The barbed-end actin capping protein FSGS3/CD2AP localizes to apical adherens junctions in polarized cells, where it promotes adhesive strength and barrier function.
Abstract
By combining in vitro reconstitution biochemistry with a cross-linking approach, we have identified focal segmental glomerulosclerosis 3/CD2-associated protein (FSGS3/CD2AP) as a novel actin barbed-end capping protein responsible for actin stability at the adherens junction. FSGS3/CD2AP colocalizes with E-cadherin and α-actinin-4 at the apical junction in polarized Madin-Darby canine kidney (MDCK) cells. Knockdown of FSGS3/CD2AP compromised actin stability and decreased actin accumulation at the adherens junction. Using a novel apparatus to apply mechanical stress to cell–cell junctions, we showed that knockdown of FSGS3/CD2AP compromised adhesive strength, resulting in tearing between cells and disruption of barrier function. Our results reveal a novel function of FSGS3/CD2AP and a previously unrecognized role of barbed-end capping in junctional actin dynamics. Our study underscores the complexity of actin regulation at cell–cell contacts that involves actin activators, inhibitors, and stabilizers to control adhesive strength, epithelial behavior, and permeability barrier integrity.
Introduction
The actin cytoskeleton participates in cell–cell adhesion and maintenance of epithelial barrier in many fundamental ways (Cavey and Lecuit, 2009; Baum and Georgiou, 2011). Actin organizes plasma membrane domains by clustering membrane adhesion molecules (Delanoë-Ayari et al., 2004; Sato et al., 2006). In addition, it provides anchorage of cell–cell adhesion molecules to the underlying cytoplasmic actin network (Lambert et al., 2002; Kametani and Takeichi, 2007; Cavey et al., 2008). Moreover, actin contributes to mechanotransduction at sites of cell–cell contacts to strengthen adhesion (Chu et al., 2004; Yonemura et al., 2010; Gomez et al., 2011; Leckband et al., 2011). Ultimately, the actin cytoskeleton determines the efficacy of cell–cell adhesion and the nature of intercellular interactions (Nishimura and Takeichi, 2009; Takeichi, 2011; Shimono et al., 2012). Therefore, elucidating the mechanism of actin regulation at cell–cell adhesive contacts is fundamental to the understanding of epithelial cell behaviors and function.
Actin could be recruited to the junctions via a capturing mechanism where actin-binding proteins located at the junction recruit preexisting filaments from the cytoplasm. Alternatively, actin filament could be synthesized de novo via a local nucleation reaction at the junctional complex. Previously, we and others have shown that monomeric actin adds directly to cell–cell contact sites (Vasioukhin et al., 2000; Kovacs et al., 2002; Verma et al., 2004; Zhang et al., 2005; Tang and Brieher, 2012). The addition of monomeric actin at the junction requires the activity of the arp2/3 nucleation-promoting complex, which indicates that actin nucleation at sites of cell–cell adhesion is the dominant reaction. Therefore, actin assembly at cell junctions can be controlled at multiple levels, including activation of arp2/3 and actin polymerization.
Several actin-binding proteins including EPLIN, N-WASP, VASP, α-catenin, vinculin, and myosin II had been shown to participate in the recruitment and organization of actin at the adherens junction (Vasioukhin et al., 2000; Baum and Perrimon, 2001; Scott et al., 2006; Smutny et al., 2010; Kovacs et al., 2011). However, how actin nucleation is coupled to polymerization, assembly, and organization at cell–cell adhesive contacts remains largely uncharacterized. In spite of the numerous cellular and genetics studies (Nandadasa et al., 2009; Morita et al., 2010; Bernadskaya et al., 2011; Johnson et al., 2011; Kovacs et al., 2011; Chu et al., 2012), elucidation of molecular mechanisms has been difficult due to a lack of a defined biochemical system. We have previously developed an in vitro functional assay that reports on actin assembly at membrane junctional complexes by combining a classic adherens junction preparation (Tsukita and Tsukita, 1989) with a fluorescent actin signal (Tang and Brieher, 2012). The reaction can be performed in the absence of cytosolic factors, which indicates that all necessary factors are on the membrane. This assay allows identification of molecular components and elucidation of mechanisms within a complex biochemical system, providing a starting point for reconstitution of actin assembly at adhesion complexes under defined conditions.
Using junction-enriched membranes, we found that focal segmental glomerulosclerosis 1 (FSGS1)/α-actinin-4, a protein that is mutated in a chronic form of kidney disease focal segmental glomerulosclerosis (FSGS), is required for arp2/3-dependent actin assembly at the adherens junction (Tang and Brieher, 2012). Here, using the same biochemical system, we discovered that FSGS3/CD2-associated protein (FSGS3/CD2AP), which is also mutated in FSGS, is required for actin stability at the adherens junction. Our results highlight the complexity of actin regulation at cell–cell adhesion and underscore the importance of in vitro biochemical dissection to reveal molecular mechanisms.
Results
Identification of two novel junctional actin-regulating activities
We have previously described a population of adherens junctional complexes that supports actin assembly in an arp2/3- and α-actinin-4–dependent fashion (Tang and Brieher, 2012). However, FSGS1/α-actinin-4 alone does not facilitate arp2/3-dependent actin nucleation under defined conditions, which suggests that additional components on the membrane are required. Moreover, junctional proteins remained clustered after stripping with high salt, which indicates that components of the complex are stably associated with each other. Thus, the adherens junction–enriched membranes can be further exploited for biochemical reconstitution study of actin assembly.
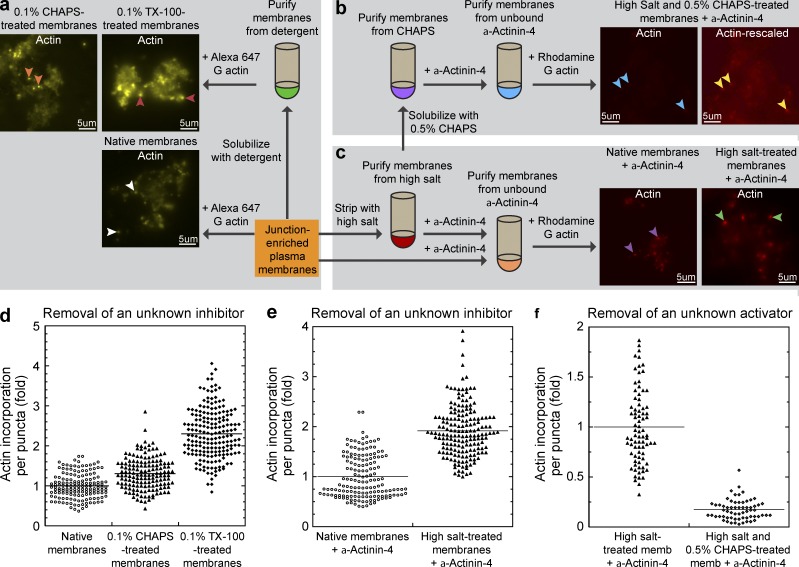
We empirically tested salt stripping and detergent solubilization conditions on purified membranes (Fig. 1, a–c). We found that treatment of membranes with 0.1% zwitterionic detergent CHAPS or nonionic detergent Triton X-100 (TX-100) yielded brighter fluorescent actin puncta upon addition of fluorescently labeled monomeric actin (Fig. 1, a and d). These results showed that actin assembly is enhanced rather than diminished after detergent treatments. In addition, the size of the junctional actin puncta also appeared larger. One possibility is that the native membrane contains endogenous inhibitory factors that can limit the amount of actin assembly at the adherens junction. Because detergents can solubilize both lipids and peripherally associated membrane proteins, the endogenous inhibitors can either be lipids or proteins. To distinguish between these possibilities, we treated the membranes with high salt, which preferentially disrupts protein–protein interactions rather than solubilizing lipids. Because high salt also removes α-actinin-4, which is an essential factor for the actin assembly reaction, we supplemented the stripped membrane assay with recombinant α-actinin-4. We found that high salt–treated membrane produced brighter actin puncta than native membranes supplemented with the same amount of recombinant α-actinin-4 (Fig. 1, c and e). These results suggest that high salt also removed inhibitory factors from the native membranes. Therefore, a peripherally associated protein is normally present on membranes to limit the amount and extent of actin assembly at the adherens junctional complexes.
Figure 1.
Identification of two actin-regulating activities at the adherens junctional complex. (a–c) Flow chart represents the overall scheme for biochemical analysis of actin-regulating factors on junction-enriched membranes. (a) Identification of a detergent extractable inhibitory factor on native membranes. Addition of Alexa Fluor 647–labeled monomeric actin to native membranes results in actin assembly associated with the junctional complexes (white arrowheads). Treatment of native membranes with zwitterionic detergent CHAPS or nonionic detergent TX-100 yielded brighter actin puncta (orange and red arrowheads, respectively). (b) Identification of a high-salt/detergent extractable activating factor on native membranes. Treatment of native membranes with high salt followed by CHAPS diminished actin incorporation at membrane puncta (blue arrowheads). Rescaling of the original image shows the presence of very dim fluorescent actin puncta (yellow arrowheads). (c) Identification of a high-salt extractable inhibitory factor on native membranes. Treatment of native membranes with high salt increased actin incorporation at membrane puncta (green arrowheads) when compared with native membranes (purple arrowheads) supplemented with α-actinin-4. (d) Quantitation of actin incorporation at junctional puncta in untreated native membranes and membranes treated with detergents CHAP (P < 0.0001) or TX-100 (P < 0.0001). (e) Quantitation of actin incorporation at junctional puncta in untreated native membranes and membranes treated with high salt (P < 0.0001). (f) Quantitation of actin incorporation at junctional puncta in high salt–treated or sequentially high salt– and CHAPS-treated membranes supplemented with α-actinin-4 (P < 0.0001).
Further perturbation of native membranes with sequential high salt and 0.5% CHAPS resulted in a dramatic decrease in actin incorporation at junctional puncta (Fig. 1, b and f). The reduction in actin incorporation is not solely due to solubilization of α-actinin-4, as the addition of excess recombinant α-actinin-4 failed to rescue the reaction. However, the number and density of actin puncta remained unchanged (Fig. 1 b, Actin-rescaled), which suggests that the membrane complexes remained largely intact.
The adherens junctional complexes are large, heterogeneous, and detergent insoluble
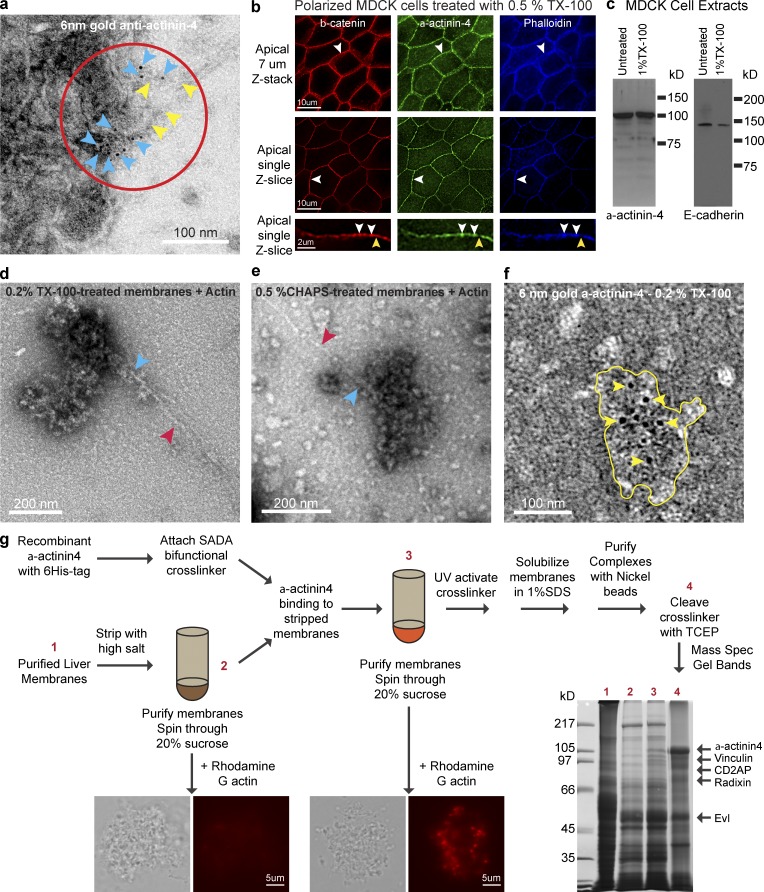
Our previous attempts to purify the membrane junctional complexes using conventional immunoprecipitation techniques had failed. Therefore, we characterized these complexes further to help guide the development of purification methods for the identification of its components. Immunogold labeling of MDCK cell plasma membranes showed that α-actinin-4 is present in clusters that span 100–200 nm in diameter at the region of actin assembly (Fig. 2 a). Immunofluorescence staining of TX-100–extracted polarized MDCK cells showed that α-actinin-4, β-catenin, and actin remained localized to the adherens junction (Fig. 2 b, white arrowheads). Although E-cadherin was partially solubilized, most α-actinin-4 was TX-100 insoluble (Fig. 2 c). These results could explain why our attempts to purify the complexes using conventional immunoprecipitation techniques had failed, which was due to the insolubilities of these large junctional complexes under conventional immunoprecipitation conditions.
