Significance
One of the central goals of developmental biology and medicine is to ascertain the relationships between the genotype and phenotype of cells. Single-cell transcriptome analysis represents a powerful strategy to reach this goal. We advance these strategies to single nuclei from neural progenitor cells and dentate gyrus tissue, from which it is very difficult to recover intact cells. This provides a unique means to carry out RNA sequencing from individual neurons that avoids requiring isolation of single-cell suspensions, eliminating potential changes in gene expression due to enzymatic-cell dissociation methods. This method will be useful for analysis of processes occurring in the nucleus and for gene-expression studies of highly interconnected cells such as neurons.
Keywords: nuclear RNA, deep sequencing, whole cell RNA
Abstract
It has recently been established that synthesis of double-stranded cDNA can be done from a single cell for use in DNA sequencing. Global gene expression can be quantified from the number of reads mapping to each gene, and mutations and mRNA splicing variants determined from the sequence reads. Here we demonstrate that this method of transcriptomic analysis can be done using the extremely low levels of mRNA in a single nucleus, isolated from a mouse neural progenitor cell line and from dissected hippocampal tissue. This method is characterized by excellent coverage and technical reproducibility. On average, more than 16,000 of the 24,057 mouse protein-coding genes were detected from single nuclei, and the amount of gene-expression variation was similar when measured between single nuclei and single cells. Several major advantages of the method exist: first, nuclei, compared with whole cells, have the advantage of being easily isolated from complex tissues and organs, such as those in the CNS. Second, the method can be widely applied to eukaryotic species, including those of different kingdoms. The method also provides insight into regulatory mechanisms specific to the nucleus. Finally, the method enables dissection of regulatory events at the single-cell level; pooling of 10 nuclei or 10 cells obscures some of the variability measured in transcript levels, implying that single nuclei and cells will be extremely useful in revealing the physiological state and interconnectedness of gene regulation in a manner that avoids the masking inherent to conventional transcriptomics using bulk cells or tissues.
Methods for measuring gene expression in single cells have been limited to reverse transcription (RT)–PCR for candidate genes (which has primer design restrictions) and to microarray analysis (which has a low dynamic range and excludes the ability to discover new transcripts or splice isoforms) (1, 2). Improvements in cDNA synthesis from single cells (3–5) have enabled RNA sequencing (RNA-seq) (6). In the protocol followed here (7–9), cDNA is synthesized from the 0.1–1.0 pg of mRNA in one cell (10, 11) by RT from poly(dT) primers, followed by second-strand cDNA synthesis by Taq DNA polymerase and PCR amplification of the cDNA to generate sufficient template for use in sequencing. Expression levels for a majority of the 10,000–20,000 genes expressed in one cell (5, 12) can be derived from read depth, and the sequences reveal genotype and splice variants.
Single-cell transcriptomics is a powerful tool to investigate gene expression at the most fundamental level of the individual cell. However, in tissues where intact cells are difficult to recover, such as highly interconnected neurons, an approach using a single nucleus becomes attractive. Previous studies demonstrated mRNA recovery from bulk nuclei (13–17). We have shown that there is sufficient mRNA in bulk nuclei (13) or in pooled nuclei isolated by fluorescence activated sorting (18) for transcript profiling using microarrays, leading to accurate identification of promoters having cell type-specific activities. Other work has shown that polyadenylated (PolyA+) transcripts within the nuclear compartment of eukaryotic cells can be measured to 5-nucleotide resolution (19). Here, we report whole transcriptome sequencing from a single nucleus. Transcript levels in nuclear and total cellular RNA from a mouse neural progenitor cell (NPC) line were generally similar, with only a small minority differing in abundance between the nucleus and the cytoplasm. Currently, single-cell transcriptomics from tissues requires proteolytic dissociation of cells at elevated temperatures which could potentially perturb transcriptional activity. Alternatively, laser capture microdissection might be used to capture single cells (20), however, mechanical damage to the cell and its dendritic and axonal processes is likely. Furthermore, tissue-fixation methods typically used would be expected to interfere with synthesis of full-length cDNAs. In contrast, RNA-seq from single nuclei will allow gene-expression studies directly from post mortem tissues that are maintained at 4 °C throughout the process.
Results
cDNA Synthesis from a Single Nucleus.
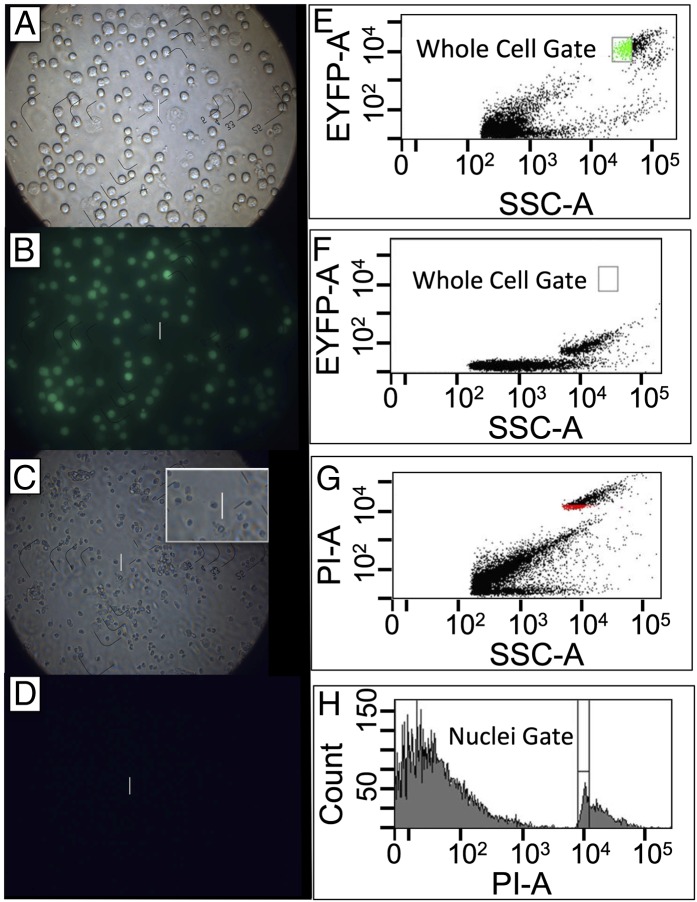
RNA-seq was done using sorted NPCs labeled with cytoplasmic Enhanced Yellow Fluorescent Protein (EYFP) (ref. 21; Methods and Fig. 1 A, B, and E), and using sorted nuclei (Fig. 1 C and D) labeled with propidium iodide (PI) (Fig. 1 G and H) but lacking EYFP fluorescence (Fig. 1F). Confirmation of the precision of sorting was also obtained using epifluorescence microscopy (SI Appendix, Fig. S1). Double-stranded cDNA was prepared (SI Appendix, Methods S2) from eight individual nuclei and eight individual whole cells. Transcript levels for five housekeeping genes [Beta-actin (Actb), glyceraldehyde-3-phosphate dehydrogenase (Gapdh), heat shock protein 90 alpha class B member 1 (HSP90ab1), and ribosomal protein L13 (Rpl13)] (22) and three NPC tissue-specific genes [fatty acid binding protein 7 (Fabp7), H2A histone family member Z (H2afz), and vimentin (Vim)] (23) were measured by TaqMan quantitative (q)PCR (SI Appendix, Methods S2). Wherever possible (Hsp90ab1, Eef2, Vim, and Fabp7), primers spanned exon–exon junctions ensuring detection was from cDNA rather than from genomic DNA.
