A subset of phagosomes in human macrophages and dendritic cells that is marked by a coat of autophagy-related proteins maintains phagocytosed antigens for prolonged presentation on MHC class II molecules.
Abstract
Antigen preservation for presentation is a hallmark of potent antigen-presenting cells. In this paper, we report that in human macrophages and dendritic cells, a subset of phagosomes gets coated with Atg8/LC3, a component of the molecular machinery of macroautophagy, and maintains phagocytosed antigens for prolonged presentation on major histocompatibility complex class II molecules. These Atg8/LC3-positive phagosomes are formed around the antigen with TLR2 agonists and require reactive oxygen species production by NOX2 for their generation. A deficiency in the NOX2-dependent formation of these antigen storage phagosomes could contribute to compromise antifungal immune control in chronic granulomatous disease patients.
Introduction
Macroautophagy is an evolutionarily conserved pathway, involving a molecular machinery of autophagy-related proteins (Atgs), that generates a double membrane–surrounded vesicle called the autophagosome. These organelles engulf cytosolic substrates to carry them to lysosomal hydrolysis (Mizushima et al., 2011). In addition to its nutrient-recycling functions during starvation, higher eukaryotes can use this pathway to restrict intracellular pathogens and present their fragments on major histocompatibility complex (MHC) class II molecules to CD4+ T cells (Münz, 2009; Levine et al., 2011).
Among the molecular machinery for macroautophagy, a core of eight Atgs (Atg3, 4, 5, 7, 8, 10, 12, and 16L1) is involved in the conjugation of Atg8/LC3 to the autophagosomal membrane, and Atg8/LC3 has been implicated in membrane elongation as well as in substrate recruitment during autophagosome formation (Weidberg et al., 2011; Randow and Münz, 2012). After autophagosome completion, the macroautophagy core machinery is removed from the outer autophagosome membrane, whereas Atg8/LC3 stays conjugated to the inner autophagosome membrane. Thus, Atg8/LC3 has been used to experimentally monitor the faith of these vesicles.
In addition to the classical function of the macroautophagy core machinery in host defense, namely delivering intracellular pathogens and antigens toward lysosomal degradation, Atgs have lately been reported to regulate phagocytosis (Sanjuan et al., 2007; Florey et al., 2011; Martinez et al., 2011). In these studies, antigens with agonists of toll-like receptors as well as live (entosis) or apoptotic cell debris were found to recruit Atg8/LC3 to phagosomes, resulting in more rapid degradation in lysosomes. More recently, Dectin-1 signaling was also reported to trigger Atg8/LC3 recruitment to phagosomes (Ma et al., 2012). In addition, this study reported that fungal antigen presentation on MHC class II molecules was compromised in the absence of LC3-associated phagocytosis (LAP). In human antigen-presenting cells (APCs), it is not yet known which receptor agonists induce LAP efficiently or which kinetics and which functional potential the Atg8/LC3-coated phagosomes have. Therefore, we set out to investigate these LAP phagosomes in human macrophages and dendritic cells (DCs).
Results and discussion
Atg8/LC3 binds to the phagosomal membrane after pattern recognition receptor (PRR) signaling in a NOX2-dependent manner
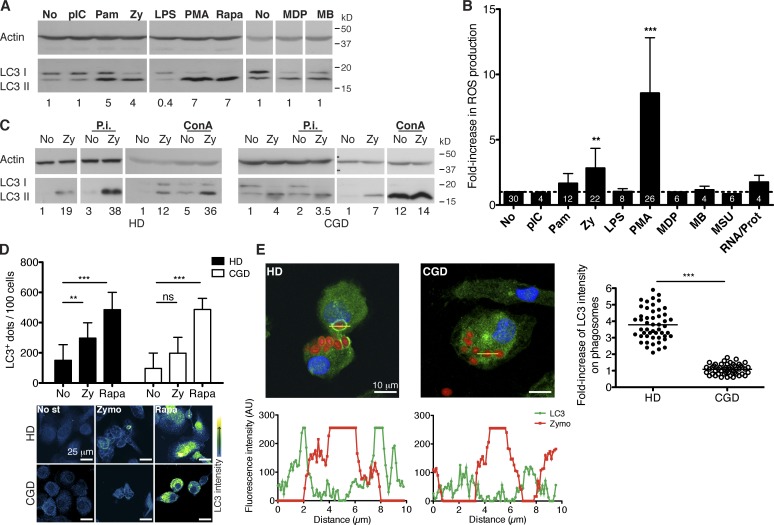
To induce Atg8/LC3-positive phagosomes in human primary monocyte-derived macrophages, we screened agonists of pathogen-associated molecular pattern (PAMP) receptors for their ability to induce macroautophagy. Even though macrophages responded with proinflammatory cytokines to all tested stimuli (Fig. S1 A), only the TLR2 (toll-like receptor 2) agonists Pam3CSK4 and zymosan significantly induced macroautophagy, as determined by the accumulation of the membrane-coupled Atg8/LC3 form (LC3-II) in whole-protein extracts (Fig. 1 A). Furthermore, the two investigated TLR2 stimuli induced reactive oxygen species (ROS) production significantly better than the other PAMP receptor agonists (Fig. 1 B). When this ROS production was abrogated by the use of NADPH oxidase inhibitors, LC3-II levels significantly decreased (Fig. S1, B and C), demonstrating a relation between pathogen recognition, ROS triggering, and up-regulation of macroautophagy, as previously reported for mouse macrophages (Huang et al., 2009). However, ROS production by NOX2 does not seem to be absolutely required for TLR2-mediated macroautophagy induction. Macrophages from chronic granulomatous disease (CGD) patients that have mutations in the NOX2 gene and thereby abrogated ROS production can still up-regulate macroautophagy upon zymosan treatment (Fig. 1, C and D). Additionally, accumulation of LC3-II in both healthy donors (HDs) and CGD macrophages was further enhanced upon lysosomal inhibition, suggesting intact autophagosome degradation. We also verified by microscopy that CGD cells are able to form autophagosomes, similar to HD cells after zymosan and rapamycin treatment (Fig. 1 D). However, CGD cells cannot recruit Atg8/LC3 to the membranes of zymosan-containing phagosomes, whereas HD cells present this marker in ∼5–10% of their phagosomes (Fig. 1 E). This Atg8/LC3 binding to phagosomes occurred also after stimulation with additional TLR2 agonists, including heat-killed Candida albicans or Pam3CSK4-coated latex beads but not when uncoated beads were phagocytosed by the cells (Fig. S1 D). To further confirm the correlation between TLR2 stimulation and Atg8/LC3 recruitment to phagosomal membranes, phagosomes isolated from HD macrophages and analyzed by flow cytometry (phago-FACS) also showed an increase in Atg8/LC3 signal (Fig. S1 G). In addition, other human phagocytes, such as human monocyte-derived DCs, formed Atg8/LC3-positive phagosomes in response to zymosan particles (Fig. S1, E and F).
Figure 1.