Figure 2.
Purification and identification of junction-associated actin-regulatory factor complexes using a target bait-based, cross-linking approach. (a) Negative staining of MDCK membranes showing 6 nm immunogold labeling for α-actinin-4 (blue arrowheads) at sites of actin incorporation (red circle) and filament association (yellow arrowheads). (b) Immunofluorescence staining of TX-100 extracted MDCK cells for β-catenin, α-actinin-4, and actin (phalloidin) showing colocalization at junctional puncta (white arrowheads). A subset of β-catenin staining has no α-actinin-4 or actin (yellow arrowheads). (c) Western blots of MDCK cell extracts for E-cadherin and α-actinin-4 showing resistance to detergent extraction by TX-100. (d) Negative staining of TX-100–treated membranes showing actin filament (red arrowhead) associated with macromolecular complexes (blue arrowhead). (e) Negative staining of CHAPS-treated membranes showing actin filament (red arrowhead) associated with macromolecular complexes (blue arrowhead). (f) Negative staining of TX-100–resistant junctional complex (yellow outlines) showing 6 nm immunogold labeling for α-actinin-4 (yellow arrowheads). (g) Addition of cross-linker–derivatized recombinant α-actinin-4 to high salt stripped membranes results in targeting of α-actinin-4 to a membrane junctional complex that supports actin assembly. Activation of a cross-linker functional group by UV results in covalent attachment of α-actinin-4 to proteins in close proximity. Non-cross-linked proteins are removed by detergent solubilization. Identification of CD2AP, EVL, radixin, and vinculin by mass spectroscopy of gel bands from cleaved α-actinin-4 cross-linked complexes is shown. Coomassie blue staining of SDS-PAGE gel shows purification steps (lanes 1–4).
To assess if we could purify the complexes based on size, we examined membranes treated with TX-100 or CHAPS using negative-stain electron microscopy. We found that the complexes are highly heterogeneous in dimension and as large as 200 nm in size even after extraction with 0.2% TX-100 or 0.5% CHAPS detergents (Fig. 2, d and e). These large detergent-insoluble complexes can assemble actin filaments when incubated with monomeric actin, which indicates that they are functionally intact. Immunogold labeling of TX-100–extracted membrane fragments confirmed that α-actinin-4 is present in complexes that are heterogeneous in shape and sizes of 100–200 nm in diameter (Fig. 2 f). Because of their heterogeneity, purification of junctional complexes by conventional size-exclusion chromatography is not suitable.
Characterization and identification of membrane junctional complexes by a target-based cross-linking approach
Based on the dimensions visualized using electron microscopy, we conservatively estimated the junctional complexes of 100 nm to consist of at least 10–15 proteins calculated from the sizes of actin at 4 × 5 nm (Aebi et al., 1981) and α-actinin dimer at 7 × 40 nm (Bennett et al., 1984). We reason that this macromolecular junctional complex consists of membrane adhesion receptors such as E-cadherin, peripherally associated proteins such as catenins, and actin regulatory proteins such as arp2/3 and α-actinin-4. We hypothesized that actin regulatory proteins would be in close proximity to α-actinin-4. Therefore, we designed a cross-linking approach that specifically targets α-actinin-4 binding partners and/or actin-regulating components at the membrane by taking advantage of the biochemical reconstitution system, where α-actinin-4 can be stripped and rebind back to membrane junctional complexes (Fig. 2 g).
We identified seven proteins and confirmed the presence of four proteins—CD2AP, radixin, vinculin, and EVL—on native membranes (Fig. S1). CD2AP and radixin are susceptible to high salt and detergent extraction, and thus are candidates for the inhibitory activity. Vinculin and EVL are mildly resistant to high salt extraction, and thus are candidates for the activating activity.
EVL and CD2AP regulate α-actinin-4–dependent actin dynamics at the membrane junctional complex
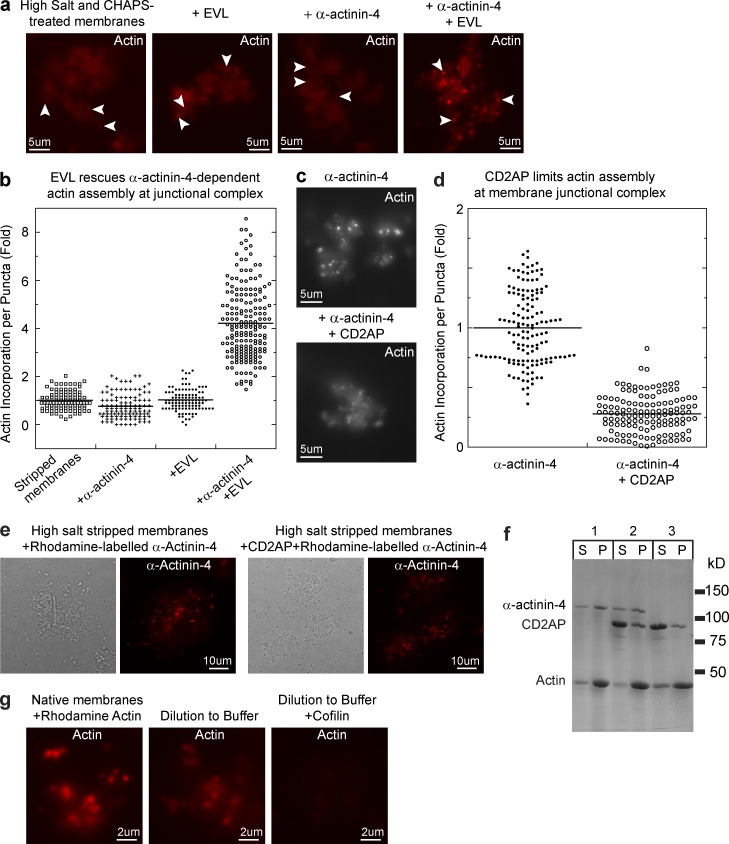
Using the membrane reconstitution assay, we confirmed that EVL and CD2AP are the activating and inhibitory activities for actin assembly at the junctional complexes (Fig. 3, a–e). Addition of EVL and α-actinin-4 to high salt/CHAPS-treated membranes restored actin puncta formation at junctional complexes (Fig. 3, a and b). However, EVL alone has no effect, which indicates that both EVL and α-actinin-4 are necessary in the same actin assembly reaction. Conversely, addition of CD2AP to high salt–treated membranes suppresses actin assembly even in the presence of α-actinin-4 (Fig. 3, c and d). However, incubation of stripped membranes with CD2AP failed to block recruitment of fluorescently labeled α-actinin-4 to junctional complexes (Fig. 3 e). CD2AP also did not inhibit α-actinin-4 from binding to F-actin in a cosedimentation assay (Fig. 3 f). Therefore, the suppression of actin assembly by CD2AP is unlikely to be caused by an interruption of α-actinin-4 function but rather by a direct modulation of actin dynamics. We found that upon actin assembly at the membrane, junctional actin puncta became very stable and would only slowly depolymerize upon dilution into buffer (Fig. 4 g). However, actin puncta could be completely disassembled by filament severing with cofilin. These observations are consistent with our findings that native membranes contain factors that can regulate actin filament dynamics at the junctional complex.
Figure 3.
Identification of CD2AP and EVL as the actin-regulating activities at the adherens junctional complex. (a) Biochemical reconstitution of actin assembly on high salt/CHAPS-treated membranes with α-actinin-4 and EVL. (b) Quantitation of actin puncta showing recovery of actin assembly on high salt/CHAPS-treated membranes (LPHSCh) supplemented with α-actinin-4 and EVL (P < 0.0001) but not with α-actinin-4 or EVL alone. (c) Biochemical reconstitution of actin assembly on high salt–treated membranes with α-actinin-4 alone or together with CD2AP. (d) Quantitation of actin puncta showing inhibition of actin assembly by CD2AP (P < 0.0001). (e) F-actin spin-down assay showing cosedimentation of α-actinin-4 and CD2AP with F-actin. (f) CD2AP did not interfere with α-actinin-4 recruitment to the membrane junctional complex. High salt–stripped membranes were incubated with rhodamine-labeled α-actinin-4 in the absence or presence of 1 µM CD2AP. (g) Actin depolymerization assay by dilution of membrane junctional actin puncta into buffer in the presence or absence of the severing protein cofilin.
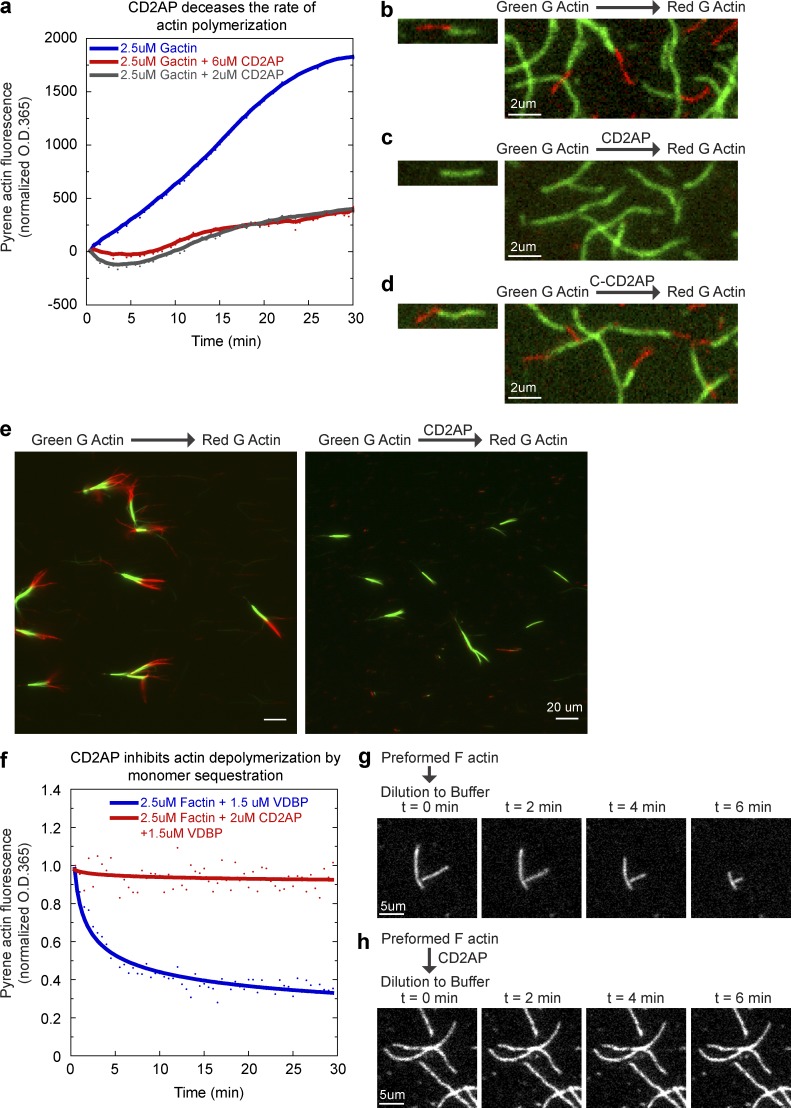
Figure 4.
CD2AP inhibits actin polymerization and depolymerization by capping actin barbed ends. (a) Pyrene-actin spectroscopy assay showing a dose-dependent decrease in spontaneous actin polymerization rate by CD2AP. Pyrene fluorescence was measured at 30-s intervals to obtain the trace. A representative trace from four experiments is shown. (b) Two-color actin single filament imaging showing addition of rhodamine-labeled (red) monomeric actin to preformed rhodamine green-labeled (green) actin filaments. (c) Two-color actin single filament imaging showing inhibition of red actin addition by preincubating green actin filaments with CD2AP. (d) Two-color actin single filament imaging showing addition of red actin after preincubating green actin filaments with the C-terminal half of CD2AP. (e) Two-color actin filament imaging of limulus parallel actin bundles showing inhibition of red actin addition to the fast-growing barbed ends by preincubating green actin filaments with CD2AP. (f) Pyrene-actin spectroscopy assay in the presence of monomer sequestration protein, vitamin D–binding protein (VDBP), showing suppression of actin depolymerization by CD2AP. Pyrene fluorescence was measured at 30-s intervals to obtain the trace. A representative trace from two experiments is shown. (g) Single filament imaging showing actin depolymerization upon dilution into buffer. (h) Single filament imaging showing inhibition of actin depolymerization by preincubating filaments with CD2AP.
