Abstract
Long non-coding RNAs (lncRNAs), transcribed from the intergenic regions of animal genomes, play important roles in key biological processes. In mice, Zdbf2linc was recently identified as an lncRNA isoform of the paternally expressed imprinted Zdbf2 gene. The functional role of Zdbf2linc remains undefined, but it may control parent-of-origin-specific expression of protein-coding neighbors through epigenetic modification in cis, similar to imprinted Nespas, Kcnq1ot1 and Airn lncRNAs. Here, we identified a novel imprinted long-range non-coding RNA, termed GPR1AS, in the human GPR1-ZDBF2 intergenic region. Although GPR1AS contains no human ZDBF2 exons, this lncRNA is transcribed in the antisense orientation from the GPR1 intron to a secondary, differentially methylated region upstream of the ZDBF2 gene (ZDBF2 DMR), similar to mouse Zdbf2linc. Interestingly, GPR1AS/Zdbf2linc is exclusively expressed in human/mouse placenta with paternal-allele-specific expression and maternal-allele-specific promoter methylation (GPR1/Gpr1 DMR). The paternal-allele specific methylation of the secondary ZDBF2 DMR was established in human placentas as well as somatic lineage. Meanwhile, the ZDBF2 gene showed stochastic paternal-allele-specific expression, possibly methylation-independent, in placental tissues. Overall, we demonstrated that epigenetic regulation mechanisms in the imprinted GPR1-GPR1AS-ZDBF2 region were well-conserved between human and mouse genomes without the high sequence conservation of the intergenic lncRNAs. Our findings also suggest that lncRNAs with highly conserved epigenetic and transcriptional regulation across species arose by divergent evolution from a common ancestor, if they do not have identical exon structures.
Keywords: lncRNA, genomic imprinting, DNA methylation, antisense RNA, placenta
Introduction
One surprising result of the human genome project is the discovery that protein coding sequences account for a very small fraction (~1%) of the genomic sequence.1 Human transcriptome analysis revealed that only one-fifth of transcription across the genome is associated with protein-coding genes.2 The untranslated transcripts are broadly referred to as ?non-coding? RNA molecules and include small RNAs and long non-coding RNAs (lncRNAs). Small RNAs (e.g., miRNA, piRNA and snoRNA) are critically important in a variety of biological processes.3 However, small RNAs represent a small portion of the non-coding genes; the majority of non-coding RNAs are lncRNAs.2 The lncRNAs are defined as non-coding RNAs with more than 200 nucleotides. Genome-wide experimental and computational studies have shown that lncRNAs are poorly conserved across species; for instance, only 14% of the mouse lncRNAs have human orthologs.4,5 Although they generally are less well conserved across species and often show high tissue specificity and low expression levels across multiple tissues and developmental stages, studies have revealed that lncRNAs also play important roles in gene regulation through chromatin remodeling, e.g., X chromosome dosage compensation or regulation of HOX genes.3,6-8 Nonetheless, most lncRNAs remain poorly understood; this class of RNA has become a new paradigm for gene regulation.
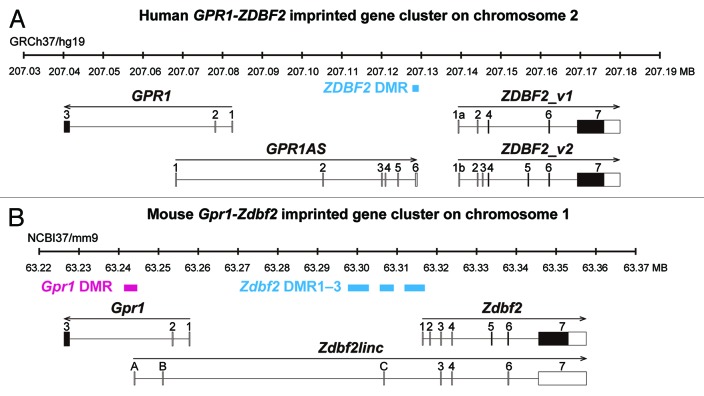
In placental mammals, the functional non-equivalence of the parental genomes is mediated by ?genomic imprinting,? an epigenetic mechanism by which imprinted genes are expressed in a parent-of-origin-specific manner.9 Approximately 150 imprinted genes have been identified so far in humans and mice, and the imprinting status of many is conserved across mammalian species (imprinted gene catalogs: www.mousebook.org/catalog.php?catalog=imprinting, igc.otago.ac.nz/home.html, www.geneimprint.com). Most of the imprinted genes occur in clusters with silencing controlled by an imprint control region (ICR) that is methylated on one parental allele, a stable epigenetic mark established during gametogenesis.10 The unmethlyated ICR acts as promoter for lncRNAs such as Nespas, Kcnq1ot1 and Airn, which then silence all imprinted genes in the cluster on that parental allele.11-17 Thus, these imprinted gene clusters provide important epigenetic models for the numerous lncRNAs mapped in the mammalian genome. We and other groups have identified an imprinted gene cluster containing the GPR1/Gpr1 (G protein-coupled receptor 1) and ZDBF2/Zdbf2 (zinc finger, DBF-type containing 2) genes on the human chromosome 2q3.33 and mouse chromosome 1C2; both are specifically expressed from the paternal allele.18-20 Zdbf2 exhibits imprinted expression in various fetal and adult tissues, whereas Gpr1 exhibits kidney-specific imprinted expression in mice. Furthermore, we identified a novel mouse Zdbf2 isoform, Zdbf2linc, a long-range (transcription across 114 kb) non-coding RNA, transcribed from Gpr1 intron 2 in the antisense orientation with respect to Gpr1 (Fig. 1).23 Zdbf2linc is also expressed in a paternal-allele-specific manner and, importantly, both the non-coding and original transcripts were controlled by maternal DNA methylation imprints via DNMT3L, a cofactor of DNA methyltransferase.24 The imprinted Gpr1-Zdbf2 locus therefore has two similarities with the above-referenced imprinted clusters: (1) two differential states of imprinted DNA methylation, namely, maternal-allele-specific methylation at ICRs (also called ?primary? differentially methylated regions or DMRs) and paternal-allele-specific methylation established after fertilization/implantation at ?secondary? DMRs, and (2) paternal specific transcription of lncRNAs from ICRs regulated by maternal methylation.23 Although little is known about the rule of these secondary DMRs, their monoallelic methylation marks may have essential regulatory function for the single or multiple imprinted neighbor genes during post-fertilization development.25 Mouse imprinted Gpr1-Zdbf2 domain harbors the germline-derived primary Gpr1 DMR at the promoter of Zdbf2linc and the post-fertilization-derived secondary Zdbf2 DMR1?3 upstream of the Zdbf2 gene (Fig. 1). In human, only ZDBF2 DMR, which exhibits paternal-allele-specific DNA methylation, was identified at the intergenic region between GPR1 and ZDBF2.21 We must elucidate the transcriptional and epigenetic profiles and structures of a gene cluster to understand the mechanisms by which imprinted expression is regulated.
