [DOI] [PubMed] [Google Scholar]
- 43.Chen CY, Schwartz RJ. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Mol Cell Biol. 1996;16(11):6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang D, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105(7):851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 45.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113(3):329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 46.Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68(3):597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- 47.Grueneberg DA, Natesan S, Alexandre C, Gilman MZ. Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor. Science. 1992;257(5073):1089–1095. doi: 10.1126/science.257.5073.1089. [DOI] [PubMed] [Google Scholar]
- 48.Dalgleish P, Sharrocks AD. The mechanism of complex formation between Fli-1 and SRF transcription factors. Nucleic Acids Res. 2000;28(2):560–569. doi: 10.1093/nar/28.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta M, et al. Physical interaction between the MADS box of serum response factor and the TEA/ATTS DNA-binding domain of transcription enhancer factor-1. J Biol Chem. 2001;276(13):10413–10422. doi: 10.1074/jbc.M008625200. [DOI] [PubMed] [Google Scholar]
- 50.Natesan S, Gilman M. YY1 facilitates the association of serum response factor with the c-fos serum response element. Mol Cell Biol. 1995;15(11):5975–5982. doi: 10.1128/mcb.15.11.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franzoso G, et al. Activation of the serum response factor by p65/NF-kappaB. EMBO J. 1996;15(13):3403–3412. [PMC free article] [PubMed] [Google Scholar]
- 52.Belaguli NS, et al. Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators. Mol Cell Biol. 2000;20(20):7550–7558. doi: 10.1128/mcb.20.20.7550-7558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang DF, et al. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev Cell. 2003;4(1):107–118. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 54.Janknecht R, Ernst WH, Pingoud V, Nordheim A. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 1993;12(13):5097–5104. doi: 10.1002/j.1460-2075.1993.tb06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miano JM. Serum response factor: Toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35(6):577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 56.Wang DZ, et al. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci USA. 2002;99(23):14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuwahara K, Barrientos T, Pipes GC, Li S, Olson EN. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol Cell Biol. 2005;25(8):3173–3181. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaromytidou AI, Miralles F, Treisman R. MAL and ternary complex factor use different mechanisms to contact a common surface on the serum response factor DNA-binding domain. Mol Cell Biol. 2006;26(11):4134–4148. doi: 10.1128/MCB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arsenian S, Weinhold B, Oelgeschläger M, Rüther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17(21):6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith-Hicks C, et al. SRF binding to SRE 6.9 in the Arc promoter is essential for LTD in cultured Purkinje cells. Nat Neurosci. 2010;13(9):1082–1089. doi: 10.1038/nn.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wickramasinghe SR, et al. Serum response factor mediates NGF-dependent target innervation by embryonic DRG sensory neurons. Neuron. 2008;58(4):532–545. doi: 10.1016/j.neuron.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramanan N, et al. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat Neurosci. 2005;8(6):759–767. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- 63.Bosch D, Saiardi A. Arginine transcriptional response does not require inositol phosphate synthesis. J Biol Chem. 2012;287(45):38347–38355. doi: 10.1074/jbc.M112.384255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Minichiello L, et al. Essential role for TrkB receptors in hippocampus-mediated learning. Neuron. 1999;24(2):401–414. doi: 10.1016/s0896-6273(00)80853-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.