Abstract
PPR proteins are a family of ubiquitous RNA-binding factors, found in all the Eukaryotic lineages, and are particularly numerous in higher plants. According to recent bioinformatic analyses, yeast genomes encode from 10 (in S. pombe) to 15 (in S. cerevisiae) PPR proteins. All of these proteins are mitochondrial and very often interact with the mitochondrial membrane. Apart from the general factors, RNA polymerase and RNase P, most yeast PPR proteins are involved in the stability and/or translation of mitochondrially encoded RNAs. At present, some information concerning the target RNA(s) of most of these proteins is available, the next challenge will be to refine our understanding of the function of the proteins and to resolve the yeast PPR-RNA-binding code, which might differ significantly from the plant PPR code.
Keywords: mitochondria, penta-tricopeptide repeat, PPR, budding yeast, S. cerevisiae, fission yeast, S. pombe, RNA metabolism, translation
Introduction
In the mitochondrial field, the budding yeast S. cerevisiae has traditionally been a leading model for the study of organelle gene expression and the biogenesis of the oxidative phosphorylation (OXPHOS) complexes. The first yeast protein to be recognized as PPR protein was Pet309 in S. cerevisiae.1 In the following years, the study of PPR proteins in plants rapidly overtook that of yeast PPR proteins, with computational analyses showing that this protein family is the largest known in land plants and proposing that PPR proteins participate actively in the very rich RNA metabolism of plant organelles, via their intrinsic RNA-binding capacity.1 Thus, the extreme importance of the PPR protein family for land plants has generated a lot of interest and much data has been gathered to list plant PPR proteins, study their role, their mechanism of action and the way they recognize their target RNA.2 In addition, it has been found that plant PPR proteins constitute an evolutionary reservoir that serves the high plasticity and adaptability of photosynthetic organisms.3 By comparison, studies focused specifically on yeast PPR proteins remained less developed until the design of a new algorithm, SCIPHER, significantly facilitated their identification.4 This has revealed that several yeast proteins already known to take part in mitochondrial gene expression are in fact PPR proteins. In addition, the initial characterization of PPR proteins from the fission yeast Schizosaccharomyces pombe has also allowed the identification of both conserved and novel PPR proteins. In consequence, considering the importance and experimental tractability of the yeast model, it seems an appropriate time to review our current knowledge of this protein family in yeasts and to discuss future directions that might be taken to understand the diverse biological functions of these proteins and the way they bind their target RNAs.
Identifying yeast PPR Proteins In Silico—Challenges and Solutions
PPR motifs, together with closely related Tetratrico-Peptide Repeats (TPR), SEL-1 like and HAT repeats, belong to a large family of solenoid repeat structures formed of α-α repeats.1,5,6 Considerable divergence and short repeat length (about 34–35 aa) present significant challenges for reliable computational identification of such sequences, and make profile-based methods, like profile hidden Markov models (pHMM),7 more suitable than pair wise methods, like BLAST. Both general pHMM approaches, for example the Pfam database,8 and more specialized tools targeted specifically toward tandem repeat families, such as TPRpred,9 allowed the identification of numerous PPR proteins, mainly encoded by plant genomes.
The abundance of the PPR proteins in plants (over 90% of the known PPR proteins, containing about 95% of the PPR motifs represented in the Pfam database come from Viridiplantae), means that the general PPR motif profiles are significantly biased, and are thus not optimal for studying sequences of non-plant origin. Before a more detailed study aimed specifically at the annotation of yeast PPR motifs had been performed by Lipinski et al.,4 TPRpred9 identified only Pet309p, Aep3p and Dmr1p as PPR proteins in S. cerevisiae, with 12, three and two PPR motif, respectively. As typical PPR proteins contain a multitude of repeats, this suggested a profound underestimation of the number of PPR motifs and proteins present in yeast, and led to the development of a phylogenetically informed pHMM method, SCIPHER, based on yeast-specific iteratively enriched profiles.4 The initial profiles were constructed from multiple sequence alignments of orthologs of known PPR proteins from 14 yeast species (13 from Saccharomycotina and one, S. pombe, representing Taphrinomycotina) and used to search all the protein sequences encoded by the genomes of these yeasts. New motifs and/or proteins uncovered in the searches were appended to the respective profiles used in subsequent rounds of searching, until saturation was reached. In this way, a total of 150 PPR proteins containing 953 repeats were identified, including 14 in S. cerevisiae and nine in S. pombe.4 The inclusion of three additional Schizosaccharomyces sp genomes10 in the analysis identified one more putative PPR protein in S. pombe (Ppr10). Additional putative PPR motifs in S. cerevisiae that failed to yield significantly positive scores using the HMM profiles were found based on similarity with identified motifs in orthologous sequences,4 bringing a total number of PPR proteins in this species to 15, with 159 repeat motifs. Following the inclusion of recently annotated PPR sequences, the current general profiles used in TPRpred recognize 11 of the 15 putative S. cerevisiae PPR proteins, with a total of 71 motifs. Table 1 presents the list of PPR proteins, with the number of predicted motifs, in the model yeasts S. cerevisiae and S. pombe.
Table 1. List of S. cerevisiae and S. pombe PPR proteins.
| Name | Generic Name | (aa) | # of motifs | Carbonate resistant (%) | Localization references |
|
|---|---|---|---|---|---|---|
|
S. cerevisiae |
Aep1/Nca1 |
Ymr064w |
504 |
4 (5)e |
ND |
77 |
| Aep2a/Atp13 |
Ymr282c |
580 |
5 (8) |
ND |
91 |
|
| Aep3 |
Ypl005w |
606 |
5 (5) |
50 |
40 |
|
| Atp22/Tcm10 |
Ydr350c |
684 |
3 (10) |
100 |
68 |
|
| Cbp1 |
Yjl209w |
654 |
5 (14) |
100 |
50 |
|
| Dmr1b/Ccm1/Rrg2 |
Ygr150c |
876 |
6 (12) |
ND |
31 |
|
| Msc6 |
Yor354c |
692 |
5 (9) |
ND |
79 |
|
| Pet111 |
Ymr257c |
800 |
7 (15) |
≈ 100 |
62 |
|
| Pet309c |
Ylr067c |
965 |
16 (22) |
100 0 |
92 54 |
|
| Rmd9 |
Ygl107c |
646 |
6 (7) |
50 |
19, 30 |
|
| Rmd9L |
Ybr238c |
731 |
4 (6) |
100 |
19 |
|
| Rpm2 |
Yml091c |
1202 |
7 (21) |
0 |
27 |
|
| Rpo41d |
Yfl036w |
1351 |
0 (5) |
ND |
79 |
|
| Sov1 |
Ymr066w |
898 |
7 (11) |
ND |
38 |
|
| Yer077c |
Yer077c |
688 |
4 (9) |
ND |
77 |
|
| Ppr1 |
Spbc1604.02c |
697 |
6 |
30 |
18 |
|
| Ppr2 |
Spbc18H10.11c |
432 |
≥3 |
80 |
18 |
|
| S. pombe | Ppr3b |
Spbc19G7.07c |
687 |
≥10 |
100 |
18 |
| Ppr4c |
Spac8C9.06c |
931 |
13 |
100 |
18 |
|
| Ppr5 |
Spac1093.01 |
1261 |
≥16 |
100 |
18 |
|
| Ppr6a |
Spcc11E10.04 |
443 |
5 |
100 |
18 |
|
| Ppr7 |
Spbc16A3.03c |
658 |
5 |
50 |
18 |
|
| Ppr8 |
Spbc1289.06c |
481 |
4 |
40 |
18 |
|
| Rpo41d |
Spac26H5.12 |
1154 |
2 |
ND |
18 |
|
| Ppr10f |
Spbc106.19 |
515 |
2 |
ND |
93 |
|
| |
|
|
|
|
|
|
All yeast PPR proteins except Ppr10 have been experimentally localized into mitochondria. They were protease protected (i.e. facing the matrix or within the matrix) whenever tested. Column 5 give the percentage that remains associated with the membrane after an alkaline carbonate treatment. Clear S. cerevisiae and S. pombe orthologs are the following: aAep2 and Ppr6; bDmr1 and Ppr3; cPet309 and Ppr4; dboth Rpo41 proteins. eThe first number is the number of PPR motifs detected as statistically significant using pHMM profiles,4 the number in parentheses is the total number of putative motifs, including those that were extrapolated based on the presence of statistically significant motifs in orthologous sequences (see text). fMitochondrial localization is predicted in silico with high probability using MitoProt.93 ND, not determined; #, number; aa, amino acids.
