Abstract
Dissolution of protein macromolecules from poly(lactic-co-glycolic acid) (PLGA) particles is a complex process and still not fully understood. As such, there are difficulties in obtaining a predictive model that could be of fundamental significance in design, development, and optimization for medical applications and toxicity evaluation of PLGA-based multiparticulate dosage form. In the present study, two models with comparable goodness of fit were proposed for the prediction of the macromolecule dissolution profile from PLGA micro- and nanoparticles. In both cases, heuristic techniques, such as artificial neural networks (ANNs), feature selection, and genetic programming were employed. Feature selection provided by fscaret package and sensitivity analysis performed by ANNs reduced the original input vector from a total of 300 input variables to 21, 17, 16, and eleven; to achieve a better insight into generalization error, two cut-off points for every method was proposed. The best ANNs model results were obtained by monotone multi-layer perceptron neural network (MON-MLP) networks with a root-mean-square error (RMSE) of 15.4, and the input vector consisted of eleven inputs. The complicated classical equation derived from a database consisting of 17 inputs was able to yield a better generalization error (RMSE) of 14.3. The equation was characterized by four parameters, thus feasible (applicable) to standard nonlinear regression techniques. Heuristic modeling led to the ANN model describing macromolecules release profiles from PLGA microspheres with good predictive efficiency. Moreover genetic programming technique resulted in classical equation with comparable predictability to the ANN model.
Keywords: poly(lactic-co-glycolic acid) (PLGA) microparticles, genetic programming, feature selection, artificial neural networks, molecular descriptors
Video abstract
Introduction
Poly(lactic-co-glycolic acid) (PLGA) microparticles play a prominent role in many drug formulations. PLGA is flexible and has a satisfactory safety profile. It also has the ability to modify the drug dissolution profile for controlled-release formulations.1 Moreover, the drug protective properties of PLGA were found to be suitable for unstable active pharmaceutical ingredients (APIs).2 Therefore, PLGA has wide applications as an excipient.
PLGA can be formulated as films, discs, microcapsules, and nano- or microspheres.3–6 The current study is focused on the formulation of PLGA nano- and microspheres. One of the most studied properties of PLGA particles over the past years is the API release mechanism. There are often contradictory results reported in the literature, which demonstrate the high level of complexity of PLGA-based dosage forms. The drug release from the PLGA matrix is mainly governed by two mechanisms: diffusion and degradation/erosion.7 The work of D’Souza and DeLuca8 and Mollo and Corrigan9 showed that the drug release profile can be divided into two stages. Initially, the release is considered to be diffusion-controlled, corresponding to the low amount of released drug. Afterwards, the drug release is controlled primarily by the degradation and erosion of the PLGA matrix. There are many factors influencing the diffusion and degradation rate of PLGA. For example, pore diameters, matrix–API interactions, API–API interactions, and formulation composition;10–13 however, there is, to date, no versatile quantitative model describing the relationships among these factors in a consistent, mathematical manner. Recently, Fredenberg et al proposed four so-called “true” drug release mechanisms from PLGA matrices: 1) diffusion through water-filled pores; 2) diffusion through the polymer; 3) osmotic pumping; and 4) polymer erosion. The release profile of large and hydrophilic molecules, such as proteins and peptides, is mostly limited by diffusion through water-filled pores.7
Over the past 2 decades, there have been several publications covering the model development of the drug release profile from PLGA microspheres. Among the most recognized are the works of Zygourakis and Markenscoff14 and Göpferich,15 in which Monte Carlo and cellular automata microscopic models were introduced. The authors used a two-dimensional model of the polymer matrix based on the physicochemical characterization to predict erosion and therefore the release rate of small molecules. Later, Siepmann et al implemented a model coupling a Monte Carlo simulator with sets of partial differential equations.16 The model described the chemical reactions and physical mass transport processes involved in erosion-controlled drug release from PLGA beads. Nevertheless, the predictability of classical models (empirical/semiempirical or mechanistic models) has not been thoroughly assessed in many studies and the versatility remains questionable.14–16 A recent study by Barat et al17 applied Göpferich’s theory15 on multi-agent systems. Good agreement between modeling and experimental results was obtained for polymer erosion coupled with a partial differential equation model of lysozyme release profiles from PLGA spheres;17 however, to the authors’ best knowledge, there is no general model able to predict the release rate of the various macromolecules from PLGA systems. Therefore, the design of the PLGA microsphere formulations loaded with macromolecules depends heavily on the trial-and-error approach.
The objective of this work was to demonstrate the cooperative use of feature selection, artificial neural networks (ANNs), and genetic programming (GP) to create a simple predictive model of the macromolecule dissolution rate from the PLGA dosage form, based on the gathered literature data.
Materials and methods
Data set
Data were collected from the literature. After careful screening of about 200 publications, release rates of 68 PLGA formulations from 18 publications were selected for data extraction (Supplementary material Table S1). Selected PLGA formulations were prepared using four methods: 1) a double-emulsion water-in-oil-in-water solvent extraction process; 2) a solid-in-oil-in-water emulsion method; 3) an oil-in-oil solvent evaporation technique; and 4) a spray-drying process. Formulations included 14 model substances: bovine serum albumin; recombinant human erythropoietin; recombinant human epidermal growth factor; lysozyme; recombinant human growth hormone; hen ovalbumin; human serum albumin; beta-amyloid; insulin; recombinant human erythropoietin coupled with human serum albumin; L-asparaginase; bovine insulin; alpha-1 antitrypsin; and chymotrypsin. The release profiles presented in the literature were manually digitized, taking mean values of the observations if possible. Standard deviations and/or errors were not included due to the manual character of the digitization and lack of this information in some sources. Overall there were 745 data records with 320 variables (Supplementary material Table S2). The independent parameters contained 319 inputs covering the formulation characteristics (PLGA inherent viscosity, PLGA molecular weight, lactide-to-glycolide ratio, inner and outer phase Polyvinyl alcohol (PVA) concentration, PVA molecular weight, inner phase volume, encapsulation rate, mean particle size, and PLGA concentration); the experimental conditions (dissolution pH, number of dissolution additives, dissolution additive concentration and production method, and dissolution time); and the molecular descriptors of the macromolecules and excipients. The molecular descriptors were computed using Marvin cxcalc plugin, UK (version 5.11; ChemAxon, Budapest, Hungary) for the drug substance as well as the excipients.18 The amount of the drug substance released (Q) was the only dependent variable. Prior to feature selection, the initially obtained 319 inputs were further reduced to 300 inputs by removing all the null and missed input during the data set preprocessing procedure. The data set was then processed in several ways:
Noise addition to prevent models from over-fitting. The noised data records were produced numerically with ±5% amplitude for each variable value and two times more records number (noted in the text as “rand”),
Data set split according to the tenfold cross-validation scheme, with the aim of excluding all the data belonging to the particular formulation, thus simulating the real application of the model forced to predict the behavior of the unknown formulation (noted in the text and tables as “10cv”),
Linear scaling using either the output range of <0.2, 0.8> or <−0.8, 0.8>, in order to match nonlinear activation functions domains were used for ANNs only (noted in text and tables as “scale”).