CD2AP inhibits barbed-end dynamics of actin filaments
To test whether CD2AP can directly regulate actin dynamics, we measured fluorescence dequenching of pyrene-actin in the presence of CD2AP (Fig. 4 a). We found that CD2AP greatly retards the rate of actin polymerization. CD2AP could inhibit actin polymerization by either binding to actin monomers or capping filament ends. To directly assess the role of CD2AP on actin filament dynamics, we imaged single filaments in perfusion chambers (Fig. 4, b–e). Perfusion chambers allow buffer exchange, and CD2AP can be washed out after incubation with actin filaments in the chamber, eliminating the possibility of monomer sequestration in subsequent reactions. We assessed filament growth by imaging the addition of rhodamine-labeled (red) monomeric actin to preformed rhodamine green-labeled single filaments (Fig. 4, b–d). In the absence of CD2AP, red actin would add to ends of green filaments (Fig. 4 b). However, preincubation of green filaments with CD2AP completely blocked the addition of red actin (Fig. 4 c). The capping activity of CD2AP requires full-length CD2AP because a truncation form of CD2AP, C-CD2AP, which retains its F-actin binding activity (Gaidos et al., 2007), failed to inhibit red actin addition (Fig. 4 d). To assess if CD2AP blocks the fast-growing barbed ends of actin filaments, we performed sequential two-color actin polymerization using limulus bundles as seeds. Limulus bundles consist of parallel actin filaments with all barbed ends of actin filaments pointing at the fast-growing end of the bundle. By marking the barbed ends with green actin, we found that preincubation with CD2AP completely blocked barbed-end addition of red actin (Fig. 4 e).
Capping actin barbed ends would affect both actin polymerization and depolymerization. To further characterize the molecular function of CD2AP, we measured depolymerization of actin filaments in the presence or absence of CD2AP. Depolymerization of actin filaments is studied by dilution of the monomer pool to below the critical concentration of actin either via addition of monomer sequestration protein vitamin D–binding protein (Fig. 4 f) or by dilution into buffer (Fig. 4, g and h). We found that CD2AP completely blocked depolymerization of pyrene-actin filaments in the presence of vitamin D–binding protein (Fig. 4 f). To assess actin filament dynamics, we performed single filament imaging and showed that preincubation of actin filaments with CD2AP completely blocked dilution-induced shrinking from an end (Fig. 4, g and h). These results showed that CD2AP is a novel capping protein that can effectively shut down fast actin dynamics at filament barbed ends.
CD2AP colocalizes with a stable pool of actin and α-actinin-4 at the adherens junction
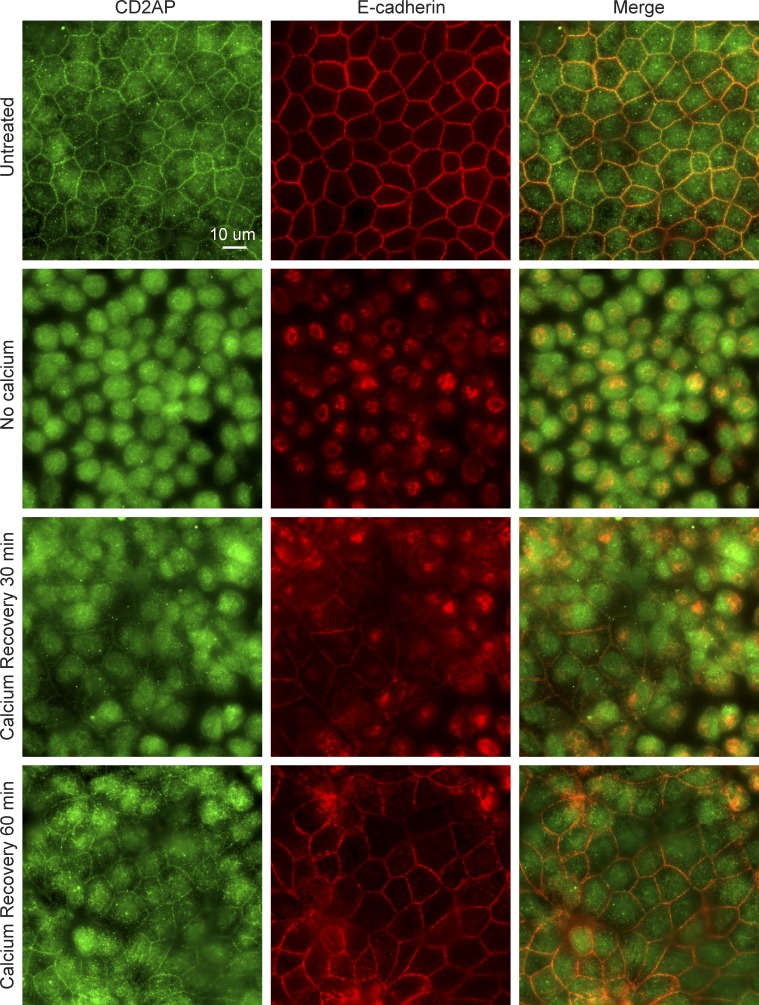
Barbed-end capping is a potential way to regulate actin assembly and disassembly at the adherens junction, which could have an important impact in the overall behaviors of intercellular interactions. We found that CD2AP is localized to the adherens junction along with E-cadherin in polarized MDCK cells (Fig. 5). Incubation of MDCK cells without calcium for 3 h results in diffuse cytoplasmic localization of CD2AP that is distinct from E-cadherin localization. Upon replenishment of calcium, CD2AP and E-cadherin reappeared at nascent cell–cell contacts within 30 min. By 1 h, most cells reformed cell–cell contacts containing E-cadherin and CD2AP.
Figure 5.
CD2AP is targeted to the adherens junction along with E-cadherin after a calcium-switch assay. Incubation of MDCK cells without calcium for 3 h results in diffuse cytoplasmic localization of CD2AP that is distinct from E-cadherin localization. Upon replenishment of calcium, CD2AP and E-cadherin appeared at nascent cell–cell contacts within 30 min. By 1 h, most cells reformed cell–cell contacts with E-cadherin and CD2AP localization at the junction.
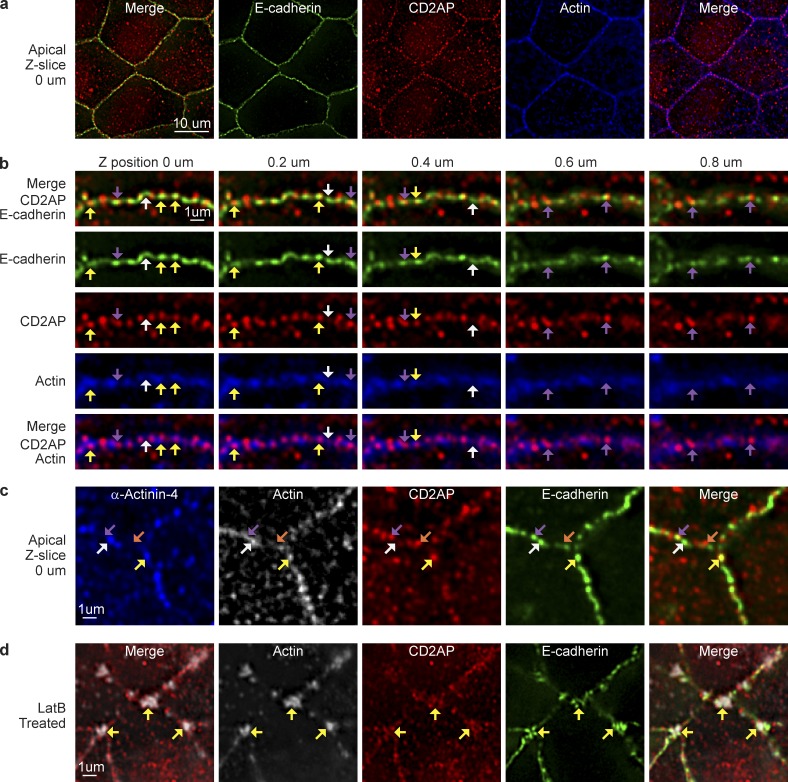
Using deconvolution microscopy, we showed that CD2AP is localized to discreet puncta along the adherens junction with actin and E-cadherin (Fig. 6 a). CD2AP is specifically colocalized to the apical (Fig. 6 b, 0–0.4 µm, yellow arrowheads) but not the lateral membrane (Fig. 6 b, 0.6–0.8 µm, purple arrowheads). Apical CD2AP puncta sometimes colocalized with α-actinin-4, E-cadherin, and actin (Fig. 6 c, yellow arrowheads). But puncta without α-actinin-4 or CD2AP are also present at the level of adherens junction (Fig. 6 c, purple and white arrowheads). These results are consistent with our electron microscopy data, which show that junctional complexes are highly heterogeneous. Previously, we and others have described a stable pool of actin at the adherens junction that is resistant to depolymerization by monomer sequestration with latrunculin (Fig. S2; Cavey et al., 2008; Tang and Brieher, 2012). Here, we found that CD2AP colocalized with α-actinin-4 and E-cadherin at latrunculin-resistant apical actin puncta (Fig. 6 d, yellow arrowheads). These observations suggest that CD2AP may have a role in stabilizing actin filaments at the adherens junction.
Figure 6.
CD2AP localizes to discreet junctional puncta in polarized MDCK cells. (a) Single deconvolved optical section showing colocalization of CD2AP with E-cadherin and actin (phalloidin). (b) Single deconvolved optical z sections scanning from the apical (0 µm) to lateral (0.8 µm) junction. Yellow arrowheads point to colocalization of CD2AP puncta with E-cadherin and actin. Purple arrowheads point to colocalization of CD2AP puncta with E-cadherin in the absence of actin. White arrowheads point to puncta with E-cadherin alone. (c) Single deconvolved optical section showing colocalization of CD2AP with α-actinin-4, E-cadherin, and actin (yellow arrowheads). A subset of E-cadherin puncta has α-actinin-4 and actin without CD2AP (white arrowheads), whereas another subset of E-cadherin puncta has CD2AP and actin without α-actinin-4 (purple arrowheads). There is also a population of CD2AP not associating with α-actinin-4, E-cadherin, or actin (orange arrowhead). (d) Single deconvolved optical section showing colocalization of CD2AP with E-cadherin at latrunculin-resistant actin clusters (yellow arrowheads).
CD2AP knockdown decreases actin accumulation at the adherens junction
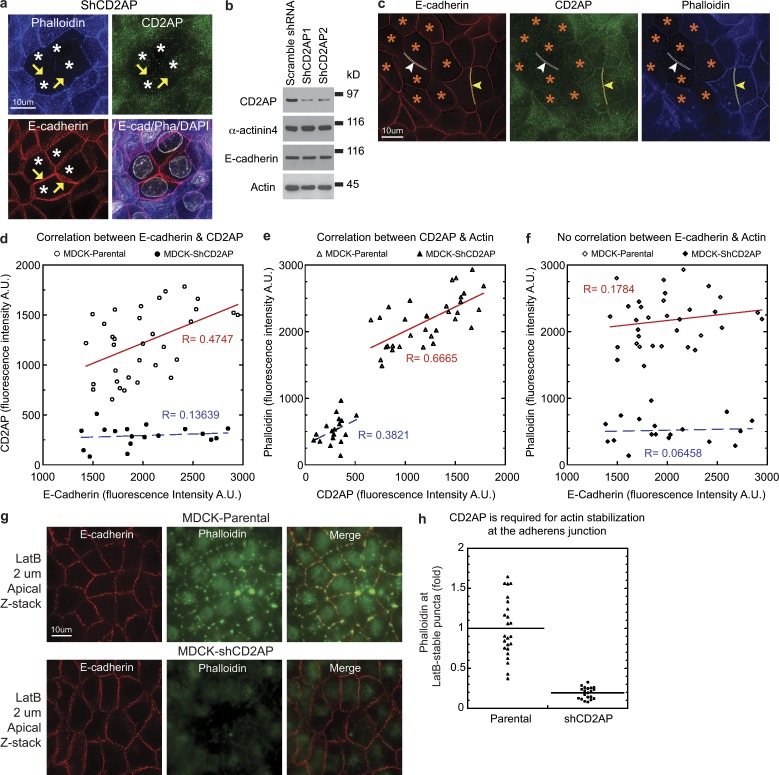
Knockdown of CD2AP (Fig. 7 a, asterisks) reduced filamentous actin without affecting the overall level of E-cadherin at the adherens junction (Fig. 7 a, arrowheads). Western blots showed no significant changes in α-actinin-4, E-cadherin, actin, vinculin, α-catenin, and ZO-2 levels in CD2AP knockdown cell lines (Fig. 7 b and Fig. S3). We compared junctional intensity of E-cadherin, CD2AP, and F-actin in single deconvoluted optical images (Fig. 7 c) and found a moderate correlation between E-cadherin and CD2AP levels (Fig. 7 d, red line), which was lost in CD2AP knockdown cells (Fig. 7 d, blue line). CD2AP levels correlated strongly with F-actin levels at the junction in both normal and knockdown cells (Fig. 7 e, red and blue lines). Surprisingly, E-cadherin levels did not correlate with F-actin accumulation, regardless of whether the cells were expressing normal or knockdown levels of CD2AP (Fig. 7 f, red and blue lines), which suggests that E-cadherin might have a more permissive role rather than an instructive role. To examine whether knockdown of CD2AP compromises actin stability, we depolymerized cellular actin by treating cells with latrunculin. We found a substantial reduction of latrunculin-resistant junctional puncta in CD2AP knockdown cells (Fig. 7, g and h). These results showed that CD2AP plays an essential role in actin stabilization and accumulation at the adherens junction.