The study of yeast PPR proteins revealed significant limitations in the pHMM-based computational approach. Distinguishing between true PPRs and related repeat motifs, like TPR or HAT repeats, is not always evident, and members of these families are often found as false positives when searching for new PPR proteins. A cursory analysis of the predicted secondary structure of known yeast PPR proteins suggests the presence of numerous additional α-helical repeat units that are not identified as statistically significant PPR motifs using the HMM profiles. Thus the numbers of PPR motifs presented in Table 1 are likely to underestimate the actual number of PPR (or PPR-like) repeats in these proteins. The high divergence of the PPR motif, and similarity to related repeat families, constitute the limitations of sequence-based methods for the identification of PPR proteins, particularly in non-plant organisms. Further refinements would require the inclusion of data on protein structure, such as the asymmetrical charge distribution and the presence of a positively charged substrate-binding surface in the PPR repeats, sub-cellular localization and biological function of the candidate proteins. Successful exhaustive identification of PPR proteins in the foreseeable future would therefore require a combination of computational approaches with experimental data and expert human input.
Finding the Target and Function of PPR Proteins in Yeast Mitochondria
Specificities of the yeast mtDNA coding capacity and expression
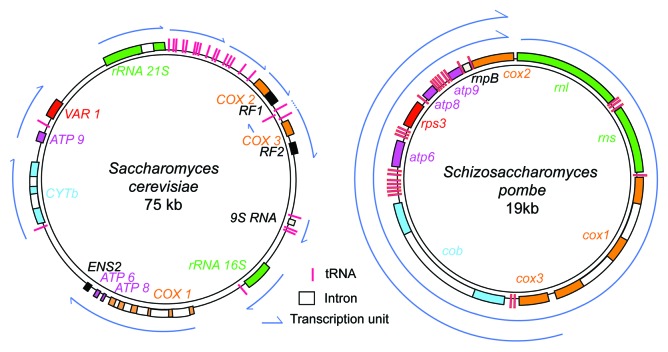
All known yeast PPR proteins have been found, or are predicted, to be in the mitochondrial matrix, sometimes associated with the inner membrane (see Table 1) and, thus, are expected to have mitochondrially encoded RNAs as their targets. The coding content of the mitochondrial genomes of S. cerevisiae and S. pombe, are similar: 24 tRNAs, two rRNAs (15S and 21S) for the small and large subunits of the mitochondrial ribosome, one RNase P component (9S in S. cerevisiae and rnpB in S. pombe) and eight protein coding genes; these produce one subunit of the respiratory complex III (Cytb), three subunits of complex IV (cytochrome c oxidase; Cox1, 2, 3), three subunits of complex V (ATP synthase; Atp6, 8, 9) and one subunit of the mitochondrial ribosome (Var1 in S. cerevisiae, Rps3 in S. pombe). Thus, all the RNAs encoded by these yeast mitochondrial genomes contribute to the formation of OXPHOS complex subunits, either by encoding structural subunits or by participating in the general translation or processing machinery (Fig. 1). The main difference in the coding capacity of these genomes is the number of introns (up to 13 in S. cerevisiae mtDNA11 and up to four in S. pombe mtDNA12), some of which encode maturases that are necessary for splicing; however, to date none of the yeast PPR proteins have been convincingly implicated in splicing per se.
Figure 1. A comparison of the mitochondrial DNA of S. cerevisiae and S. pombe showing their transcriptional units. The color code for the different classes of genes is: tRNA, fuchsia; RRNA, green; other non-coding RNA: gray; ATP synthase subunit genes, magenta; complex III subunit genes, blue; complex IV subunit genes, orange; ribosomal subunit genes, red; unknown ORFs (S. cerevisiae): black. Transcription units and introns are indicated. The intron content corresponds to D273-10B for S. cerevisiae and 972h- for S. pombe.
Another important difference regarding RNA metabolism in S. pombe and S. cerevisiae is the number of transcription units. In S. pombe only two major RNAs are produced, one corresponds to the whole genome and the other to half of the genome (Fig. 1). These large primary transcripts from S. pombe are then processed, largely due to a tRNA punctuation system, in which the 5′ ends of mitochondrial RNAs are mostly produced by the excision of the tRNAs, or in two cases, directly by the initiation of transcription. In addition, most 3′ ends are generated after cleavage at a C-rich motif located shortly after the stop codon13 and recognized by a helicase and RNase complex.14 In one case, rnpB, the 3′ end is generated by tRNA excision and for the 21S rRNA, the 3′ processing signal is unknown. Consequently, there is little need for additional RNA-specific processing factors in S. pombe. However, as the amount of individual proteins that are ultimately derived from these polycistronic transcripts can vary by a factor of 10 (e.g., Atp9 and Atp6), factors involved in post-transcriptional regulation must play an important role in mitochondrial gene expression in S. pombe.
In S. cerevisiae, the mitochondrial genome is expressed as 11 transcription units (Fig. 1), and many more specific processing factors are involved because tRNA excision is not sufficient to produce the 5′ ends of mt-mRNAs. In addition, the factor(s) responsible for the generation of the 3′ ends of the mt-mRNAs by cleavage at the conserved dodecamer sequence are not known. So it is possible that in S. cerevisiae, some PPR proteins may play a role in processing in addition to RNase P (see below § 3.2).
Specific strategies used to delineate the role of a PPR protein in yeast
In yeast, the classic strategy to find the RNA target of a PPR protein is to analyze the consequences of a mutation in the corresponding gene (either a deletion or a point mutation) on OXPHOS complex formation. It is important to determine first whether the effect is general (e.g., a reduction in the de novo translation of all mt-mRNAs) or specific (e.g., the destabilization of a specific mt-mRNA). Unfortunately, this is easier said than done. A brief survey of the literature for the most studied PPR proteins will often suggest that they are involved in many different steps of mitochondrial gene expression. While it is quite possible for a single protein to be involved in more than one step, the extremely integrated nature of mitochondrial gene expression often means that the interpretation of the experimental data are difficult. More specific details will be presented later in this review, but it has long been accepted that mitochondrial ribosomes are associated with the inner mitochondrial membrane and some assembly factors to facilitate the translation and membrane insertion of the very hydrophobic mitochondrial proteins. There is also evidence that several mt-mRNA-specific translation factors are membrane associated and interact with the ribosome, and there is some evidence that the mitochondrial RNA polymerase interacts with the mitochondrial ribosome.15,16 Taken together, this suggests that essentially all the steps of mitochondrial gene expression occur at, or near, the inner membrane; thus mitochondrial gene expression is essentially a two-dimensional process. This has several important consequences, first there will be considerable steric constraints, for example, in such a system it is difficult to envisage several ribosomes simultaneously translating the same mt-mRNA, second, each step will probably be much slower than the cytoplasmic equivalent and third, disruption of any step in the process will often have secondary effects elsewhere and distinguishing between the primary and secondary effects can be complicated.