Fig. 1.
Fluorescence-activated sorting of whole cells and nuclei. (A) NPCs visualized by phase-contrast microscopy (100×). (B) NPCs visualized by epifluorescence microscopy. (C) NPC nuclei visualized by phase-contrast microscopy (100×); demonstrates expected size and morphology. (D) NPC nuclei, examined by epifluorescence microscopy, lack EYFP fluorescence. The white line is a 25-µm calibration ruler. (E) Biparametric flow cytometric analysis of EYFP fluorescence, detected using a 525-nm/25-nm band pass filter, versus side scatter (area signals). The intact cells form a discrete cluster, well separated from cellular debris. The gate (green) designates the region used as the sort window for isolation of single cells. Intact cells lacked red fluorescence, detected using a 620-nm/40-nm band pass filter, when PI was included in the medium. (F) Biparametric flow analysis of PI-stained nuclei using the same instrument settings as in E. The nuclei form a discrete cluster lacking EYFP fluorescence. (G) Biparametric-flow cytometric analysis of PI-stained nuclei, examining red fluorescence, detected using a 620-nm/40-nm band pass filter, versus forward-angle light scatter. The nuclei form a discrete cluster, well separated from cellular debris. (H) Uniparametric display of the 620-nm fluorescence emission from PI-stained nuclei. The window used for sorting single nuclei surrounds the major peak within the nuclear distribution to exclude nuclear aggregates (doublets, triplets, etc.).
Single nuclei contained sufficient mRNA for analysis although the amounts were less than for whole cells based on qPCR (Fig. 2 and SI Appendix, Fig. S2). Controls established that cDNA was not derived from contaminating free nucleic acids present in the NPC culture media; polystyrene fluorescent microspheres were added to either the whole cell or the nuclear preparations and then recovered by FACS (SI Appendix, Fig. S3). The milieu isolated along with the microspheres did not support cDNA synthesis (SI Appendix, Table S1).
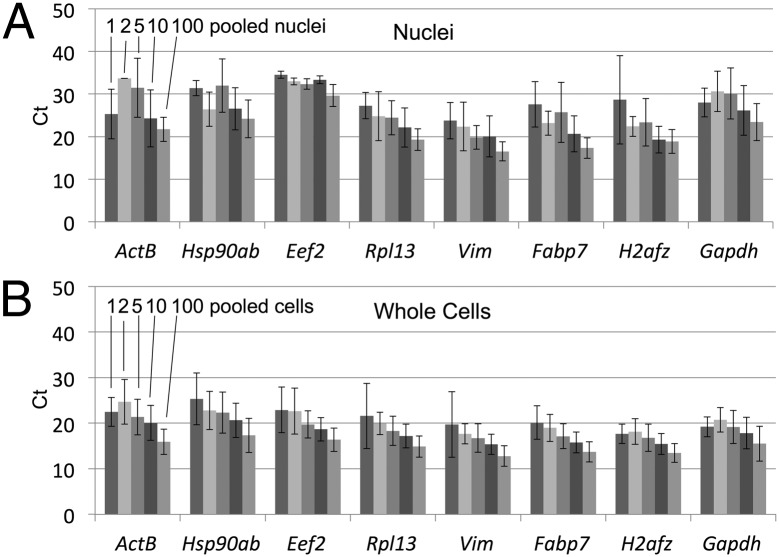
Fig. 2.