Figure 7.
CD2AP is required for actin stability at the adherens junction of polarized MDCK epithelial cells. (a) Projection of 40 deconvolved optical z slices spanning the apical 8 µm of cells showing decreased actin accumulation (yellow arrowheads) at cell–cell contacts in CD2AP knockdown cells (white asterisks) stained for E-cadherin, CD2AP, actin (phalloidin), and DNA (DAPI). (b) Western blots of total cell extracts showing normal levels of E-cadherin, α-actinin-4, and actin in stable CD2AP knockdown cell lines, ShCD2AP1 and ShCD2AP2. Molecular weight markers are 150 kD for CD2AP, α-actinin-4, and E-cadherin blots and 50 kD for the actin blot. (c) Single deconvolved optical z slice at the apical region of cells showing knockdown of CD2AP (white arrowheads and outlines) within a monolayer of cells expressing normal levels of CD2AP (yellow arrowheads and outlines). (d) Quantitation of junctional staining of E-cadherin and CD2AP showing a correlation between CD2AP levels and E-cadherin levels in parental but not CD2AP knockdown cells. R is the correlation coefficient. Representative data from four separate experiments are shown. (e) Quantitation of junctional staining of CD2AP and actin (phalloidin) showing a correlation between CD2AP levels and actin levels in both parental and CD2AP knockdown MDCK cells. Representative data from four separate experiments (n = 4) are shown. (f) Quantitation of junctional staining of E-cadherin and actin (phalloidin) showing a lack of correlation between E-cadherin levels and actin levels in both parental and CD2AP knockdown MDCK cells. Representative data from four separate experiments (n = 4) are shown. (g) Projection of 10 deconvolved optical z slices spanning the apical 2 µm of cells showing decreased actin levels at latrunculin-resistant puncta in CD2AP knockdown cells. (h) Quantitation of actin levels at latrunculin-resistant puncta in parental and CD2AP knockdown cells (P < 0.0001). Representative data from four separate experiments (n = 4) are shown.
CD2AP knockdown compromises intercellular adhesive strength and epithelial barrier function
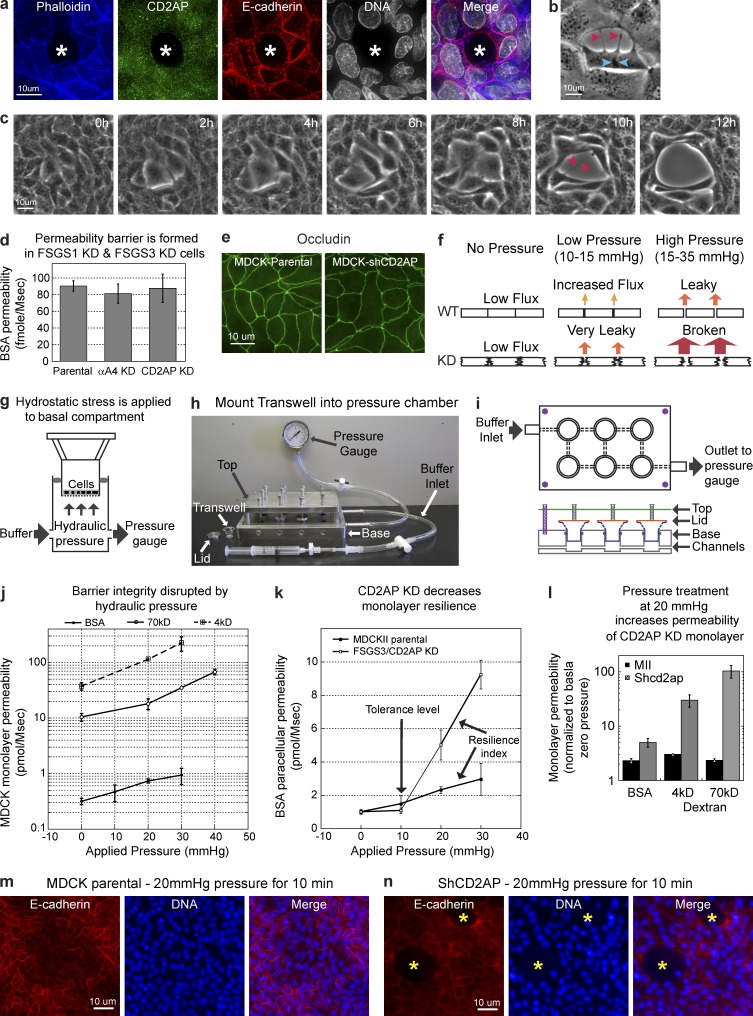
CD2AP knockdown cells spontaneously detach from their neighbors in growing monolayers, resulting in holes (Fig. 8 a, asterisks) and tearing between cells (Fig. 8, b and c). We hypothesized that CD2AP-dependent actin accumulation is essential for stability of cell–cell adhesion. However, we did not observe any holes or tearing between CD2AP knockdown cells after they formed polarized monolayers.
Figure 8.
CD2AP is required for the epithelial monolayer to withstand mechanical stress. (a) Projection of 70 deconvolved optical z slices of the entire 14-µm height of polarized MDCK cells showing a missing cell (white asterisks) within a cell monolayer transfected with CD2AP shRNA. (b) Phase-contrast microscopy showing a cell in mitosis (chromosome condensation, blue arrowheads) detaching from neighboring cells (red arrowheads) within a monolayer of cells transfected with CD2AP shRNA. (c) Time-lapse images of phase-contrast microscopy showing cells tearing away (red arrowheads) from each other within a monolayer of cells transfected with CD2AP shRNA. (d) BSA flux assays showing similar monolayer permeability between parental, CD2AP knockdown, and α-actinin-4 knockdown cells. (e) Occludin localization at the tight junction is normal in CD2AP knockdown cells. (f) Logistics of applying mechanical stress at intercellular junctions for the analysis of epithelial cohesion. (g) A pressure chamber apparatus that can generate hydraulic pressure across a cell monolayer to induce stress at intercellular junctions. (h) Pressure chamber apparatus is attached to a syringe for pumping fluid and a pressure gauge to monitor applied pressure. Transwell supports were held in position during pressure application by lids and screws at the top. (i) Schematics for the operation of the pressure chamber apparatus. Buffer is pumped through channels connecting the basal chambers to produce hydrostatic pressure. (j) Permeability assays showing increased tracer flux with increasing applied hydrostatic pressure to MDCK monolayers. (k) Permeability assays showing a low tolerance level and decreased resilience to mechanical stress in CD2AP knockdown cells. (l) Permeability assays showing greater increases in tracer permeability in CD2AP knockdown cells than in parental cells after mechanical stress. (m) E-cadherin localization at the cell–cell junction of MDCK cells showing intact monolayer after mechanical stress. (n) E-cadherin localization at the cell–cell junction of CD2AP knockdown cells showing holes (asterisks) in the monolayer after mechanical stress. All error bars indicate standard errors.
To directly assess the integrity of cell monolayers, we measured the permeability of cell monolayer to extracellular tracer BSA. We found that CD2AP knockdown did not compromised the barrier function of the monolayer (Fig. 8 d). In addition, localization of the tight junction protein, occludin (Furuse et al., 1993; Wong and Gumbiner, 1997), was unchanged (Fig. 8 e).
We hypothesized that reduction of junctional actin could compromise cell–cell adhesive strength rather than formation of intercellular junctions. To directly test our hypothesis, we assessed the strength of cell–cell cohesion by testing the ability of cells to resist external mechanical stress and preserve epithelial permeability barrier (Fig. 8 e).
Until now, there is no cell culture model to study the causative role of mechanical stress in the disruption of cell–cell adhesion and permeability barriers. Therefore, we designed and built a new apparatus that can generate perpendicular hydraulic pressure to study the strength of cell–cell adhesion (Fig. 8, f–i). To measure the ability of cell monolayer to withstand mechanical stress, hydraulic pressure was applied to cell monolayers grown on Transwell filters mounted onto a sealed fluid-filled basal chamber. The hydrostatic pressure in the basal chamber was monitored continuously using a pressure gauge during pressure treatment. Cells were subjected to physiological (0–15 mmHg) and pathological (15–35 mmHg) tissue pressures (Kinoshita and Knox, 1989; Shiotsu et al., 1995). The ability of the cell monolayer to withstand mechanical stress and preserve epithelial barrier function was monitored by the permeability of the cell monolayer.
Using the pressure chamber, we found that mechanical load caused a dose-dependent increase in MDCK monolayer permeability to extracellular tracers (Fig. 8 j). Comparison of parental and CD2AP knockdown cells showed a dramatic difference in their response to mechanical stress (Fig. 8 k). At low hydrostatic pressure, CD2AP knockdown cells behaved similar to parental cells. However, at higher hydrostatic pressure, CD2AP knockdown cells became substantially leakier to all three extracellular tracers, with molecular mass ranging from 4 to 70 kD (Fig. 8 l). In addition, mechanical stress also induced tearing, resulting in holes within the CD2AP knockdown cell monolayer (Fig. 8, m and n). These results showed that strengthening of cell–cell adhesion and generation of epithelial resilience to mechanical stress requires actin accumulation and stabilization of the adherens junction supported by CD2AP.
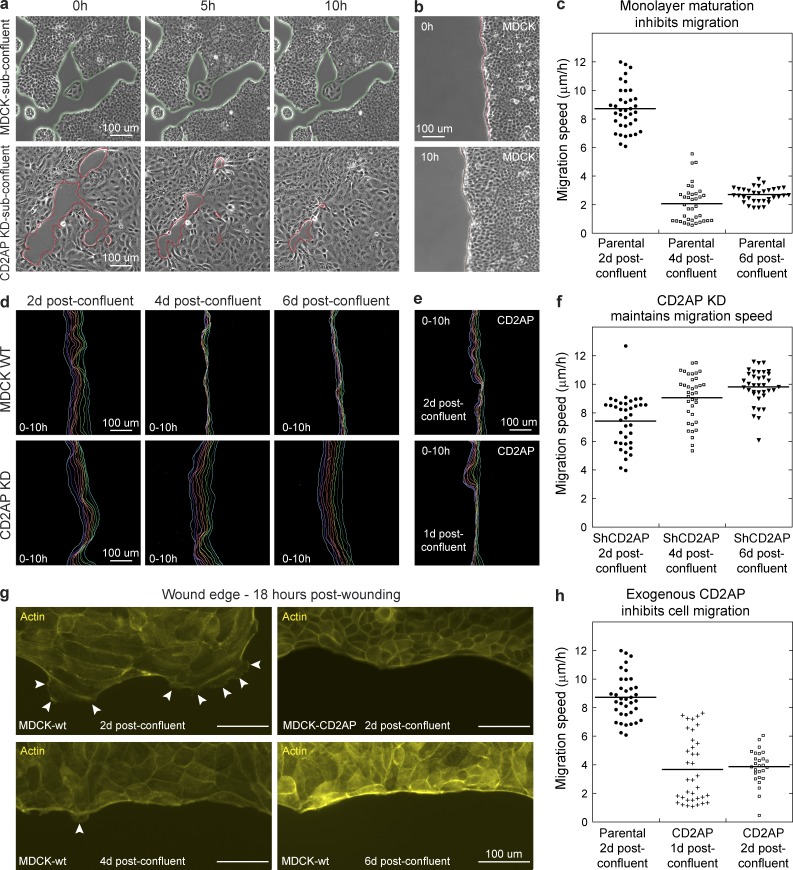
CD2AP knockdown compromises junction maturation and increases cell motility
Maturation and strengthening of cell–cell adhesion in epithelial cells has important physiological implications, including suppression of cell motility (Takai et al., 2008; Mayor and Carmona-Fontaine, 2010; Etienne-Manneville, 2012). In subconfluent culture, MDCK cells were relatively stationary, whereas CD2AP knockdown cells would move into free space (Fig. 9 a). However, MDCK cells can be induced to migrate when wounded. To assess the role of CD2AP in wound-induced migration, we measured wound edge progression in MDCK monolayers (Fig. 9 b). We found that the rate of wound invasion depends on the age of the confluent monolayer, which suggests that the MDCK monolayer matures over time (Fig. 9, c and d). Knockdown of CD2AP prevented this maturation process, and the rate of wound invasion did not slow down over time (Fig. 9, d and f). Conversely, overexpression of exogenous CD2AP accelerated junction maturation and suppressed wound-induced migration (Fig. 9, e and h). Examination of the monolayer wound edge showed a correlation between the cell migration rate and the formation of protrusive structures (Fig. 9 g, white arrowheads). These results indicate that actin accumulation supported by CD2AP directly participates in maturation and stabilization of the adherens junctions, contributing to the overall behaviors of epithelial cells.