Figure 1. Schematic physical maps of human (top) and mouse (bottom) orthologous imprinted GPR1-GPR1AS-ZDBF2/Gpr1-Zdbf2linc-Zdbf2 clusters. Imprinted genes are represented as follows: Open and filled boxes represent untranslated regions and ORFs, respectively; arrows indicate transcription orientation. Human GPR1AS does not contain any significant ORFs. ZDBF2_v2 codes for an ORF similar to the original ZDBF2 protein sequence encoded by ZDBF2_v1 (see Fig. S1). Magenta and cyan bars represent maternally or paternally methylated DMRs in mice. The extent of each DMR was determined in previous DNA methylome studies.20-22
In this study, we identified a novel, long (>1.5 kb, transcription across 60 kb) imprinted, non-coding gene transcribed from GPR1 intron 2 in the human genome. The novel lncRNA gene does not include any human ZDBF2 exons (meaning that the transcript is not an isoform of human ZDBF2), but its expression profile is very similar to that of mouse Zdbf2linc. Our findings strongly indicate that epigenetic and transcriptional regulation mechanisms of the GPR1-ZDBF2 locus are well-conserved between human and mouse genomes and that this lncRNA might play a crucial role in local gene regulation.
Results
Identification of novel long-range RNA transcript, GPR1AS, at the human GPR1-ZDBF2 locus
We mapped the 5? end of the human ZDBF2 transcript using rapid amplification of cDNA ends (RACE) analysis of human embryonic stem (ES) cells to identify a human ZDBF2 splicing variant homologous to the mouse Zdbf2linc. The original and new variants were termed ZDBF2_v1 and ZDBF2_v2. ZDBF2_v1 contains 5 original exons, renamed exon 1a, 2, 4, 6 and 7 (RefSeq database: exon 1?5). ZDBF2_v2 contains an alternative first exon, exon 1b, located between the first and second exons of ZDBF2_v1; 4 exons match NCBI exons (RefSeq database: exon 2?5); extra exons (exon 3 and 5) are located between the second/third and third/fourth exons of ZDBF2_v1 (Fig. 1; Table S1). ZDBF2_v2 has a large open reading frame (ORF, 2352 amino acids), similar to the ZDBF2 protein encoded by ZDBF2_v1 (Fig. S1). It is unclear whether the newly identified ZDBF2_v2 is the true coding or non-coding RNA; however, we identified no large intergenic isoform of ZDBF2 transcribed from the GPR1 gene (as well as mouse Zdbf2linc) in this approach.
Next, we focused on an EST clone, BE734382 (914 bp without the poly-A tail), which is an antisense transcript transcribed from GPR1 intron 2 to the ZDBF2 gene, just like the mouse Zdbf2linc (Fig. S2). To determine the full length of the antisense GPR1 gene transcript, we therefore performed 5?- and 3?-RACE using human ES cells. The RACE analyses showed the antisense transcript encodes a 1505-nt RNA molecule and contains 6 exons; the first exon is located at GPR1 intron 2 and the remaining 5 exons are located in the intergenic region between GPR1 and ZDBF2 (Fig. 1; Fig. S3). This spliced antisense RNA is transcribed through the 60-kb genomic region but does not include any ZDBF2 exons and contains no large ORFs encoding larger than 100 amino acids. Interestingly, the transcription termination site of this novel ?lncRNA? reaches and overlaps the human ZDBF2 DMR, the known DMR upstream of the ZDBF2 gene.21 We named this novel gene encoding a long antisense non-coding RNA transcript GPR1-AntiSense (GPR1AS).
GPR1AS is a novel paternally expressed imprinted gene in the human genome
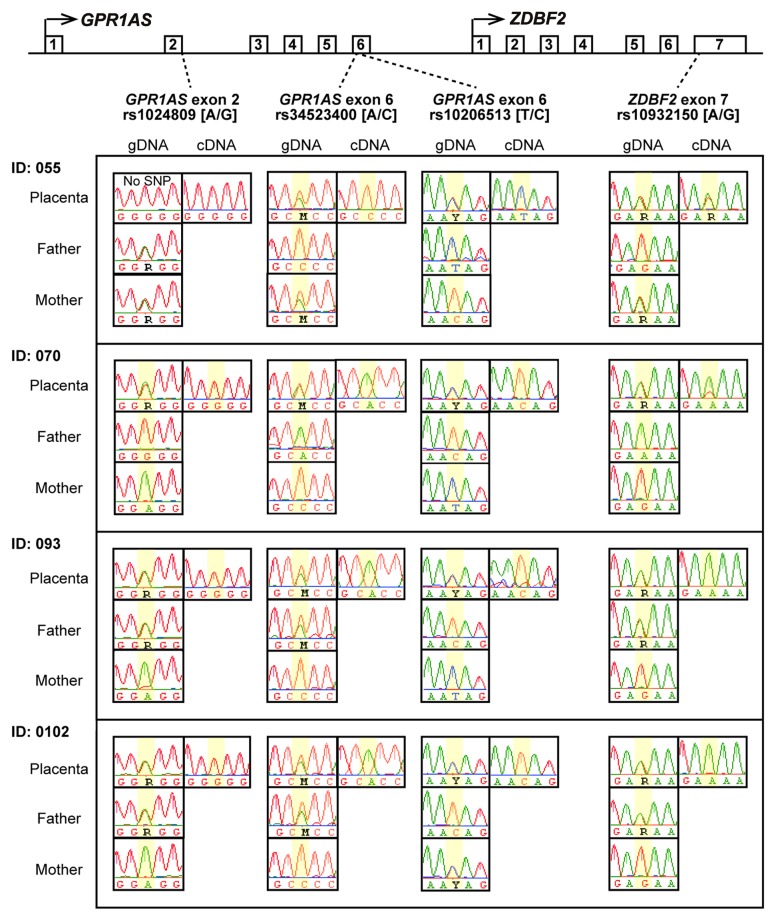
Next, we characterized GPR1AS/Zdbf2linc expression profiles in fetal and adult human and mouse tissues by relative quantification in real-time RT-PCR. Therefore, we demonstrated that GPR1AS/Zdbf2linc is exclusively and strongly expressed in the placenta and adult testis, and Zdbf2linc is expressed weakly in mouse brain; however, GPR1AS/Zdbf2linc transcript levels were very low or almost undetectable in other tissues (Fig. 2). Notably, this strong placental expression of GPR1AS is reminiscent of the original BE734382 EST clone obtained from human placenta. We also performed allelic expression profiling of human placentas using single nucleotide polymorphism (SNP). Three SNPs have been identified in GPR1AS exons; we designed RT-PCR primers within these exons to detect these SNPs (reference SNP ID: rs1024809; at exon 2, rs34523400 and rs10932150; at exon 6). Allele-specific expression analysis was performed by direct sequencing of RT-PCR products of these exons in informative cases: four human placental tissues from a child heterozygous for individual SNP sites, and only the paternal allele was detected (Fig. 3). Thus, we demonstrated that GPR1AS is a novel imprinted gene that is expressed from a paternal allele in the human placenta.