Quantification of transcript levels in isolated cells and nuclei. Average cycle threshold (Ct) values for eight genes for mouse NPC nuclei (A) and whole cells (B) measured by TaqMan qPCR. Eight replicates of 1, 2, 5, 10, or 100 cells (indicated only for the ActB gene in this figure) were FACS sorted into individual wells in a 384-well plate and used for cDNA synthesis and amplification by PCR. The PCR products were diluted 10-fold and tested for expression of five housekeeping genes (Actb, Eef2, Gapdh, Hsp90ab1, and Rpl13) and three NPC tissue-specific genes (Fabp7, H2afz, and Vim). Ct values of less than 50 were recorded. The number detected for each set of 8 replicate nuclei or cells (error bars) was as follows: (nuclei) Actb—3, 1, 7, 7, and 8 (for 1 nucleus, 2 nuclei, 5 nuclei, 10 nuclei, and 100 nuclei, respectively); Hsp90ab1—4, 2, 7, 8, and 8; Rpl13—7, 8, 8, 8, and 8; Eef2—2, 3, 4, 4, and 8; Vim—4, 6, 7, 8, and 8; Fabp7—5, 4, 8, 8, and 8; H2afz—2, 3, 5, 8, and 8; and Gapdh—6, 5, 7, 8, and 8. (Whole cells) Actb—5, 8, 8, 8, and 8 (for 1 cell, 2 cells, 5 cells, 10 cells, and 100 cells, respectively); Hsp90ab1—6, 8, 8, 8, and 8; Rpl13—6, 8, 8, 8, and 8; Eef2—6, 7, 8, 8, and 8; Vim—7, 6, 8, 8, and 8; Fabp7—6, 8, 8, 8, and 8; H2afz—5, 8, 8, 8, and 8; and Gapdh—5, 8, 8, 8, and 8. The error bars represent the SD of the average Ct measured for each gene locus. The presence of some cells lacking expression for these genes is expected, based on the stochastic activation of gene expression and the current state of knowledge concerning transcriptional bursting within single cells.
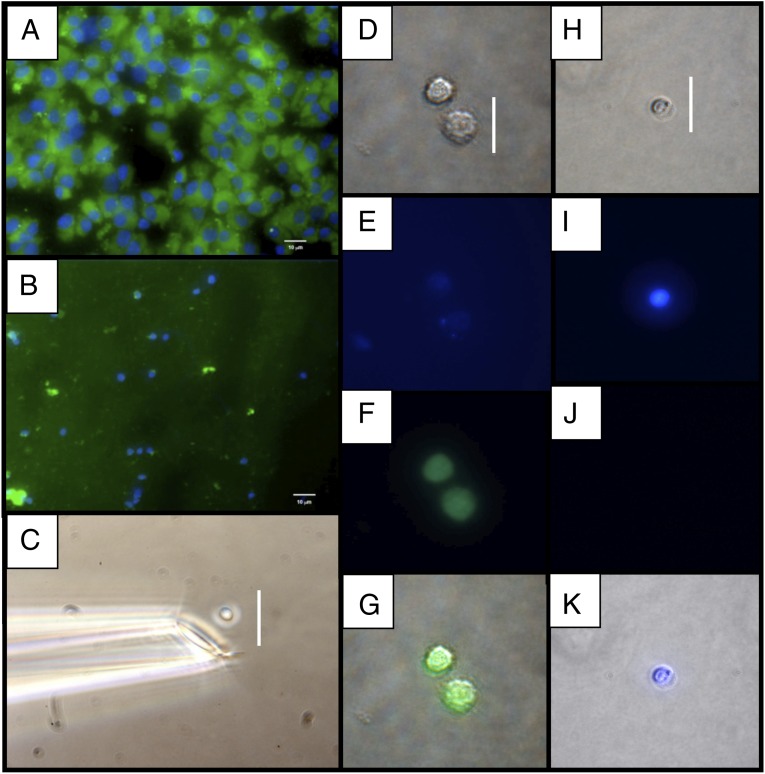
Single NPC whole cells (Fig. 3 A and D–G) and nuclei (Fig. 3 B and H–K) were also isolated by micromanipulation (Fig. 3C). Double-stranded cDNA was prepared from three individual nuclei, three individual cells, and pools of 10 and 100 flow-sorted cells to investigate the effect of averaging transcriptional profiles. Based on qPCR for the Gapdh transcript, cDNA was synthesized from all cells and nuclei, but not from the final PBS wash used to remove contaminating mRNA or DNA (SI Appendix, Fig. S4).
Fig. 3.
Isolation of mouse NPCs and nuclei by micromanipulation. (A) Dispersed NPC whole cells. All cells have an intense EYFP signal in the cytoplasm and a DAPI-stained nucleus. (B) Nuclei purified by density-gradient centrifugation. (C) Micromanipulation of a nucleus with a glass capillary. Before micromanipulation, cells and nuclei were visualized by phase-contrast microscopy, and the DAPI and EYFP signals determined by epifluorescence microscopy. (D–G) Whole cells. (H–K) Nuclei. (D and H) Phase contrast. (E and I) DAPI epifluorescence. (F and J) EYFP epifluorescence. (G and K) Stacked phase contrast plus epifluorescence images. (Scale bars: 10 μm in A and B and 25 μm in C, D, and H.)
RNA-Seq from Single Nuclei and Single Cells.
The cDNA preparations (SI Appendix, Fig. S4) generated 942 million total sequencing reads (SI Appendix, Methods S3) from 18 samples (SI Appendix, Table S2) with single nuclei and single cells averaging 47 and 56 million reads, respectively. An average of 47% of the total reads uniquely mapped to the Mus musculus genome [assembly MGSCv37 (mm9), to which the EYFP transgene transcript sequence was added], and 46% mapped to exonic regions (SI Appendix, Fig. S5), similar to published data (5). For 50 NPC markers and housekeeping transcripts analyzed in detail (SI Appendix, Table S3), only exons were detected (for example, Vim and Eef2 in SI Appendix, Fig. S6), demonstrating that most or all of the nuclear transcripts are rapidly spliced before cDNA synthesis, and that the intronic RNA reads detected (SI Appendix, Fig. S5) do not simply represent nonspecific transcriptional noise which would be expected to occur as a background across all genes. Although we lack a full understanding of the intronic reads, we conclude that all genes with deep exon coverage and lacking introns can be reliably analyzed. An average of 27% unique reads mapped to intronic regions, possibly identifying previously unknown splice isoforms or transcribed noncoding RNA (ncRNA) (24). Alternatively, these may result from poly(dT) priming from poly(dA) tracts in genomic DNA (SI Appendix, Fig. S7), but can be identified bioinformatically. An average of 20,065 and 20,243 unique transcripts were detected in the cell and nucleus samples, respectively, which is comparable to previous results for single cells (5). Unique mapping of 10 million, 50-base reads is sufficient to account for the majority of nonsplice variants (5, 8). Others (25) have shown that sequencing to a depth of 3 million reads was sufficient to identify the majority of genes in various human and mouse tissues, and sequencing to greater depth did not increase transcript detection. Another study (6) indicated that 10–30 million 25-bp reads are sufficient to map unique sites in the mouse genome. The transcripts detected in our study represent ∼78% of the manually curated 27,553 unique protein-coding transcripts in the NCBI Reference Sequence Database (RefSeq). 23% of unique reads mapped to published (9) intergenic sequences found in single cells (SI Appendix, Fig. S5). Possibly these regions are transcriptionally active (24), or the reads may represent artifacts of RT priming from genomic DNA.