Atg8/LC3 binding to the phagosomal membrane depends on PRR signaling and on NOX2 activity. (A) Human macrophages were treated with the indicated stimuli for 8 h, and LC3-II levels were assessed by WB. Numbers below the blot represent the fold increase of LC3-II compared with the nonstimulated control cells after normalization to the corresponding actin levels. (B) ROS production was determined after 3 h of PRR stimulation (for PMA stimulation after 30 min), by the increase in fluorescence of the probe Amplex UltraRed. Data are expressed as the means ± SD; paired t test; **, P < 0.01; ***, P < 0.001. Numbers inside the bars correspond to the n values. No increase is indicated as a dashed line. (C) LC3-II protein levels increase both in the presence and in the absence of lysosomal inhibitors (P.i., protease inhibitors; ConA, concanamycin A) for the indicated donors. (D) Quantification of Atg8/LC3+ dots on cells of nine HD and nine CGD patients, treated with the indicated stimuli for 6 h. (bottom) Representative confocal images showing Atg8/LC3 signal intensity. Data are expressed as the means ± SD; **, P < 0.01; ***, P < 0.001. No st, no stimulus; Zymo, zymosan. (E) Macrophages were treated with red-labeled zymosan and stained with Atg8/LC3 antibody and DNA content (DAPI). Profile plots of the fluorescence intensity across the indicated trajectories are shown below the pictures. The graph on the right represents the fold increase of Atg8/LC3 intensity on phagosomes (n = 50) compared with the cytosol signal in HD and CGD cells; mean values are represented as horizontal lines. Mann–Whitney test; ***, P < 0.001. AU, arbitrary unit; pIC, polyinosinic/polycytidylic acid; Pam, Pam3CSK4; Zy, zymosan; LPS, lipopolysaccharide; Rapa, rapamycin; MDP, muramyl dipeptide; MB, murabutide; MSU, monosodium urate crystals; RNA/Prot, RNA/protamine.
Atg8/LC3-positive phagosomes do not contain other classical autophagy markers
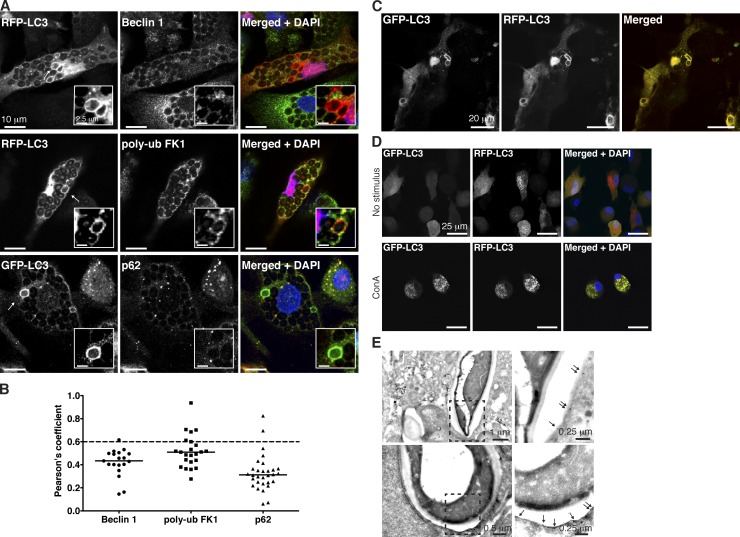
Next, we investigated whether Atg8/LC3 recruitment resulted from autophagosome fusion with phagosomes. Classical markers for macroautophagy, such as Atg6/Beclin-1, p62/SQSTM1, or polyubiquitinated substrates did not colocalize with the Atg8/LC3-positive phagosomes (Fig. 2, A and B). These findings suggest that Atg8/LC3-positive phagosomes do not result from autophagosome fusion with phagosomes but rather from a direct Atg8/LC3 lipidation of phagosomes. To further address this topic, GFP-Atg8/LC3–decorated phagosomes were examined by immunoelectron microscopy. Anti-GFP gold labeling was found on single membranes surrounding heat-killed C. albicans (Fig. 2 E). This indicates direct Atg8/LC3 binding to the phagosomal membrane. For additional characterization of this process, we investigated whether Atg8/LC3 was anchored to the membrane from the luminal or cytosolic side. For this purpose, we used tandem fluorescent-tagged Atg8/LC3 (Kimura et al., 2007), a lentiviral construct encoding both mRFP and GFP fused to Atg8/LC3’s N terminus, whereby GFP fluorescence is lost in the transition from neutral to acidic pH (Fig. 2 D). Because the phagosomal lumen acidifies with maturation, mRFP-GFP-Atg8/LC3 would lose its GFP fluorescence on the luminal side but maintain it at the cytosolic side during acidification. As shown in Fig. 2 C and Video 1, Atg8/LC3-positive phagosomes preserve an intense yellow signal at all times.
Figure 2.
Classical autophagy markers are absent from PRR-induced Atg8/LC3-positive phagosomes. (A) Macrophages transduced with lentiviral constructs of fluorescent-tagged Atg8/LC3 were stained for the indicated proteins and DAPI. Arrows point to Atg8/LC3-positive phagosomes. Insets show a zoomed-in view of the phagosomes of interest. (B) Pearson’s coefficient values were calculated for each marker in Atg8/LC3-positive phagosomes (n ≥ 20). Mean values are represented as horizontal lines. A Pearson score >0.6 is indicated in each graph as a dashed line. poly-ub, polyubiquitin. (C) Zymosan-containing phagosomes in macrophages transduced with the tandem fluorescent-tagged Atg8/LC3 construct and treated with zymosan for 6 h. (D) Macrophages transduced with tandem fluorescent Atg8/LC3 and treated or not treated with concanamycin A (ConA). (E) Representative GFP immunogold labeling in electron micrographs of phagosomes harboring heat-killed C. albicans in GFP-Atg8/LC3–transduced macrophages. The areas highlighted by dashed line boxes are shown with higher magnification on the right. Arrows point to the single membrane structure labeled with gold particles.
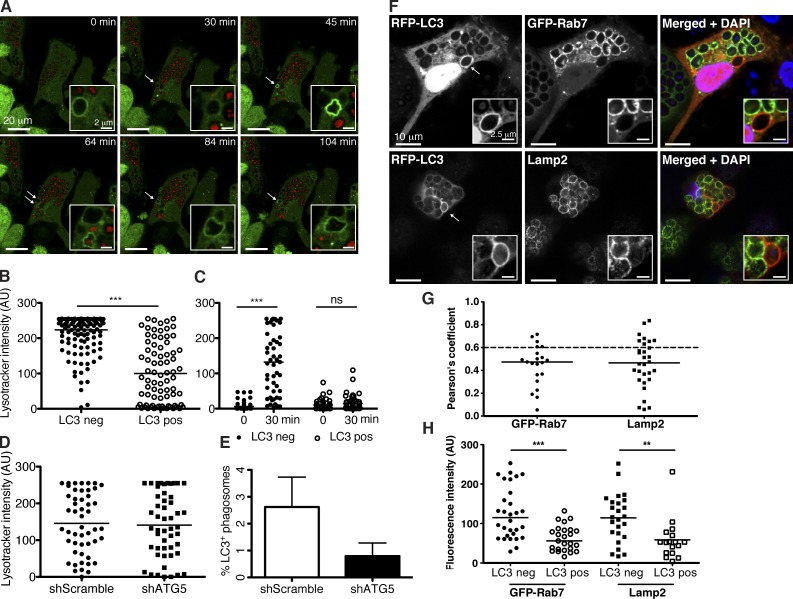
To further analyze fusion of Atg8/LC3-positive phagosomes with lysosomes, we followed these vesicles by live-cell imaging of macrophages. Lysosomes of these cells were labeled with the LysoTracker fluorescent dye. Of note, Atg8/LC3-positive phagosomes were found to be less often positive for this lysosome marker (Fig. 3, A–C; and Video 3). Nevertheless, LysoTracker and Atg8/LC3 double-positive phagosomes maintained their GFP fluorescence (Fig. 3 B). Together, these data strongly argue that in human macrophages, a subset of TLR2 agonist-carrying phagosomes are labeled with Atg8/LC3 on the cytosolic side of their single membrane and do not contain autophagosome substrates.