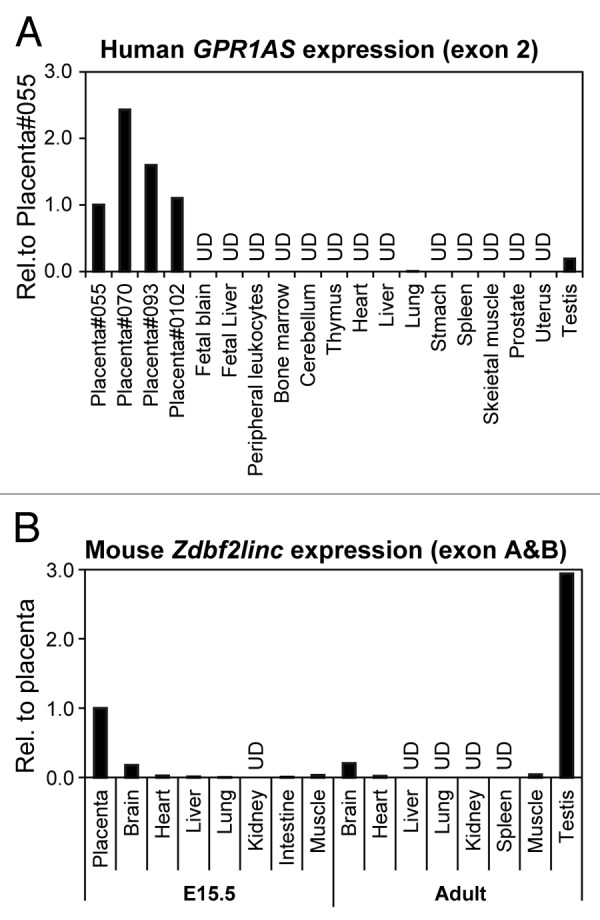
Figure 2. Relative expression of GPR1AS/Zdbf2linc in multiple human and mouse tissues (qRT-PCR). Expression in the human placenta (Family ID: 055) or mouse placenta at E15.5 is set as 1 for each gene. UD means undetectable after 40 cycles of amplification.
Figure 3. Allele-specific RT-PCR sequencing of GPR1AS and ZDBF2 in human placentas. Four heterozygous genotypes were used to distinguish between maternal and paternal alleles in human placental samples from four Japanese patients (National Center for Child Health and Development). SNP positions are highlighted in yellow.
Our previous study in mice demonstrated that Zdbf2 and Zdbf2linc transcripts were controlled by maternal methylation imprinting, most likely in the Gpr1 DMR at the promoter region of Zdbf2linc.23 Therefore, we investigated the methylation status of the promoter region of GPR1AS in human placental tissues. Three SNPs have been identified upstream of the first GPR1AS exon (reference SNP ID: rs16838070, rs1683074 and rs130009940). Of these, rs16838070 and rs1683074 were observed in the Japanese population, and were in strong linkage disequilibrium; rs130009940 is rare Japanese (data not shown). Therefore, we designed primer sets for bisulfite sequencing within the promoter region that includes rs1683074. Four informative cases of placentas and umbilical cord blood lymphocytes were investigated. In all cases, the GPR1AS promoter was hypomethylated (methylation levels, ?25%) on the active paternal allele and hypermethylated (methylation levels, ?75%) on the repressed maternal allele in placenta; the promoter was hypermethylated on both parental alleles in blood lymphocytes (Fig. 4). Thus, a novel maternally methylated ?human GPR1 DMR? was identified in a human region orthologous to the mouse Gpr1 DMR.
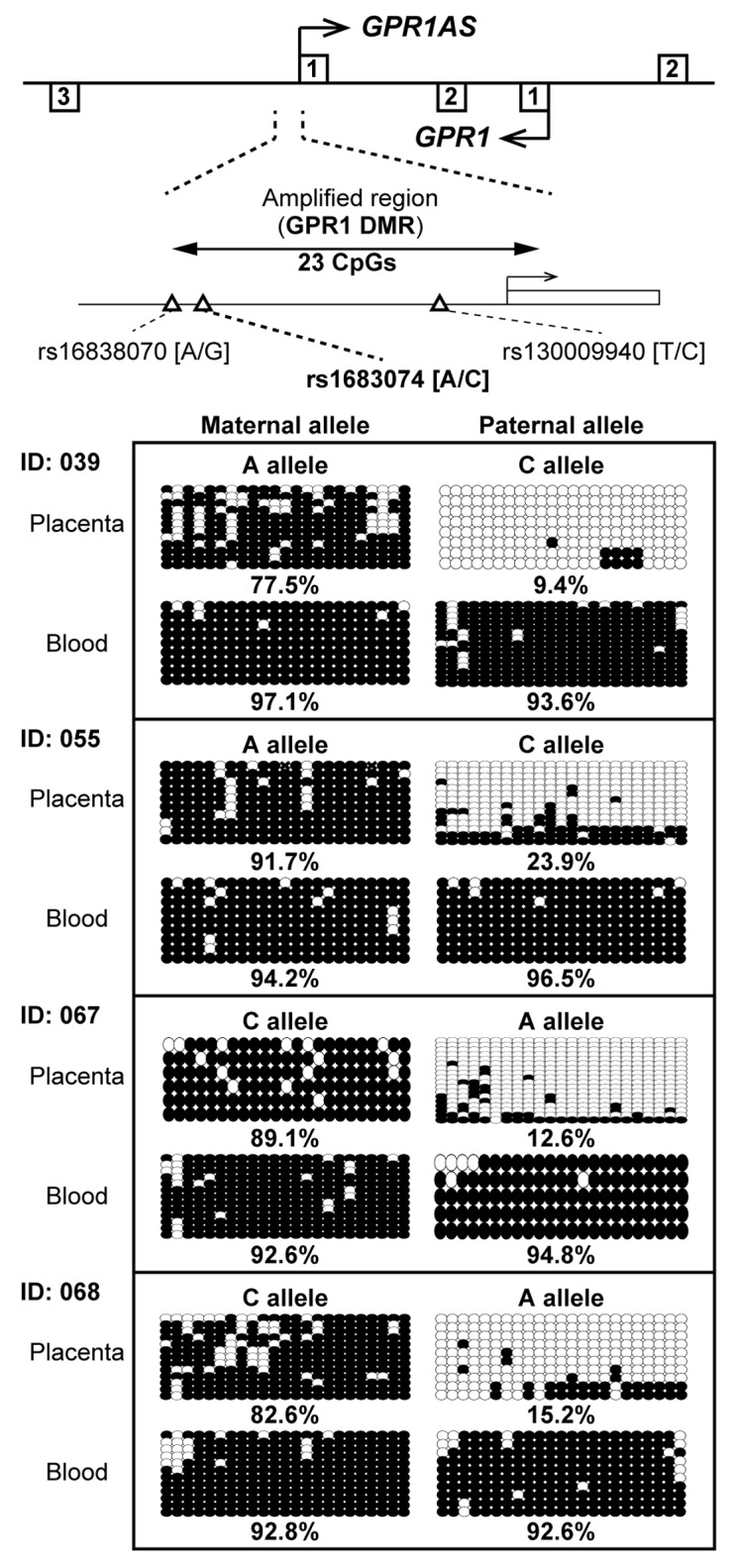
Figure 4. Bisulfite sequencing of the GPR1AS promoter region in human placenta and umbilical blood samples. Each row represents the results of an independent sequencing reaction. Open and closed circles denote unmethylated and methylated CpGs. Calculated methylation levels are shown below each graph. Heterozygous SNP rs1683074 was used to distinguish between maternal and paternal alleles in human samples. One of case (Family ID: 055) was also investigated by allelic expression analysis in Figure 3.