The distribution of reads-per-kilobase-of-transcript-per-million mapped (RPKM) values was determined for a series of bins between 0 and 100,000 (SI Appendix, Fig. S8). For both nuclei and cells, almost all of the mapped exons have an RPKM (6) value between 0.1 and 1,000, a dynamic range of four logarithms, and sensitivity approaching a single transcript per cell. The coverage for introns is higher in nuclei relative to cells (possibly due to unprocessed mRNA in the nucleus) with 40% mapped around 1 and 0.1 RPKM, respectively, consistent with the Encyclopedia of DNA Elements (ENCODE) bulk RNA data (19), where 34% of the total nuclear transcriptome mapped to intronic regions compared with 22% in the cytoplasm. The intron- and intergenic-coverage distribution in single cells are similar suggesting that the RPKM intervals of high confidence lie at a threshold of 0.1. Further, the percent of transcripts in the lowest bin of expression (RPKM of 0.1–1.0) is not increased as more cells or nuclei are pooled, indicating that detection sensitivity is sufficient for single cells or single nuclei, and that ∼50 million reads is adequate to uniquely map to almost all of the available reference transcripts.
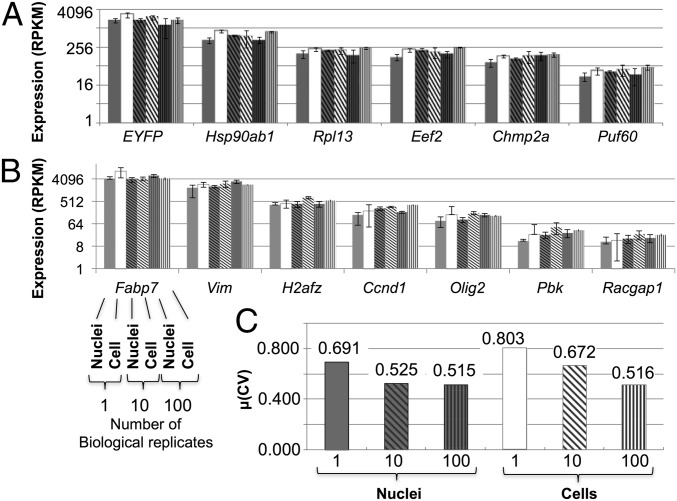
One concern was that use of nuclei might introduce sources of experimental variation, such as increased mRNA degradation accompanying cell homogenization or mRNA loss during purification of the nuclei. However, the RPKM values were similar for nuclei and cells over a range of approximately three orders of magnitude (Fig. 4 A and B), based on transcripts of five housekeeping genes [Hsp20ab1, Rpl13, Eef2, Chmp2a, and poly-U binding splicing factor 60 (Puf60)] (22), five NPC-specific genes (Vim, Olig2, Fabp7, Racgap1, and H2afz) (23), and the EYFP transgene. Analysis of variance (ANOVA) applied to all samples indicated only a 3% difference between nuclei and cells, and the transcript levels for housekeeping genes remained within a 3.5-fold range across all samples. NPC-specific transcripts were also at similar levels across all libraries, as would be expected. The neuronal cell marker, Vim, was highly expressed with an average RPKM value of 2,368. Global gene-expression variation [mean values for the coefficients of variation (CVs) of the transcript levels for each gene] across three replicate nuclei was not higher than in cells (69.1% and 80.3%, respectively) (Fig. 4C). Variation in global gene expression for bulk samples of 100 cells and 100 nuclei was almost identical (51.6% and 51.5%, respectively). Comparison of RNA-seq data from these NPC samples to recently published data (26) provides external confirmation of cell-type specificity (SI Appendix, Fig. S9).
Fig. 4.
Transcript levels and variation are the same in nuclei and whole cells after sequencing. Expression (RPKM) values for housekeeping genes (A) and NPC-specific genes (B) were used to compare sequenced cells and nuclei. For each gene (x axis), sets of six bars represent the six samples of various numbers of pooled biological triplicates and are in the following order: 1 nucleus, 1 cell, 10 nuclei, 10 cells, 100 nuclei, and 100 cells (indicated for the Fabp7 gene only in B). Error bars denote 1 SD. The y axis is log2 scaled. (C) Measure of dispersion for each group of samples for single and bulk (10 and 100) nuclei and cells. Dispersion (statistical variability) is calculated as the mean of the CV of each gene across biological triplicates.
Another concern was that low-copy transcripts might not be detected from single nuclei. However, single-nucleus RPKM values ranged from 1.00 (Gm71) to 5,016.78 (Tmsb4x). Because an RPKM value of 0.2 corresponds to about one transcript per cell (9), the dynamic range appears to span from 5 to about 25,083 transcript copies. Furthermore, expression variation and expression level are not positively correlated (for example, compare SDs of Puf60 with Hsp90ab1, Fig. 4A). This implies that detection biases are not increased for low-copy relative to high-copy transcripts. Global transcriptional data (27) have previously shown that genes with an RPKM of at least 1 (about five transcripts per cell) can be analyzed in this cell type under similar conditions. Consistent with their observation, our analysis indicated that about 85% and 100% of the transcripts with at least 1 RPKM and 0.1 RPKM, were detected with less than 10% variation when 10 and 45 million reads were mapped, respectively (SI Appendix, Fig. S10). These results suggest that whereas at the lower end of the read mapping density (10 million reads), transcripts with 1 RPKM or greater constitute the high confidence set, those expressed at lower RPKM levels of at least 0.1 can be identified with greater reproducibility and confidence with an increasing number of mapped reads up to ∼45 million.