In yeast, general effects on the transcription, processing, stability or translation of mitochondrial RNAs can profoundly modify the mitochondrial physiology and the generation of the mitochondrial membrane potential, since all OXPHOS complexes will be affected. In S. cerevisiae, which is petite-positive, this will result in a partial to complete instability of the mitochondrial genome. To allow the study of such mutants, unstable mtDNA in S. cerevisiae can be counteracted, at least to some extent, by growing the cells on minimal medium and a high glucose concentration (10%), generating heterozygotes or conditional mutants, and using strains containing deleted mitochondrial genomes called rho-, which can be stabilized and are transcribed in a background where the full mtDNA would be lost. This is an important issue since several PPR proteins from S. cerevisiae appear to have a role that is sufficiently general to compromise mtDNA stability (Rpo41, Rpm2, Dmr1, Rmd9 and Rmd9L in combination with Rmd9).
In S. pombe, a petite-negative yeast, the absence of some mitochondrial functions leads to mtDNA loss which is lethal, this lethality can be suppressed by mutations in two unidentified genes, ptp1 and ptp2, which make S. pombe petite-positive.17 To date, only one PPR protein from S. pombe has such a general function, the mitochondrial RNA polymerase Rpo41, otherwise, all other deletions of genes encoding PPR proteins are viable in a classical wild-type background18 indicating that the corresponding factors all have either specific target(s), or only partial effects on general steps of mitochondrial gene expression.
For mutants which do not have a general effect on mitochondrial gene expression, the classical investigation involves the analysis of growth on non-fermentable medium and/or in the presence of drugs targeting the respiratory chain, the recording of cytochrome spectra, the radioactive labeling of newly synthesized mitochondrial proteins in presence of cycloheximide (which blocks cytoplasmic protein synthesis) and the analysis of mitochondrial RNA by northern blot and RT-PCR (for example, see refs. 18 and 19). In addition, in S. cerevisiae, the translation of individual mt-mRNAs can be monitored by growth analysis. 35S labeling or western blot analysis of strains containing translational fusions of mitochondrial genes with a mitochondrial reporter, ARG8m,20 integrated into the mtDNA after biolistic transformation of the fusion constructions.21 Finally, the powerful genetic systems of S. cerevisiae and S. pombe mean that screens to find suppressors of a respiratory deficient phenotype, either genetic or multi-copy suppressors, can be performed easily (see Table 2). This allows the identification of genetic partners, which may uncover direct physical interactions, or by-pass pathways.
Table 2. Genetic and physical interactions of S. cerevisiae PPR protein-coding genes .
| PPR gene | Variant, mutant, dosage or tag | Type of interaction | Interacting gene/factor | Function | Variant, mutant, dosage or tag | Refs |
|---|---|---|---|---|---|---|
|
AEP2 |
Mutation ND |
Suppressor of |
ATP9 |
Complex V F0 subunit |
+T at -87 in 5’ UTR |
44 |
| L413P |
Suppressed by |
ATP9 |
Complex V F0 subunit |
T-16C (5’ UTR) |
||
|
AEP3 |
Y305N |
Synthetic respiratory defect |
FMT1 |
Formyl methionine transferase |
∆fmt1 |
41 |
|
MIS1 |
Mt tetrahydrofolate synthase |
∆mis1 |
||||
| Aep3-HA |
Co-IP |
mtIF2 |
Mitochondrial initiation factor 2 |
Overproduced mtIF2 |
||
| |
Purified Aep3-MBP |
MBP pulldown assay |
Purified mtIF2 |
|||
|
ATP22 |
High copy |
Suppressor of |
ATP6 |
Complex V F0 subunit |
5’ UTR-atp6::ARG8m ∆atp12 |
70 |
| High copy |
Suppressor of |
ATP8 |
Complex V F0 subunit |
5’ UTR-atp8::ARG8m ∆atp12 |
||
|
∆atp22 |
Suppressed by |
ATP6 |
Complex V F0 subunit |
5’ UTR-COX1::ATP6 |
69 |
|
|
CBP1 |
G454(f.s.) |
Suppressor of |
COX18 |
Cox2 C-tail translocase |
∆cox18 |
94 |
| K205R, E241G, I249T, N281D, S289G, I293L, S330R, Q358K, Q358R, L489W, K532M, D533Y or I638M |
Suppressor of |
CYTb |
Complex III subunit |
C-944 to A or G-942 to U |
4849 |
|
|
∆cbp1 |
Partially suppressed by |
PET127 |
Processing degradation |
∆pet127 |
47 |
|
| Several cbp1-ts alleles |
Suppressed by |
SOC1/ CBT1 |
Processing/stabilization CYTb |
? |
46 |
|
| Cbp1-Bio |
Affinity puri-fication & 2D gels |
Pet309 |
COX1 stability and translation |
Pet309-HA |
50 |
|
|
DMR1 |
∆dmr1 |
Partially suppressed by |
PET127 |
Processing degradation |
∆pet127 |
31 |
| D785V |
Suppressed by |
RMD9 |
Processing stability |
High copy |
P.G. |
|
|
PET111 |
High copy |
Suppressor of |
COX2 |
Complex IV subunit |
- AUG->AUA - Many mutations lowering translation |
95, 96 64 |
| A652T (PET111-20) |
Suppressor of |
COX2 |
Complex IV subunit |
Deletion of base -24 in the 5’ UTR |
97 |
|
|
∆pet111 PET111 |
Suppressor of |
COX18 |
Cox2 C-tail translocase |
∆cox18 COX18 |
94 |
|
|
∆pet111 |
Suppressed by |
COX2 |
Complex IV subunit |
5’ UTR-COX3::COX2 |
63 |
|
| High copy |
Suppressed by |
COX1 |
Complex IV subunit |
5’ UTR-cox2::COX1 |
65 |
|
|
MSS51 |
COX1 translation |
High copy |
||||
|
PET309 |
COX1 translation |
High copy |
||||
| ∆37aa N-ter |
2 hybrid |
Pet54 Pet494 Nam1 |
COX3 translational activators Stability factor |
Full length ∆146aa N-ter Full length |
80 |
|
| Pet111-cMyc |
Co-IP |
Pet494 |
COX3 translation |
Pet494-HA |
80 |
|
|
PET309 |
L318Stop |
Suppressor of |
COX18 |
Cox2 C-tail translocase |
∆cox18 |
94 |
| S35Stop | ||||||
|
∆pet309 |
Suppressor of H202 sensitivity |
COX11 |
Copper assembly |
∆cox11 |
98 |
|
|
∆pet309 |
Suppressed by |
COX1 |
Complex IV subunit |
5’ UTR-CYTb::COX1 |
53 |
|
|
∆pet309 PET309 |
Synthetic [Arg] growth defect |
MSS51 |
COX1 translation |
∆mss51 MSS51 5’ UTR- cox1::ARG8m |
99 |
|
| Full length |
2 hybrid |
Pet54 |
COX3 translational activators |
Full length |
80 |
|
| Pet122 | ||||||
| Nam1 |
Stability factor |
|||||
| Pet309-cMyc |
Co-IP |
Pet494 |
COX3 translation |
Pet494-HA |
80 |
|
| Nam1 |
Stability factor |
Endogenous |
||||
| Pet309-HA |
Affinity purification & 2D gels |
Cbp1 |
COX1 stability & translation |
Cbp1-Bio |
50 |
|
| Endogenousa |
Pull down assay |
Rpo41 |
Mitochondrial transcription |
Rpo41-TAP |
16 |
|
|
RMD9 |
High copy |
Suppressor of |
DMR1 |
15S stability |
D785V |
P.G. |
| High copy |
Suppressor of |
OXA1 |
Insertion/assembly |
E65G-F229S |
19 |
|
| V363I |
Synthetic respiratory defect |
Rsm28 |
Ribosomal Protein (small subunit) |
∆rsm28 |
30 |
|
|
RPM2a |
High copy |
Suppressor of |
TOM40 |
Mitochondrial import |
ts, tom40-3 |
27 |
| Low copy of ∆736-1202 |
Suppressor of |
TOM40 |
Mitochondrial import |
ts, tom40-3 |
28 |
|
|
∆rpm2 |
Suppressed by |
PRE4 |
20S Proteasome subunit |
V12F |
100 |
|
|
∆aa146-246 |
Suppressed by |
DHH1 |