Imprinted GPR1AS/Zdbf2linc gene expression with parent-of-origin-specific methylation was retained in extra-embryonic lineages
Placental imprinted expression of human GPR1AS suggests mouse Zdbf2linc may also be expressed in an allele-specific manner in placental tissues. Previously, we demonstrated that Zdbf2linc expression is paternally expressed during the blastula to gastrula stages in mouse embryos, but is rarely expressed post-gastrulation, with bi-allelic complete methylation at the Gpr1 DMR.23 However, the expression profiles were not investigated in extra-embryonic tissues. Therefore, we re-analyzed Zdbf2linc expression profiles in mouse E3.5 to E9.5 embryonic tissues, including extra-embryonic tissues, by quantitative real-time RT-PCR (qRT-PCR). The accumulation of Zdbf2linc mRNA in whole embryos or embryonic and extra-embryonic tissues was clearly detected during blastula (E3.5) and gastrula (E5.5 to E7.5) stages; however, although Zdbf2linc mRNA levels were very low or undetectable in embryos, it remained detectable in ectoplacental cones (placental precursors) post-gastrulation (E9.5) (Fig. 5A). We performed allele-specific expression analysis of mouse tissues, including placental tissues, in which Zdbf2linc expression was detectable, in a C57BL/6N × JF1 genetic background (designated B6 and JF) using SNP information. Predictably, Zdbf2linc showed paternal-allele-specific expression in placental tissues at E9.5 and E15.5 (Fig. 5B). Thus, as with human GPR1AS, imprinted expression of Zdbf2linc appeared in placental tissues. Meanwhile, paternal-allele-specific expression was also observed in the fetal brain at E9.5, whereas bi-parental expression was observed in adult testis (Fig. S4). Testis-specific escape from imprinting has been also observed for Zdbf2 exon 7.19
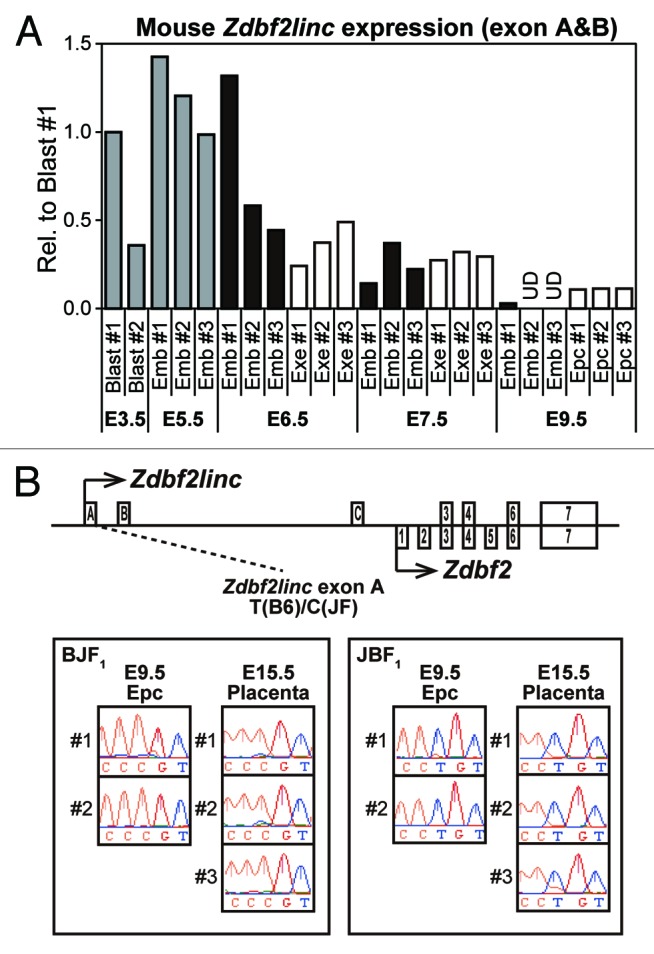
Figure 5. (A) Embryonic and extra-embryonic expression of Zdbf2linc in pre- and post-gastrulation mouse embryo (at E3.5 to E9.5). Abbreviations: E3.5 Blast, pools of 5 blastocysts (gray bars); E5.5 Emb, whole embryos (gray bars), E6.5-E9.5, dissected embryos free of extra-embryonic tissues (black bars); E6.5-E7.5 Exe, extra-embryonic tissues (white bars); E9.5 Epc, ectoplacental cones dissected from the remaining extra-embryonic tissues (white bars). After normalization against the housekeeping Actb gene, relative expression in mouse blast #1 is set as 1. UD indicates undetected mRNA expression. (B) Allele-specific RT-PCR sequencing of Zdbf2linc-specific exon (left) and common Zdbf2 exon (right) in mouse ectoplacental cones at E9.5 and placentas at E15.5. We prepared 2 parental mouse strains, C57BL/6N (B6) and JF1/Msf (JF), and reciprocal F1 hybrids (BJF1; F1 hybrid derived from mating between B6 female and JF male mice, JBF1; the reciprocal cross of JF female with B6 male mice)
To characterize the correlation between imprinted expression of Zdbf2linc and methylation status of the promoter region of Zdbf2linc (Gpr1 DMR), we compared the methylation status of Gpr1 DMR in mouse embryonic and placental tissues at E9.5. Interestingly, the placental tissues showed maternal-allele-specific methylation, whereas embryonic tissues showed hypermethylation of both parental alleles in the Gpr1 DMR (Fig. 6). Our previous study showed the Gpr1 DMR exhibits maternal-allele-specific methylation during blastula to gastrula stages in mice. Thus, our previous and current data showed that GPR1AS/Zdbf2linc is rarely expressed throughout post-gastrulation development and into somatic tissues due to bi-allelic hypermethylation at their transcription start sites.
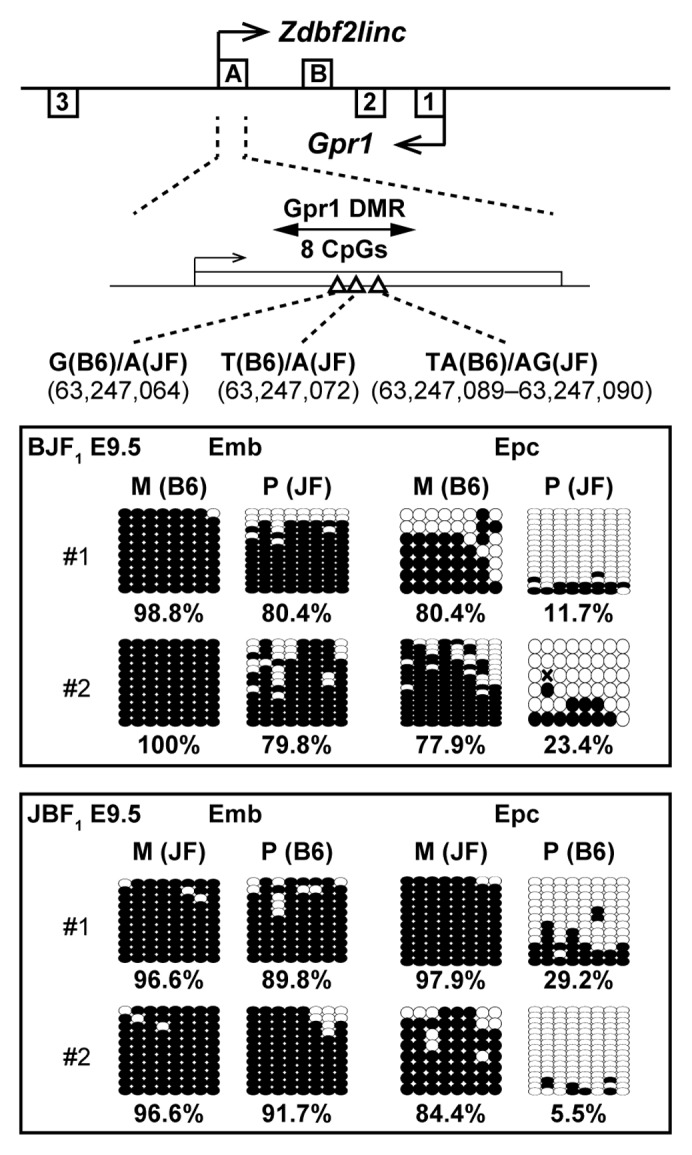
Figure 6. Bisulfite sequencing of Gpr1 DMR in mouse BJF1/JBF1 embryos and placental tissues at E9.5. M and P indicate maternally and paternally inherited alleles. Both alleles were discriminated by more than one polymorphism.