Figure 3.
Phagosomes harboring Atg8/LC3 have delayed maturation. (A) GFP-Atg8/LC3–transduced macrophages were preloaded with red LysoTracker dye for 1 h and incubated with zymosan. Atg8/LC3 recruitment to the phagosomes (white arrows) was followed at 5-min intervals (see Video 3) and LysoTracker signal intensity registered. Insets show a zoomed-in view of the phagosomes of interest. Representative time-lapse images are shown. (B) Lysosomal dye intensity was compared in phagosomes with (LC3 positive [pos]) and without (LC3 negative [neg]) GFP-Atg8/LC3 signal (n ≥ 80); Mann–Whitney test; ***, P < 0.001. (C) Recruitment of the lysosomal dye was followed over time, and signal intensity was registered immediately (0 min) or after 30 min of zymosan uptake within the same phagosome (n = 50); paired t test; ***, P < 0.001. (D) LysoTracker signal intensity was compared in macrophage phagosomes transduced with scramble shRNA versus atg5 shRNA lentiviruses. Graph shows phagosomes (n = 50) analyzed after 2 h of zymosan uptake. (E) Atg8/LC3 recruitment to phagosomes was compared in macrophages transduced with scramble shRNA versus atg5 shRNA lentivirus. Percentage of Atg8/LC3-positive phagosomes is calculated from a total of 50 phagosomes counted in each condition after 5 h of zymosan engulfment. Data are pooled from three different donors. Error bars show SDs. (F) Macrophages transduced with RFP-Atg8/LC3 lentiviral construct were stained for Lamp2 and DAPI or double transduced with the GFP-Rab7 construct. Arrows point to Atg8/LC3-positive phagosomes. Insets show a zoomed-in view of the phagosomes of interest. (G) Pearson’s coefficient values were calculated for each marker in Atg8/LC3-positive phagosomes (n ≥ 20). A Pearson score >0.6 is indicated in each graph as a dashed line. (H) Indicated marker signal intensity was compared in Atg8/LC3-positive versus negative phagosomes (n ≥ 25). Mean values are represented as horizontal lines. **, P < 0.01; ***, P < 0.001. AU, arbitrary unit.
Phagosomes decorated with Atg8/LC3 show delayed maturation
The low frequency of LysoTracker-labeled Atg8/LC3-positive phagosomes led us to compare the kinetics of lysosome fusion of Atg8/LC3-positive phagosomes versus Atg8/LC3-negative phagosomes. We imaged live GFP-Atg8/LC3–transduced cells and found that the majority of Atg8/LC3-positive phagosomes were long lived and could be observed for >4 h (Video 2). In addition, late endosome and lysosome markers, such as Rab7 and Lamp2, were tested for their colocalization with zymosan-containing phagosomes (Fig. 3, F–H). The majority of Atg8/LC3-positive phagosomes were negative for Rab7 and Lamp2, further confirming the delay of maturation for Atg8/LC3-positive phagosomes. This could also be demonstrated by live-cell imaging using LysoTracker-loaded cells (Fig. 3 C). Atg8/LC3-negative vesicles acquired the lysosomal dye much faster than Atg8/LC3-positive vesicles. In addition, we used GFP–C. albicans to infect RFP-Atg8/LC3–transduced cells. In this experimental setup, the maintenance of GFP fluorescence within the phagosomes can be used to monitor phagolysosomal fusion. The longer GFP fluorescence is preserved, the less acidification occurs. Our results show that in Atg8/LC3-positive vesicles, GFP fluorescence is not affected during 5 h, whereas Atg8/LC3-negative vesicles decrease cargo GFP fluorescence intensity considerably in the same time period (Fig. S2, B and C). Moreover, acidification of Atg8/LC3-positive phagosomes occurred primarily after uncoupling of Atg8/LC3 from the phagosome membrane (Fig. S2 A and Video 4). We, therefore, suggest that Atg8/LC3-positive phagosomes are long-lived endosomal compartments, which mostly fuse with lysosomes after Atg8/LC3 is cleaved off the phagosomal membrane.
Atg8/LC3-positive phagosomes enable prolonged antigen presentation by MHC class II molecules
To analyze whether the endocytosed cargo preservation could result in prolonged antigen processing and thereby sustained MHC class II presentation, we isolated C. albicans–specific CD4+ T cells with an IL-17 secretion assay (Fig. S3, A and B). Further characterization of these T cell clones showed that the restriction element was human leukocyte antigen (HLA)–DR4 (DRB1*0401; Fig. S3 C).
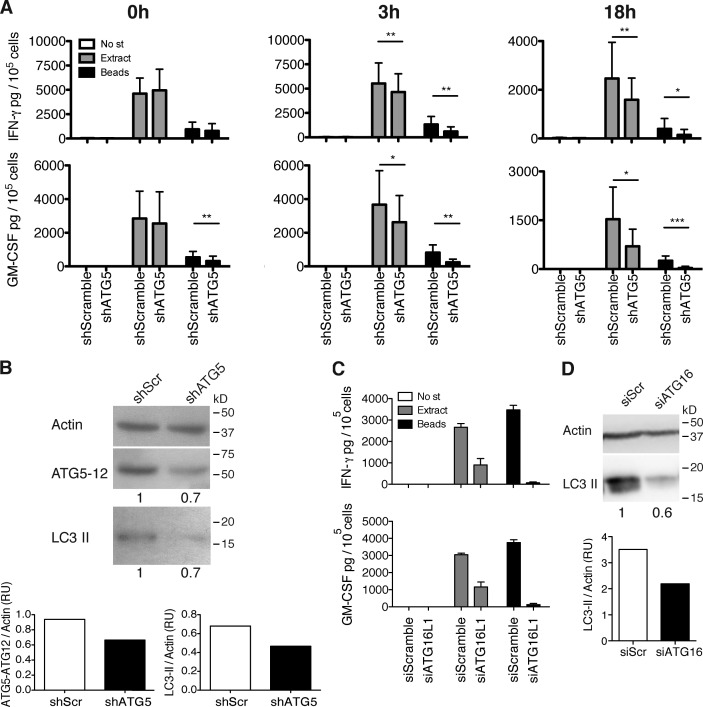
To evaluate whether C. albicans–specific CD4+ T cell recognition of antigen-pulsed macrophages would differ upon macroautophagy inhibition, HLA-DR4–matched donor cells were used as targets after transduction with lentiviral anti-atg5 shRNA. The lentiviral shRNA resulted in a decrease of ∼30–60% of both Atg5–Atg12 complexes and LC3-II levels, when compared with control transduced cells (Fig. 4 B, shScramble). Importantly, RNA silencing of atg5 in lentivirally transduced human macrophages had no effect on their survival (Fig. S3, F and G), cell activation (Fig. S3 H), or overall phagosome maturation (Fig. 3 D) but significantly reduced the percentage of cells with Atg8/LC3-positive phagosomes (Fig. 3 E).