Gene Expression Is More Variable for Single Cells or Single Nuclei than for Bulk Samples.
A goal in single-cell analysis is to unmask biological interactions that are obscured using multiple cells or cell types (28). Principal component analysis (PCA) (SI Appendix, Fig. S11A) and hierarchical clustering (SI Appendix, Fig. S16A) confirmed that single-cell and -nucleus transcriptomes appear more variable than the pools of 10 and 100 with higher CVs (Fig. 4C) and lower correlation coefficients (Rs) (SI Appendix, Figs. S11B and S12 A and B) for biological replicates. The highly variable transcripts in single cells and single nuclei were strongly reduced in the pooled samples (SI Appendix, Fig. S13), underscoring the potential of single-cell and -nucleus analysis to provide resolution not achievable using bulk samples. Single-nuclei transcriptome variability appears to be lower than in the single cells. A larger sample size would allow a more thorough investigation of transcripts that are consistently highly variable.
Nuclear and Whole-Cell Transcriptomes Are Similar, with Notable Exceptions.
Bulk mRNA contains some transcripts that are differently represented between cells and nuclei (13). Differential expression analysis of the entire data set (SI Appendix, Table S1; single, 10, and 100 nuclei and cells; n = 9 for nuclei and n = 9 for whole cells) indicated a subset of the transcriptome was enriched within the nuclei compared with the cells. Based on a one-way ANOVA, 26,167 (98.3%) transcripts were equally represented in the two groups (P ≤ 0.05), similar to previous studies (13–15, 19), and confirming that use of nuclei as the mRNA source does not introduce gross perturbations to gene-expression measurements. Microarray analysis on bulk human cells (19) found 96.5% of genes equally represented in nuclei and cytoplasm. Only 3.5% of the genes (735) displayed differential transcript accumulation. We also observed a minor proportion of transcripts (438 or 2.0%) at least threefold accumulated either within the nucleus or the whole cell (SI Appendix, Table S4), 80.4% (352) of which was enriched 3- to 30-fold in the nucleus. Gene Ontology (GO) analysis identified nuclear transcripts for several protein families with nuclear functions involving the cell cycle, mitosis, and transcription (29). Likewise, transcripts overrepresented within the NPC nuclei are significantly enriched (P ≤ 0.05) for biological processes, including regulation of transcription (32 transcripts; GO:0006355) and regulation of RNA metabolic processes (32 transcripts; GO:0051252) (SI Appendix, Table S5), supporting earlier conclusions from bulk RNA (13–15, 19). Interestingly, 7 of the 352 nucleus-enriched transcripts represent long intergenic ncRNAs (lincRNAs) and other ncRNAs, such as Xist, the product of which is a cis-acting regulator and triggers the recruitment of chromatin modifying factors (30).
ncRNA.
Many ncRNAs function in regulatory pathways, including regulation of proliferation, pluripotency, and development (24). Single nuclei should be ideal for analysis of polyA+ ncRNAs. Many examples of ncRNAs were identified from the NPC samples (SI Appendix, Table S6), including lincRNAs [which function in chromatin remodeling (31–33) and in the regulation of pluripotency and differentiation (34)] and microRNA (miRNA) [which regulate of a diverse set of genes in eukaryotes (24, 35)]. We detected about 14% of all lincRNAs annotated in the Mouse Genome Database at the Mouse Genome Informatics Web site (36): from all nuclei samples, 146 of 1053 were detected, and 147 in whole cells. Likewise, 90 (12.3%) and 103 (14.1%) miRNAs were detected in nuclei and cells, respectively, of 729 miRNAs described in the database. One hundred twenty lincRNAs (of 239) and 57 (of 293) miRNAs were expressed in both whole cells and nuclei. In nuclei, we detected 97 of the 330 miRNAs reported in an RNA-seq study of single whole cells (8). The miRNAs detected are expected to be in the unprocessed, primary miRNA (pri-miRNA) form which are contained in the nucleus and have poly(A) tails (37). Full-length pri-miRNA cDNAs are indicated that result from poly-T priming at the 3′ polyA tail and complete RT-extension, giving deep coverage of the 5′ end (SI Appendix, Fig. S14). We cannot formally disprove that clipped species could contribute 5′ reads (see the figure legend of SI Appendix, Fig. S14), however, the kinetics of processing might be investigated either from full-length or clipped species. Overall, this initial study suggests that single nuclei will be an important source of information for ncRNAs and that the number detected is about the same for whole cells and nuclei. About 50% of all transcribed sequences (many ncRNAs) have been found only in the nucleus (19). Whereas these nuclear RNAs will also be detected in the whole-cell analysis, the nuclei should be a more direct source that is free of other RNA forms present in the cytoplasm.
Applying Single-Nucleus Sequencing to Brain Tissue.
Use of single nuclei is of particular importance where interconnected cells are difficult to isolate, as is the case for the CNS. RNA-seq is demonstrated using hippocampus dentate gyrus (DG) tissue dissected from a mouse engineered with a Prox1-promoter-driven GFP for identification of DG neuronal cells (SI Appendix, Fig. S15). Two biological replicates of single DG nuclei yielded more than a combined 123 million reads, with 82% of these mapping to the mouse genome (SI Appendix, Table S2). Hierarchical clustering and PCA showed that the replicate DG nuclei profiles both shared similarities and were distinct from the bulk stromal control and replicate NPC profiles (SI Appendix, Fig. S16). Based on this small dataset, tissue-derived nuclei perform as well in RNA-seq as cultured cells and nuclei. The high number of detectable gene transcripts (∼17,000 gene loci) and efficiency of read mapping and the observation of tissue-specific profiles indicate the feasibility of single nuclei transcriptomics from neuronal tissue.