Stochastic paternal-allele-specific expression of ZDBF2/Zdbf2 in placental lineages
In mice, Zdbf2 transcription is positively correlated with methylation of the Zdbf2 DMRs (Zdbf2 DMR1?3), established on the paternal allele in the post-implantation embryo, upstream of Zdbf2.23 In human, as previously mentioned, paternally methylated DMR (ZDBF2 DMR) was also identified at intergenic region between GPR1 and ZDBF2.21 We investigated the methylation status of the ZDBF2 DMR in human placental tissues and umbilical cord blood lymphocytes. A SNP has been identified at ZDBF2 DMR (reference SNP ID: rs10206513), but any SNP has not been found around the first exon of ZDBF2. Therefore, we designed primer sets for bisulfite sequencing within the ZDBF2 DMR, which includes rs10206513, and the ZDBF2 promoter region. Four informative cases of placentas and lymphocytes were investigated; these cases have also been investigated in Figure 3. Interestingly, both placental tissues and lymphocytes showed clear maternal-allele-specific hypermethylation in the ZDBF2 DMR and almost complete hypomethylation in the ZDBF2 promoter (Fig. 7).
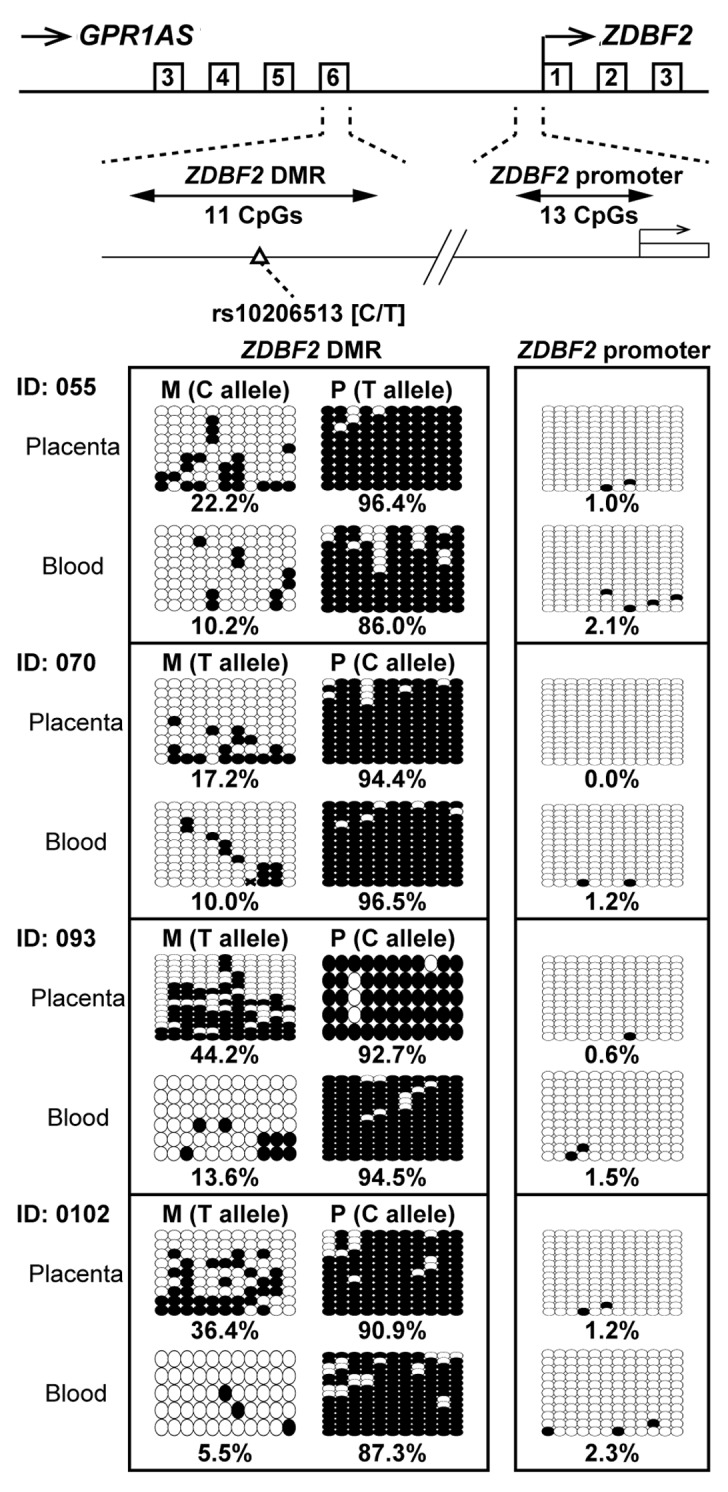
Figure 7. Bisulfite sequencing of ZDBF2 DMR and ZDBF2 promoter in human placenta and umbilical blood samples. Calculated methylation levels are shown below the graphs. Heterozygous SNP rs10206513 was used to distinguish between maternal and paternal alleles in human samples (all cases were tested in Figure 3).
We previously described bi-allelic ZDBF2 expression in human placental tissues after allelic expression profiling of coding exon (exon 7); however, the study included only one human placenta.19 Here, we re-analyzed allelic the expression profiles of ZDBF2 in human placental tissues. Similar to the GPR1AS gene, we performed allele-specific expression analysis of ZDBF2 in human placentas (reference SNP ID: rs10932150, at ZDBF2 exon 7). Four placentas were investigated and we were surprised to find only one case (Family ID: 055) showed bi-allelic expression; the other three cases showed paternal-allele-specific expression (Fig. 3). Although it is possible that maternal contamination of fetal tissue obscures parental expression bias of ZDBF2 in some placental cases (e.g., Family ID: 055 in human case),26,27 our results indicated stochastic, paternal-allele-specific ZDBF2 expression in placental tissues and that imprinting might be independent of DNA methylation at the ZDBF2 DMR or ZDBF2 promoter.
Mouse germ cell DNA methylome profiling in the Gpr1-Zdbf2 locus
We previously performed whole-genome shotgun bisulfite sequencing of samples from various stages of mouse germ cells, blastocysts and ES cells.22,28 To clear the hotspot for DNA methylation, we generated and investigated the DNA methylome map of mouse Gpr1-Zdbf2 (Fig. S5). In sperm cells, although almost CpGs including Zdbf2 DMRs were hypermethylated, all promoters and CpG islands (including Gpr1 DMR, Gpr1 promoter and Zdbf2 CpG island promoter) were hypomethylated. The paternally methylated Zdbf2 DMRs were demethylated (probably biallelically) in blastocysts. This is consistent with the fact that Zdbf2 DMRs were secondary DMRs, in which paternal-allele-specific methylation was re-established during post-implantation fetal development.23 In oocytes, the intragenic region of Gpr1, including the Gpr1 DMR, was hypermethylated, whereas the other intergenic and intragenic regions were hypomethylated. All CpG islands except Gpr1 DMR remained hypomethylated during early embryo development and gametogenesis. Complete hypomethylation of the Zdbf2linc promoter (Gpr1 DMR) might result in bi-allelic expression of Zdbf2linc in adult testes containing spermatogenetic cells.