Figure 4.
Components of the molecular machinery of macroautophagy can stabilize pathogen-containing phagosomes for prolonged antigen processing onto MHC class II molecules. (A) IFN-γ and GM-CSF ELISAs on supernatants of the C. albicans–specific T cell clone, co-cultured with cognate macrophages transduced with lentiviral atg5-specific or control shRNA, and pulsed or not pulsed with C. albicans extract or C. albicans–coated beads. Time indicated above each graph corresponds to the time point at which the T cells were added for co-culture, after the initial 4 h of pulsing. Bars represent means ± SD of data pooled from three independent experiments; paired t test; *, P < 0.5; **, P < 0.01; ***, P < 0.001. No st, no stimulus. (B) atg5 silencing levels were controlled by WB for LC3-II and Atg5–Atg12 complexes on day 3 after transduction and quantified by densitometry. Numbers below the blot represent the fold increase of LC3-II or Atg5–Atg12 complexes compared with the control cells after normalization to the corresponding actin levels. The results are representative of three assays, corresponding to the experiments shown in A. shScr, scrambled shRNA. (C) Same as in A using macrophages transfected with atg16L1 siRNA or control siRNA. After 4 h of pulsing, cells were washed and incubated in new medium for an additional 18 h before the T cells were added. Error bars show SDs. (D) atg16L1 silencing levels were controlled by WB for LC3-II on day 2 after transduction and quantified by densitometry. Numbers below the blot represent the fold increase of LC3-II compared with the control cells after normalization to the corresponding actin levels. One representative blot out of two performed is shown. RU, relative units; SiScr, scrambled siRNA.
The requirement for Atg5 in fungal antigen presentation was assessed by cytokine production of the specific T cell clones that were stimulated with antigen-pulsed macrophages. For this purpose, C. albicans extract was added to human macrophages either directly or coupled to latex beads before incubation with the C. albicans–specific T cells. Atg5-deficient macrophages were significantly inhibited in presenting extracellular antigen onto MHC class II molecules. This inhibition was pronounced at later time points after antigen pulsing with bead-coupled C. albicans antigens (Fig. 4 A). When fungal extract was processed via phagocytosis, the resulting MHC class II antigen presentation was inhibited up to sevenfold 18 h after antigen processing. In contrast, processing of soluble C. albicans extract for CD4+ T cell activation was only affected up to twofold in atg5-silenced macrophages (Fig. 4 A). siRNA targeting of atg16L1 in macrophages or shRNA atg5 silencing in DCs resulted in similarly reduced responses by C. albicans–specific CD4+ T lymphocytes (Fig. 4, C and D; and Fig. S3, D and E). These findings suggest that Atg8/LC3-positive phagosomes store antigen for prolonged processing onto MHC class II molecules. Consistent with this model, MHC class II molecules and Lamp2, hallmarks of the MHC class II–loading compartment, do not colocalize with Atg8/LC3-positive phagosomes but with each other (Fig. 3, F–H; and Fig. S2, D–F). Only after uncoupling of Atg8/LC3 from the phagosomal membrane, fusion with lysosomes occurs, and the stored antigen is delivered for MHC class II processing. To support the increased immunogenic capacity of zymosan-containing phagosomes, we purified phagosomal fractions of macrophages treated with uncoated beads (control) or with zymosan (to induce LAP) by density gradient centrifugation. The resulting fractions were blotted for Atg8/LC3 and Lamp2, and double-positive fractions were selected for mass spectrometry (MS) analysis. Fig. S2 G confirms up-regulation of Atg8/LC3 in zymosan-containing phagosomes compared with those without. In addition, we could detect enrichment of HLA molecules and of other relevant markers, such as redox regulators or Rab proteins (Fig. S2 H).
Our experiments indicate that human phagocytes generate a phagosome subset with Atg8/LC3 coupled to the cytosolic side of their membranes. This subset is preferentially induced by engulfment of TLR2 ligand-carrying cargo. In these phagosomes, the antigen is preserved for longer periods of time for continuous loading onto MHC class II molecules and thereby CD4+ T cell activation. We also show that ROS production by NOX2 is required for the generation of these antigen storage vesicles. In contrast to previous studies (Sanjuan et al., 2007; Florey et al., 2011; Martinez et al., 2011), our data suggest that coupling of Atg8/LC3 to phagosomes hinders their maturation. The use of different species or other cellular context might account for the different observations. Indeed, it has been recently shown that LAP in human plasmacytoid DCs reroutes phagocytosed immune complexes to TLR9-containing compartments, rather than simply degrading it (Henault et al., 2012).
Antigen preservation for prolonged MHC class II loading has recently emerged as a hallmark of potent APCs, such as DCs, which are able to limit degradation in their endocytic compartment for efficient antigen processing (Delamarre et al., 2005; Savina et al., 2006). These studies suggest that macrophages degrade endocytosed material rapidly, whereas DCs maintain antigens longer and therefore sustain antigen presentation on MHC class II. If lysosomal processing is attenuated by increasing lysosomal pH, macrophages become more potent APCs (Delamarre et al., 2006). Our results now extend these findings by characterizing a phagosomal subset in macrophages and DCs that allows antigenic peptides to be conserved for many hours. Inhibition of the formation of these phagosomes compromises endocytosed antigen processing for MHC class II presentation. These findings might also explain recent observations in macroautophagy-deficient mouse models in vivo. DCs from such mice were found to process extracellular antigen for MHC class II presentation less efficient in vivo, probably contributing to decreased herpes simplex virus–specific CD4+ T cell responses (Lee et al., 2010). Although the authors suggest that this was caused by decreased recruitment of lysosomal proteases to phagosomes, they did not distinguish between phagosomes that simply degrade extracellular antigens and those that deliver these antigens for MHC class II presentation. Another study in murine DCs in vitro showed that Atg8/LC3 recruitment to phagosomes containing β-glucan particles influences the efficiency of MHC class II antigen presentation (Ma et al., 2012). In this study, membrane-bound Atg8/LC3 seemed to colocalize with MHC class II molecules, increasing T cell activation. Contrary to what we observe in human APCs, murine cells display Atg8/LC3-positive phagosomes only for 60 min. Our experimental approach of live-cell imaging and immunoelectron microscopy allowed us to determine the long half-life of Atg8/LC3-associated phagosomes in human phagocytes. We could demonstrate that Atg8/LC3 is coupled to the single phagosomal membrane most likely on the outside of these vesicles and that this subset of phagosomes fuses with lysosomes only after Atg8/LC3 uncoupling. Also, we observed that the molecular machinery of macroautophagy seems to modify a subset of phagosomes to retain antigen in macrophages and DCs for enhanced processing toward MHC presentation. However, the majority of phagosomes will not recruit Atg8/LC3 and will keep their proteolytic potential. Macroautophagy components then seem to regulate phagocytosis by modifying the fate of a subgroup of phagocytic vesicles. This might contribute to an optimization of resources because upon the same stimulus, phagosomes may be directed to phagolysosome formation to rapidly degrade the engulfed material or, alternatively, preserve antigen for presentation.