Discussion
Transcriptional profiling of multiple single cells has recently emerged as a productive approach to define cell types and states. We extend use of this approach to single nuclei, which were unambiguously distinguished from whole cells by cytoplasmic expression of EYFP. Purification of the flow-sorted nuclei and cells was confirmed by epifluorescence microscopy (SI Appendix, Fig. S1). Overrepresentation of transcripts related to regulation of transcription and RNA metabolic processes (SI Appendix, Table S5) was also consistent with nuclei being the source of mRNA. Neither the final rinse buffer for micromanipulated nuclei (SI Appendix, Figs. S2 and S4) nor flow-sorted microspheres (SI Appendix, Table S2) supported cDNA amplification, demonstrating the absence of contaminating free nucleic acids. Thus, either a cell or a nucleus must be present within the sample to obtain cDNA. With our method, single nuclei could be isolated by either flow sorting or by micromanipulation. Another method for cell type-specific isolation of bulk nuclei has recently been described (16), but this approach is limited to transgenic cell lines and has not been shown to be feasible using single nuclei.
A single nucleus generated less cDNA than the corresponding cell (Fig. 2), presumably reflecting the 10- to 100-fold lower content of polyA+ transcripts, however, similar numbers of transcripts were detectable (on average, 20,065 unique transcripts from single cells and 20,243 from single nuclei). Ninety-eight percent of the transcripts were found at similar levels between nuclei and whole cells (having a less-than-threefold difference in average transcript levels), and differences in transcript levels across genes were also similar (Fig. 4). We conclude that general-expression differences between genes are conserved between nuclei and cells. A more detailed analysis will be required to determine whether smaller changes in transcript levels can be detected with equal accuracy across nuclei and cells. The global expression patterns among single nuclei or single cells were more variable than for the pooled 10- and 100-sample replicates (SI Appendix, Fig. S12), consistent with earlier studies on cell-to-cell variation, including use of genetically identical cells (38–43), and recognizing that different genes differ in their degree of cell-to-cell variability (42, 44). The observed variability between replicate single cells and between single nuclei (SI Appendix, Figs. S11A and S12 A and B) was similar to that described from the RNA-seq of single cells (45). The average number of genes that are similarly expressed in single nucleus replicates was 96.8% and for single cells, 85.7%. Single-nuclei transcriptome variability appears to be lower than in the single cells. Possibly, this reflects the compartmentalized mechanisms of mRNA decay leading to the differential turnover of transcripts in the two locations. Where fewer mechanisms of nuclear mRNA decay are known, several exist in the cytoplasm, potentially leading to a higher rate of mRNA decay (46, 47). Isolation of nuclei by tissue homogenization at 4 °C, thereby arresting gene expression, is preferable to enzymatic dissociation of whole cells (which has the potential to perturb gene expression) (48), and to mechanical separation of cells [micromanipulation or laser capture microdissection, which sever dendrites and axons]. A detailed comparison of the transcriptional profiles of whole neurons and their nuclei will be needed to further explore the effects of cellular stress on gene expression and the advantages of using nuclei. Use of nuclear transcripts is particularly attractive where isolation of intact single cells from complex tissues is difficult, for example, in the CNS. Single DG nuclei from tissues yielded whole-transcriptome data comparable to data from cultured NPC nuclei and whole cells (SI Appendix, Table S2).
Several classes of regulatory ncRNAs were detected in single nuclei, including lincRNA and miRNA (SI Appendix, Table S6). Nuclei should also be ideal for investigating transcripts that are selectively accumulated within the nucleus. According to ENCODE data (19), half of all transcribed sequences are found only in the nucleus and are mostly unannotated. Single nuclei contained a higher percentage of intergenic and intronic sequences than single cells (SI Appendix, Fig. S5), suggesting that nuclei will be an ideal source for discovery of new unannotated ncRNAs and will facilitate studies of some RNA processing events, including processing of pri-miRNAs.
Further improvements in methods for cDNA synthesis from single cells and single nuclei for use in RNA-seq analyses will emerge. Two recent methods appear limited by selective 3′ (49) or 5′ (50) end-tagging strategies for measuring mRNA abundance. Another protocol, published during the course of our investigation [switching mechanism at 5′ end of RNA template or Smart-seq (51)] demonstrates a reduced 5′ attenuation of the transcript signal, although it does not entirely eliminate it. Additional validation studies will be required to fully compare the performance of the available single-cell protocols. However, we expect that single nuclei will be a suitable source of mRNA for most protocols.
Methods
NPC Derivation and Culture.
Mouse embryonic stem cells expressing EYFP and LacZ transgenes (ROSA26 loci), with tamoxifen-inducible CRE recombinase from untargeted viral integration, were maintained on a mouse embryonic fibroblast feeder layer by daily replacement of mES media [KO-DMEM (Invitrogen); 15% (vol/vol) knock-out serum (Sigma), 1X glutaMAX (Gibco), 1 × nonessential amino acids (Gibco), 55 µm 2-mercaptoethanol (Gibco), and 1,000 U/mL mLIF (Millipore)]. Cells were a gift from A. McMahon (Keck School of Medicine of University of Southern California, Los Angeles) (52).
Differentiation was initiated by withdrawal of mLIF and transfer to low adherence (i.e., 3262 polystyrene) culture dishes (Sigma). Embryoid bodies (EBs) were observed after overnight culture, aspirated, filtered (70 µm) to improve size homogeneity, and then replated in N2 media [DMEM/F12 (Invitrogen), 1X B27 (Gibco), 1X N2 (Invitrogen)] supplemented with 500 ng/mL Noggin (Peprotech). Media was changed every other day. After 5 d, EBs were collected and dissociated with papain (Worthington Biochemical Corp.) for subsequent NPC culture.