Comparison of human and mouse GPR1-ZDBF2/Gpr1-Zdbf2 loci
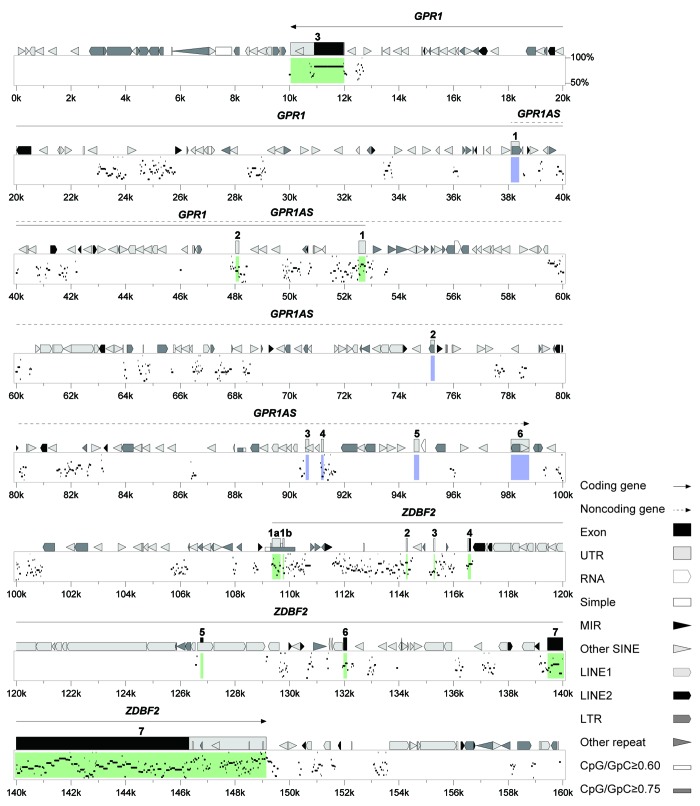
These results indicated that epigenetic and transcriptional profiles of human GPR1AS and mouse Zdbf2linc were similar. Our findings and those of previous studies demonstrated that the regulatory mechanism of the imprinted GPR1-ZDBF2 cluster was well-conserved between human and mouse. Therefore, we used the advanced PipMaker program to perform a detailed comparison between mouse and human genomic sequences. This analysis is shown graphically in the percent identity plot (PIP) in Figure 8 for the protein-coding exons of GPR1 and ZDBF2. Overall, the PIP revealed strong conservation of GPR1/Gpr1 and ZDBF2/Zdbf2 between the human and mouse genomes and poor conservation of GPR1AS lncRNAs, with a number of transposable elements (Fig. S6). Evolutionary conservation is also supported by certain similarities between their human/mouse gene products (GPR1 proteins, 94% similarity; ZDBF2 proteins, 76% similarity) (Fig. S7). In all our comparisons, the exon sequences and structures of human GPR1AS were very different from the mouse Zdbf2linc transcript.
Figure 8. Human and mouse sequences of the imprinted GPR1-ZDBF2 domain. Percent Identity Plot (PIP, a scale of 50% to 100% conservation, on the y-axis) showing order and alignment of the entire imprinted domain on human chromosome 2q33.3 (chr2: 207,030,001?207,190,000, on the x-axis) and the orthologous region on mouse chromosome 1C2. Detected sequence homology (>50% identity) is plotted by position along the test sequence and level of sequence similarity. Structural features in the human ortholog, including exons, repeats and CpG-rich regions, are shown above the top line. The novel imprinted lncRNA gene, GPR1AS, is shown in blue, and known imprinted protein-coding genes, GPR1 and ZDBF2, are shown in green.
Discussion
We identified a novel antisense lncRNA gene, GPR1AS, at the imprinted GPR1-ZDBF2 domain on human chromosome 2q33.3. GPR1AS is specifically expressed in placenta and testis; in the placenta, the paternal allele is exclusively expressed. We identified several similarities between human GPR1AS and mouse Zdbf2linc lncRNAs: (1) antisense transcription from GPR1/Gpr1 intron 2; (2) long intergenic transcripts (human GPR1AS, 60 kb; mouse Zdbf2linc, 114 kb) throughout the secondary ZDBF2/Zdbf2 DMRs; (3) specific expression in ES cells, placental tissues and testis; (4) transcript expression from the paternal allele with silencing of the maternal allele by promoter DNA hypermethylation in placental tissues and; and (5) complete biallelic promoter methylation and silencing in fetal and adult tissues (Fig. 9). These facts strongly suggested that GPR1AS is a human ortholog of mouse Zdbf2linc. Furthermore, previous studies have revealed that neighbor genes, GPR1/Gpr1 and ZDBF2/Zdbf2, are also commonly imprinted with maternal and paternal DMRs (GPR1/Gpr1 and ZDBF2/Zdbf2 DMRs) in human and mouse. These and previous findings demonstrated that epigenetic and transcriptional regulatory mechanisms of the imprinted GPR1-ZDBF2 cluster were well conserved between human and mouse genomes. Some structural differences were also identified: (1) no ZDBF2 exon in the GPR1AS transcript and (2) no significant similarity between lncRNA sequences. However, the conservation of epigenetic and transcriptional traits between GPR1AS and Zdbf2linc can be assumed to have arisen by divergent evolution from a common ancestor, in spite of their differing sequences, and are likely functional.
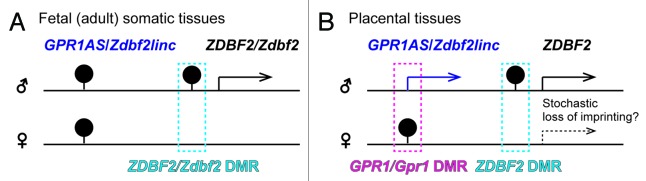
Figure 9. Summary of allele-specific epigenetic and gene expression differences within the GPR1AS-ZDBF2 (Zdbf2linc-Zdbf2) locus during fetal (A) and extra-embryonic (B) development. Arrows represent transcriptional orientation of individual imprinted genes. DMR positions are indicated by filled pins on the methylated alleles.
Imprinted gene clusters in the eutherian genome contain many lncRNA genes, most of which are imprinted. These non-coding RNAs are often closely linked to the regulation of imprinted expression in cis. Perhaps the best studied of such molecules are the imprinted Nespas, Kcnq1ot1 and Airn lncRNAs. These lncRNAs induce parental-specific silencing and suggest their transcription is more important than their products.13-15,17 The mono-allelic transcription of GPR1AS/Zdbf2linc lncRNAs may have a similar cis-regulating effect on flanking coding genes because these lncRNAs were transcribed from an unmethylated ICR candidate (maternally methylated primary GPR1/Gpr1 DMR: There is no direct evidence that this region actually acts as an ICR because there is no report of a knockout mouse) as well as the three functional imprinted lncRNAs. Some studies have suggested that transcription across DMRs is associated with epigenetic changes and acquisition of genomic imprinting.13,29-31 For example, Williamson et al. demonstrated that paternally expressed Nespas lncRNA transcription is required for demethylation of trimethylated histone H3 lysine 4, followed by DNA methylation, at the secondary Nesp DMR.13 Despite the differing exon structures of GPR1AS and Zdbf2linc, both lncRNA transcripts reached the secondary ZDBF2/Zdbf2 DMRs (Fig. 1; Fig. 9). It is also possible that monoallelic epigenetic modification (e.g., paternal-allele-specific methylation of ZDBF2/Zdbf2 DMRs) is established via long-range transcription of GPR1AS and Zdbf2linc during the early embryonic stage, and such epigenetic modification might stably control the parental-allele expression of ZDBF2/Zdbf2 in fetal and adult somatic tissues, even after the disappearance of lncRNA transcript (Fig. 9).23 Although paternal-allele-specific methylation at ZDBF2 DMRs was established in all tested cases, stochastic non-imprinted expression of ZDBF2 was observed in placental tissues. Perhaps for this reason, imprinted expression of ZDBF2 may be independent of DNA methylation in placental lineages, similar to the imprinted Kcnq1 gene clusters.32 In both human and mice, promoter regions of ZDBF2/Zdbf2 were completely hypomethylated whereas their paternal-allele-specific expression (Fig. 7; Fig. S5). Thus, the promoter might be marked monoallelically by other epigenetic modification (e.g., histone modifications). In mice, we previously described bi-allelic Zdbf2 expression in (E15.5) placentas by expression profiling of exon 7 (18); however, this exon is common between Zdbf2 and Zdbf2linc. Unfortunately, between B6 and JF mice, no SNPs were detectable in coding Zdbf2-specific exons 1, 2 and 5 (data not shown); therefore, further analysis is required to determine whether coding Zdbf2 variant is expressed in a parent-of-origin-specific manner in placental tissues, e.g., using other mouse strains.