Finally, we have now shown for human APCs that LAP is dependent on NOX2 activity similarly to murine cells (Huang et al., 2009; Ma et al., 2012). The lack of ROS-dependent formation of this antigen storage compartment could also contribute to the susceptibility of CGD patients to fungal infection (Seger, 2010; Kuijpers and Lutter, 2012). Thus, our findings propose that diminished MHC class II antigen processing could impair antifungal CD4+ T cell responses, resulting in a compromised immune control of these pathogens.
Materials and methods
Antibodies and cell dyes
Antibody anti-Atg8/LC3 (clone 5F10) and -Atg5 (clone 7C6) were purchased from Nanotools for Western blotting (WB). Directly labeled anti–β-actin (HRP) antibody was obtained from Abcam. For immunofluorescence stainings, anti-Atg8/LC3 was received from Medical and Biological Laboratories, anti-Beclin1 was purchased from Novus Biologicals, antipolyubiquitinylated proteins (clone FK1) were obtained from Enzo Life Sciences, anti-p62 was obtained from American Research Products, anti-Lamp2 (clone H4B4) was obtained from SouthernBiotech or directly labeled to Alexa Fluor 647 from eBioscience, and anti–HLA-DR (clone L243) was purchased from BioLegend. For EM, anti-GFP was provided by EMD Millipore. Blocking antibodies, anti–HLA-DR and IgG isotype control, were purchased from BioLegend. For flow cytometric staining, directly labeled anti-CD3 (clone UCHT1), anti-CD4 (clone Sk3), anti-CD8 (clone RPA-T8), and anti–IFN-γ (clone BSB3) were obtained from BioLegend. Anti–IL-17A (clone DEC1) was provided by eBioscience. LIVE/DEAD Fixable Aqua was purchased from Invitrogen, Annexin V Alexa Fluor 647 and 7-AAD were received from BioLegend. For ELISA, anti–IFN-γ and anti–granulocyte–macrophage colony-stimulating factor (GM-CSF) duosets were obtained from Mabtech, and anti–IL-17A was purchased from BioLegend. The lysosomal dye LysoTracker red DND-99 and the redox-sensitive probe Amplex UltraRed were provided by Invitrogen.
Cell preparation and stimulation
Peripheral blood mononuclear cells (PBMCs) were isolated from leukocyte concentrates (Zürich Blood Center) or from whole blood (CGD patients) by density-gradient centrifugation on Ficoll/Hypaque. CD14+ cells were isolated from PBMCs by positive magnetic cell separation (Miltenyi Biotec), and macrophages were generated in D10 (DMEM + 10% heat-inactivated FBS + penicillin/streptomycin + nonessential amino acids). Recombinant human GM-CSF (1,000 U/ml) was added at days 0, 3, and 5. Differentiated macrophages were used between day 6 and 8 of culture. For generation of immature DCs, CD14+ were cultured for 5 d in R2 (RPMI 1640 + 2% heat-inactivated pooled human antibody serum + gentamycin), adding 500 U/ml recombinant human IL-4 and 500 U/ml recombinant human GM-CSF on days 0 and 3. For stimulation of macrophages with PAMPs and other compounds, the following concentrations were used: No in the figures indicates medium alone, 25 µg/ml polyinosinic/polycytidylic acid, 100 µg/ml Pam3CSK4, 100 µg/ml zymosan, 0.5 µg/ml lipopolysaccharide, 1 µg/ml PMA, 25 µM rapamycin, 20 µg/ml muramyl dipeptide, 20 µg/ml murabutide, 150 mg/ml monosodium urate crystals, 2 µg RNA/protamine, extract is 20 µg/ml C. albicans protein extract, 1:5 heat-killed C. albicans/macrophages ratio, 56 µM E64d, 37 µM Pepstatin A, 84 µM leupeptin, and 2 nM concanamycin A. For adsorption of Pam3CSK4 and C. albicans protein extract to polystyrene beads (Bangs Laboratories), 50 µg of the respective agonist was added to 108 beads in PBS buffer, pH 7.4. After incubation at RT for 1 h with constant mixing, beads were left at 4°C on in a rotator, washed extensively with PBS, counted, and used immediately (at 1:1 macrophage/bead ratio).
Subcellular fractionation and immunoblotting
To obtain protein extracts, cells were collected at the indicated times, washed once in PBS, resuspended in ice-cold lysis buffer (1% NP-40 with complete protease inhibitor cocktail obtained from Roche), vortexed for 10 s, and incubated on ice for 10 min. Cell lysates were centrifuged for 10 min at 4°C at 20,000 g, and protein concentrations were quantified with the bicinchoninic acid protein assay. For all samples, 25–50 µg of total protein extracts were boiled for 5 min in the presence of SDS-PAGE loading buffer, resolved by SDS-PAGE, and transferred onto polyvinylidene difluoride membranes. For detection of primary antibodies, HRP-conjugated secondary antibodies and the ECL Plus detection system were used. For quantification of protein levels, densitometry was performed on films using the ImageJ software (National Institutes of Health), always on pictures without pixel saturation.
Quantitative PCR
TNF mRNA quantitation was achieved from cDNA generated using reverse transcription (GoScript; Promega). cDNA was used as a template for quantitative PCR on a detection system (MyiQ; Bio-Rad Laboratories) with Platinum Quantitative PCR SuperMix-UDG (Life Technologies). Primers used for quantification of TNF were 5′-GCCGCATCGCCGTCTCCTAC-3′ and 5′-AGCGCTGAGTCGGTCACCCT-3′. Expression levels of TNF were normalized to the expression of GAPDH. GAPDH-detecting primers were 5′-ATGGGGAAGGTGAAGGTCG-3′ and 5′-GGGTCATTGATGGCAACAATATC-3′.
Measurement of ROS
Production of ROS by macrophages was assessed with the cell-permeable fluorescent probe Amplex UltraRed. Macrophage monolayers were cultured in 48-well plates and stimulated with the indicated compounds for 30 min or 3 h in phenol red–free D10 medium. Cells were then incubated with a mixture of 25 µM Amplex UltraRed and 0.5 U/ml HRP for 30 min at 37°C and analyzed in a fluorometer (excitation of 530 nm, emission of 580 nm, and cutoff of 550 nm; Infinite; Tecan).