Feature selection
It is considered state of the art to implement the feature selection technique prior to the actual modeling stage when dealing with multidimensional data sets. Two variable selection techniques were used to reduce redundant or irrelevant information gathered during data set preparation: based on the methodology proposed by Żurada et al19 with further modification by Mendyk and Jachowicz,20 which uses ANNs trained with the back propagation algorithm as the modeling tools; and feature ranking created by the fscaret package of the R environment (The R Foundation for Statistical Computing, Vienna, Austria).21,22
The main parameters of the applied techniques are listed in Table 1. The resulting feature rankings were then inspected on the substantial decrease of variable significance; two cutoff points were chosen for each method.
Table 1.
Feature selection parameters used in the assay according to applied methods for artificial neural networks and fscaret
| ANNs | fscaret |
|---|---|
| Feature selection technique applied | |
| Whole data set. | Whole data set. |
| Trained and tested over 170 ANN architectures. 5M iterations. |
Time-limiting function was set to 2 hours for single model development. |
| Final ensemble based on the goodness of fit criterion: up to 200% of minimum error achieved. | “PreprocessData” function was off. The feature ranking was created according to the results of 58 models. |
Abbreviations: M, millions; ANN, artificial neural network.
ANNs
Based on reduced vectors of inputs, two types of ANN model were obtained, namely, multi-layer perceptron artificial neural networks (MLP-ANNs) and monotone multi-layer perceptron artificial neural networks (MON-MLP) networks. The tools used during modeling were the Nets2012 ANN23 simulator (developed in-house, Aleksander Mendyk, Jagiellonian University, Krakow, Poland), and R environment with the monmlp24 package for MON-MLP networks.23,24 MLP-ANNs and MON-MLP were trained on reduced vectors after feature selection procedure.
MLP-ANNs
MLP-ANNs used the backpropagation training algorithm and were tested with activation functions such as linear (“lin”), logistic (“sigma”), hyperbolic tangent (“tanh”), and logarithmic function (“fsr”). The architecture of MLP-ANNs comprised one to seven hidden layers. In addition to the MLP-ANNs, neuro-fuzzy systems (NFs) were employed by adapting the simplest Mamdani type and contained from five to 100 nodes in the hidden layer. All models were multiple input/single output (MISO) type.25 Considering a tenfold cross-validation, a total of 3,680 models were trained in a single run. Each model was trained up to 10,000,000 epochs with several stop points. The generalization error was assessed in order to find the most optimal training conditions. The stop points were selected as 100,000, 200,000, 500,000, 1,000,000, 1,500,000, 2,000,000, 3,000,000, 5,000,000, and 10,000,000 epochs. The epoch size was equal to 1. Other parameters were as follows:
momentum technique, with the momentum factor 0.3,
delta-bar-delta algorithm, with the initial learning factor 0.65,
jog-of-weights technique designed to prevent getting stuck in the local minima of the cost function; a simple noise addition to the weights was performed when the ANN was not improving its efficiency during the 100,000 epochs (the patience criterion).
The root-mean-square error (RMSE) was used to measure the goodness of fit and was calculated according to Equation 1; additionally, relative RMSEs (relRMSEs) for each dissolution profile were calculated (Equation 2).
| [1] |
where obsi, predi = observed and predicted values respectively, i = data record number, and n = total number of records.
| [2] |
where RMSEi = RMSE for formulation i and Qmax and Qmin = maximum and minimum percentage of released molecule, respectively.
MON-MLP
MON-MLP networks were created and trained by the “monmlp.fit” function of the monmlp R package.24 Bootstrap aggregation was used to avoid over-fitting of models. All models consisted of two hidden layers with three to 32 nodes per layer. Two transfer functions for hidden layer modification of hyperbolic tangent (“tansig”) and for output layer linear function were applied in each model. The prepared ensemble system consisted of ten or 20 neural networks. Performance of the models was assessed using RMSE. Other parameters were as follows:
“trials” function, set to 5, was used to avoid local minima in ensemble,
Iteration number for learning models was set to 100, 500, 1,000, or 2,000.
GP and symbolic regression
Mathematical equations were produced by means of GP and the symbolic regression mode available in the rgp package of the R environment.26 Direct mapping of the input and output variables was applied. GP was performed on reduced input vectors after the feature selection procedure.
The parameters of the function “symbolicRegression” of the rgp package were set as follows:
“individualSizeLimit” (the chromosome length) varied from 10 to 300 and was the subject of the optimization in regard to the RMSE (Equation 1),
“populationSize” was set to 10,000,
“myfunctionSet,” a set of prototype functions, was restricted to the simplest arithmetic operators like addition, subtraction, multiplication, and division, together with power, natural logarithm, square root, and exponent function,
the algorithm stop condition makeFitnessStopCondition (RMSE) was equal 1.0,
“makeTimeStopCondition” was set to 1 hour for the indirect and 120 hours for the direct modeling mode.
The direct mapping approach applied GP on the original data set to create functional relationships between the amount of the macromolecule released (Q) and the vector of parameters containing the time variable (t), formulation characteristics, and molecular descriptors.
Although GP provides the model with optimized values of its parameters, to achieve better generalization, the multivariate optimization of equation parameters was applied using the optimx package (The R Foundation for Statistical Computing).27
Parameters used for the “optimx” function were:
“par” – initial value for the parameters was set to 0.1,
“all.methods” was set to “true”, which means that all available (and suitable) methods were used,
“follow.on” was set to “false.” All optimization methods were used separately.
Fitting was performed in the mode of tenfold cross-validation. The latter procedure was done with the same training/testing data sets as for MLP and MON-MLP ANNs in order to compare the two systems’ generalization abilities. The selection criterion for the parameters was the model goodness of fit expressed as the RMSE obtained on the training data set.