In mammals, over a thousand evolutionarily conserved long-intergenic non-coding RNAs (lincRNAs: lncRNAs overlapping intergenic regions) have been identified by their chromatin structures.33 Although it is now estimated that the human genome produces over 8000 lincRNAs, only ~12% of human lincRNAs have orthologous transcripts in other species and only a few of them have been functionally characterized.34 These functional lincRNAs play essential roles in the regulation of epigenetic marks and gene expression in trans or cis. For example, HOTAIR lincRNA silences transcription across 40 kb of the HOXD locus by inducing a repressive chromatin state in trans,35 and the XIST, TSIX, JPX and XACT lincRNAs are key players in X-chromosome inactivation (XCI) in cis.36-39 The XCI and genomic imprinting represent prominent examples of the recruitment of chromatin-modifying activities by lncRNAs and provides a prototype model for a cis localization mechanism. Our findings suggested the most important cue for cis-acting lncRNA might be transcription through a particular region (e.g., cis-regulating elements, such as promoter, silencer and insulator) via specific interaction with chromatin to mediate targeted recruitment of repressive epigenetic-modifying activities to silence transcription. Thus, the lncRNA sequences and structures might not be important. Our results suggest many more homologous lncRNAs could be defined using transcriptional and epigenetic profiles including DNA methylation. lncRNAs represent a largely unexplored world of genetic/epigenetic variation and may provide mechanistic insight into transcriptional regulation.
Materials and Methods
Human samples
All human studies were approved by the Institutional Review Board Committee at the National Center for Child Health and Development (NCCHD) and performed after obtaining written informed consent. The derivation and cultivation of human ES Cell (hESC) lines was performed in full compliance with ?The Guidelines on the Derivation and Distribution of Human Embryonic Stem Cells? of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (Notification No.156 of 2009). We used MEXT-registered hESC lines, SEES1 and SEES3.Genomic DNA from human placentas was isolated with a standard proteinase K-phenol/chloroform procedure. Genomic DNA from human lymphocytes was isolated with the QIAamp DNA Blood Midi Kit (Qiagen). Total RNA from human placentas was extracted with Sepasol-RNA I Super G (Nacalai Tesque); total RNA from hESCs was extracted with the Allprep DNR/RNA Mini Kit (Qiagen). RNA was treated with RQ1 RNase-free DNase (Promega) and purified by phenol/chloroform extraction. Total RNA from other human normal tissues were purchased from TaKaRa Bio.
Mouse samples
Mouse embryos, placental tissues, fetal tissues, and adult tissues were prepared as described previously.19,23,40 To use DNA polymorphisms for allele discrimination, reciprocal F1 hybrids were obtained by crossing C57BL/6N (B6; Clea Japan) and JF1/Msf (JF; National Institute of Genetics) mice. Total RNA from E3.5?7.5 embryos and extra-embryonic tissues were extracted with the RNeasy Mini Kit. Genomic DNA and Total RNA were isolated from E9.5 embryos and ectoplacental cones (yolk sacs and amnions removed) with the Allprep DNR/RNA Mini Kit (Qiagen). Total RNA from E15.5 placental and fetal tissues and adult tissues from 8-week-old male mice were isolated with TRIzol reagent (Invitrogen). The extracted RNA was treated with RQ1 RNase-free DNase (Promega) and purified by phenol/chloroform extraction.
RACE analysis
The 5?- and 3?-regions of the Human ZDBF2 and GPR1AS gene was obtained using the GeneRacer Kit (Invitrogen). Genomic DNA-free total RNA was prepared from human ES cells and 2 rounds of PCR were performed using TaKaRa EX Taq (TaKaRa Bio) under the following conditions: 5 cycles of 30 sec at 94 °C, 1 min at 72 °C, 5 cycles of 30 sec at 94 °C, 1 min at 70 °C, 20 cycles of 30 sec at 94 °C, 30 sec at 64 °C, 1 min at 68 °C and 10 min final elongation at 68 °C for the first PCR; and 20 cycles of 30 sec at 94 °C, 30 sec at 65 °C, 2 min at 68 °C and 10 min final elongation at 68 °C for the second PCR. The Zdbf2 gene-specific primers for the nested PCR were as follows: anti-sense ZDBF2 5RA1: 5?-GCATTACAAATCACTGGGGGGCGTACCA-3? for the first PCR and anti-sense ZDBF2 5RA2: 5?-CTGGGGGGCGTACCAAGTTGCTTCTA-3? for the second PCR for 5?RACE. The BE734382 (GPR1AS gene) primer sets nested PCR were as follows: anti-sense BE734382_5RA1: 5?-TACTACAAGGGCCAGAACTCAGGAAGAG-3? for the first PCR and anti-sense BE734382_5RA2: 5?-TGGAAGCGATGCACGGGACAAGGTCT-3? for the second PCR for 5?RACE, and sense BE734382_3RA1: 5?-AGGAGCCCCAGAGGGCCATTGTAATTC-3? for the first PCR and sense BE734382_3RA2: 5?-TCAAGATCTTCAGTAAACTTCTGCCGC-3? for the second PCR for 3?RACE. Spliced form of GPR1AS was obtained by PCR using two sets of primers; BE734382_F1: 5?-GTCCCGTGCATCGCTTCCAT-3? and BE734382_R1: 5?-CCACCTTTTTCCAAGTGGCC-3?, and BE734382_F2: 5?-ATCCGGCTCTTCCTGAGTTC-3? and BE734382_R2: 5?-CGGCAGAAGTTTACTGAAGA-3?. The amplified products were sequenced directly, after purification.
Real-time RT-PCR and allelic expression analysis
Genomic DNA-free total RNA samples from human and mouse fetal samples were reverse transcribed to cDNA with SuperScript III (Invitrogen). Quantitative expression analysis of human GPR1AS and mouse Zdbf2linc was performed in a 7500 Real-time PCR system (Applied Biosystems) with SYBR Green PCR Master Mix (Applied Biosystems). Relative expression of each gene was calculated by the ??Ct method and normalized to human ACTB or mouse Actb housekeeping genes. RT-PCR and direct sequencing of GPR1AS/Zdbf2linc and ZDBF2/Zdbf2 exons were performed using an ABI PRISM 3730xl genetic analyzer (Applied Biosystems) as described previously.19 Primer sequences and PCR cycling conditions are listed in Table S2.
