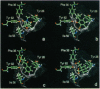
Abstract
The search for novel leads is a critical step in the drug discovery process. Computational approaches to identify new lead molecules have focused on discovering complete ligands by evaluating the binding affinity of a large number of candidates, a task of considerable complexity. A new computational method is introduced in this work based on the premise that the primary molecular recognition event in the protein binding site may be accomplished by small core fragments that serve as molecular anchors, providing a structurally stable platform that can be subsequently tailored into complete ligands. To fulfill its role, we show that an effective molecular anchor must meet both the thermodynamic requirement of relative energetic stability of a single binding mode and its consistent kinetic accessibility, which may be measured by the structural consensus of multiple docking simulations. From a large number of candidates, this technique is able to identify known core fragments responsible for primary recognition by the FK506 binding protein (FKBP-12), along with a diverse repertoire of novel molecular cores. By contrast, absolute energetic criteria for selecting molecular anchors are found to be promiscuous. A relationship between a minimum frustration principle of binding energy landscapes and receptor-specific molecular anchors in their role as "recognition nuclei" is established, thereby unraveling a mechanism of lead discovery and providing a practical route to receptor-biased computational combinatorial chemistry.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abkevich V. I., Gutin A. M., Shakhnovich E. I. Specific nucleus as the transition state for protein folding: evidence from the lattice model. Biochemistry. 1994 Aug 23;33(33):10026–10036. doi: 10.1021/bi00199a029. [DOI] [PubMed] [Google Scholar]
- Bryngelson J. D., Onuchic J. N., Socci N. D., Wolynes P. G. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995 Mar;21(3):167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- Bryngelson J. D., Wolynes P. G. Spin glasses and the statistical mechanics of protein folding. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caflisch A., Miranker A., Karplus M. Multiple copy simultaneous search and construction of ligands in binding sites: application to inhibitors of HIV-1 aspartic proteinase. J Med Chem. 1993 Jul 23;36(15):2142–2167. doi: 10.1021/jm00067a013. [DOI] [PubMed] [Google Scholar]
- Clackson T., Wells J. A. A hot spot of binding energy in a hormone-receptor interface. Science. 1995 Jan 20;267(5196):383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- Devlin J. J., Panganiban L. C., Devlin P. E. Random peptide libraries: a source of specific protein binding molecules. Science. 1990 Jul 27;249(4967):404–406. doi: 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- Ebeling M., Nadler W. On constructing folding heteropolymers. Proc Natl Acad Sci U S A. 1995 Sep 12;92(19):8798–8802. doi: 10.1073/pnas.92.19.8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M. B., Wiley D. C., Karplus M., Hubbard R. E. HOOK: a program for finding novel molecular architectures that satisfy the chemical and steric requirements of a macromolecule binding site. Proteins. 1994 Jul;19(3):199–221. doi: 10.1002/prot.340190305. [DOI] [PubMed] [Google Scholar]
- Feng S., Chen J. K., Yu H., Simon J. A., Schreiber S. L. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994 Nov 18;266(5188):1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- Fremont D. H., Matsumura M., Stura E. A., Peterson P. A., Wilson I. A. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science. 1992 Aug 14;257(5072):919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- Friedman A. R., Roberts V. A., Tainer J. A. Predicting molecular interactions and inducible complementarity: fragment docking of Fab-peptide complexes. Proteins. 1994 Sep;20(1):15–24. doi: 10.1002/prot.340200104. [DOI] [PubMed] [Google Scholar]
- Fukugita M., Lancaster D., Mitchard M. G. Kinematics and thermodynamics of a folding heteropolymer. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6365–6368. doi: 10.1073/pnas.90.13.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop M. A., Barrett R. W., Dower W. J., Fodor S. P., Gordon E. M. Applications of combinatorial technologies to drug discovery. 1. Background and peptide combinatorial libraries. J Med Chem. 1994 Apr 29;37(9):1233–1251. doi: 10.1021/jm00035a001. [DOI] [PubMed] [Google Scholar]
- Garcia K. C., Ronco P. M., Verroust P. J., Brünger A. T., Amzel L. M. Three-dimensional structure of an angiotensin II-Fab complex at 3 A: hormone recognition by an anti-idiotypic antibody. Science. 1992 Jul 24;257(5069):502–507. doi: 10.1126/science.1636085. [DOI] [PubMed] [Google Scholar]
- Gordon E. M., Barrett R. W., Dower W. J., Fodor S. P., Gallop M. A. Applications of combinatorial technologies to drug discovery. 2. Combinatorial organic synthesis, library screening strategies, and future directions. J Med Chem. 1994 May 13;37(10):1385–1401. doi: 10.1021/jm00036a001. [DOI] [PubMed] [Google Scholar]
- Gutin A. M., Abkevich V. I., Shakhnovich E. I. Evolution-like selection of fast-folding model proteins. Proc Natl Acad Sci U S A. 1995 Feb 28;92(5):1282–1286. doi: 10.1073/pnas.92.5.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmirski S. L., Alonso D. O., Cohen F. E., Prusiner S. B., Daggett V. Theoretical studies of sequence effects on the conformational properties of a fragment of the prion protein: implications for scrapie formation. Chem Biol. 1995 May;2(5):305–315. doi: 10.1016/1074-5521(95)90049-7. [DOI] [PubMed] [Google Scholar]