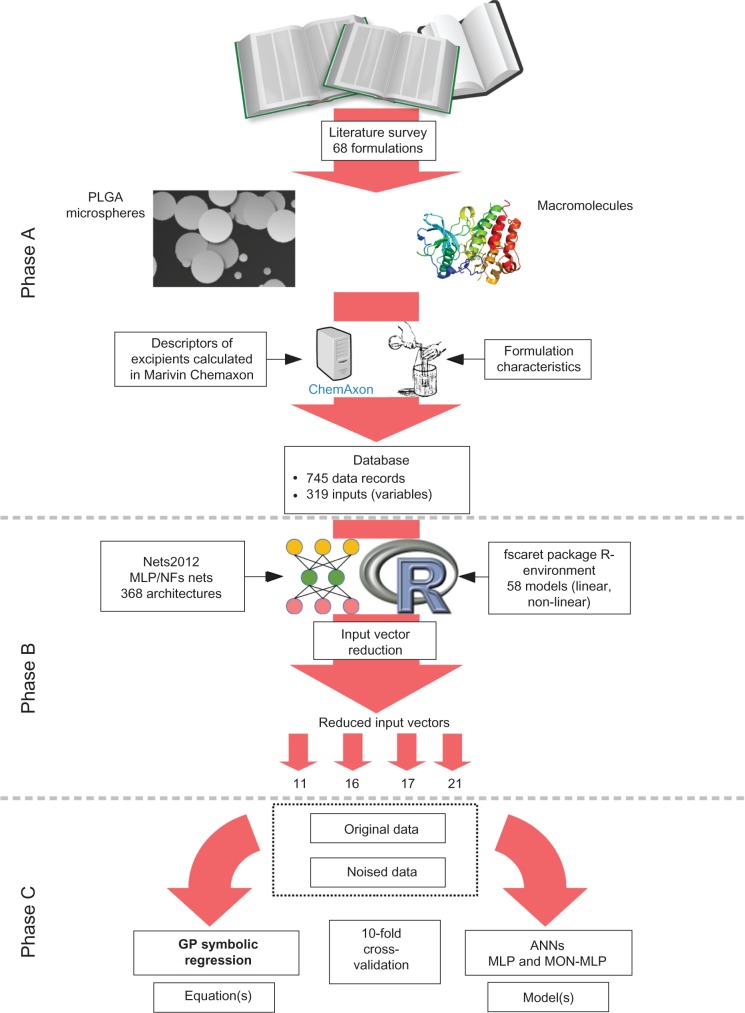
The outline of the workflow, in conjunction with some of the results of feature selection, is presented in Figure 1.
Figure 1.
Schematic representation of the experimental setup.
Notes: Data acquisition (Phase A). Data preprocessing (Phase B). Modeling (Phase C).
Abbreviations: ANNs, artificial neural networks; GP, genetic programming; MLP, multi-layer perceptron artificial neural networks; MON-MLP, monotone multi-layer perceptron artificial neural networks; NFs, neuro-fuzzy systems; PLGA, poly(lactic-co-glycolic acid).
Hardware environment
All calculations were performed using 21 professional workstations equipped mainly with Xeon central processing units (Intel Corporation, Santa Clara, CA, USA) with a total of 120 cores and at least 16 GB of RAM operating on the openSUSE 12.3 (x86_64) operating system (SUSE, Nürnberg, Germany).
Results and discussion
Feature selection
According to the feature ranking created by ANNs and fscaret, four reduced input vectors containing 21, 17, 16, and eleven independent variables were selected. The resulting input vectors for 17 and eleven independent variables, which yielded the best results for GP and ANNs, respectively, are shown in Tables 2 and 3. They originate from two different feature-ranking methods and gave the lowest RMSE when applied to ANNs and GP. Evaluation of the input vector was based on the generalization error (RMSE). A substantial reduction of RMSE from 22.8% (ANN models trained on tenfold cross-validation with all 300 inputs) to 15%–18% was obtained for all four reduced input vectors (Tables 4 and 5). Further attribute reduction applied to reduced vectors was unsuccessful, giving about 2%–3% higher generalization errors in each case.
Table 2.
Reduction to 17-variable input vector
| Variable name | Description | Group |
|---|---|---|
| In1 | Hyper-Wiener index | Macromolecule descriptors |
| In2 | Szeged index | |
| In3 | PLGA inherent viscosity (dL/g) | Formulation characteristic |
| In4 | PLGA molecular weight (Da) | |
| In5 | PVA inner phase concentration (%) | |
| In6 | PVA outer phase concentration (%) | |
| In7 | Encapsulation rate (%) | |
| In8 | Mean particle size (μm) | |
| In9 | PLGA-to-plasticizer ratio | |
| In10 | Dissolution pH | |
| In11 | Production method: 1) w/o/w; 2) s/o/w; 3) s/o/o; 4) spray-dried | |
| In12 | Hyper-Wiener index | Plasticizer descriptors |
| In13 | log D at pH 0 | |
| In14 | log D at pH 1 | |
| In15 | log D at pH 14 | |
| In16 | Basic pKa1 | |
| In17 | Time (days) | Formulation characteristic |
| Q | % of macromolecule dissolved | Output |
Abbreviations: PLGA, poly(lactic-co-glycolic acid); PVA, Poly(vinyl alcohol); s/o/o, oil-in-oil solvent; s/o/w, solid-in-oil-in-water; w/o/w, water-in-oil-in-water.
Table 3.
Reduction to eleven-variable input vector
| Variable name | Description | Group |
|---|---|---|
| In1 | Szeged index | Macromolecule descriptors |
| In2 | pI | |
| In3 | Quaternary structure of macromolecule: 1) monomer; 2) dimer | |
| In4 | Lactide-to-glycolide in polymer ratio | Formulation characteristics |
| In5 | PVA inner phase concentration (%) | |
| In6 | PVA outer phase concentration (%) | |
| In7 | Encapsulation rate (%) | |
| In8 | Mean particle size (μm) | |
| In9 | Dissolution pH | |
| In10 | Production method: 1) w/o/w; 2) s/o/w; 3) s/o/o; 4) spray-dried | |
| In11 | Time (days) | |
| Q | % of macromolecule dissolved | Output |
Abbreviations: PVA, Poly(vinyl alcohol); s/o/o, oil-in-oil solvent; s/o/w, solid-in-oil-in-water; w/o/w, water-in-oil-in-water; pI, isoelectric point.
Table 4.
Artificial neural network–MLP results, according to the lowest root-mean-square error SE obtained
| Input vector | Architecture | Iterations | Relative RMSE (%) |
|---|---|---|---|
| 21in | 120_50_30_20_10_tanh_scale | 3,000,000 | 16.9 |
| 17in | 7_5_3_fsr_rand_scale | 10,000,000 | 18.0 |
| 16in | 200_80_40_20_tanh_rand | 3,000,000 | 17.9 |
| 11in | 60_20_8_tanh_rand | 5,000,000 | 16.2 |
Abbreviation: MLP, multi-layer perceptron; RMSE, root-mean-square error.
Table 5.
MON-MLP results, according to the lowest root-mean-square error SE obtained
| Input vector | Architecture | Ensemble | Iterations | Relative RMSE (%) |
|---|---|---|---|---|
| 21in | 8_6_trials_5_orig | 20 | 1,000 | 17.7 |
| 17in | 12_6_trials_5_orig | 20 | 2,000 | 17.1 |
| 16in | 12_6_trials_5_orig | 10 | 2,000 | 16.4 |
| 11in | 8_6_trials_5_orig | 20 | 500 | 15.4 |
Abbreviations: MON-MLP, monotone multi-layer perceptron artificial neural networks; RMSE, root-mean-square error.