Dead-Box helicase |
High copy |
29 |
|
|
∆aa146-246 ∆rpm2 |
Suppressed by |
PAB1 |
Poly-A binding protein |
High copy |
29 |
|
|
∆rpm2 |
Suppressed by |
SEF1 |
Unknown |
High copy |
101 |
|
|
∆rpm2 |
Suppressed by |
UMP1 |
Chaperone 20S proteasome |
Stop at 10 Stop at 36 |
100 |
|
| ∆1-41 |
2 hybrid |
Dcp2 |
mRNA decapping enzyme |
? |
29 |
|
|
RPO41b |
E543G |
Suppressor of |
SUV3 |
RNA degradation |
Δsuv3 |
102 |
| V978F | ||||||
|
∆aa27-117 |
Suppressed by |
NAM1 |
Stability factor |
High copy V5-tagged |
81 |
|
| R129D |
Suppressed by |
SLS1 |
mRNA delivery/translation |
High copy |
103 |
|
| E543G |
Suppressor of |
SUV3 |
RNA degradation |
Δsuv3 |
102 |
|
| V978F | ||||||
| N-ter (27-392) |
2 hybrid | Nam1 | Stability factor | Full length | 81 |
Because of frequent high backgrounds of false-positives, data from high-throughput studies have not been included in this compilation of the genetic and physical interactions of the S. cerevisiae PPR proteins. aRpm2 is associated to a large so-called RMT (RNA processing, metabolism, translation) complex,104 which components are not listed here. bRpo41-TAP can be used to pull down a number of mitochondrial proteins not listed here, including Pet309, although this interaction is probably indirect through Nam1.16 a.a., amino-acid; f.s., frame shift; ND, not determined.
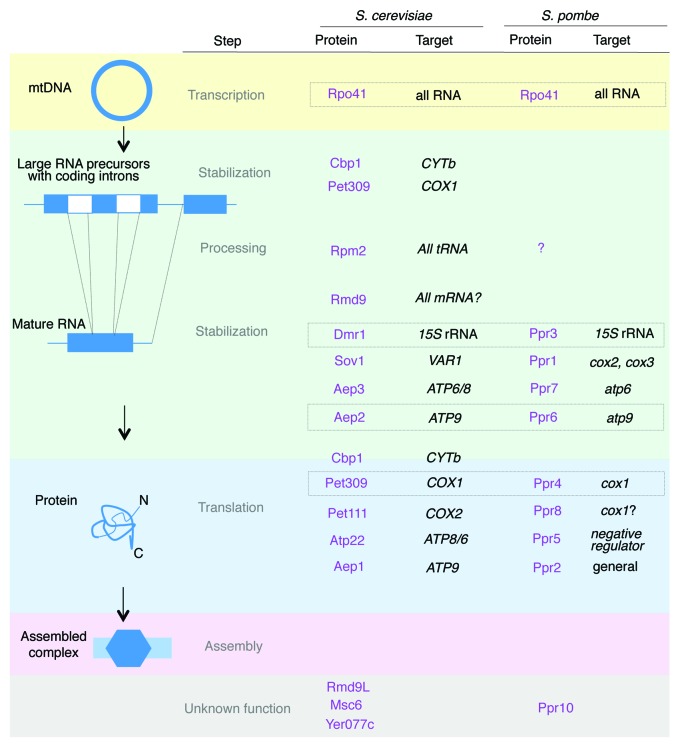
These types of studies have been undertaken for most of the genes encoding PPR proteins in S. cerevisiae and S. pombe and a summary of the data gathered so far is presented below; we have tried to classify all the genes according to the step of mitochondrial gene expression that they control (Fig. 2). As mentioned above, PPR proteins are most abundant and have been extensively studied in land plants. Most of the techniques used in yeasts are also used in plants, although the mitochondrial genome is essential in plants and the possibility of manipulating the mitochondrial genome is very restricted, especially compared with S. cerevisiae. One of the principal advantages of the yeast models is the facility with which genetic interactions can be investigated. This perhaps explains why in vitro experiments examining RNA-protein interactions are more advanced in plants. At present, direct biochemical analysis of the protein-RNA interactions is just starting for yeast PPR proteins and their targets, but this will obviously be an important step in the future characterization of the proteins.
Figure 2. Summary of the main functions of the S. cerevisiae and S. pombe PPR proteins. The different S. cerevisiae and S. pombe proteins are given in purple with their probable targets in black. Proposed S. cerevisiae and S. pombe orthologs are boxed. The different steps of OXPHOS complex formation are differentiated by colors, although they are generally interdependent. Some factors have been proposed to have dual functions and are marked at several steps; however, the data are discussed in detail in the text.
Principal Steps of Mitochondrial Gene Expression Affected by Yeast PPR Proteins
It is generally accepted that the abundance of PPR proteins in land plants is largely explained by the rich RNA metabolism of their mitochondria and chloroplasts. In yeast, where there is no poly-A addition, editing or trans-splicing in mitochondria and the coding capacity of the mitochondrial genome is limited, only a small number of PPR proteins are present. Despite the presence of mitochondrial introns in yeast, none of the yeast PPR proteins are directly required for splicing (see the discussion concerning Dmr1 § 3.d). In fact, most of the yeast PPR proteins appear to be involved in stability and/or translation, most often of specific mt-mRNAs. Thus, the majority of yeast PPR proteins are regulators of mitochondrial gene expression and only two have been clearly demonstrated to have a general role in mitochondrial RNA formation: the mitochondrial RNA polymerase and RNase P. This section will classify step by step the functions of the different budding and fission yeast PPR proteins.
Transcription requires the mitochondrial polymerase, a PPR protein with only a few motifs
The mitochondrial RNA polymerase is of the viral T7 type (reviewed in ref. 22), it is a multi-domain polypeptide that contains two to several PPR domains, depending on the organism, located in an N-terminal extension of non-viral origin. The structure of the human mitochondrial RNA polymerase (mtRNAP or POLRMT) has been determined and this is the first crystal structure of a PPR protein.23 However, POLRMT is not a typical PPR protein, so it is not possible to extrapolate the information from this structure to other PPR proteins and the function of POLRMT’s two PPR motifs is not known. One possibility is that these PPR domains could help to channel the newly synthesized RNA strand, but they have also been proposed to be involved in protein-protein interactions like TPR domains. The SCIPHER algorithm predicts two PPR motifs for the S. pombe Rpo41 and five for the S. cerevisiae protein. As the function of these motifs is unclear it is difficult to speculate on significance in difference in the number of motifs. However, whatever their function, these motifs appear to be important as they have been conserved during evolution from yeasts to humans and a form of POLRMT with an N-terminal deletion that removes the PPR motifs can no longer initiate transcription in vitro.23
As mentioned above, the PPR motifs are located in the N-terminal region of the mitochondrial RNA polymerase. In S. cerevisiae, mutations in this domain strongly decrease the de novo synthesis of mitochondrially-encoded proteins. In addition, two-hybrid studies have shown that this domain is able to interact with the mtRNA stability factor Nam1 (Table 2). Overexpression of NAM1 can suppress a partial deletion in the N-terminal domain of Rpo41 and overexpression of SLS1, a membrane protein required for mitochondrial translation, can also compensate for a point mutation in the N terminus (Table 2). These data suggest that the N-terminal domain of Rpo41 could couple several factors and activities involved in mitochondrial gene expression directly to the transcription machinery, in order to coordinate transcription and translation at specific sites on the inner membrane; whether the PPR motifs are important for this remains to be elucidated.
tRNA excision is mediated by the PPR riboprotein complex RNase P
RNase P is the enzyme that excises mitochondrial tRNAs from their precursor transcripts. As well as liberating tRNAs from the primary mitochondrial transcripts this often generates the extremities of other RNAs present on the same co-transcript, thus RNase P plays a crucial and general role in mitochondrial RNA processing.