Figure 9.
CD2AP suppresses cell motility and wound-induced cell migration. (a) Time-lapse phase-contrast microscopy showing migration of CD2AP knockdown cells into free space, whereas parental MDCK cells are relatively stationary. (b) Time-lapse phase-contrast microscopy showing wound-induced migration of 2 d postconfluent MDCK cell monolayer. (c) Quantitation of migration speed in MDCK cells showing decreased migration rate as a function of time (P < 0.0001). Representative data from three separate experiments (n = 3) are shown. (d) Line tracings of wound edge at 1-h intervals showing a decrease in migration rates of parental but not CD2AP knockdown cells at 4 and 6 d postconfluency (4 d and 6 d). (e) Line tracings of wound edge at 1-h intervals showing suppression of wound-induced migration of cells expressing exogenous CD2AP at 1 and 2 d postconfluency. (f) Quantitation of migration speed in CD2AP knockdown cells showing a lack of suppression in wound-induced migration after confluency. Representative data from three separate experiments (n = 3) are shown. (g) Actin (phalloidin) staining showing correlation of cell migration with wound-edge protrusions (white arrowheads). Protrusions were suppressed in 4 d and 6 d postconfluent parental cells and cells expressing exogenous CD2AP. (h) Quantitation of migration speed showing suppression of wound-induced migration in 1 d and 2 d postconfluent cells expressing exogenous CD2AP (P < 0.0001). Representative data from three separate experiments (n = 3) are shown.
Discussion
Dissecting the molecular mechanism of actin regulation at the cell–cell adhesive junction is fundamental to the understanding of intercellular interactions. We have developed a novel target-based, two-step cross-linking approach to identify junctional components associated with α-actinin-4–dependent actin assembly. Using this approach, we discovered two new opposing activities that regulate actin dynamics at the adherens junction; an activating activity by EVL and a capping activity by FSGS3/CD2AP. Furthermore, we have designed and built a new apparatus that can apply tangential mechanical stress to a cell–cell junction to study how actin regulation contributes to epithelial integrity and cohesion.
We showed that EVL is required for actin assembly at adherens junctional complexes in an arp2/3- and α-actinin-4–dependent manner. EVL belongs to the Ena/VASP family of actin elongation factors (Dominguez, 2009), which play roles in actin accumulation (Baum and Perrimon, 2001) and organization at cadherin–adhesive contacts (Vasioukhin et al., 2000; Scott et al., 2006). However, EVL and α-actinin-4 do not directly stimulate arp2/3 nucleation. Therefore, additional unknown factors at the membrane junctional complexes are required for actin assembly. Recently, knockdown of WAVE2, an arp2/3 activator that localizes to the adherens junction, has been shown to decrease actin polymerization and reduce actin accumulation at the adherens junction (Verma et al., 2012). Because WAVE can activate arp2/3 in pure solution (Chen et al., 2010), it could be involved in actin assembly in our reconstituted system. Future experiments to directly address the involvement of auxiliary factors in actin assembly will provide deeper understanding of actin nucleation and elongation at membrane junctional complexes.
Our findings reveal a new function for CD2AP in actin barbed-end capping, which inhibits both actin polymerization and depolymerization. CD2AP was originally identified as mesenchyme-to-epithelium transition protein with SH3 domains 1 (METS-1) in a screen for RNA transcripts that are up-regulated during mesenchyme-to-epithelium transition (Jansson et al., 1997; Lehtonen et al., 2000). CD2AP has been shown to organize the actin cytoskeleton at immunological synapses (Dustin et al., 1998) and podocyte slit diaphragms (Yaddanapudi et al., 2011; Zhao et al., 2013). However, the mechanism by which CD2AP regulates membrane actin dynamics is unknown. Previous experiments demonstrated that a peptide from the C terminus of CD2AP would uncap the barbed-end capping protein CapZ from filament ends to allow actin polymerization (Bruck et al., 2006; Hernandez-Valladares et al., 2010). However, we found that the C-terminal half of CD2AP, which contains the CapZ unbinding peptide sequence, was unable to block barbed-end dynamics. In addition, molecular genetic studies showed that a 4% truncation from the C terminus of CD2AP protein was able to cause FSGS (Löwik et al., 2007). Thus, the physiological function of CD2AP appears to require full-length CD2AP. Nevertheless, CD2AP could also be regulated in vivo to perform additional functions including uncapping CapZ to allow filament elongation (Bruck et al., 2006; Hernandez-Valladares et al., 2010).
Despite an almost complete inhibition of barbed-end dynamics in pyrene and single filament assays in vitro, endogenous CD2AP did not completely block actin assembly on purified native membranes. Preincubation of stripped membranes with excess CD2AP only resulted in a modest decrease in actin assembly. It is possible that CD2AP, when tethered to the membrane, is not in the proximity of actin barbed ends. Another possibility is that actin barbed ends are protected during polymerization by the large membrane junctional complex. In addition, other actin regulators such as the capping protein CapZ can modulate actin dynamics on the membrane (Cooper and Sept, 2008). Although CD2AP is expressed at a significantly higher level than CapZ in MDCK cells (Fig. S4), CD2AP and CapZ could have overlapping and distinct roles in regulating actin barbed-end dynamics. A future direction for research will be to understand the role of different actin regulators, including barbed-end binding proteins such as CapZ and pointed-end binding proteins such as tropomodulin (Yamashiro et al., 2012), in actin dynamics at the adherens junction.
Although both FSGS1/α-actinin-4 and FSGS3/CD2AP knockdown decreased actin accumulation at the adherens junction, their mechanisms of action are entirely different. Knockdown of FSGS1/α-actinin-4 disrupted actin assembly without affecting junctional actin stability (Tang and Brieher, 2012). However, knockdown of FSGS3/CD2AP substantially compromised actin stability. Therefore, actin accumulation at the adherens junction can be a consequence of either actin assembly or filament stabilization. Our study highlights the importance of molecular dissection and biochemical characterization to complement cellular and animal studies and in the interpretation of cellular phenotypes and disease states.
We showed that CD2AP is not required for the formation of epithelial intercellular junctions. Knockdown of CD2AP did not affect the levels or localization of junctional components. In addition, CD2AP knockdown did not compromise the permeability barrier function of polarized MDCK cell monolayer. Previous studies found that CD2AP depletion perturbed the reformation of transepithelial resistance in a calcium switch assay, which suggests that CD2AP may be involved in junction dynamics (Maun et al., 1996). However, CD2AP is not necessary for the maintenance of cell junctions upon polarization of the epithelial monolayer. In agreement with our findings, homozygous and heterozygous CD2AP knockout mice show no gross morphological defects at birth, which indicates that the formation of cell junctions and development of epithelial polarity is normal in the absence of CD2AP (Shih et al., 1999; Huber et al., 2006). In this study, we found that CD2AP is targeted along with E-cadherin to nascent junctions after calcium switch. Our preliminary experiments indicate that the actin-binding domain of CD2AP alone is not sufficient for targeting to cell–cell junctions (Fig. S5). A goal for future research will be to understand how CD2AP is recruited to junctions and whether targeting to the adherens junction requires known interacting proteins such as cortactin (Lynch et al., 2003) and Rac (van Duijn et al., 2010) or novel binding partners.
Despite having normal permeability barrier function, CD2AP knockdown cells would become very leaky when challenged with physiological hydraulic pressures similar to fluid filtration pressures in the kidney (Kinoshita and Knox, 1989; Shiotsu et al., 1995). These observations are consistent with the findings that CD2AP knockout mice eventually develop proteinuria due to loss of glomerular filtration barrier (Shih et al., 1999; Kim et al., 2003; Huber et al., 2006). Therefore, stabilization of actin at the adherens junction by CD2AP plays an important role in epithelial cohesion and cell–cell adhesive strengthening to withstand normal mechanical stress. Our data provide a plausible molecular explanation for the disruption of barrier function and development of proteinuria and kidney disease reminiscent of FSGS in CD2AP knockout mice.
There are precedents that the adherens junction contributes to the barrier function of epithelial and endothelial cells in knockout mouse models. Knockout of E-cadherin in the skin had been shown to result in the loss of permeability barrier of the epidermis, leading to dehydration of the animal (Tunggal et al., 2005). In addition, triple knockout of the EVL family members, which regulate actin polymerization at the adherens junction, led to a dramatic leakage of the endothelial barrier of the blood vessel (Furman et al., 2007). Our study provides further evidence that the adherens junction and its associating actin dynamics play very important roles in the maintenance of epithelial tissue integrity to defend the permeability barrier.
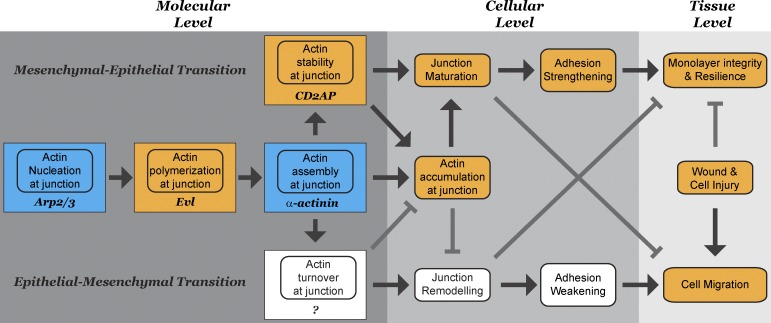
Our results implicate CD2AP in the maturation of epithelial junction, which is consistent with the findings that CD2AP expression is up-regulated during mesenchyme-to-epithelium transitions (Jansson et al., 1997). By stabilizing actin at the adherens junction, CD2AP plays an important role not only in cell–cell adhesion but also in the development of epithelial characteristics such as suppression of cell migration. As stabilization of actin at the adherens junction suppresses cell migration, destabilization of actin is associated with increased actin dynamics, resulting in enhanced cell migration during epithelium-to-mesenchyme transition and cancer metastasis (Ghosh et al., 2004; Wang et al., 2007). Therefore, regulating actin stability at the adherens junction is a potential way to modulate the strength of cell–cell adhesion as well as epithelial cell behaviors without destroying the entire cell–cell contact. Our observation is consistent with others that CD2AP regulates junctions and cytoskeleton dynamics during tissue patterning (Johnson et al., 2008). The rich interplay of actin regulators at the adherens junction reflects the complexity of epithelial dynamics (Fig. 10). Regulation of actin polymerization, stability, and depolymerization locally at the adherens junction dramatically influences junction dynamics, which ultimately dictates the strength of adhesion, influences cell motility, and contributes to the ability of epithelial cells to withstand mechanical stress and defend tissue integrity and barrier function.
Figure 10.
A relationship between actin accumulation at the molecular level, adhesion strengthening at the cellular level, and epithelial cohesion at the tissue level. Actin accumulation at the adherens junction is a balance of actin polymerization, stabilization, and depolymerization. Actin polymerization at the adherens junction requires the coupling of actin nucleation by the arp2/3 complex, actin elongation by EVL, and actin assembly by α-actinin-4. Thus, junction actin assembly is not simply a product of actin polymerization but resulted from the coordinated spatial and temporal coupling between an actin assembly process and a nucleation/polymerization reaction at the adherens junction. After actin is assembled, filament barbed ends are capped by CD2AP. Stabilization of actin by CD2AP contributes to strengthening and maturation of adherens junctions, leading to suppression of cell motility and increase in resilience of epithelial sheet to mechanical stress. Orange boxes represent this work. Red boxes represent work using the same biochemical reconstitution system.