Microscopy and live-cell imaging
For immunofluorescence stainings, cells on 8-well chamber slides (Thermo Fisher Scientific) were fixed in 3% PFA for 20 min at 4°C, permeabilized with 0.01% Triton X-100 for 1 min at RT, and then incubated with the signal enhancer (Image-iT FX; Invitrogen) and stained with the indicated antibodies followed by the appropriate secondary reagent. All washes were performed in PBS supplemented with 1% fish skin gelatin and 0.02% saponin. Slides were counterstained with DAPI and mounted with ProLong gold antifade reagent (Invitrogen). Cells were visualized through a 63×, 1.4 NA oil immersion lens with an inverted confocal laser scanning microscope (SP2 or SP5; Leica). To determine colocalization, Pearson’s correlation coefficient values were calculated using JACoP (Just Another Colocalization Plugin) in ImageJ software for the region surrounding Atg8/LC3-positive phagosomes or for the area of interest. Data are expressed as dot plots with median values displayed as horizontal lines. A Pearson score >0.6 indicates a statistically relevant colocalization of the signals and is indicated in each graph as a dashed line. Intensity values for the different markers were given by Imaris (Bitplane) for the correspondent region of interest (usually, part of the phagosomal membrane). Data are expressed as dot plots with median values displayed as horizontal lines. For live-cell imaging, macrophages were cultured in 8-well chamber slides obtained from ibidi in phenol red–free D10 medium. Cells were visualized through a 63×, 1.4 NA oil immersion lens with an inverted confocal microscope (SP5) at 37°C and 5% CO2 atmosphere. Live-imaging analysis was performed using Imaris 7 software. All pictures shown represent one representative picture out of at least three independent experiments.
Immunoelectron microscopy
Macrophages infected or not infected with heat-killed C. albicans were fixed for 2 h at RT with 2% formaldehyde and 0.1% glutaraldehyde in 0.1 M phosphate buffer containing 50 mM NaCl, pH 7.4. Samples were prepared as previously described (Zuber et al., 2000) and analyzed in an electron microscope (CM100; Philips), and pictures were taken with a charge-coupled device camera (ORIUS; Gatan). In brief, cell pellets were immersed in 2 M sucrose containing 15% polyvinyl pyrrolidone, enclosed in 2% agarose (FMC Bioproducts), mounted on aluminum pins, and frozen and stored in liquid nitrogen. Frozen ultrathin sections were prepared by using an ultramicrotome (Ultracut S; Reichert) equipped with a cryochamber (FCS; Reichert), picked up on nickel grids, and stored overnight on gelatin at 4°C. Before immunolabeling, gelatin was liquefied at 37°C, and nickels grids were removed and washed by floating them on droplets of PBS, pH 7.4. For immunolabeling, grids with the attached thin sections were conditioned on droplets of PBS containing 1% BSA, 0.01% Triton X-100, and 0.01% Tween 20 for 10 min at ambient temperature. Grids were then transferred to droplets of primary antibody (dilution 1:50) diluted in conditioning buffer for 2 h at ambient temperature, rinsed on droplets of PBS, and incubated with gold-labeled secondary antibody. Finally, grids with the attached thin sections were rinsed in PBS, fixed with 2% glutaraldehyde in PBS for 10–20 min, rinsed with PBS and distilled water, and embedded and stained with methylcellulose and uranyl acetate.
Lentiviral transduction and shRNA silencing
The GFP-tagged Atg8/LC3 was obtained as described in Gannagé et al. (2009) by PCR amplification of the cDNA of human MAP1LC3B sequence (available from GenBank under accession no. NM_022818) from a human B-LCL and cloning into the multiple cloning site of the mammalian expression vector pEGFP-C2 (Takara Bio Inc.). The mRFP-tagged Atg8/LC3 was cloned out of plasmid 21075 p.m.RFP-Atg8/LC3 (Addgene) and reintroduced into the BamHI–NotI sites of the pHR-SIN-CSGWΔNotI (pCSGW) vector. The mRFP-GFP-Atg8/LC3 vector was a gift from T. Yoshimori (University of Osaka, Osaka, Japan). For silencing atg5, shRNA expression plasmid pLKO.1-puromycin shRNA scramble (5′-TCCTAAGGTTAAGTCGCCCTCG-3′) and shRNA atg5 (5′-CCTGAACAGAATCATCCTTAA-3′) were gifts from J. Tschopp (University of Lausanne, Lausanne, Switzerland). For these constructs, the puromycin gene was substituted by the GFP gene (from lentivirus-GFP; Addgene) using BamHI–KpnI. Lentiviral construct for GFP-Rab7 was obtained by subcloning the Rab7 gene from plasmid 12605 (Addgene) into BamHI sites of pENTR4-GFP-C1 (Addgene). The tagged Rab7 gene was then introduced into the pLenti-CMV-puromycin DEST vector (Addgene) using the cloning kit (Gateway; Invitrogen). Production of lentiviral particles was performed as described in Schmid et al. (2007). In brief, lentiviral vectors were cotransfected with the helper plasmids pCMVΔR8.91 and pMDG into 293T cells by calcium phosphate transfection. Culture supernatants containing recombinant viral particles were harvested on day 2 after transfection, filtered through a 0.22-µm filter, and frozen at −80°C. For transduction of primary human macrophages, cells were used between day 5 and 7, incubated with viruslike particles for 3 h at 37°C, infected with the correspondent lentiviruses at an MOI of 5, and centrifuged at 1,900 g for 45 min at 37°C. For expression vectors, 6 µg/ml polybrene (EMD Millipore) was added together with the lentiviruses, and 24 h after, culture medium was replaced, and cells were incubated at 37°C 5% CO2 for an additional 24 h before visualization. For silencing with shRNA constructs, no polybrene was added, and cells were incubated with the lentiviruses for a total of 3 or 5 d before use of macrophages or immature DCs, respectively.
siRNA silencing
Macrophages were plated in 96-well plates in Opti-MEM medium. The cells were transfected with siRNA formulated into liposomes (Lipofectamine 2000; Invitrogen) according to the manufacturer’s instructions. siRNA atg16L1 (5′-GAGUUGUCUUCAGCCCUGAUGGCAG-3′) and siRNA scramble (Stealth RNAi siRNA negative control Hi GC by Invitrogen) concentration was 15 pmol/2 × 105 cells. The final volume of culture medium was 115 µl/well. After incubation for 48 h, cells were used for co-culture with CD4+ T cell clones.