In budding yeast, like most other organisms, RNase P is a riboprotein complex, composed of a mitochondrially encoded RNA called 9S or RPM1, and a nuclear-encoded protein, Rpm2. Recently, it has been shown that the mitochondrial RNase P from humans and plants are protein complexes that contain at least one PPR protein, but have no RNA component; the structure of the A. thaliana PRORP1 has been solved24 and in this case the PPR motifs are involved in substrate binding. However, when RNA is present in the enzyme, it is the RNA that is responsible for the catalytic activity.25 In addition to mt-tRNA processing, budding yeast RNase P is responsible for the maturation of its own RNA component.26 Rpm2 is a PPR protein, but it is not known if the function of the PPR motifs it to bind the catalytic 9S RNA and/or the pre-tRNA substrates. Since Rpm2 is also required for growth on fermentable medium, it must have another function other than the processing of mitochondrial RNA.27 Rpm2 has been shown to be present in the nucleus and cytoplasmic P bodies28,29; its precise function is not clear, but it seems to be involved in mRNA transcription/turn-over, in particular for components of the mitochondrial import system (see Table 2). Thus, Rpm2 is a rare PPR protein that has a role outside an organelle, although this function seems to be related to the function of the organelle.
Surprisingly, no clear equivalent of Rpm2 has been detected in silico in S. pombe, but a possible homolog of the 9S RNA, rnpB, is present in the mitochondrial genome.12 Thus, we would expect to find a PPR protein that would interact with rnpB to form the S. pombe RNase P. The newly identified Ppr10 is a possible candidate, it is a much smaller protein than Rpm2 from S. cerevisiae, but this might be because it is only involved in mitochondrial RNA processing. Alternatively, another as yet unidentified PPR protein could be the bona fide partner of rnpB.
The stability and processing of several mt-RNAs are dependent on Rmd9
In S. cerevisiae, Rmd9 encodes a PPR protein and the gene was isolated in two independent screens: as a multi-copy suppressor of point mutations in Oxa1, which encodes an inner membrane insertase for newly translated mitochondrial proteins (Table 219), and as a synthetic respiratory defective mutant of the deletion of a small ribosomal subunit gene, Rsm28 (∆rsm28 alone only leads to a partial respiratory deficiency) (Table 230). It was also shown that a small fraction of Rmd9 co-sediments with the small subunit of the mitochondrial ribosome in sucrose gradients. Thus, Rmd9 interacts with the ribosome but is not a ribosomal subunit per se.30
The loss of Rmd9 affects many mitochondrial RNAs, which are either highly unstable (CYTb, COX2, COX3) or not correctly processed (COX1, ATP8/6, ATP9); thus, all the mRNAs encoding respiratory complex subunits appear strongly affected in some way.19 However, CYTb is transcribed as a co-transcript with the Glu-tRNA (Fig. 1) and the level of this mitochondrial tRNA in a ∆rmd9 strain is normal, indicating that transcription per se is not affected. During respiratory growth, the ∆rmd9 strain produces cytoplasmic petites, possibly because of the reduced level of the small mito-ribosomal rRNA. Rmd9 is a peripheral membrane protein facing the mitochondrial matrix and it has a homolog, Rmd9L, which shows a stronger association to the membrane.19 As yet, no clear phenotype has been associated with the deletion of RMD9L, except that when combined with ∆rmd9, it increases the instability of the mtDNA. Overexpression of Rmd9 increases the steady-state level of the mitochondrial RNAs encoding respiratory complex subunits in an oxa1 mutant (see Table 2). Thus, the insertion defect caused by the oxa1 mutation is probably compensated for by an elevated level of mt-mRNAs due to Rmd9 overexpression, possibly leading to an increase in translation facilitating insertion in the presence of the oxa1 mutation.
Rmd9 is one of the rare PPR proteins that has a very general effect, it may be involved in stabilizing/protecting mt-mRNAs at sites specific for the translation of respiratory subunits via an interaction with the small subunit of the ribosome and possibly with other factors.
A conserved PPR protein is a rRNA stability factor
In S. cerevisiae, Dmr1 was one of the first three yeast proteins to be classified as PPR proteins, together with Pet309 and Aep3 (see § 1). Cells devoid of Dmr1 rapidly and irreversibly accumulate deletions of the mtDNA, whatever the mitochondrial intron content, suggesting that Dmr1 is involved in a general step of mitochondrial gene expression. Using a rho- mtDNA that can be stably maintained in ∆dmr1 cells, it has been shown that the main target of Dmr1 is the 15S rRNA, which is unstable in the absence of Dmr1 and shows discrete bands of higher mobility probably corresponding to degradation intermediates.31 This suggests that the 15S rRNA is stabilized by Dmr1 binding. In vitro, it has been found that purified Dmr1 binds to several sequences derived from the 15S rRNA.
It has also been reported that Dmr1 (also called Ccm1) could be a splicing factor for the fourth introns of the CYTb and COX1 genes.32,33 However, it is well documented that the splicing of these introns is dependent on the translation of the maturase encoded by the fourth intron of the CYTb gene,34 which complicates the interpretation of the Dmr1 data. Upon extinction of DMR1, translation will be impaired leading to a progressive loss of the mitochondrial genome (accumulation of rho- and rho° cells). Concomitant with this will be a lack of translation of the intron-encoded maturases leading to the accumulation of some precursors. Also, transcription, but not translation, still occurs in rho- cells, and some of the rho- cells generated upon the extinction of DMR1 will retain parts of the genome that encode the introns bI4 and aI4, and the surrounding sequences. Thus, we would expect intron-containing precursors to be detectable by RT-PCR. Therefore, all the consequences of DMR1 loss described by32,33 can be explained by its activity as a stability factor of the 15S rRNA without the need to invoke any additional activity as a specific splicing factor.
In S. pombe, Dmr1 has a clear sequence homolog, Ppr3. The major effect of Ppr3 is on the stability of the 15S rRNA.18 A reduction in the steady-state level of the 15S rRNA is observed in a ∆ppr3 strain, associated with the appearance of a shorter discrete band, which is probably a degradation product, similar to that seen in the S. cerevisiae ∆dmr1 strain. At 36°C, the deletion of ppr3 is lethal, suggesting that all the 15S rRNA must then become fully unstable. Thus, Dmr1 and Ppr3 appear to be bona fide orthologs, required for the 15S rRNA stability.