Materials and methods
Antibodies and reagents
Antibodies to α-actinin-4, radixin, and EVL were raised in-house against 6-His–tagged recombinant full-length human proteins expressed in Rosetta cells. Antibodies to E-cadherin were made in-house from hybridoma RR1. Antibodies to α-actinin-4 (catalog no. 49333), β-catenin (catalog no. 7963), CD2AP (catalog no. 9137), and vinculin (catalog no. 7649) were purchased from Santa Cruz Biotechnology, Inc. Secondary antibodies were obtained from Bio-Rad Laboratories (HRP anti–rabbit), Invitrogen (FITC and Cy3 anti–mouse, FITC and Cy3 anti–rabbit, Alexa Fluor 488 anti–mouse, Alexa Fluor 568 anti–rabbit, and Alex Fluor 647 anti–goat), and Electron Microscopy Sciences (colloidal 6 nm gold anti–rabbit). Tetramethyl-rhodamine N-succinimidyl ester, rhodamine green N-succinimidyl ester, tetramethyl-rhodamine maleimide, and pyrene maleimide were purchased from Invitrogen. Cross-linker sulfo-NHS-SS-Diazirine (sulfo-SDAD) was purchased from Thermo Fisher Scientific. Reducing agent Tris(2-carboxyetheyl)-phosphine hydrochloride (TCEP) was purchased from Thermo Fisher Scientific. Vitamin D–binding protein (GC-globin) was purchased from EMD Millipore. Alexa Fluor 647, Alexa Fluor 350–phalloidin, and Alexa Fluor 647–phalloidin were purchased from Invitrogen. FITC-labeled dextrans (4 kD and 70 kD) were purchased from Sigma-Aldrich. Leupeptin, Pefabloc, E-64, antipain, aprotinin, bestatin, and calpain inhibitors I and II were purchased from A.G. Scientific, Inc.
DNA constructs
Full-length human α-actinin-4 tagged with 6-His at the N terminus cloned into pET30a+ bacterial expression vector was used for protein expression as described previously (Tang and Brieher, 2012). The coding sequences of human CD2AP, C-CD2AP truncation mutant (aa 351–639), human radixin, and EVL were subcloned into the bacterial expression vector pET30a+ (EMD) for expression in Rosetta cells (EMD). Full-length CD2AP was subcloned with G418-selectable mammalian expression vector pNTAPB (Agilent Technologies) with a streptavidin-binding peptide tag at the N terminus for expression in MDCK cells. shRNAs for CD2AP (5′-TCAACACCTTCAAGTGCTTCG-3′) and scramble shRNA were synthesized and subcloned into puromycin-selectable pRS vector (OriGene).
Protein purification
Recombinant α-actinin-4 was prepared as described previously (Tang and Brieher, 2012). In brief, for expression of recombinant 6-His–tagged α-actinin-4, actin-binding domain, and K255E, Rosetta cells were induced with 500 µM isopropyl β-d-1-thiogalactopyranoside for 8 h at 25°C. Cells were centrifuged at 6,000 g for 15 min and resuspended in 20 mM NaCl, 20 mM Hepes, pH 7.8, in the presence of 5 mg/ml of lysozymes. After a freeze/thaw cycle, lysed cells were centrifuged at 20,000 g for 30 min. The supernatant was loaded onto a nickel column (QIAGEN). The column was washed with 20 bed volumes of 500 mM NaCl, 25 mM Imidazole, and 20 mM Hepes, pH 7.8. The recombinant proteins were eluted with 10 bed volumes of 500 mM NaCl, 500 mM Imidazole, and 20 mM Hepes, pH 7.8. Full-length proteins were concentrated using centricon (EMD Millipore) and purified by gel filtration in 150 NaCl, 20 mM Hepes, and 10 mM β-mercaptoethanol. For expression of recombinant 6-His–tagged CD2AP, C-CD2AP, EVL, and radixin, Rosetta cells were induced with 500 µM isopropyl β-d-1-thiogalactopyranoside for 4–8 h at 25°C and lysed by a freeze/thaw cycle. Recombinant proteins were purified using nickel resin followed by Superdex 200 gel filtration (CD2AP and C-CD2AP), Q HP ion exchange (radixin), and S ion exchange (EVL) columns. Actin was labeled on lysine residues using N-hydroxysuccinimide–activated fluorophores (Invitrogen). α-Actinin-4 was labeled on cysteine using maleimide-activated fluorophores or derivatized with sulfo-SDAD. Labeled proteins were separated from free dyes or cross-linker by gel filtration using Superdex 200 (GE Healthcare). Recombinant cofilin was purified as described previously (Maciver et al., 1998). In brief, BL21-DE3 cells were induced with 500 µM isopropyl β-d-1-thiogalactopyranoside for 3 h at 25°C and lysed with 1 mg/ml lysozyme and a freeze/thaw cycle. Recombinant proteins were purified using DEAE followed by Q HP ion exchange columns. Recombinant human CapZ (α and β) was made in Rosetta cells by induction with 500 µM isopropyl β-d-1-thiogalactopyranoside for 3 h at 25°C followed by lysing the cells and a freeze/thaw cycle. Recombinant proteins were purified using S HP followed by Sephadex gel filtration columns (Sigma-Aldrich).
Actin assembly and reconstitution assays
Purification of junction-enriched membranes was prepared as described previously (Tang and Brieher, 2012). In brief, frozen rat livers (Pel-Freez) were thawed in 2 volumes of 10 mM Hepes, pH 8.5/10 mM DTT. Protease inhibitors (see above) were added and the livers were briefly blended in a blender (Waring; 5 × 15 s). The liver slush was filtered through four layers of cheesecloth to obtain the total liver homogenate. Total liver homogenate was centrifuged at 1,000 g for 30 min. The pellet was homogenized in 10 mM Hepes, pH 8.5/10 mM DTT in a homogenizer (Dounce) and centrifuged at 100 g for 30 min. The supernatant was collected and centrifuged at 1,000 g for 30 min. The membrane pellet contains the majority of actin assembly activity and is used for all membrane experiments.
Actin assembly assays were performed in actin assembly buffer (50 mM KCl, 2 mM EGTA, 2 mM MgCl2, and 100 mM Hepes, pH 7.8) supplemented with 2 mM of buffered ATP, pH 8. In brief, a 20-µl reaction consisting of ∼15 µg of total proteins from the membrane fraction and 0.5 µM of fluorescently labeled monomeric actin was allowed to carry out at room temperature for 30 min. For reconstitution assays, purified membranes were stripped with high salt (500 mM NaCl, 2 mM MgCl2, 2 mM EGTA, 20 mM Hepes, pH 7.8, and 10 mM DTT), TX-100, or CHAPS (in 50 mM NaCl, 2 mM MgCl2, 2 mM EGTA, 20 mM Hepes, pH 7.8, and 10 mM DTT) on ice. Stripped membranes were collected by centrifugation through a 20% sucrose cushion at 10,000 g for 10 min. Purified proteins (CD2AP, EVL, and α-actinin-4) were allowed to bind to native or stripped membranes for 1 h at room temperature. Unbound proteins were removed by spinning membranes through a 20% wt/wt sucrose cushion at 10,000 g for 10 min. The final reconstitution reaction consists of ∼8 µg of total protein from stripped membranes, 0.5 µM fluorescently labeled actin, and purified proteins (CD2AP, EVL, and α-actinin-4), and was performed at room temperature for 30 min. The membranes were imaged using a light microscope (Axio Imager) with the Colibri illumination system (both from Carl Zeiss) using a 63× objective lens (NA 1.4) attached to a 1,000 × 1,000 charge-coupled device camera (ORCA-ER; Hamamatsu Photonics). Quantitation of actin puncta intensity was performed in ImageJ (National Institutes of Health) using unprocessed original images. A defined area (6 × 6 pixels) was used to measure the signal intensity of actin fluorescence. The measured intensities were subtracted from background before being used for calculation of the intensity ratios.
Negative-stain electron microscopy and immunogold labeling
For visualizing actin assembly, membrane reactions were performed as above before processing for electron microscopy. For immunogold labeling of MDCK plasma membranes, cell homogenates were prepared as described previously (Tang and Brieher, 2012). In brief, MDCK cells were rinsed and hypo-osmotically shocked on dish in 10 mM Hepes, pH 8.5, for 60 min at 4°C. Cells were scraped and homogenized on ice through a 25G needle in 10 mM Hepes, pH 8.5, in the presence of 10 mM DTT and protease inhibitors (10 µg/ml Leupeptin, 1 mg/ml Pefabloc, 10 µg/ml E-64, 2 µg/ml antipain, 2 µg/ml aprotinin, 50 µg/ml bestatin, 20 µg/ml calpain inhibitors I, and 10 µg/ml calpain inhibitor II). Actin assembly reactions were allowed to continue for 10 min before incubation with anti–α-actinin-4 antibodies for 2 h on ice. For immunogold labeling of junction-enriched membrane, membranes were treated with 0.2% TX-100 in actin assembly buffer on ice for 1 h, spun through a 20% sucrose cushion before incubation with anti–α-actinin-4 antibodies for 2 h on ice. Unbound antibodies were removed by spinning membranes through a 20% sucrose cushion before incubation with 6 nm colloidal gold anti–rabbit antibodies for 2 h. Unbound gold antibodies were removed by spinning membranes through a 20% sucrose cushion before processing for electron microscopy. Membrane reactions were put onto glow-discharged carbon-coated grids for 10 min, washed three times with assembly buffer, and stained with 2% uranyl acetate. Images were collected with a microscope (2100EX; JEOL) at 120 kV using a 2,000 × 2,000 charge-coupled device camera (UltraScan; Gatan, Inc.). For figure generation, images were cropped, contrasted, and scaled using Photoshop software (Adobe) before importing into Illustrator (Adobe).
Bait-based cross-linking and mass spectroscopy
Cross-linking was performed with a heterobifunctional cross-linker, succinimidyl-diazirine, that allows a controlled two-step cross-linking reaction using succinimidyl-based chemistry combined with diazirine-based photochemistry. We covalently attached the cross-linker to recombinant 6-His–tagged α-actinin-4 by selectively activating the succinimidyl functional group. The derivatized α-actinin-4 was used as “bait” for binding to membranes that had been pretreated with high salt to remove endogenous α-actinin-4. Unbound α-actinin-4 was removed by spinning the membranes through a 20% sucrose cushion at 10,000 g for 10 min. The membranes were resuspended in actin assembly buffer and placed under UV to activate the diazirine functional group. The cross-linked α-actinin-4 complexes were solubilized in boiling 2% SDS and centrifuged at 16,000 g to remove insoluble materials. The supernatant was diluted to a final concentration of 0.2% SDS in 1% TX-100 and incubated overnight with nickel resin (QIAGEN). The nickel resin was washed with 20 bed volumes of 0.5% TX-100 in 20 mM Hepes, pH 7.8, and the cross-linked α-actinin-4 complexes were eluted with 5 bed volumes of 0.5% TX-100 in 500 mM imidazole and 20 mM Hepes, pH 7.8. The cross-linker was cleaved by dialysis of the eluate against TCEP for 24 h at 37°C. Cleaved products were separated by SDS-PAGE. Distinct protein bands were excised from a Coomassie-stained gel and submitted for mass spectroscopy analysis at the University of Illinois Roy J. Carver Biotechnology Center.
Pyrene-actin polymerization assay
Pyrene-actin polymerization/depolymerization was monitored in 96-well plates by fluorescence dequenching/quenching of pyrene (excitation 365 nm; emission 410 nm) upon initiation of actin polymerization/depolymerization in the absence or presence of arp2/3, actA, EVL, CD2AP, and vitamin D–binding protein with SpectraMax M2 (Molecular Devices).