Reduced input vectors mostly consisted of formulation characteristics, which is consistent with previous publications stating that the method of preparation and parameters describing the particles, influence in great extent the drug dissolution rate from the PLGA particles.1,4,7 Sandor et al stated that the release rate of proteins is dependent on the protein molecular weight;28 however, distance-based geometry properties coded as hyper-Wiener index and Szeged index were not discussed in the literature.
ANNs
The best result obtained using the ANN modeling tools had a relRMSE of 15.4% for MON-MLP networks with a vector consisting of eleven inputs (Tables 3 and 5).
Although the generalization error of the ANN models was about 15%–18%, the relRMSE (related to single formulation) of the best MON-MLP model range was from 1.75% to 36.03% between formulations, indicating that there is good potential for further improvements. To accomplish this task, the data set should be enriched with other crucial parameters influencing the dissolution, such as the porosity of particles, pore diameters, chemical descriptors of polymer, and particle size of the excipients. Unfortunately, data were not available or scant in the selected publications, therefore not useful for systematic quantitative analysis. As a consequence, the model has inferior performance when compared to classical models, although it is important to note that all the predictive model generalization errors were a result of the tenfold cross-validation procedure, which is not the case in classical approaches.7–9,14,15 Moreover, our models are universal in terms of the drug encapsulated in PLGA microspheres, whereas most of the classical models are focused on a limited and/or predefined number of drugs.1,4,5,7,9
Symbolic regression
Obtained using GP classical equation, though complicated and derived from a database consisting of 17 inputs, not symbolic regression but equation yielded better generalization error (14.3%) and narrower relRMSE, ranging from 4.76% to 32.83% between formulations, in comparison to the MON-MLP model. The equation was characterized by four parameters (Equation 3). Moreover, during the evolution process, the GP algorithm further reduced the number of necessary variables by eliminating input numbers 2, 4, 9, 10, 12, 13, 15, and 16. The simplified mathematical model still retained all the types of input variables (with one exception of the production method), yet its predictive performance was comparable to the more complex MON-MLP model (Tables 6 and Supplementary material Table S3 [full results]).
Table 6.
The results of the formulation-to-formulation relRMSE (%)
| Descriptive statistics | relRMSE (%)
|
|
|---|---|---|
| MON-MLP model | GP model (Eq 3) | |
| Min | 1.75 | 4.76 |
| Max | 36.03 | 32.83 |
| Mean | 14.39 | 13.75 |
Abbreviations: GP, genetic programming; MLP, multi-layer perceptron artificial neural networks; MON-MLP, monotone multi-layer perceptron artifical neural networks; relRMSE, relative root-mean-square error.
| [3] |
in which In1–In17 correspond to labels in Table 2 and C1–C4 = constants. Equation parameters: C1 =0.1382595046751, C2 =0.1063092562504, C3 =4.3209748739209, and C4 =0.3010676275288.
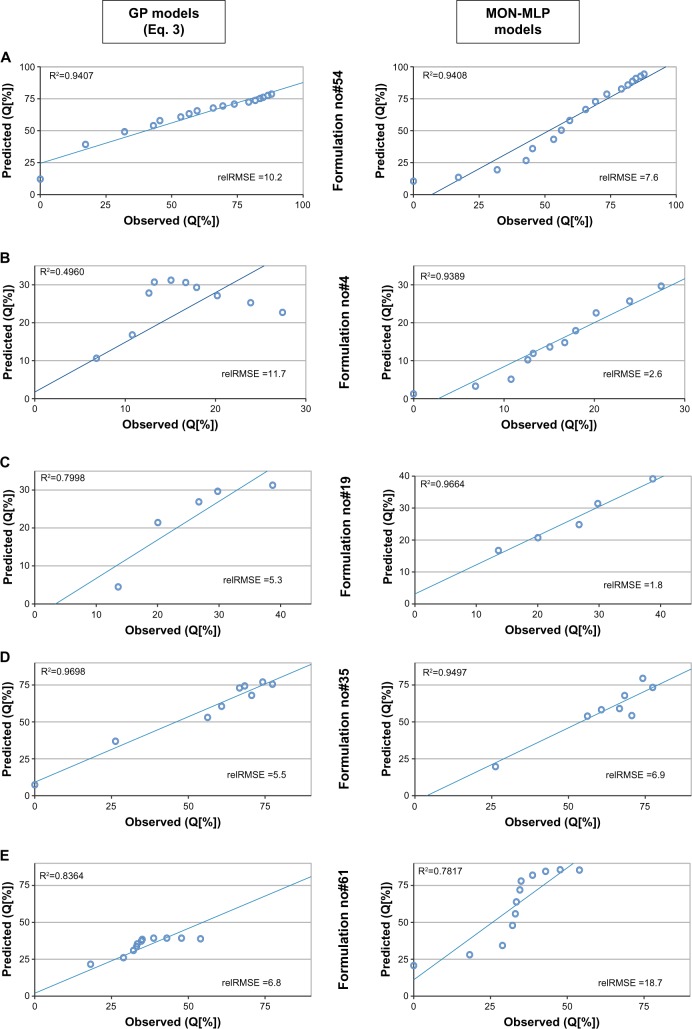
Due to the lack of data, only 68 formulations and 14 model substances were included. Nonetheless, since our model is based on the molecular descriptors, it is able to predict the behavior of a completely unknown drug substance. An example of such accurate prediction is formulation 54 (MON-MLP model relRMSE 7.6%, GP relRMSE 10.2%, molecule recombinant human erythropoietin coupled with human serum albumin [Figure 2A]).
Figure 2.
Comparison of predicted versus observed values for two best models (MON-MLP and classical equation).
Note: Data from the gathered database (Table S2).
Abbreviations: GP, genetic programming; MLP, multi-layer perceptron artifical neural networks; MON-MLP, monotone multi-layer perceptron neural networks; relRMSE, relative root-mean-square error.
In general, when considering the relRMSE of formulations, almost 60% of the release profiles had a relRMSE less than 15% when the MON-MLP model was applied. Moreover, the equation obtained by GP yielded 64% of profiles with a rel-RMSE less than 15%. Formulation-to-formulation comparison indicates that ten formulations yielded a relRMSE below 10%. It is important to note that those formulations consisted of medium-to-large molecules with a molecular weight ranging from 14.2 kDa (lysozyme) to 66.5 kDa (bovine serum albumin). Formulations that have a relRMSE of more than 30% could be perceived as outliers, with two formulations consisting of a relatively small molecule of bovine insulin (5.8 kDa).