PTCD3 is required for the stability of the 12S rRNA35 and, thus, appears to be the human mitochondrial Dmr1 and Ppr3 ortholog (and not a Pet309 homolog, as proposed in the summary of ref. 36); PTCD3 has been identified as a small ribosomal subunit that cross-links to the initiation factor IF3.36 It will be interesting to know whether interactions can be found between the yeast Dmr1 or Ppr3 and the corresponding yeast IF3 factors, and whether Dmr1 and Ppr3 co-sediment with the small ribosomal subunit in sucrose gradients of mitochondrial extracts.
Several PPR proteins act mainly as stability, and/or translation factors
When the absence of a PPR protein leads to both the destabilization of a given mt-mRNA and a defect in the accumulation of the corresponding protein, further experiments are necessary to determine whether (1) a primary RNA stability defect causes a secondary translation deficiency, (2) a primary defect in translation destabilizes the mt-mRNA or (3) the factor has a dual role in stability and in translation, as shown for example for PPR10 in Arabidopsis,37 although in this case the RNA stability and translation effects concern different chloroplastic mRNAs. For several of the yeast PPR proteins whose deletion has such a dual effect, this analysis has not yet been conducted, and for this review they will be classified as stability and/or translation factors. To date, six factors belong to this class: S. cerevisiae Sov1, Aep3 and Aep2 and S. pombe Ppr1, Ppr6 and Ppr7; the available data suggest that Aep2 and Ppr6 are orthologs.
Sov1
The original sov1 (synthesis of Var1) mutants were isolated in a screen for mutants unable to synthesize a mitochondrial reporter protein, Arg8m, when it was fused to the VAR1 5′ UTR.38 In this screen, the Var1 ribosomal protein was encoded in the nucleus by a relocated construct producing a mitochondrially targeted version of Var1 that could replace the endogenous mitochondrial Var1 and maintain mitochondrial translation.39 However, the sov1 mutants were not only unable to synthesize Arg8m from the VAR1 5′ UTR, they also failed to accumulate the chimeric VAR1::ARG8m mRNA as well as the wild-type VAR1 mt-mRNA. In these strains, the tRNASer-VAR1 precursor level was wild-type, so Sov1 appears to be required for stability of the mature VAR1 mt-mRNA rather than processing of the precursor. Finally, overexpression of Sov1 only slightly stabilizes the VAR1 and VAR1::ARG8m mt-mRNAs but leads to a significant overproduction of the Var1 protein, suggesting that Sov1 could have an effect on Var1 that is independent of the stabilization of the VAR1 mt-mRNA.
Aep3
Aep3 is a peripheral membrane protein facing the mitochondrial matrix.40 A deletion of the gene prevents respiratory growth and partly destabilizes the mtDNA, leading to an accumulation of about 50% of rho- cells. Further molecular analysis showed that Aep3 targets the ATP8/6 bi-cistronic transcript.40 The ATP8/6 mt-mRNA is transcribed together with COX1 upstream and ENS2 downstream (Fig. 1), and its 5′ end is processed in two steps to generate a long and a short transcript. In a ∆aep3 mitochondrial intron-less strain, the short transcript was undetectable while the long transcript was reduced by 60%, and the COX1-ATP8/6 precursor slightly accumulated. 35S labeling of de novo mitochondrial translation products showed that the Atp6 and Atp8 proteins were almost undetectable. Taken together, these data suggested that Aep3 is primarily involved in the stability of the ATP8/6 transcript, although an effect on processing and on translation could not be excluded.40
In addition, it has been reported that Aep3 is an accessory factor in mitochondrial translation initiation, which interacts physically with the initiation factor mt-IF2, i.e. the main GTPase that brings the initiator fMet-mt-tRNA to the mitochondrial ribosome. Aep3 would allow the use of unformylated methionine as initiator amino acid by promoting the binding of unformylated Met-mt-tRNA to mt-IF2.41 Whether this additional function is linked to the presence of the PPR motifs or rather to an additional domain of the protein has not yet been investigated.
Aep2/Ppr6
Mutants of Aep2 (also known as Atp13) were devoid of subunit 9 of ATP synthase and showed strongly decreased level of mature mt-mRNA but normal to higher steady-state levels of the precursor RNA. Thus, it has been proposed that Aep2 could be involved in the processing and/or stability and/or translation of ATP9.42,43
A suppressor analysis has reinforced the relationship between Aep2 and ATP9, as a point mutation, Aep2-P413L, can be suppressed by a T-to-C transition in the ATP9 5′ UTR, at position -16 (Table 244). Even though current HMM profiles do not detect a PPR motif around residue 413,4 secondary structure modeling indicates that it is in a region containing multiple tandem α helices, superficially resembling PPR repeats, which may be extremely divergent variants of the motif. This suggests a hypothesis where the Aep2-P413L mutation changes the binding specificity of Aep2 and that the T-to-C transition is a compensatory mutation that restores binding. If this were the case, it would tend to argue in favor of a role for Aep2 in translation as the T-to-C transition is close to the ATP9 translation start and far from the processing sites. However, in the absence of experimental data, this remains speculative. In addition, the suppression of an ATP9 5′ UTR mutation at position -87 by a mutation in Aep2 has been cited in a discussion but never fully documented (Table 244). Thus, while it seems highly probable that Aep2 interacts with the ATP9 5′ UTR, the precise role of this interaction remains uncertain.
Aep2 appears conserved in other yeasts including S. pombe,4 and its fission yeast homolog, Ppr6, is similarly important for the stability of the atp9 mRNA and for Atp9 synthesis.18 As in S. cerevisiae, whether both defects reflect a simultaneous a dual role of Ppr6 or are a consequence of one another is not yet understood.
Finally, AEP2 is transcribed as two transcripts using different polyadenylation sites, one within the ORF and one after the end of the ORF, and the shorter transcript, which is unable to produce a functional Aep2 protein, is more abundant during the switch to non-fermentable conditions.45 The function of this surprising modulation of expression is not understood but might somehow regulate the adaptation to respiratory conditions. A similar regulation exists for another PPR protein, Cbp1 (see § 3. f. 1).
Ppr7
Ppr7 is the second fission yeast PPR protein that is involved in the biogenesis of ATP synthase, as shown by the sensitivity of ∆ppr7 cells to antimycin. In mutants lacking Ppr7, the main effect on RNA was a strong decrease of the atp6 mRNA.18 In S. cerevisiae, ATP synthase F0 mutants often have secondary effects on protein synthesis and destabilize the mtDNA. In the absence of S. pombe, Ppr7 de novo mitochondrial translation is also severely reduced. In particular, Atp6 is lacking and another faster migrating protein appears, but it is not known if this is related to Atp6. The simplest synthesis of these data are that the primary defect in ∆ppr7 cells is a reduction in the level of the atp6 mRNA, which leads to various secondary effects due to the complex V deficiency. However, at present, a direct role for Ppr7 in translation and/or the stability of other mt-mRNAs cannot be excluded.
Ppr1
The ∆ppr1 mutant compromises the stability of the cox2 and cox3 mRNAs, which become barely detectable, and the Cox2 and Cox3 proteins are not detected by 35S labeling as well as western blot for Cox2 18. Thus, even though Ppr1 is definitely required for the stability of two mt-mRNAs, an effect in translation cannot be excluded. Interestingly, cox3 and cox2 are located at opposite ends of the S. pombe minor transcript (cox3 at the beginning and cox2 at the end), which corresponds to about half of the mitochondrial genome (Fig. 1). A tempting hypothesis is that Ppr1 stabilizes the extremities of this large co-transcript. However, RNA-circularization experiments followed by PCR amplification and sequencing of the junction do not reveal any obvious degradation events at the 5′ end of the cox3 mRNA or the 3′ end of cox2 mRNA (C.J.H., unpublished results). Also, the 3′ UTRs of fission yeast mt-mRNAs are very short and cannot include obvious binding sites for a specific factor, since they already contain a C-rich motif that has been proposed to be the target of a general 3′ processing complex.14 The Ppr1 binding sites within the cox2 and cox3 mRNAs might correspond to a common or rather similar sequence and the identification of this sequence would be a significant step toward understanding the binding parameters of fission yeast PPR proteins.