T cell clones
C. albicans–specific CD4+ T cells were prepared from HLA-DRB1*0401 HD PBMCs. After 16 h of incubation of PBMCs with 20 µg/ml C. albicans protein extract, IL-17–producing T cells were collected by the IL-17 secretion assay (Miltenyi Biotec), according to the manufacturer’s protocol. T cells were plated in U-bottom 96-well plates with irradiated feeder cells (105 allogenic PBMCs and 2 × 104 B-LCL cells/well), at concentrations of 10, 1, or 0.3 T cells/well. Stimulatory medium consisted of RPMI 1640, 5% heat-inactivated pooled human AB serum, gentamycin, 150 U/ml IL-2, and 1 µg/ml phytohemagglutinin-L (Sigma-Aldrich). Further culturing was performed as previously described (Fonteneau et al., 2001); after 5 d, half of the volume of medium (75 µl/well) was replaced with culture medium containing 5% pooled human serum and 150 U/ml IL-2 without phytohemagglutinin-L. After 8 or 9 d, proliferating clones could be seen by microscopy and were maintained with <1.5 × 106 T cells/ml. After 2–3 wk, clones were tested for specificity by IFN-γ ELISA or intracellular cytokine staining (B3 CD4+ T cell clone was selected based on highest specificity and cytokine production). Initially, the resulting T cell clones (T cell clone B3) secreted both IL-17 and IFN-γ in response to C. albicans protein extracts. However, after further expansion of the cells, IL-17 production ceased, and only IFN-γ and GM-CSF secretions were maintained. These cytokines could be used as an indication for specific antigen recognition by T cells. For antigen presentation, macrophages and DCs were pulsed with C. albicans protein extract–coated beads (1:1 ratio) or 20 µg/ml of soluble C. albicans protein extract for 4 h. After pulsing, cells were washed, and new medium was added (RPMI 1640 + 5% heat-inactivated FBS + gentamycin). After 0, 3, and 18 h, B3 CD4+ T cells were added for co-culture with the macrophages at a 1:1 ratio. After 0, 18, and 38 h, B3 CD4+ T cells were added for co-culture with the DCs at a 1:1 ratio.
ELISA assays
For cytokine detection, supernatants were harvested after 18 h of co-culture of the T cell clone B3 with the target cells (1:1), diluted, and plated directly onto previously coated 96-well ELISA plates (Nunc-Immuno MaxiSorp; Thermo Fisher Scientific). Cytokines were then detected with the biotinylated specific antibodies and streptavidin-HRP, using the peroxidase substrate tetramethylbenzidine. Recombinant human IFN-γ, GM-CSF, and IL-17A (15–1,000 pg/ml) were used as standards.
HLA-DR blocking experiment
Target cells were plated in 96-well flat-bottom plates and incubated with 10 µg/ml anti–HLA-DR or isotype control for 1.5 h at 4°C. After this time, 20 µg/ml C. albicans protein extract was added to the cells together with the T cell clone B3 at a 1:1 ratio, and cells were incubated at 37°C and 5% CO2 for 18–20 h. Co-culture supernatants were analyzed for the presence of IFN-γ and IL-17 by ELISA.
Phago-FACS
Recruitment of Atg8/LC3 to phagosomal membranes was analyzed by flow cytometry in isolated phagosomes as previously described (Savina et al., 2010). In brief, cells were disrupted by passing them through a 24-gauge needle in homogenization buffer (8% sucrose, 3 mM imidazole, and 1 mM DTT in PBS). Afterwards, lysates were centrifuged at 100 g for 4 min, and the supernatant was collected (phagosomes). The desired stainings were performed in this enriched phagosomal fractions with primary and secondary antibodies diluted in PBS for 30 min at 4°C. Every subsequent centrifugation step was performed at 1,000 g for 4 min. Phagosome pellets were resuspended in 150 µl PBS for acquisition at the flow cytometer (LSRFortessa; BD).
Phagosome purification and MS
Phagosome purification was modified from the protocol described in Dengjel et al. (2012). The cell lysate was spun at 100 g for 4 min at 4°C to collect the supernatant containing the phagosomes. This supernatant was further spun for 10 min at 1,000 g at 4°C, and the pellet containing the vesicular fraction was resuspended in 1 ml homogenization medium (0.25 M sucrose, 1 mM EDTA, and 20 mM Hepes, pH 7.4). The suspension was loaded on top of an iodixanol gradient and centrifuged for 17 h at 100,000 g at 6°C. For the gradient, iodixanol was mixed with homogenization medium in solutions of 5, 10, 16, 24, and 30%. Gradient steps consist in total of 1.6 ml. The stacked gradients were allowed to diffuse for 4–5 h in a horizontal position at RT. Mass spectrometry sample preparation and analysis were performed according to (Dengjel et al., 2012); therefore, samples for liquid chromatography–MS/MS were fractionated by nanoscale HPLC on either a 1200 (Agilent Technologies) or a NanoLC Ultra (Eksigent) connected online to a mass spectrometer (LTQ Orbitrap XL; Thermo Fisher Scientific). Peptides were separated over a linear gradient from 10–30% acetonitrile in 0.5% acetic acid with a flow rate of 250 nl/min. All full-scan acquisition was performed in the Fourier transform MS part of the mass spectrometers in the range from mass/charge (m/z) 350 to 2,000 with an automatic gain control target value of 106 and at resolution 60,000 at m/z 400. MS acquisition was performed in data-dependent mode to sequentially perform MS/MS on the five (first replicate) or 10 (second replicate) most intense ions in the full scan (top 10) in the LTQ using the following parameters: automatic gain control target value of 5,000, ion selection thresholds of 1,000 counts (first replicate) or 100 (second replicate), and a maximum fill time of 100 ms. Wide-band activation was enabled with an activation q = 0.25 applied for 30 ms at a normalized collision energy of 35%. Singly charged and ions with unassigned charge state were excluded from MS/MS. Dynamic exclusion was applied to reject ions from repeated MS/MS selection for 45 s. All MS/MS scans were recorded with multistage activation enabled with a neutral loss mass list of m/z 195.95, 97.97, 48.99, and 32.66. All recorded liquid chromatography–MS/MS raw files (359) were processed together in MaxQuant26 version 1.2.2.5 with default parameters using the March 2012 UniProt database, which contains 81,470 protein sequences. For database searching, parameters were mass accuracy thresholds of 0.5 (MS/MS) and 6 ppm (precursor), maximum two missed cleavages, carbamidomethylation as fixed modification, and deamidation (NQ), oxidation (M), phosphorylation (STY), and protein N-terminal acetylation as variable modifications. MaxQuant was used to filter the identifications for a false discovery rate <1% for peptides, sites, and proteins using forward-decoy searching. Match between runs were enabled with a retention time window of 2 min.
Statistical analysis
Where indicated, Mann–Whitney or t tests were performed using the Prism software (version 5.0a; GraphPad Software).
Online supplemental material
Fig. S1 shows that Atg8/LC3 binding to the phagosomal membrane depends on PRR signaling. Fig. S2 shows the characterization of Atg8/LC3-positive phagosomes. Fig. S3 shows the generation and characterization of C. albicans–specific T cells, antigen presentation assay after silencing of autophagy-related genes, and survival and activation patterns of macrophages with atg5 silencing. Video 1 shows that the RFP-GFP-Atg8/LC3 tandem reporter construct is coupled to and uncoupled from phagosomes without ever switching from yellow RFP/GFP composite fluorescence to red RFP only fluorescence, indicating that the Atg8/LC3 is never exposed to a low pH during LAP. Video 2 shows the longevity of LC3-associated phagosomes. Video 3 demonstrates that LysoTracker and Atg8/LC3 fluorescence is found on separate vesicles. Video 4 shows that LC3-associated phagosomes only acidify and become LysoTracker positive after Atg8/LC3 uncoupling. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201308173/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201308173.dv.