Single filament imaging
Perfusion chambers coated with filamin and blocked with casein were used to capture actin filaments for imaging. For two-color imaging, rhodamine actin was flushed into the perfusion chamber, allowed to polymerize for 30 s, and flushed out with actin assembly buffer. Samples were then incubated with or without 1 µM CD2AP or C-CD2AP for 90 s and rinsed twice with actin assembly buffer, followed by flushing in rhodamine green–actin and polymerization for 30 s. Unpolymerized actin was flushed out, and a photo buffer (Brieher et al., 2006) was flushed into the chamber before imaging. For actin depolymerization imaging, rhodamine-actin was allowed to polymerize for 90 s in the perfusion chamber, rinsed twice in actin assembly buffer, incubated with or without 1 µM CD2AP, and rinsed twice with actin assembly buffer, followed by imaging in photo buffer. Images were collected at room temperature with a light microscope (Axio Imager) using AxioVision release 4.7 software with the Colibri illumination system using a 63× objective lens (NA 1.4) attached to a 1,000 × 1,000 charge-coupled device camera (ORCA-ER; Hamamatsu Photonics). For figure generation, images were cropped, contrasted, and scaled using Photoshop software before importing into Illustrator.
F-actin spin down
10 µM actin was polymerized for 2 h in the absence or presence of 5 µM CD2AP and 1 µM α-actinin-4. F-actin was centrifuged at 200,000 g for 30 min.
Cell culture and transfection
MDCK cells were maintained in MEM/Earle’s balanced salt solution supplemented with 25 mM Hepes and 10% FBS. For transfection, MDCK cells were incubated in Opti-MEM (Invitrogen) with a 1:1 mixture of DNA/polyethylenimine and selected for 10 d using puromycin. Total cell lysates were obtained by solubilization of cells directly in SDS-PAGE sample buffer containing 25 mM dithiothreitol, 2% SDS, 50 mM Tris-Cl, 5% glycerol, pH 8.8, and protease inhibitors (10 µg/ml Leupeptin, 1 mg/ml Pefabloc, 10 µg/ml E-64, 2 µg/ml antipain, 2 µg/ml aprotinin, 50 µg/ml bestatin, 20 µg/ml calpain inhibitors I, and 10 µg/ml calpain inhibitor II).
Immunofluorescence and quantitation of junctional staining
MDCK cells grown on Transwell-Clear inserts (Corning) for 10 d were rinsed twice in Earle’s Balanced Salt Solution and fixed in 1% formaldehyde/150 mM NaCl/20 mM Hepes, pH 7.8, at 4°C for 2 h. The reaction was quenched with 50 mM Tris in staining buffer (0.1% TX-100/100 mM NaCl/20 mM Hepes, pH 7.8) for 1 h. After rinsing in staining buffer, the cells were incubated with primary antibodies in staining buffer overnight. After rinsing in staining buffer three times, the cells were incubated in secondary antibodies for 90 min. The cells were rinsed again three times, and poststain fixed with 1% formaldehyde in staining buffer. Finally, the cells were incubated with fluorescently labeled phalloidin for 60 min. Transwell filters were excised and mounted on glass slides using ProLong Gold antifade reagent (Invitrogen).
Optical z slices in 200-nm steps were collected using a 60× objective (NA 1.42), with a 1.6× auxiliary magnification, with an inverted microscope (1X71; Olympus) attached to a 1,000 × 1,000 charge-coupled device camera (CoolSNAP HQ; Applied Precision). All images were obtained at room temperature using SoftWoRx DMS software (Applied Precision) and deconvolved by enhanced ratio deconvolution using 10 iteration cycles (Applied Precision). Z stack projections were generated from deconvolved slices using the maximum intensity criteria. Composite images were generated using ImageJ software (National Institutes of Health). Quantitation of immunofluorescence intensity was performed in ImageJ using unprocessed original single optical z slice images taken at the level of adherens junction. A defined junctional area was used to compare the fluorescence intensity of actin (phalloidin), E-cadherin (immunofluorescence using RR1 antibodies), and CD2AP (immunofluorescence using N-terminal antibodies). The measured intensities were subtracted from background (cytoplasm) before being used for calculating the intensity ratios. For figure generation, images were cropped, contrasted, and scaled using Photoshop software (Adobe) before importing into Illustrator (Adobe). Correlation coefficient R was calculated using KaleidaGraph software (Synergy Software).
Permeability measurement
FITC-dextrans 70 and 4 kD were purchased from Molecular Probes. BSA was labeled on lysine residues using N-hydroxysuccinimide–activated rhodamine (Invitrogen) and purified by gel filtration using Sephadex S-200. Extracellular tracers (rhodamine-BSA, FITC-dextran 70 and 4 kD) were kept frozen until used. Thawed tracers were purified away from residual free dye using Sephadex 25G spin columns immediately before use. Permeability assays were performed as described in detail previously (Tang and Goodenough, 2003). In brief, 12-mm Transwell supports were rinsed with 50 ml of prewarmed flux buffer (145 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, and 10 mM Hepes, pH 7.4). The apical well was filled with 0.5 ml of flux buffer and the Transwell supports were placed in a 12-well plate with 1 ml of flux media containing 3 µM of labeled extracellular tracers. Flux was performed at 37°C for 2 h. At the end of the flux experiment, the apical solutions were collected and fluorescence was measured using a fluorimeter. Permeability is the amount of tracer in the apical bathing solution (in micromoles) per unit driving force (basal molar concentration of tracer), per unit time (hours), per area of the monolayer (cubic centimeters).
Hydraulic pressure application to cell monolayers
Pressure chambers were fabricated from medical-grade stainless steel by the mechanical engineering machine shop at the University of Illinois at Urbana-Champaign. Transwell filter cups were mounted onto pressure chamber, held in place against the lid using a screw and a top. Hydraulic pressure was applied to the basal compartment by pumping buffer into an O-ring sealed fluid-filled basal chamber while the apical compartment is exposed to ambient pressure. Sustained pressure was applied to cell monolayers for 5 min and monitored with a low-pressure gauge attached to the basal compartment.
Cell motility assay
For wound migration rates, cells were plated at confluent density on glass coverslips. Wounds were made at 1, 2, 4, and 6 d after plating with a razor blade. Wound-induced cell migration was imaged in a sealed chamber containing normal media every 5 min for 12–18 h. Wound edge progression was measured at 20-µm intervals along the perpendicular axis of wound. Images were collected at room temperature with a light microscope (Axio Imager) using AxioVision release 4.7 with the Colibri illumination system using a 10× objective lens (NA 0.25) attached to a 1,000 × 1,000 charge-coupled device camera (ORCA-ER; Hamamatsu Photonics). For figure generation, images were cropped, contrasted, and scaled using Photoshop software (Adobe) before importing into Illustrator (Adobe). Wound edges were measured at 20 µm intervals along the perpendicular axis of the wound using ImageJ software (NIH).
Online supplemental material
Fig. S1 shows Western blots of junction-enriched liver membranes untreated or treated with 0.1% TX-100 or 1 M KCl, showing differential extraction of CD2AP, vinculin, radixin, and EVL. Fig. S2 shows that actin dynamics at the adherens junction involve complex barbed-end regulation. Fig. S3 shows Western blots of total MDCK lysate showing vinculin, α-catenin, and ZO-2 levels unchanged in CD2AP knockdown cells. Fig. S4 shows that MDCK cells express higher levels of CD2AP than CapZ. Fig. S5 shows that exogenous streptavidin-binding protein (SBP)-tagged full-length CD2AP is localized with total CD2AP at the junction. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201304143/DC1.
Supplementary Material
Acknowledgments
We thank Stephanie Sams and Sarvanand Patel for assistance with cell culture and protein purification. We thank Ambika Nadkarni and Hui-Chia Yu for the purification of EVL and CapZ proteins. The pressure chamber was built by the mechanical engineering machine shop at the University of Illinois, Urbana-Champaign. Deconvolution microscopy was performed at the Microscopy Facility in the School of Molecular and Cellular Biology at the University of Illinois, Urbana-Champaign. Mass Spectrometry and spectroscopy were performed at the Roy J. Carver Biotechnology Center at the University of Illinois, Urbana-Champaign. Electron microscopy was carried out at the University of Illinois Frederick Seitz Materials Research Laboratory central facilities.
This work is funded by the the National Institutes of Health (R01-DK098398-01 to V.W. Tang and R01-GM106106-01 to W.M. Brieher) and a March of Dimes Basil O’Connor award (5FY11).
Footnotes
Abbreviation used in this paper:
- FSGS
- focal segmental glomerulosclerosis
- TX-100
- Triton X-100
References
- Aebi U., Fowler W.E., Isenberg G., Pollard T.D., Smith P.R. 1981. Crystalline actin sheets: their structure and polymorphism. J. Cell Biol. 91:340–351 10.1083/jcb.91.2.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B., Georgiou M. 2011. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J. Cell Biol. 192:907–917 10.1083/jcb.201009141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B., Perrimon N. 2001. Spatial control of the actin cytoskeleton in Drosophila epithelial cells. Nat. Cell Biol. 3:883–890 10.1038/ncb1001-883 [DOI] [PubMed] [Google Scholar]
- Bennett J.P., Zaner K.S., Stossel T.P. 1984. Isolation and some properties of macrophage alpha-actinin: evidence that it is not an actin gelling protein. Biochemistry. 23:5081–5086 10.1021/bi00316a039 [DOI] [PubMed] [Google Scholar]
- Bernadskaya Y.Y., Patel F.B., Hsu H.T., Soto M.C. 2011. Arp2/3 promotes junction formation and maintenance in the Caenorhabditis elegans intestine by regulating membrane association of apical proteins. Mol. Biol. Cell. 22:2886–2899 10.1091/mbc.E10-10-0862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieher W.M., Kueh H.Y., Ballif B.A., Mitchison T.J. 2006. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J. Cell Biol. 175:315–324 10.1083/jcb.200603149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck S., Huber T.B., Ingham R.J., Kim K., Niederstrasser H., Allen P.M., Pawson T., Cooper J.A., Shaw A.S. 2006. Identification of a novel inhibitory actin-capping protein binding motif in CD2-associated protein. J. Biol. Chem. 281:19196–19203 10.1074/jbc.M600166200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey M., Lecuit T. 2009. Molecular bases of cell-cell junctions stability and dynamics. Cold Spring Harb. Perspect. Biol. 1:a002998 10.1101/cshperspect.a002998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey M., Rauzi M., Lenne P.F., Lecuit T. 2008. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature. 453:751–756 10.1038/nature06953 [DOI] [PubMed] [Google Scholar]
- Chen Z., Borek D., Padrick S.B., Gomez T.S., Metlagel Z., Ismail A.M., Umetani J., Billadeau D.D., Otwinowski Z., Rosen M.K. 2010. Structure and control of the actin regulatory WAVE complex. Nature. 468:533–538 10.1038/nature09623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y.S., Thomas W.A., Eder O., Pincet F., Perez E., Thiery J.P., Dufour S. 2004. Force measurements in E-cadherin-mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. J. Cell Biol. 167:1183–1194 10.1083/jcb.200403043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D., Pan H., Wan P., Wu J., Luo J., Zhu H., Chen J. 2012. AIP1 acts with cofilin to control actin dynamics during epithelial morphogenesis. Development. 139:3561–3571 10.1242/dev.079491 [DOI] [PubMed] [Google Scholar]
- Cooper J.A., Sept D. 2008. New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell Mol. Biol. 267:183–206 10.1016/S1937-6448(08)00604-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoë-Ayari H., Al Kurdi R., Vallade M., Gulino-Debrac D., Riveline D. 2004. Membrane and acto-myosin tension promote clustering of adhesion proteins. Proc. Natl. Acad. Sci. USA. 101:2229–2234 10.1073/pnas.0304297101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. 2009. Actin filament nucleation and elongation factors—structure-function relationships. Crit. Rev. Biochem. Mol. Biol. 44:351–366 10.3109/10409230903277340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin M.L., Olszowy M.W., Holdorf A.D., Li J., Bromley S., Desai N., Widder P., Rosenberger F., van der Merwe P.A., Allen P.M., Shaw A.S. 1998. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell. 94:667–677 10.1016/S0092-8674(00)81608-6 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. 2012. Adherens junctions during cell migration. Subcell. Biochem. 60:225–249 10.1007/978-94-007-4186-7_10 [DOI] [PubMed] [Google Scholar]