Two PPR proteins may affect both the translation and stability of their target RNA
Cbp1
In S. cerevisiae, the CYTb mt-mRNA is produced as a co-transcript with the Glu-tRNA (Fig. 1) as mentioned in §3.c. First the Glu-tRNA is processed to yield a precursor CYTb mRNA, this is further processed at the 5′ end to produce the mature CYTb 5′ UTR of 954 or 955 nucleotides. A mutant lacking Cbp1 has been implicated in all these steps apart from the primary excision of the tRNA Glu,46 and is proposed to protect the CYTb mRNA and promote its processing. Further evidence supporting a role for Cbp1 in the stability of the CYTb mRNA is that thermosensitive mutants of CBP1 are suppressed by a mutation in the SOC1/CBT1 gene, involved in the stability of the CYTb mature mRNA,46 and the combination of the deletion CBP1 with that of PET127, involved in mitochondrial RNA processing and decay, stabilizes the mature CYTb mRNA (Table 247). In this latter case, the mature CYTb mRNA is stabilized, but without restoring synthesis of the Cytb protein. Thus, in addition to its stability function, Cbp1 plays a role in CYTb translation, at least in a pet127-deficient background.
A 64 bp sequence from nucleotide -961 to -898 of the precursor CYTb mRNA appears to be sufficient for Cbp1-mediated stabilization of the CYTb mRNA. Within this otherwise AU-rich element, an 11 nucleotide sequence (-948 to -938) is particularly important, especially the bases CCG (-944 to -942).48 Mutation of in any of these three nucleotides destabilizes the CYTb transcripts. However, mutations of the CCG sequence to ACG or CCU are suppressed by many single substitutions in Cbp1 (Table 2)48,49). All of these mutations fall within the PPR motifs predicted by ref. 4, but they occur between residues 205 and 638 of the protein so they are not restricted to adjacent PPR motifs. Thus, the position and the number of the amino acid substitutions means that they cannot be simple compensatory mutations in the PPR motifs that “read” the CCG sequence. It is always possible that they are responsible for long-range modifications of the protein structure, but this would seem unusual in an RNA binding protein with a repeated domain structure.
The role of Cbp1 appears to be complicated; it is clearly required for the stability of the CYTb mRNA and appears to have some function in translation. However, it is not clear if this is a direct role in translation per se, or an indirect role, possibly via interactions with other translational activators. This would be consistent with the observation that Cbp1 interacts with the COX1 translational activator Pet309 (Table 250).
Like AEP2, CBP1 is transcribed as a full-length transcript able to encode the normal Cbp1 protein, and a truncated transcript that cannot produce Cbp1.51 The level of the short transcript increases over the full-length transcript during the switch to a non-fermentable carbon source, while the overall transcript level remains the same.45 As for AEP2, this regulation is surprising and not fully understood because if the protein level follows the transcript level the cell would appear to be reducing the production of the full-length protein at the very moment it was needed.45 It would be interesting to know if this type of regulation is unique to CBP1 and AEP2, or if it also occurs for other PPR proteins.
Pet309 and Ppr4
The pet309 mutant was initially characterized by a striking deficiency of both mature and precursor COX1 RNA, encoding subunit 1 of complex IV. The maturation of the COX1 mRNA is complex in S. cerevisiae, because it is derived from a primary transcript that also contains ATP8/6 and ENS2 (Fig. 1). In addition, the large COX1-ATP8/6-ENS2 co-transcript is even longer if COX1 contains introns (up to seven in some mitochondrial genomes11). Finally, some of these COX1 introns are involved in their own excision as they encode maturases, so translation is necessary for pre-COX1 splicing.52
An analysis of pre-COX1 mRNA from strains with different intron content shows that in the absence of Pet309 the stability of the COX1 mRNA and of the large precursor co-transcript varied inversely with the number of COX1 introns.53 Thus, Pet309 probably plays a role in the stability of the intron-containing COX1-ATP8/6-ENS2 precursor, although an involvement in transcription has not been formerly eliminated.53 Crucially, a strain devoid of mitochondrial introns accumulates the mature COX1 mRNA but is unable to translate this mRNA in the absence of Pet309. In such an intron-less context, translation of COX1 can be restored by a mitochondrial rearrangement placing the 5′ UTR of the CYTb gene in front of the COX1 ORF. This rearrangement places COX1 translation under the control of CYTb translational activators, and coexists in a heteroplasmic state with the normal wild-type mtDNA (Table 253). The interpretation of such rearrangements is that they bypass the requirement for Pet309 by replacing its RNA-binding site by that of another translational activator. Taken together, these results clearly demonstrate that Pet309 is required for the translation of the COX1 mRNA.
The requirement of Pet309 for the translation of the COX1 mRNA is further demonstrated by experiments where a partial deletion of some of the predicted Pet309 motifs abolished COX1 translation.54 However, if the Pet309 deletion constructions were overexpressed, the COX1 transcripts accumulated. In general, the interpretation of the data concerning Pet309 and the stabilization of the large COX1 precursors is complicated, because with the combinations of introns that were examined it is possible that the absence of synthesis of the Cox1 maturase(s) is the primary cause of the destabilization.53 Pet309 binds to the 5′ UTR of the large COX1-ATP8/6-ENS2 co-transcript, in the absence of Pet309 many of the introns in the COX1 pre-mRNA will not be spliced resulting in the accumulation of large transcripts. It is not unreasonable to assume that large transcripts will be less stable than small transcripts, so in the absence of Pet309 the production of mature ATP8/6 and ENS2 mRNAs and the COX1 pre-mRNA will reflect the equilibrium between degradation and processing. Thus, while it is clear that Pet309 plays an active, primary role in the translation of the COX1 mRNA, the stabilizing effect of Pet309 on the long co-transcripts might be considered secondary.
In S. pombe, Ppr4 is the ortholog of Pet309;18 deletion of the gene prevents Cox1 synthesis even in an intron-less background whereas mRNA accumulation is not significantly affected. In an intron containing background (i.e. with two introns in COX1), the COX1 precursor RNA is detected albeit at a low level. Thus, whereas Ppr4 is not absolutely required to accumulate intron-containing cox1 mRNA in S. pombe, its absence nevertheless lowers its stability. Clearly there is no role in transcription since the other individual mRNAs derived from the same large co-transcript are not affected.
An ortholog of Pet309 has also been studied in Neurospora crassa (Cya-555), and in humans, LRPPRC was initially proposed to be a homolog of Pet309 since mutations in patients affected by the French-Canadian Leigh syndrome led to a lower synthesis of the COXI protein.56 However, COXIII synthesis is also affected57 and subsequent studies have shown that LRPPRC is actually a much more general factor than previously thought, with multiple functions (refs. 58 and 59 and references within), and that the specific effect observed on COXI and COXIII reflects an acute sensitivity of these two mRNAs to LRPPRC mutations. It should be also noted, that due to the high divergence and repetitive nature of the PPR sequences, their orthology cannot be reliably inferred based on sequence similarity in organisms as distant as yeasts and vertebrates (see Section 5 below).
The example of Pet309 and its orthologs clearly illustrates the difficulty of assigning primary function in such an integrated system as mitochondrial gene expression, but it could be argued that distinctions of primary and secondary function are of more importance to mitochondrial research workers than they are to mitochondria.