It has been observed that, in some cases, the GP model yielded better predictions than MON-MLP, and vice versa (Figure 2B and C). The GP model had over- or underestimated the initial burst of molecule release (Figure 2D and E) and the end-point release (Figure 2D), while the midpoints of release profiles were in good accordance with the observed data. In contrast, for the MON-MLP models, even if it yielded worse generalization error, the shapes of the predicted profiles were preserved for most formulations (Figure 2B–E).
Based on the exploratory analysis of the modeling results, it could be concluded that the GP models are inferior in the presence of recombinant human epidermal growth factor and human serum albumin, showing much higher errors (Supplementary material Tables S1 and S2), whereas the MON-MLP model responded poorly to the presence of beta-amyloid, L-asparaginase and bovine insulin (Figure 2C). Moreover, if the value of the variable “mean particle size” is above 20 μm, the GP model predictions are most likely poor (Figure 2B) and, in the case of the MON-MLP model, the values below 1 μm yielded worse predictions. The above presents limitations of our model, which will be a subject of its development in the future. Due to the heuristic nature of the models, it is difficult to explain all the unexpected behaviors of the models, such as the erratic profile seen in Figure 2E, where the MON-MLP model creates a completely opposite profile at the time of 20 days. Thus we wanted to present our analysis of the models and their advantages (generic for the molecule) versus disadvantages (limitations of predictions) in order to allow every user to understand their applicability. The predicted versus observed release profiles of discussed formulations are presented in Figure 2, with the “observed” data derived from the literature.29–33
Conclusion
In this paper, a new approach was presented for modeling the dissolution of macromolecules from PLGA particles. It was also shown which aspects of the method of production coupled with molecular description of the formulation have the most impact on the release rate. Heuristic modeling techniques showed their usefulness in the complex and still not fully understood process of dissolution from PLGA particles. Moreover, they led to the mathematical formula (Equation 3) describing the process with predictive efficiency comparable to the ANN models.
With predictive modeling, knowledge extraction was performed simultaneously as a part of the modeling protocol. The major procedure employed feature selection techniques for reduction of input vector. A successful reduction from an initial 300 to 17 and eleven provided both information about the analyzed problem and appropriate generalization ability of MON-MLP and mathematical model, avoiding the well-known problem of “curse of dimensionality.” Analysis of crucial variables confirmed the dominating role of PLGA formulation characteristics; however, geometric characteristics of protein molecules and plasticizer ionic descriptor were also found to be important features. Analysis of the selected variables is a direct source of knowledge and a possible hint for creating a more general theory of how macromolecules are released from the PLGA particles.
This work was possible thanks to the application of two key elements:
Marvin, a cheminformatic tool, which allowed direct computation of macromolecule descriptors – an ability not present in every cheminformatic package;18 and
heuristic modeling tools for feature selection and predictive modeling integrating various types of information into the single model.
The resulting models are versatile in terms of prediction of the unknown macromolecule release profiles from the PLGA particles, thus it might be useful for the future development of PLGA microparticles as a decision support system.
Supplementary material
List of publications used in data extraction
| 1. Kang F, Singh J. Effect of additives on the release of a model protein from PLGA microspheres. AAPS PharmSciTech. 2001;2(4):30. |
| 2. Zhou XL, He JT, Du HJ, et al. Pharmacokinetic and pharmacodynamic profiles of recombinant human erythropoietin-loaded poly(lactic-co-glycolic acid) microspheres in rats. Acta Pharmacol Sin. 2012;33(1):137–44. |
| 3. Fan D, De Rosa E, Murphy MB, et al. Mesoporous silicon-PLGA composite microspheres for the double controlled release of biomolecules for orthopedic tissue engineering. Adv Funct Mater. 2012;22(2):282–293. |
| 4. Kim TH, Lee H, Park TG. Pegylated recombinant human epidermal growth factor (rhEGF) for sustained release from biodegradable PLGA microspheres. Biomaterials. 2002;23(11):2311–2317. |
| 5. Blanco D, Alonso MJ. Protein encapsulation and release from poly(lactide-co-glycolide) microspheres: effect of the protein and polymer properties and of the co-encapsulation of surfactants. Eur J Pharm Biopharm. 1998;45(3):285–294. |
| 6. Mok H, Park TG. Water-free microencapsulation of proteins within PLGA microparticles by spray drying using PEG-assisted protein solubilization technique in organic solvent. Eur J Pharm Biopharm. 2008;70(1):137–144. |
| 7. Buske J, König C, Bassarab S, Lamprecht A, Mühlau S, Wagner KG. Influence of PEG in PEG-PLGA microspheres on particle properties and protein release. Eur J Pharm Biopharm. 2012;81(1):57–63. |
| 8. Corrigan OI, Li X. Quantifying drug release from PLGA nanoparticulates. Eur J Pharm Sci. 2009;37(3–4):477–485. |
| 9. Puras G, Salvador A, Igartua M, Hernández RM, Pedraz JL. Encapsulation of Aβ(1–15) in PLGA microparticles enhances serum antibody response in mice immunized by subcutaneous and intranasal routes. Eur J Pharm Sci. 2011;44(3):200–206. |