NPC cultures were initiated by plating dissociated EB/neural rosettes (200,000 per mL) on laminin-coated plates in N2 media with 20 ng/mL EGF (StemGent), 20 ng/mL FGF (Peprotech), and 0.2% FBS (Atlanta Biologicals). NPC cultures were maintained by feeding every other day in N2 media with 20 ng/mL EGF, 20 ng/mL FGF, and 1 μg/mL laminin (Invitrogen). Splits (1:3) were performed at confluence (circa a weekly basis) using TrypLE (Gibco).
Isolation and Staining of Whole Cells and Nuclei from NPC Cultures.
NPCs were grown and harvested, as for passaging, and kept on ice in HBSS (Gibco). One-third of this population was used for whole-cell micromanipulation or sorting. Whole cells expressing EYFP were chosen for downstream micromanipulation and cDNA synthesis. The rest (two-thirds) was transferred to nuclei isolation media [(NIM) 250 mM sucrose, 25 mM KCl, 5 mM MgCl2, 10 mM Tris], aspirated three times through a 27-gauge needle, and centrifuged at 1,200 × g for 8 min. Nuclei were further purified using a 29% iodixanol cushion and centrifuged at 10,300 × g for 20 min. An aliquot was observed by fluorescence microscopy to confirm the absence of EYFP signal. A candidate single cell or single nucleus was selected from the population and serially washed in cold PBS to remove potential nucleic acid contaminants from the sample. Nuclei were stained by addition either of DAPI (20 µg/mL) or PI (50 µg/mL), as previously described (18). RNA-seq was performed using single nuclei from which the cytoplasm had been removed.
Cell Staining and Nuclei Isolation from Hippocampal DG.
All protocols were approved by the Salk Institute's Institutional Animal Care and Use Committee. Prox1-GFP mice (MMRRC, stock no. 031006-UCD) express (53) GFP in DG neurons. Mice were anesthetized with a ketamine/xylazine mixture and transcardially perfused with 0.9% NaCl solution followed by 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer, pH 7.4. Brain tissue was postfixed overnight in 4% PFA at 4 °C and then transferred to a 30% sucrose solution. Forty-micron coronal sections were cut on a sliding microtome and all immunohistochemistry was performed on free-floating sections. Tissue from animals was incubated with a primary antibody [rabbit anti-Prox1 (1:250, Covance)]. for 48 h at 4 °C followed by incubation with a secondary antibody. Images were taken using a confocal microscope (Radiance 2100; Bio-Rad). A transgenic mouse expressing GFP under the Prox1 promoter enabled the observation of endogenous Prox1 expression in vivo. The DG was isolated by dissection as before (54). Nuclei were obtained from freshly dissected tissue using a Polytron (Kinematica, Inc.), and dounce homogenization in NIM + 0.5% triton. Purification of nuclei was performed as for NPCs.
Flow Cytometry and FACS Sorting of Single Nuclei.
A FACS Aria II flow sorter (Becton Dickinson, San Jose, CA), (argon laser, 100 mW at 488 nm), used a custom forward scatter photomultiplier for high-sensitivity small-particle detection. An aliquot of the purified nuclei (Methods, Cell Staining and Nuclei Isolation from Hippocampal DG) stained with propidium iodide (PI, 20 μg/mL final concentration) lacked EYFP. Sorting gates were based on flow analysis of events (cells, nuclei), and validated by sorting onto glass slides, and examination via phase contrast and fluorescence microscopy. Samples were sorted at a rate of 50 events per second, based on side scatter (threshold value >200). Fluorescence detection used a 510-nm dichroic longpass beam splitter, and a 525-nm/25-nm-band pass barrier filter for EYFP, and a 620-nm/40-nm-band pass filter for PI. Biparametric histograms of light scatter versus fluorescence (with log scaling) were collected for a total count of at least 50,000 events. The sequenced 10 and 100 cells and nuclei were isolated using FACS, whereas the single samples were isolated via micromanipulation.
For micromanipulation of single cells and single nuclei, see SI Appendix, Methods S1; for cDNA synthesis, amplification, and TaqMan analysis, see SI Appendix, Methods S2; for SOLiD (Life Technologies) sequencing, mapping, and error correction, see SI Appendix, Methods S3; for bioinformatics analysis, see SI Appendix, Methods S4; and for GO analysis, see SI Appendix, Methods S5.
Supplementary Material
Acknowledgments
We thank Jan Bruder, Dane Clemenson, and Diana Yu for help with fluorescence imaging; Katherina Psathaki for discussions on NPC and DG heterogeneity; and Gene Yeo for bioinformatics discussions. R.S.L. was funded by National Institutes of Health (NIH)-1 RC1 HG005471 and NIH 2R01 HG003647; R.S.L. and F.H.G. by TRO1MH095741; F.H.G. by the JPB Foundation, the Mathers Foundation, the McDonnell Foundation, and the National Institute of Mental Health; and D.W.G. and G.M.L. acknowledge support from the College of Agriculture and Life Sciences, University of Arizona, and NIH-NCI R21-CA164690.
Footnotes
The authors declare no conflict of interest.
Data deposition: The RNA sequencing reads and quality files reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA), http://www.ncbi.nlm.nih.gov/sra (SRA accession nos. SRX255934, SRX255935, SRX255938, SRX256509, SRX256510, SRX256516, SRX256805, SRX256806, SRX256809–SRX256812, SRX256825–SRX256829, and SRX256832–SRX256835, and found in SRA study SRP020097).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319700110/-/DCSupplemental.