Supplementary Material
Acknowledgments
This work was supported by the National Cancer Institute (R01CA108609), the Sassella Foundation (10/02), Cancer Research Switzerland (KFS-02652-08-2010), the Association for International Cancer Research (11-0516), the Vontobel Foundation, the Baugarten Foundation, the EMDO Foundation, Novartis, the Gebert Rüf Stiftung (Program Rare Diseases–New Approaches; GRS-046/10), the European Union Seventh Framework Programme Cell Primary Immune Deficiencies program, and the Swiss National Science Foundation (310030_126995). S. Romao was a recipient of a Fundação para a Ciência e Tecnologia postdoctoral fellowship (SFRH/BPD/63459/2009). J. Dengjel is funded by the Excellence Initiative of the German federal and state governments (Freiburg Institute for Advanced Studies and Centre for Biological Signalling Studies) and the Deutsche Forschungsgemeinschaft (GZ DE1757/2-1).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used in this paper:
- APC
- antigen-presenting cell
- Atg
- autophagy-related protein
- CGD
- chronic granulomatous disease
- DC
- dendritic cell
- GM-CSF
- granulocyte–macrophage colony-stimulating factor
- HD
- healthy donor
- HLA
- human leukocyte antigen
- LAP
- LC3-associated phagocytosis
- MHC
- major histocompatibility complex
- MS
- mass spectrometry
- m/z
- mass/charge
- PAMP
- pathogen-associated molecular pattern
- PBMC
- peripheral blood mononuclear cell
- PRR
- pattern recognition receptor
- ROS
- reactive oxygen species
- WB
- Western blotting
References
- Delamarre L., Pack M., Chang H., Mellman I., Trombetta E.S. 2005. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science. 307:1630–1634 10.1126/science.1108003 [DOI] [PubMed] [Google Scholar]
- Delamarre L., Couture R., Mellman I., Trombetta E.S. 2006. Enhancing immunogenicity by limiting susceptibility to lysosomal proteolysis. J. Exp. Med. 203:2049–2055 10.1084/jem.20052442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengjel J., Høyer-Hansen M., Nielsen M.O., Eisenberg T., Harder L.M., Schandorff S., Farkas T., Kirkegaard T., Becker A.C., Schroeder S., et al. 2012. Identification of autophagosome-associated proteins and regulators by quantitative proteomic analysis and genetic screens. Mol. Cell. Proteomics. 11:M111.014035 10.1074/mcp.M111.014035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O., Kim S.E., Sandoval C.P., Haynes C.M., Overholtzer M. 2011. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol. 13:1335–1343 10.1038/ncb2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonteneau J.F., Larsson M., Somersan S., Sanders C., Münz C., Kwok W.W., Bhardwaj N., Jotereau F. 2001. Generation of high quantities of viral and tumor-specific human CD4+ and CD8+ T-cell clones using peptide pulsed mature dendritic cells. J. Immunol. Methods. 258:111–126 10.1016/S0022-1759(01)00477-X [DOI] [PubMed] [Google Scholar]
- Gannagé M., Dormann D., Albrecht R., Dengjel J., Torossi T., Rämer P.C., Lee M., Strowig T., Arrey F., Conenello G., et al. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 6:367–380 10.1016/j.chom.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henault J., Martinez J., Riggs J.M., Tian J., Mehta P., Clarke L., Sasai M., Latz E., Brinkmann M.M., Iwasaki A., et al. 2012. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 37:986–997 10.1016/j.immuni.2012.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Canadien V., Lam G.Y., Steinberg B.E., Dinauer M.C., Magalhaes M.A., Glogauer M., Grinstein S., Brumell J.H. 2009. Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. USA. 106:6226–6231 10.1073/pnas.0811045106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460 [DOI] [PubMed] [Google Scholar]
- Kuijpers T., Lutter R. 2012. Inflammation and repeated infections in CGD: two sides of a coin. Cell. Mol. Life Sci. 69:7–15 10.1007/s00018-011-0834-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Mattei L.M., Steinberg B.E., Alberts P., Lee Y.H., Chervonsky A., Mizushima N., Grinstein S., Iwasaki A. 2010. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 32:227–239 10.1016/j.immuni.2009.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Mizushima N., Virgin H.W. 2011. Autophagy in immunity and inflammation. Nature. 469:323–335 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Becker C., Lowell C.A., Underhill D.M. 2012. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J. Biol. Chem. 287:34149–34156 10.1074/jbc.M112.382812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Almendinger J., Oberst A., Ness R., Dillon C.P., Fitzgerald P., Hengartner M.O., Green D.R. 2011. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. USA. 108:17396–17401 10.1073/pnas.1113421108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Ohsumi Y. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27:107–132 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Münz C. 2009. Enhancing immunity through autophagy. Annu. Rev. Immunol. 27:423–449 10.1146/annurev.immunol.021908.132537 [DOI] [PubMed] [Google Scholar]
- Randow F., Münz C. 2012. Autophagy in the regulation of pathogen replication and adaptive immunity. Trends Immunol. 33:475–487 10.1016/j.it.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan M.A., Dillon C.P., Tait S.W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J.L., Withoff S., Green D.R. 2007. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 450:1253–1257 10.1038/nature06421 [DOI] [PubMed] [Google Scholar]
- Savina A., Jancic C., Hugues S., Guermonprez P., Vargas P., Moura I.C., Lennon-Duménil A.M., Seabra M.C., Raposo G., Amigorena S. 2006. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 126:205–218 10.1016/j.cell.2006.05.035 [DOI] [PubMed] [Google Scholar]
- Savina A., Vargas P., Guermonprez P., Lennon A.M., Amigorena S. 2010. Measuring pH, ROS production, maturation, and degradation in dendritic cell phagosomes using cytofluorometry-based assays. Methods Mol. Biol. 595:383–402 10.1007/978-1-60761-421-0_25 [DOI] [PubMed] [Google Scholar]
- Schmid D., Pypaert M., Münz C. 2007. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 26:79–92 10.1016/j.immuni.2006.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger R.A. 2010. Chronic granulomatous disease: recent advances in pathophysiology and treatment. Neth. J. Med. 68:334–340 [PubMed] [Google Scholar]
- Weidberg H., Shvets E., Elazar Z. 2011. Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80:125–156 10.1146/annurev-biochem-052709-094552 [DOI] [PubMed] [Google Scholar]
- Zuber C., Spiro M.J., Guhl B., Spiro R.G., Roth J. 2000. Golgi apparatus immunolocalization of endomannosidase suggests post-endoplasmic reticulum glucose trimming: implications for quality control. Mol. Biol. Cell. 11:4227–4240 10.1091/mbc.11.12.4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.