| 10. Kim HK, Park TG. Microencapsulation of dissociable human growth hormone aggregates within poly(D,L-lactic-co-glycolic acid) microparticles for sustained release. Int J Pharm. 2001;229(1–2):107–116. |
| 11. Han Y, Tian H, He P, Chen X, Jing X. Insulin nanoparticle preparation and encapsulation into poly(lactic-co-glycolic acid) microspheres by using an anhydrous system. Int J Pharm. 2009;378(1–2):159–166. |
| 12. He J, Feng M, Zhou X, et al. Stabilization and encapsulation of recombinant human erythropoietin into PLGA microspheres using human serum albumin as a stabilizer. Int J Pharm. 2011;416(1):69–76. |
| 13. Gasper MM, Blanco D, Cruz ME, Alonso MJ. Formulation of L-asparaginase-loaded poly(lactide-co-glycolide) nanoparticles: influence of polymer properties on enzyme loading, activity and in vitro release. J Control Release. 1998;52(1–2):53–62. |
| 14. Kawashima Y, Yamamoto H, Takeuchi H, Fujioka S, Hino T. Pulmonary delivery of insulin with nebulized DL-lactide/glycolide copolymer (PLGA) nanospheres to prolong hypoglycemic effect. J Control Release. 1999;62(1–2):279–287. |
| 15. Ungaro F, d’Emmanuele di Villa Bianca R, Giovino C, et al. Insulin-loaded PLGA/cyclodextrin large porous particles with improved aerosolization properties: in vivo deposition and hypoglycaemic activity after delivery to rat lungs. J Control Release. 2009;135(1):25–34. |
| 16. Jiang HL, Jin JF, Hu YQ, Zhu KJ. Improvement of protein loading and modulation of protein release from poly(lactide-co-glycolide) microspheres by complexation of proteins with polyanions. J Microencapsul. 2004;21(6):615–624. |
| 17. Pirooznia N, Hasannia S, Lotfi AS, Ghanei M. Encapsulation of alpha-1 antitrypsin in PLGA nanoparticles: in vitro characterization as an effective aerosol formulation in pulmonary diseases. J Nanobiotechnology. 2012;10:20. |
| 18. Castellanos IJ, Flores G, Griebenow K. Effect of cyclodextrins on alpha-chymotrypsin stability and loading in PLGA microspheres upon S/O/W encapsulation. J Pharm Sci. 2006;95(4):849–858. |
Full data base used in the study
| The full data base of macromolecules release from PLGA microparticles http://nigella.farmacja.cm-uj.krakow.pl/~kuba/Int_J_Nanomedicine/Supplementary_material_S2.xlsx |
The results of the formulation-to-formulation relRMSE (%)
| Formulation no | relRMSE (%)
|
|
|---|---|---|
| MON-MLP model | GP model (Equation 2) | |
| 1 | 9.56 | 14.43 |
| 2 | 10.29 | 11.70 |
| 3 | 22.14 | 18.00 |
| 4 | 2.56 | 11.74 |
| 5 | 22.63 | 14.75 |
| 6 | 12.01 | 19.47 |
| 7 | 10.72 | 13.99 |
| 8 | 10.12 | 13.87 |
| 9 | 11.42 | 12.84 |
| 10 | 20.92 | 21.92 |
| 11 | 17.73 | 5.25 |
| 12 | 7.48 | 8.78 |
| 13 | 17.47 | 8.94 |
| 14 | 12.79 | 23.37 |
| 15 | 3.12 | 5.35 |
| 16 | 1.75 | 9.75 |
| 17 | 16.10 | 6.76 |
| 18 | 13.47 | 8.76 |
| 19 | 1.83 | 5.27 |
| 20 | 4.31 | 8.38 |
| 21 | 12.97 | 15.05 |
| 22 | 3.87 | 7.57 |
| 23 | 6.26 | 12.34 |
| 24 | 6.55 | 6.99 |
| 25 | 5.35 | 12.21 |
| 26 | 25.05 | 13.12 |
| 27 | 14.48 | 8.83 |
| 28 | 17.05 | 14.67 |
| 29 | 11.20 | 9.26 |
| 30 | 15.05 | 7.01 |
| 31 | 36.03 | 27.25 |
| 32 | 34.23 | 26.38 |
| 33 | 8.21 | 10.49 |
| 34 | 26.99 | 25.33 |
| 35 | 6.85 | 5.46 |
| 36 | 26.82 | 22.38 |
| 37 | 28.55 | 22.87 |
| 38 | 19.27 | 24.45 |
| 39 | 8.78 | 11.05 |
| 40 | 9.20 | 9.04 |
| 41 | 18.09 | 17.55 |
| 42 | 16.36 | 15.15 |
| 43 | 10.71 | 5.39 |
| 44 | 7.15 | 10.71 |
| 45 | 20.51 | 7.39 |
| 46 | 8.75 | 32.83 |
| 47 | 15.23 | 23.02 |
| 48 | 21.05 | 20.61 |
| 49 | 4.70 | 17.83 |
| 50 | 11.18 | 6.56 |
| 51 | 10.90 | 16.59 |
| 52 | 11.20 | 18.85 |
| 53 | 10.85 | 19.87 |
| 54 | 7.63 | 10.20 |
| 55 | 25.31 | 10.20 |
| 57 | 19.16 | 24.17 |
| 58 | 24.05 | 21.14 |
| 59 | 31.39 | 29.31 |
| 60 | 28.53 | 22.90 |
| 61 | 30.81 | 5.76 |
| 62 | 18.73 | 6.79 |
| 63 | 7.59 | 8.59 |
| 64 | 13.37 | 6.16 |
| 65 | 8.93 | 7.73 |
| 66 | 4.61 | 10.34 |
| 67 | 11.72 | 4.76 |
| 68 | 14.25 | 5.85 |
| Min | 1.75 | 4.76 |
| Max | 36.03 | 32.83 |
| Mean | 14.39 | 13.75 |
Acknowledgments
This work was funded by Poland-Singapore bilateral cooperation project no 2/3/POL-SIN/2012 and IPROCOM Marie Curie Initial Training Network project no 316555.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Makadia HK, Siegel SJ. Poly lactic-co-glycolic acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers (Basel) 2011;3:1377–1397. doi: 10.3390/polym3031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Préat V. PLGA-based nanoparticles: an overview of biomedical applications. J Control Release. 2012;161(2):505–522. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 3.Houchin ML, Topp EM. Physical properties of PLGA films during polymer degradation. J Appl Polym Sci. 2009;114:2848–2854. [Google Scholar]
- 4.Wang J, Ng CW, Win KY, et al. Release of paclitaxel from polylactide-co-glycolide (PLGA) microparticles and discs under irradiation. J Microencapsul. 2003;20(3):317–327. doi: 10.1080/0265204021000058401. [DOI] [PubMed] [Google Scholar]
- 5.Hamishehkar H, Emami J, Najafabadi AR, et al. Pharmacokinetics and pharmacodynamics of controlled release insulin loaded PLGA microcapsules using dry powder inhaler in diabetic rats. Biopharm Drug Dispos. 2010;31(2–3):189–201. doi: 10.1002/bdd.702. [DOI] [PubMed] [Google Scholar]
- 6.De S, Robinson DH. Particle size and temperature effect on the physical stability of PLGA nanospheres and microspheres containing Bodipy. AAPS PharmSciTech. 2004;5(4):e53. doi: 10.1208/pt050453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fredenberg S, Wahlgren M, Reslow M, Axelsson A. The mechanisms of drug release in poly(lactic-co-glycolic acid)-based drug delivery systems – a review. Int J Pharm. 2011;415:34–52. doi: 10.1016/j.ijpharm.2011.05.049. [DOI] [PubMed] [Google Scholar]