- Furman C., Sieminski A.L., Kwiatkowski A.V., Rubinson D.A., Vasile E., Bronson R.T., Fässler R., Gertler F.B. 2007. Ena/VASP is required for endothelial barrier function in vivo. J. Cell Biol. 179:761–775 10.1083/jcb.200705002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Hirase T., Itoh M., Nagafuchi A., Yonemura S., Tsukita S., Tsukita S. 1993. Occludin: a novel integral membrane protein localizing at tight junctions. J. Cell Biol. 123:1777–1788 10.1083/jcb.123.6.1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidos G., Soni S., Oswald D.J., Toselli P.A., Kirsch K.H. 2007. Structure and function analysis of the CMS/CIN85 protein family identifies actin-bundling properties and heterotypic-complex formation. J. Cell Sci. 120:2366–2377 10.1242/jcs.004333 [DOI] [PubMed] [Google Scholar]
- Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D.S., Condeelis J.S. 2004. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 304:743–746 10.1126/science.1094561 [DOI] [PubMed] [Google Scholar]
- Gomez G.A., McLachlan R.W., Yap A.S. 2011. Productive tension: force-sensing and homeostasis of cell-cell junctions. Trends Cell Biol. 21:499–505 10.1016/j.tcb.2011.05.006 [DOI] [PubMed] [Google Scholar]
- Hernandez-Valladares M., Kim T., Kannan B., Tung A., Aguda A.H., Larsson M., Cooper J.A., Robinson R.C. 2010. Structural characterization of a capping protein interaction motif defines a family of actin filament regulators. Nat. Struct. Mol. Biol. 17:497–503 10.1038/nsmb.1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber T.B., Kwoh C., Wu H., Asanuma K., Gödel M., Hartleben B., Blumer K.J., Miner J.H., Mundel P., Shaw A.S. 2006. Bigenic mouse models of focal segmental glomerulosclerosis involving pairwise interaction of CD2AP, Fyn, and synaptopodin. J. Clin. Invest. 116:1337–1345 10.1172/JCI27400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S., Olkkonen V., Martin-Parras L., Chavrier P., Stapleton M., Zerial M., Lehtonen E. 1997. Mouse metanephric kidney as a model system for identifying developmentally regulated genes. J. Cell. Physiol. 173:147–151 [DOI] [PubMed] [Google Scholar]
- Johnson R.I., Seppa M.J., Cagan R.L. 2008. The Drosophila CD2AP/CIN85 orthologue Cindr regulates junctions and cytoskeleton dynamics during tissue patterning. J. Cell Biol. 180:1191–1204 10.1083/jcb.200706108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.I., Sedgwick A., D’Souza-Schorey C., Cagan R.L. 2011. Role for a Cindr-Arf6 axis in patterning emerging epithelia. Mol. Biol. Cell. 22:4513–4526 10.1091/mbc.E11-04-0305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametani Y., Takeichi M. 2007. Basal-to-apical cadherin flow at cell junctions. Nat. Cell Biol. 9:92–98 10.1038/ncb1520 [DOI] [PubMed] [Google Scholar]
- Kim J.M., Wu H., Green G., Winkler C.A., Kopp J.B., Miner J.H., Unanue E.R., Shaw A.S. 2003. CD2-associated protein haploinsufficiency is linked to glomerular disease susceptibility. Science. 300:1298–1300 10.1126/science.1081068 [DOI] [PubMed] [Google Scholar]
- Kinoshita Y., Knox F.G. 1989. Role of prostaglandins in proximal tubule sodium reabsorption: response to elevated renal interstitial hydrostatic pressure. Circ. Res. 64:1013–1018 10.1161/01.RES.64.5.1013 [DOI] [PubMed] [Google Scholar]
- Kovacs E.M., Goodwin M., Ali R.G., Paterson A.D., Yap A.S. 2002. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr. Biol. 12:379–382 10.1016/S0960-9822(02)00661-9 [DOI] [PubMed] [Google Scholar]
- Kovacs E.M., Verma S., Ali R.G., Ratheesh A., Hamilton N.A., Akhmanova A., Yap A.S. 2011. N-WASP regulates the epithelial junctional actin cytoskeleton through a non-canonical post-nucleation pathway. Nat. Cell Biol. 13:934–943 10.1038/ncb2290 [DOI] [PubMed] [Google Scholar]
- Lambert M., Choquet D., Mège R.M. 2002. Dynamics of ligand-induced, Rac1-dependent anchoring of cadherins to the actin cytoskeleton. J. Cell Biol. 157:469–479 10.1083/jcb.200107104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckband D.E., le Duc Q., Wang N., de Rooij J. 2011. Mechanotransduction at cadherin-mediated adhesions. Curr. Opin. Cell Biol. 23:523–530 10.1016/j.ceb.2011.08.003 [DOI] [PubMed] [Google Scholar]
- Lehtonen S., Ora A., Olkkonen V.M., Geng L., Zerial M., Somlo S., Lehtonen E. 2000. In vivo interaction of the adapter protein CD2-associated protein with the type 2 polycystic kidney disease protein, polycystin-2. J. Biol. Chem. 275:32888–32893 10.1074/jbc.M006624200 [DOI] [PubMed] [Google Scholar]
- Löwik M.M., Groenen P.J., Pronk I., Lilien M.R., Goldschmeding R., Dijkman H.B., Levtchenko E.N., Monnens L.A., van den Heuvel L.P. 2007. Focal segmental glomerulosclerosis in a patient homozygous for a CD2AP mutation. Kidney Int. 72:1198–1203 10.1038/sj.ki.5002469 [DOI] [PubMed] [Google Scholar]
- Lynch D.K., Winata S.C., Lyons R.J., Hughes W.E., Lehrbach G.M., Wasinger V., Corthals G., Cordwell S., Daly R.J. 2003. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J. Biol. Chem. 278:21805–21813 10.1074/jbc.M211407200 [DOI] [PubMed] [Google Scholar]
- Maciver S.K., Pope B.J., Whytock S., Weeds A.G. 1998. The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur. J. Biochem. 256:388–397 10.1046/j.1432-1327.1998.2560388.x [DOI] [PubMed] [Google Scholar]
- Maun N.A., Speicher D.W., DiNubile M.J., Southwick F.S. 1996. Purification and properties of a Ca(2+)-independent barbed-end actin filament capping protein, CapZ, from human polymorphonuclear leukocytes. Biochemistry. 35:3518–3524 10.1021/bi952470p [DOI] [PubMed] [Google Scholar]
- Mayor R., Carmona-Fontaine C. 2010. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 20:319–328 10.1016/j.tcb.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita H., Nandadasa S., Yamamoto T.S., Terasaka-Iioka C., Wylie C., Ueno N. 2010. Nectin-2 and N-cadherin interact through extracellular domains and induce apical accumulation of F-actin in apical constriction of Xenopus neural tube morphogenesis. Development. 137:1315–1325 10.1242/dev.043190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandadasa S., Tao Q., Menon N.R., Heasman J., Wylie C. 2009. N- and E-cadherins in Xenopus are specifically required in the neural and non-neural ectoderm, respectively, for F-actin assembly and morphogenetic movements. Development. 136:1327–1338 10.1242/dev.031203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Takeichi M. 2009. Remodeling of the adherens junctions during morphogenesis. Curr. Top. Dev. Biol. 89:33–54 10.1016/S0070-2153(09)89002-9 [DOI] [PubMed] [Google Scholar]
- Sato T., Fujita N., Yamada A., Ooshio T., Okamoto R., Irie K., Takai Y. 2006. Regulation of the assembly and adhesion activity of E-cadherin by nectin and afadin for the formation of adherens junctions in Madin-Darby canine kidney cells. J. Biol. Chem. 281:5288–5299 10.1074/jbc.M510070200 [DOI] [PubMed] [Google Scholar]
- Scott J.A., Shewan A.M., den Elzen N.R., Loureiro J.J., Gertler F.B., Yap A.S. 2006. Ena/VASP proteins can regulate distinct modes of actin organization at cadherin-adhesive contacts. Mol. Biol. Cell. 17:1085–1095 10.1091/mbc.E05-07-0644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih N.Y., Li J., Karpitskii V., Nguyen A., Dustin M.L., Kanagawa O., Miner J.H., Shaw A.S. 1999. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science. 286:312–315 10.1126/science.286.5438.312 [DOI] [PubMed] [Google Scholar]
- Shimono Y., Rikitake Y., Mandai K., Mori M., Takai Y. 2012. Immunoglobulin superfamily receptors and adherens junctions. Subcell. Biochem. 60:137–170 10.1007/978-94-007-4186-7_7 [DOI] [PubMed] [Google Scholar]
- Shiotsu T., Yamamoto A., Kagawa S., Tamaki T., Abe Y., Reid I.A. 1995. Effect of moderately increased intrapelvic pressure on renal tissue pressure and vasopressin release in rabbits. Hypertens. Res. 18:197–202 10.1291/hypres.18.197 [DOI] [PubMed] [Google Scholar]
- Smutny M., Cox H.L., Leerberg J.M., Kovacs E.M., Conti M.A., Ferguson C., Hamilton N.A., Parton R.G., Adelstein R.S., Yap A.S. 2010. Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat. Cell Biol. 12:696–702 10.1038/ncb2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y., Miyoshi J., Ikeda W., Ogita H. 2008. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell Biol. 9:603–615 10.1038/nrm2457 [DOI] [PubMed] [Google Scholar]
- Takeichi M. 2011. Self-organization of animal tissues: cadherin-mediated processes. Dev. Cell. 21:24–26 10.1016/j.devcel.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Tang V.W., Brieher W.M. 2012. α-Actinin-4/FSGS1 is required for Arp2/3-dependent actin assembly at the adherens junction. J. Cell Biol. 196:115–130 10.1083/jcb.201103116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang V.W., Goodenough D.A. 2003. Paracellular ion channel at the tight junction. Biophys. J. 84:1660–1673 10.1016/S0006-3495(03)74975-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S., Tsukita S. 1989. Isolation of cell-to-cell adherens junctions from rat liver. J. Cell Biol. 108:31–41 10.1083/jcb.108.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunggal J.A., Helfrich I., Schmitz A., Schwarz H., Günzel D., Fromm M., Kemler R., Krieg T., Niessen C.M. 2005. E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions. EMBO J. 24:1146–1156 10.1038/sj.emboj.7600605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijn T.J., Anthony E.C., Hensbergen P.J., Deelder A.M., Hordijk P.L. 2010. Rac1 recruits the adapter protein CMS/CD2AP to cell-cell contacts. J. Biol. Chem. 285:20137–20146 10.1074/jbc.M109.099481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Yin M., Fuchs E. 2000. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 100:209–219 10.1016/S0092-8674(00)81559-7 [DOI] [PubMed] [Google Scholar]
- Verma S., Shewan A.M., Scott J.A., Helwani F.M., den Elzen N.R., Miki H., Takenawa T., Yap A.S. 2004. Arp2/3 activity is necessary for efficient formation of E-cadherin adhesive contacts. J. Biol. Chem. 279:34062–34070 10.1074/jbc.M404814200 [DOI] [PubMed] [Google Scholar]
- Verma S., Han S.P., Michael M., Gomez G.A., Yang Z., Teasdale R.D., Ratheesh A., Kovacs E.M., Ali R.G., Yap A.S. 2012. A WAVE2-Arp2/3 actin nucleator apparatus supports junctional tension at the epithelial zonula adherens. Mol. Biol. Cell. 23:4601–4610 10.1091/mbc.E12-08-0574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Eddy R., Condeelis J. 2007. The cofilin pathway in breast cancer invasion and metastasis. Nat. Rev. Cancer. 7:429–440 10.1038/nrc2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong V., Gumbiner B.M. 1997. A synthetic peptide corresponding to the extracellular domain of occludin perturbs the tight junction permeability barrier. J. Cell Biol. 136:399–409 10.1083/jcb.136.2.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaddanapudi S., Altintas M.M., Kistler A.D., Fernandez I., Möller C.C., Wei C., Peev V., Flesche J.B., Forst A.L., Li J., et al. 2011. CD2AP in mouse and human podocytes controls a proteolytic program that regulates cytoskeletal structure and cellular survival. J. Clin. Invest. 121:3965–3980 10.1172/JCI58552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S., Gokhin D.S., Kimura S., Nowak R.B., Fowler V.M. 2012. Tropomodulins: pointed-end capping proteins that regulate actin filament architecture in diverse cell types. Cytoskeleton (Hoboken). 69:337–370 10.1002/cm.21031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Wada Y., Watanabe T., Nagafuchi A., Shibata M. 2010. alpha-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12:533–542 10.1038/ncb2055 [DOI] [PubMed] [Google Scholar]
- Zhang J., Betson M., Erasmus J., Zeikos K., Bailly M., Cramer L.P., Braga V.M. 2005. Actin at cell-cell junctions is composed of two dynamic and functional populations. J. Cell Sci. 118:5549–5562 10.1242/jcs.02639 [DOI] [PubMed] [Google Scholar]
- Zhao J., Bruck S., Cemerski S., Zhang L., Butler B., Dani A., Cooper J.A., Shaw A.S. 2013. CD2AP links cortactin and capping protein at the cell periphery to facilitate formation of lamellipodia. Mol. Cell. Biol. 33:38–47 10.1128/MCB.00734-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.