Bisulfite sequencing
Genomic DNA samples were isolated from human samples and mouse E9.5 embryos and placental tissues from an F1 hybrid of B6 and JF. Genomic DNA (100 ng per sample) was treated with sodium bisulfite using the MethylCode Bisulfite Conversion Kit (Invitrogen). The bisulfite-treated DNA was PCR-amplified with TaKaRa EpiTaq HS (TaKaRa Bio) with primers specific for human GPR1 DMR, ZDBF2 DMR, ZDBF2 promoter and mouse Gpr1 DMR. Primers and PCR cycling conditions are listed in Table S2. Subcloning and sequencing were performed as described.19 Visualization and quantification of bisulfite sequence data for CpG methylation was performed using the QUMA web-based tool.41
Comparative in silico analysis
The genomic alignments of GPR1 and ZDBF2 were obtained from the UCSC Genome Browser database (genome.ucsc.edu/) for human (GRCh37/hg19) and mouse (NCBI37/mm9) genomes. The Advanced PipMaker program (pipmaker.bx.psu.edu/cgi-bin/pipmaker?advanced) was used to compare the sequences of the genomic regions surrounding human GPR1 and ZDBF2 (hg19, chr2: 207,030,001?207,190,000) and mouse Gpr1 and Zdbf2 (mm9, chr1: 63,160,001?63,380,000). Repetitive DNA sequences were classified using RepeatMasker (www.repeatmasker.org). The nucleotide and amino acid sequences were aligned by Genetyx software (Genetyx Corporation).
Accession numbers
Full-length sequences of human GPR1AS and ZDBF2 and mouse Zdbf2linc have been deposited DNA Data Bank of Japan (DDBJ) under accession numbers AB774455, AB775780, AB775781 and AB777270.
Supplementary Material
Acknowledgments
We would like to thank Dr. Akihiro Umezawa and other members of NCCHD for providing hESCs and for many helpful discussions. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (Grant No. 222228004) awarded to T.K., including the MEXT-Supported Program for the Strategic Research Foundation at Private Universities (Grant No. S0801025). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
Supplementary materials may be found here:
http://www.landesbioscience.com/journals/epigenetics/article/24887
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/24887
References
- 1.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, et al. ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children?s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–23. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 4.Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, et al. Mouse Genome Sequencing Consortium Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–21. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–40. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 11.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 12.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–82. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson CM, Ball ST, Dawson C, Mehta S, Beechey CV, Fray M, et al. Uncoupling antisense-mediated silencing and DNA methylation in the imprinted Gnas cluster. PLoS Genet. 2011;7:e1001347. doi: 10.1371/journal.pgen.1001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Latos PA, Pauler FM, Koerner MV, ?energin HB, Hudson QJ, Stocsits RR, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–72. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 15.Mohammad F, Pandey GK, Mondal T, Enroth S, Redrup L, Gyllensten U, et al. Long noncoding RNA-mediated maintenance of DNA methylation and transcriptional gene silencing. Development. 2012;139:2792–803. doi: 10.1242/dev.079566. [DOI] [PubMed] [Google Scholar]
- 16.Williamson CM, Turner MD, Ball ST, Nottingham WT, Glenister P, Fray M, et al. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet. 2006;38:350–5. doi: 10.1038/ng1731. [DOI] [PubMed] [Google Scholar]
- 17.Korostowski L, Sedlak N, Engel N. The Kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart. PLoS Genet. 2012;8:e1002956. doi: 10.1371/journal.pgen.1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babak T, Deveale B, Armour C, Raymond C, Cleary MA, van der Kooy D, et al. Global survey of genomic imprinting by transcriptome sequencing. Curr Biol. 2008;18:1735–41. doi: 10.1016/j.cub.2008.09.044. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi H, Yamada K, Morita S, Hiura H, Fukuda A, Kagami M, et al. Identification of the mouse paternally expressed imprinted gene Zdbf2 on chromosome 1 and its imprinted human homolog ZDBF2 on chromosome 2. Genomics. 2009;93:461–72. doi: 10.1016/j.ygeno.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Hiura H, Sugawara A, Ogawa H, John RM, Miyauchi N, Miyanari Y, et al. A tripartite paternally methylated region within the Gpr1-Zdbf2 imprinted domain on mouse chromosome 1 identified by meDIP-on-chip. Nucleic Acids Res. 2010;38:4929–45. doi: 10.1093/nar/gkq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang F, Hodges E, Molaro A, Dean M, Hannon GJ, Smith AD. Genomic landscape of human allele-specific DNA methylation. Proc Natl Acad Sci U S A. 2012;109:7332–7. doi: 10.1073/pnas.1201310109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8:e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi H, Sakurai T, Sato S, Nakabayashi K, Hata K, Kono T. Imprinted DNA methylation reprogramming during early mouse embryogenesis at the Gpr1-Zdbf2 locus is linked to long cis-intergenic transcription. FEBS Lett. 2012;586:827–33. doi: 10.1016/j.febslet.2012.01.059. [DOI] [PubMed] [Google Scholar]
- 24.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–93. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 25.Kagami M, O?Sullivan MJ, Green AJ, Watabe Y, Arisaka O, Masawa N, et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet. 2010;6:e1000992. doi: 10.1371/journal.pgen.1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson QJ, Seidl CI, Kulinski TM, Huang R, Warczok KE, Bittner R, et al. Extra-embryonic-specific imprinted expression is restricted to defined lineages in the post-implantation embryo. Dev Biol. 2011;353:420–31. doi: 10.1016/j.ydbio.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okae H, Hiura H, Nishida Y, Funayama R, Tanaka S, Chiba H, et al. Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum Mol Genet. 2012;21:548–58. doi: 10.1093/hmg/ddr488. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi H, Sakurai T, Miura F, Imai M, Mochiduki K, Yanagisawa E, et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 2013;23:616–27. doi: 10.1101/gr.148023.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, et al. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009;23:105–17. doi: 10.1101/gad.495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henckel A, Chebli K, Kota SK, Arnaud P, Feil R. Transcription and histone methylation changes correlate with imprint acquisition in male germ cells. EMBO J. 2012;31:606–15. doi: 10.1038/emboj.2011.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith EY, Futtner CR, Chamberlain SJ, Johnstone KA, Resnick JL. Transcription is required to establish maternal imprinting at the Prader-Willi syndrome and Angelman syndrome locus. PLoS Genet. 2011;7:e1002422. doi: 10.1371/journal.pgen.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, et al. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–5. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- 33.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sado T, Brockdorff N. Advances in understanding chromosome silencing by the long non-coding RNA Xist. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110325. doi: 10.1098/rstb.2011.0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohhata T, Senner CE, Hemberger M, Wutz A. Lineage-specific function of the noncoding Tsix RNA for Xist repression and Xi reactivation in mice. Genes Dev. 2011;25:1702–15. doi: 10.1101/gad.16997911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallot C, Huret C, Lesecque Y, Resch A, Oudrhiri N, Bennaceur-Griscelli A, et al. XACT, a long noncoding transcript coating the active X chromosome in human pluripotent cells. Nat Genet. 2013;45:239–41. doi: 10.1038/ng.2530. [DOI] [PubMed] [Google Scholar]
- 40.Sato S, Yoshida W, Soejima H, Nakabayashi K, Hata K. Methylation dynamics of IG-DMR and Gtl2-DMR during murine embryonic and placental development. Genomics. 2011;98:120–7. doi: 10.1016/j.ygeno.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36(Web Server issue):W170-5. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.