- Kick E. K., Ellman J. A. Expedient method for the solid-phase synthesis of aspartic acid protease inhibitors directed toward the generation of libraries. J Med Chem. 1995 Apr 28;38(9):1427–1430. doi: 10.1021/jm00009a002. [DOI] [PubMed] [Google Scholar]
- Koshland D. E. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc Natl Acad Sci U S A. 1958 Feb;44(2):98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz I. D. Structure-based strategies for drug design and discovery. Science. 1992 Aug 21;257(5073):1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- Li Z., Chen X., Davidson E., Zwang O., Mendis C., Ring C. S., Roush W. R., Fegley G., Li R., Rosenthal P. J. Anti-malarial drug development using models of enzyme structure. Chem Biol. 1994 Sep;1(1):31–37. doi: 10.1016/1074-5521(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Richards F. M., Fox R. O. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature. 1994 Nov 24;372(6504):375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- Madden D. R., Garboczi D. N., Wiley D. C. The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell. 1993 Nov 19;75(4):693–708. doi: 10.1016/0092-8674(93)90490-h. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Fremont D. H., Peterson P. A., Wilson I. A. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992 Aug 14;257(5072):927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- McDonald I. K., Thornton J. M. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994 May 20;238(5):777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Miyamoto S., Kollman P. A. What determines the strength of noncovalent association of ligands to proteins in aqueous solution? Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8402–8406. doi: 10.1073/pnas.90.18.8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro C. M., Kuntz I. D., Dixon J. S. Flexible ligand docking using a genetic algorithm. J Comput Aided Mol Des. 1995 Apr;9(2):113–130. doi: 10.1007/BF00124402. [DOI] [PubMed] [Google Scholar]
- Pande V. S., Grosberg A. Y., Tanaka T. Thermodynamic procedure to synthesize heteropolymers that can renature to recognize a given target molecule. Proc Natl Acad Sci U S A. 1994 Dec 20;91(26):12976–12979. doi: 10.1073/pnas.91.26.12976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring C. S., Sun E., McKerrow J. H., Lee G. K., Rosenthal P. J., Kuntz I. D., Cohen F. E. Structure-based inhibitor design by using protein models for the development of antiparasitic agents. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3583–3587. doi: 10.1073/pnas.90.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rini J. M., Schulze-Gahmen U., Wilson I. A. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science. 1992 Feb 21;255(5047):959–965. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- Sali A., Shakhnovich E., Karplus M. Kinetics of protein folding. A lattice model study of the requirements for folding to the native state. J Mol Biol. 1994 Feb 4;235(5):1614–1636. doi: 10.1006/jmbi.1994.1110. [DOI] [PubMed] [Google Scholar]
- Shoichet B. K., Kuntz I. D. Matching chemistry and shape in molecular docking. Protein Eng. 1993 Sep;6(7):723–732. doi: 10.1093/protein/6.7.723. [DOI] [PubMed] [Google Scholar]
- Shoichet B. K., Kuntz I. D. Protein docking and complementarity. J Mol Biol. 1991 Sep 5;221(1):327–346. doi: 10.1016/0022-2836(91)80222-g. [DOI] [PubMed] [Google Scholar]
- Shoichet B. K., Stroud R. M., Santi D. V., Kuntz I. D., Perry K. M. Structure-based discovery of inhibitors of thymidylate synthase. Science. 1993 Mar 5;259(5100):1445–1450. doi: 10.1126/science.8451640. [DOI] [PubMed] [Google Scholar]
- Stanfield R. L., Fieser T. M., Lerner R. A., Wilson I. A. Crystal structures of an antibody to a peptide and its complex with peptide antigen at 2.8 A. Science. 1990 May 11;248(4956):712–719. doi: 10.1126/science.2333521. [DOI] [PubMed] [Google Scholar]
- Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science. 1991 May 10;252(5007):839–842. doi: 10.1126/science.1709302. [DOI] [PubMed] [Google Scholar]
- Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993 Jan 5;229(1):105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- Verkhivker G. M., Rejto P. A. A mean field model of ligand-protein interactions: implications for the structural assessment of human immunodeficiency virus type 1 protease complexes and receptor-specific binding. Proc Natl Acad Sci U S A. 1996 Jan 9;93(1):60–64. doi: 10.1073/pnas.93.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Pantoliano M. W., Thompson L. D. Crystal structure and ligand-binding studies of a screened peptide complexed with streptavidin. Biochemistry. 1992 Oct 6;31(39):9350–9354. doi: 10.1021/bi00154a004. [DOI] [PubMed] [Google Scholar]
- Weiss G. A., Collins E. J., Garboczi D. N., Wiley D. C., Schreiber S. L. A tricyclic ring system replaces the variable regions of peptides presented by three alleles of human MHC class I molecules. Chem Biol. 1995 Jun;2(6):401–407. doi: 10.1016/1074-5521(95)90221-x. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Erickson J. W. Structure-based inhibitors of HIV-1 protease. Annu Rev Biochem. 1993;62:543–585. doi: 10.1146/annurev.bi.62.070193.002551. [DOI] [PubMed] [Google Scholar]
- Yue K., Dill K. A. Folding proteins with a simple energy function and extensive conformational searching. Protein Sci. 1996 Feb;5(2):254–261. doi: 10.1002/pro.5560050209. [DOI] [PMC free article] [PubMed] [Google Scholar]