- 8.D’Souza SS, DeLuca PP. Development of a dialysis in vitro release method for biodegradable microspheres. AAPS PharmSciTech. 2005;6(2):E323–E328. doi: 10.1208/pt060242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mollo R, Corrigan OI. An investigation of the mechanism of release of the amphoteric drug amoxycillin from poly(D,L-lactide-co-glycolide) matrices. Pharm Dev Technol. 2002;7:333–343. doi: 10.1081/pdt-120005730. [DOI] [PubMed] [Google Scholar]
- 10.Kang J, Schwendeman SP. Pore closing and opening in biodegradable polymers and their effect on the controlled release of proteins. Mol Pharm. 2007;4(1):104–18. doi: 10.1021/mp060041n. [DOI] [PubMed] [Google Scholar]
- 11.Blanco MD, Alonso MJ. Development and characterization of protein-loaded poly(lactide-co-glycolide) nanospheres. Eur J Pharm Biopharm. 1997;43(3):287–294. [Google Scholar]
- 12.Kang J, Lambert O, Ausborn M, Schwendeman SP. Stability of proteins encapsulated in injectable and biodegradable poly(lactide-co-glycolide)-glucose millicylinders. Int J Pharm. 2008;357(1–2):235–243. doi: 10.1016/j.ijpharm.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mainardes RM, Evangelista RC. PLGA nanoparticles containing praziquantel: effect of formulation variables on size distribution. Int J Pharm. 2005;290(1–2):137–144. doi: 10.1016/j.ijpharm.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 14.Zygourakis K, Markenscoff PA. Computer-aided design of bioerodible devices with optimal release characteristics: a cellular automata approach. Biomaterials. 1996;17(2):125–135. doi: 10.1016/0142-9612(96)85757-7. [DOI] [PubMed] [Google Scholar]
- 15.Göpferich A. Mechanisms of polymer degradation and erosion. Biomaterials. 1996;17(2):103–114. doi: 10.1016/0142-9612(96)85755-3. [DOI] [PubMed] [Google Scholar]
- 16.Siepmann J, Faisant N, Benoit JP. A new mathematical model quantifying drug release from bioerodible microparticles using Monte Carlo simulations. Pharm Res. 2002;19:1885–1893. doi: 10.1023/a:1021457911533. [DOI] [PubMed] [Google Scholar]
- 17.Barat A, Crane M, Ruskin HJ. Quantitative multi-agent models for simulating protein release from PLGA bioerodible nano- and microspheres. J Pharm Biomed Anal. 2008;48(2):361–368. doi: 10.1016/j.jpba.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 18.Marvin ChemAxon [Accessed 10, Jan 2013]. ( http://www.chemaxon.com)
- 19.Żurada JM, Malinowski A, Usuib S. Perturbation method for deleting redundant inputs of perceptron networks. Neurocomputing. 1997;14:177–193. [Google Scholar]
- 20.Mendyk A, Jachowicz R. Unified methodology of neural analysis in decision support systems built for pharmaceutical technology. Expert Syst Appl. 2007;32:1124–1131. [Google Scholar]
- 21.Szlęk J. The fscaret. Automated feature selection using variety of models provided by caret package. [Accessed Jun 2013]. Available from: http://cran.r-project.org/web/packages/fscaret/index.html.
- 22.R Core Team R: a language and environment for statistical computing. Vienna: The R Foundation for Statistical Computing. [Accessed 1, Jun 2013]. Available from: http://www.R-project.org/
- 23.Mendyk A, Tuszyński PK, Polak S, Jachowicz R. Generalized in vitro-in vivo relationship (IVIVR) model based on artificial neural networks. Drug Des Devel Ther. 2013;7:223–232. doi: 10.2147/DDDT.S41401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cannon AJ. monmlp: Monotone multi-layer perceptron neural network [webpage on the Internet] [Accessed Jun 2012]. Available from: http://cran.r-project.org/web/packages/monmlp/index.html.
- 25.Yager RR, Filev DP. Essentials of Fuzzy Modeling and Control. NY: John Wiley & Sons; 1994. [Google Scholar]
- 26.Flasch O, Mersmann O, Bartz-Beielstein T, Stork J, Zaefferer M. rgp: R genetic programming framework [webpage on the Internet] [Accessed 1, Jun 2013]. Available from: http://CRAN.R-project.org/package=rgp.
- 27.Nash JC, Varadhan R. Unifying optimization algorithms to aid software system users: optimx for R. J Stat Softw. 2011;43(9):1–14. [Google Scholar]
- 28.Sandor M, Enscore D, Weston P, Mathiowitz E. Effect of protein molecular weight on release from micron-sized PLGA microspheres. J Control Release. 2001;76(3):297–311. doi: 10.1016/s0168-3659(01)00446-1. [DOI] [PubMed] [Google Scholar]
- 29.He J, Feng M, Zhou X, et al. Stabilization and encapsulation of recombinant human erythropoietin into PLGA microspheres using human serum albumin as a stabilizer. Int J Pharm. 2011;416(1):69–76. doi: 10.1016/j.ijpharm.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Kang F, Singh J. Effect of additives on the release of a model protein from PLGA microspheres. AAPS PharmSciTech. 2001;2(4):30. doi: 10.1007/BF02830570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanco D, Alonso MJ. Protein encapsulation and release from poly(lactide-co-glycolide) microspheres: effect of the protein and polymer properties and of the co-encapsulation of surfactants. Eur J Pharm Biopharm. 1998;45(3):285–294. doi: 10.1016/s0939-6411(98)00011-3. [DOI] [PubMed] [Google Scholar]
- 32.Buske J, König C, Bassarab S, Lamprecht A, Mühlau S, Wagner KG. Influence of PEG in PEG-PLGA microspheres on particle properties and protein release. Eur J Pharm Biopharm. 2012;81(1):57–63. doi: 10.1016/j.ejpb.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 33.Jiang HL, Jin JF, Hu YQ, Zhu KJ. Improvement of protein loading and modulation of protein release from poly(lactide-co-glycolide) microspheres by complexation of proteins with polyanions. J Microencapsul. 2004;21(6):615–624. doi: 10.1080/02652040400000538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of publications used in data extraction