References
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okoniewski MJ, Miller CJ. Hybridization interactions between probesets in short oligo microarrays lead to spurious correlations. BMC Bioinformatics. 2006;7:276. doi: 10.1186/1471-2105-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurimoto K, Yabuta Y, Ohinata Y, Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc. 2007;2(3):739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- 4.Lao KQ, et al. mRNA-sequencing whole transcriptome analysis of a single cell on the SOLiD system. J Biomol Tech. 2009;20(5):266–271. [PMC free article] [PubMed] [Google Scholar]
- 5.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 6.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 7.Tang F, et al. Deterministic and stochastic allele specific gene expression in single mouse blastomeres. PLoS ONE. 2011;6(6):e21208. doi: 10.1371/journal.pone.0021208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang F, et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6(5):468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang F, et al. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc. 2010;5(3):516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uemura E. Age-related changes in neuronal RNA content in rhesus monkeys (Macaca mulatta) Brain Res Bull. 1980;5(2):117–119. doi: 10.1016/0361-9230(80)90182-3. [DOI] [PubMed] [Google Scholar]
- 11.Roozemond RC. Ultramicrochemical determination of nucleic acids in individual cells using the Zeiss UMSP-I microspectrophotometer. Application to isolated rat hepatocytes of different ploidy classes. Histochem J. 1976;8(6):625–638. doi: 10.1007/BF01003963. [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki ES. Microarrays and the gene expression profile of a single cell. Ann N Y Acad Sci. 2004;1020:92–100. doi: 10.1196/annals.1310.010. [DOI] [PubMed] [Google Scholar]
- 13.Barthelson RA, Lambert GM, Vanier C, Lynch RM, Galbraith DW. Comparison of the contributions of the nuclear and cytoplasmic compartments to global gene expression in human cells. BMC Genomics. 2007;8:340. doi: 10.1186/1471-2164-8-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trask HW, et al. Microarray analysis of cytoplasmic versus whole cell RNA reveals a considerable number of missed and false positive mRNAs. RNA. 2009;15(10):1917–1928. doi: 10.1261/rna.1677409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwanekamp JA, et al. Genome-wide analyses show that nuclear and cytoplasmic RNA levels are differentially affected by dioxin. Biochim Biophys Acta. 2006;1759(8-9):388–402. doi: 10.1016/j.bbaexp.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Deal RB, Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc. 2011;6(1):56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tilgner H, et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22(9):1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Barthelson RA, Lambert GM, Galbraith DW. Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 2008;147(1):30–40. doi: 10.1104/pp.107.115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308(5725):1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 20.Morris J, Singh JM, Eberwine JH. Transcriptome analysis of single cells. J Vis Exp. 2011;(50):2634. doi: 10.3791/2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hadjantonakis AK, Nagy A. The color of mice: In the light of GFP-variant reporters. Histochem Cell Biol. 2001;115(1):49–58. doi: 10.1007/s004180000233. [DOI] [PubMed] [Google Scholar]
- 22.Kouadjo KE, Nishida Y, Cadrin-Girard JF, Yoshioka M, St-Amand J. Housekeeping and tissue-specific genes in mouse tissues. BMC Genomics. 2007;8:127. doi: 10.1186/1471-2164-8-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurok U, et al. Gene expression changes in the course of neural progenitor cell differentiation. J Neurosci. 2004;24(26):5982–6002. doi: 10.1523/JNEUROSCI.0809-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5(4):e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramsköld D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLOS Comput Biol. 2009;5(12):e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergsland M, et al. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25(23):2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Łabaj PP, et al. Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling. Bioinformatics. 2011;27(13):i383–i391. doi: 10.1093/bioinformatics/btr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ståhlberg A, Bengtsson M. Single-cell gene expression profiling using reverse transcription quantitative real-time PCR. Methods. 2010;50(4):282–288. doi: 10.1016/j.ymeth.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Yeom YI, Ha HS, Balling R, Schöler HR, Artzt K. Structure, expression and chromosomal location of the Oct-4 gene. Mech Dev. 1991;35(3):171–179. doi: 10.1016/0925-4773(91)90016-y. [DOI] [PubMed] [Google Scholar]
- 30.Senner CE, et al. Disruption of a conserved region of Xist exon 1 impairs Xist RNA localisation and X-linked gene silencing during random and imprinted X chromosome inactivation. Development. 2011;138(8):1541–1550. doi: 10.1242/dev.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr Opin Genet Dev. 2010;20(2):142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gursanscky NR, Searle IR, Carroll BJ. Mobile microRNAs hit the target. Traffic. 2011;12(11):1475–1482. doi: 10.1111/j.1600-0854.2011.01253.x. Accessed October 10, 2013. [DOI] [PubMed] [Google Scholar]
- 36. The Jackson Laboratory. (2013) Mouse Genome Informatics. Available at www.informatics.jax.org.
- 37.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 39.Blake WJ, KAErn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422(6932):633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 40.Dunlop MJ, Cox RS, 3rd, Levine JH, Murray RM, Elowitz MB. Regulatory activity revealed by dynamic correlations in gene expression noise. Nat Genet. 2008;40(12):1493–1498. doi: 10.1038/ng.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raj A, van Oudenaarden A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell. 2008;135(2):216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen KD, Wu Z, Irizarry RA, Leek JT. Sequencing technology does not eliminate biological variability. Nat Biotechnol. 2011;29(7):572–573. doi: 10.1038/nbt.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suter DM, et al. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332(6028):472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 44.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci USA. 2006;103(47):17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Islam S, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21(7):1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 47.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13(4):246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang HL, et al. Trypsin-induced proteome alteration during cell subculture in mammalian cells. J Biomed Sci. 2010;17:36. doi: 10.1186/1423-0127-17-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: Single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2(3):666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 50.Islam S, et al. Highly multiplexed and strand-specific single-cell RNA 5′ end sequencing. Nat Protoc. 2012;7(5):813–828. doi: 10.1038/nprot.2012.022. [DOI] [PubMed] [Google Scholar]
- 51.Ramsköld D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244(2):305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 53.François M, et al. Sox18 induces development of the lymphatic vasculature in mice. Nature. 2008;456(7222):643–647. doi: 10.1038/nature07391. [DOI] [PubMed] [Google Scholar]
- 54.Lein ES, Zhao X, Gage FH. Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci. 2004;24(15):3879–3889. doi: 10.1523/JNEUROSCI.4710-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.