| 1. Kang F, Singh J. Effect of additives on the release of a model protein from PLGA microspheres. AAPS PharmSciTech. 2001;2(4):30. |
| 2. Zhou XL, He JT, Du HJ, et al. Pharmacokinetic and pharmacodynamic profiles of recombinant human erythropoietin-loaded poly(lactic-co-glycolic acid) microspheres in rats. Acta Pharmacol Sin. 2012;33(1):137–44. |
| 3. Fan D, De Rosa E, Murphy MB, et al. Mesoporous silicon-PLGA composite microspheres for the double controlled release of biomolecules for orthopedic tissue engineering. Adv Funct Mater. 2012;22(2):282–293. |
| 4. Kim TH, Lee H, Park TG. Pegylated recombinant human epidermal growth factor (rhEGF) for sustained release from biodegradable PLGA microspheres. Biomaterials. 2002;23(11):2311–2317. |
| 5. Blanco D, Alonso MJ. Protein encapsulation and release from poly(lactide-co-glycolide) microspheres: effect of the protein and polymer properties and of the co-encapsulation of surfactants. Eur J Pharm Biopharm. 1998;45(3):285–294. |
| 6. Mok H, Park TG. Water-free microencapsulation of proteins within PLGA microparticles by spray drying using PEG-assisted protein solubilization technique in organic solvent. Eur J Pharm Biopharm. 2008;70(1):137–144. |
| 7. Buske J, König C, Bassarab S, Lamprecht A, Mühlau S, Wagner KG. Influence of PEG in PEG-PLGA microspheres on particle properties and protein release. Eur J Pharm Biopharm. 2012;81(1):57–63. |
| 8. Corrigan OI, Li X. Quantifying drug release from PLGA nanoparticulates. Eur J Pharm Sci. 2009;37(3–4):477–485. |
| 9. Puras G, Salvador A, Igartua M, Hernández RM, Pedraz JL. Encapsulation of Aβ(1–15) in PLGA microparticles enhances serum antibody response in mice immunized by subcutaneous and intranasal routes. Eur J Pharm Sci. 2011;44(3):200–206. |
| 10. Kim HK, Park TG. Microencapsulation of dissociable human growth hormone aggregates within poly(D,L-lactic-co-glycolic acid) microparticles for sustained release. Int J Pharm. 2001;229(1–2):107–116. |
| 11. Han Y, Tian H, He P, Chen X, Jing X. Insulin nanoparticle preparation and encapsulation into poly(lactic-co-glycolic acid) microspheres by using an anhydrous system. Int J Pharm. 2009;378(1–2):159–166. |
| 12. He J, Feng M, Zhou X, et al. Stabilization and encapsulation of recombinant human erythropoietin into PLGA microspheres using human serum albumin as a stabilizer. Int J Pharm. 2011;416(1):69–76. |
| 13. Gasper MM, Blanco D, Cruz ME, Alonso MJ. Formulation of L-asparaginase-loaded poly(lactide-co-glycolide) nanoparticles: influence of polymer properties on enzyme loading, activity and in vitro release. J Control Release. 1998;52(1–2):53–62. |
| 14. Kawashima Y, Yamamoto H, Takeuchi H, Fujioka S, Hino T. Pulmonary delivery of insulin with nebulized DL-lactide/glycolide copolymer (PLGA) nanospheres to prolong hypoglycemic effect. J Control Release. 1999;62(1–2):279–287. |
| 15. Ungaro F, d’Emmanuele di Villa Bianca R, Giovino C, et al. Insulin-loaded PLGA/cyclodextrin large porous particles with improved aerosolization properties: in vivo deposition and hypoglycaemic activity after delivery to rat lungs. J Control Release. 2009;135(1):25–34. |
| 16. Jiang HL, Jin JF, Hu YQ, Zhu KJ. Improvement of protein loading and modulation of protein release from poly(lactide-co-glycolide) microspheres by complexation of proteins with polyanions. J Microencapsul. 2004;21(6):615–624. |
| 17. Pirooznia N, Hasannia S, Lotfi AS, Ghanei M. Encapsulation of alpha-1 antitrypsin in PLGA nanoparticles: in vitro characterization as an effective aerosol formulation in pulmonary diseases. J Nanobiotechnology. 2012;10:20. |
| 18. Castellanos IJ, Flores G, Griebenow K. Effect of cyclodextrins on alpha-chymotrypsin stability and loading in PLGA microspheres upon S/O/W encapsulation. J Pharm Sci. 2006;95(4):849–858. |
Full data base used in the study
| The full data base of macromolecules release from PLGA microparticles http://nigella.farmacja.cm-uj.krakow.pl/~kuba/Int_J_Nanomedicine/Supplementary_material_S2.xlsx |
The results of the formulation-to-formulation relRMSE (%)
| Formulation no | relRMSE (%)
|
|
|---|---|---|
| MON-MLP model | GP model (Equation 2) | |
| 1 | 9.56 | 14.43 |
| 2 | 10.29 | 11.70 |
| 3 | 22.14 | 18.00 |
| 4 | 2.56 | 11.74 |
| 5 | 22.63 | 14.75 |
| 6 | 12.01 | 19.47 |
| 7 | 10.72 | 13.99 |
| 8 | 10.12 | 13.87 |
| 9 | 11.42 | 12.84 |
| 10 | 20.92 | 21.92 |
| 11 | 17.73 | 5.25 |
| 12 | 7.48 | 8.78 |
| 13 | 17.47 | 8.94 |
| 14 | 12.79 | 23.37 |
| 15 | 3.12 | 5.35 |
| 16 | 1.75 | 9.75 |
| 17 | 16.10 | 6.76 |
| 18 | 13.47 | 8.76 |
| 19 | 1.83 | 5.27 |
| 20 | 4.31 | 8.38 |
| 21 | 12.97 | 15.05 |
| 22 | 3.87 | 7.57 |
| 23 | 6.26 | 12.34 |
| 24 | 6.55 | 6.99 |
| 25 | 5.35 | 12.21 |
| 26 | 25.05 | 13.12 |
| 27 | 14.48 | 8.83 |
| 28 | 17.05 | 14.67 |
| 29 | 11.20 | 9.26 |
| 30 | 15.05 | 7.01 |
| 31 | 36.03 | 27.25 |
| 32 | 34.23 | 26.38 |
| 33 | 8.21 | 10.49 |
| 34 | 26.99 | 25.33 |
| 35 | 6.85 | 5.46 |
| 36 | 26.82 | 22.38 |
| 37 | 28.55 | 22.87 |
| 38 | 19.27 | 24.45 |
| 39 | 8.78 | 11.05 |
| 40 | 9.20 | 9.04 |
| 41 | 18.09 | 17.55 |
| 42 | 16.36 | 15.15 |
| 43 | 10.71 | 5.39 |
| 44 | 7.15 | 10.71 |
| 45 | 20.51 | 7.39 |
| 46 | 8.75 | 32.83 |
| 47 | 15.23 | 23.02 |
| 48 | 21.05 | 20.61 |
| 49 | 4.70 | 17.83 |
| 50 | 11.18 | 6.56 |
| 51 | 10.90 | 16.59 |
| 52 | 11.20 | 18.85 |
| 53 | 10.85 | 19.87 |
| 54 | 7.63 | 10.20 |
| 55 | 25.31 | 10.20 |
| 57 | 19.16 | 24.17 |
| 58 | 24.05 | 21.14 |
| 59 | 31.39 | 29.31 |
| 60 | 28.53 | 22.90 |
| 61 | 30.81 | 5.76 |
| 62 | 18.73 | 6.79 |
| 63 | 7.59 | 8.59 |
| 64 | 13.37 | 6.16 |
| 65 | 8.93 | 7.73 |
| 66 | 4.61 | 10.34 |
| 67 | 11.72 | 4.76 |
| 68 | 14.25 | 5.85 |
| Min | 1.75 | 4.76 |
| Max | 36.03 | 32.83 |
| Mean | 14.39 | 13.75 |