Several PPR proteins function uniquely in translation
Long 5′ UTRs containing sites recognized by proteins that are specific translation activators are a unique feature of the mitochondrial genetic system of S. cerevisiae and other closely related species. Some of these budding yeast proteins that are known, or proposed, to be mitochondrial mRNA-specific translational activators, like Pet309, but with no effect on the stability of their target mt-mRNA are also PPR proteins: Pet111 for COX2, Atp22 for ATP8/6 and Aep1 for ATP9.
mt-mRNA-specific translation factors: Pet111, Atp22, Aep1
Pet111
Pet111, with Pet309, is the best studied of the S. cerevisiae PPR proteins acting as translational activators.60 Like Pet309, it is absolutely required for the translation of its target mRNA, COX2, but a noticeable difference with Pet309 is that Pet111 does not appear to stabilize the COX2 mRNA. However, unlike COX1, COX2 is a short transcript that never contains introns in S. cerevisiae. In addition, Pet111 function appears less conserved since homologs can be found only in closely related yeasts.61
As is usual for S. cerevisiae mitochondrial translational activators, Pet111 levels in the cell are limiting, as shown by quantitative immunodetection studies62 and overexpression analyses.63-65 Interestingly, the PET111 mRNA has an extended 5′ UTR, which contains four short ORFs that overlap with each other and with the PET111 ORF.66 It is possible that these ORFs participate in the regulation of the translation of the PET111 mRNA. Thus, since their quantity in the cell is limiting, S. cerevisiae translational activators can also be viewed as means to tightly control the translation of their target mRNAs, which are usually abundantly transcribed and poorly regulated at the level of transcription. In addition, Pet111 is associated with the membrane and interacts with other translational activators and post-transcriptional factors, so it could participate in the spatial localization of Cox subunit synthesis at specific sites of the inner membrane (see § 4).
The construction of rearranged mitochondrial genes introduced into mitochondria by biolistic transformation has shown that Pet111 acts solely on the short 54nt 5′ UTR of COX263: when most of the COX3-5′ UTR is fused to the COX2 ORF, translation becomes independent of Pet111, whereas replacement of the COX3 5′ UTR with the 54nt COX2 5′ UTR confers Pet111-dependent translational activation to the COX3 mRNA (see Table 2). Further evidence for the interaction between the COX2 5′ UTR and Pet111 has been obtained through mutant and suppressor analyses. For example, the isolation of suppressors of cox2-11, a one base deletion at position -24 of the COX2 5′ UTR, identified a dominant variant called Pet111-20, carrying the substitution A652T, which is located in the middle of one of the PPR motifs listed by.4 Pet111-20 activated COX2 translation more efficiently than overexpressed wild type Pet111, irrespective of whether the COX2 5′ UTR was wild type, or carried the original cox2-11 base deletion (Table 263); thus, PET111-20 is probably not a direct compensatory mutation of cox2-11 but rather a variant of Pet111 that increases binding to the wild type RNA target within COX2.
Extensive analysis of the COX2 5′ UTR has shown that the region -54 to -16 is sufficient for a good level of translational activation by Pet111, and that this domain contains a stem-loop structure, important for Pet111 binding.67 It seems unlikely that Pet111 recognizes the loop sequence itself since some mutations that profoundly alter this structure still allow translational activation by Pet111. In addition, several nucleotides surrounding the stem-loop structure appear crucial for translational activation, suggesting that they might belong to the target sequence. Thus, numerous data have already been gathered to delineate the Pet111 recognition site and in the future it would be interesting to complete this study and correlate it to a thorough analysis of Pet111 PPR motifs in order to try and elucidate the binding code of this yeast PPR protein.
Atp22
Atp22 was initially described to be necessary at a post-translational level for the assembly of the F0 section of ATP synthase,68 but it is now known that the mutant used in this study was still partially functional. The complete loss of Atp22 function prevented the synthesis of Atp6, although the mRNA was still present.69 A bi-cistronic mRNA, ATP8/6, encodes Atp6 and the ∆atp22 strain is still able to produce Atp8; thus, Atp22 is a translational activator that is specific for the second ORF of a bi-cistronic transcript.
Three mitochondrial revertants of ∆atp22 all corresponded to a rearrangement that fuses the 5′ UTR, first exon and first intron of COX1 to the fourth codon of ATP6; this rearranged genome coexists with a normal wild-type mitochondrial genome (Table 269). Thus, the predicted protein encoded by the rearranged DNA should be 6 kDa longer than the wild-type Atp6. However, western blot and 35S labeling revealed an apparently wild-type Atp6 in the revertants. Since Atp6 is formed as a precursor containing a 10 amino-acid pre-sequence that is removed upon insertion, the fusion protein synthesized in the revertant appeared to be efficiently processed to yield the wild-type mature Atp6. The ability to bypass the requirement for Atp22 by replacing the ATP6 5′ UTR with that of another gene strongly suggests that like other classical translational activators, Atp22 target sequence is located within 5′ UTR of the gene.
Further work showed that Atp22 plays a role in ATP synthase biogenesis.70 It had been noticed that several mutants that compromise the assembly of the F1 section of the ATP synthase also fail to synthesize Atp6 and Atp8, or a reporter gene replacing the ATP6 or ATP8 ORFs, despite normal mRNA levels. Controlling the synthesis of Atp6 appears to be important when F1 assembly is impaired, probably to prevent the formation of an Atp6-Atp9 complex that would dissipate the membrane potential of mitochondria. Overexpression of ATP22 could relieve the translation inhibition linked to the F1 assembly defect, not only for ATP6, but also for ATP8, albeit to a lower extent.70 Accordingly, ATP22 overexpression stimulated Arg+ growth of an 5′ UTR-ATP6::ARG8m mutant but also of a 5′ UTR-ATP8::ARG8m fusion, but again to a lower extent. Thus, Atp22 appears to be an efficient translational activator of ATP6 that can have a weak effect on the translation of ATP8 when overexpressed. Whether Atp22 plays a specific role in this feedback effect of sub complex F1 assembly onto ATP6 and ATP8 translation, like Mss51 for COX171 or Cbp6 and Cbp3 for CYTb72 has not been elucidated.
Aep1
AEP1 has also been studied and published under the name NCA1. In both cases, temperature conditional mutants were studied, and both types of mutants failed to synthesize Atp9. However, although the nca1 mutant did not accumulate the ATP9 mRNA,73 the temperature-conditional aep1-ts1860 contained an almost normal level of mature ATP9 mRNA.42,74 Thus, the primary role of Aep1 appears to be in ATP9 translation, although it is possible that there is some role in ATP9 mRNA stability.
A general translation factor
Ppr2 is one of the most enigmatic of the S. pombe PPR proteins since no effect of its absence could be observed on mitochondrial mRNAs or rRNAs; the Ppr2 mutant is respiratory deficient but shows some leaky growth on non-fermentable medium upon extended incubation.18 De novo synthesis of all the mitochondrially encoded proteins is very low in a deleted strain, although all the proteins were in fact still produced and detectable after long exposure of the labeled protein gels. The proteins produced might be unstable as Cytb and Cox2 are under the detection limit in western blots performed on purified mitochondria. The current hypothesis is that Ppr2 might be associated with the small subunit of the ribosome (L. Dujeancourt, unpublished data), and could be involved in translation efficiency, or the initiation of translation.
Translation inhibitor
Ppr5 is perhaps the most distinct yeast PPR protein, since its deletion stimulates respiratory growth, whereas the lack of most other PPR proteins is deleterious to respiratory growth. At the molecular level, the deletion of Ppr5 increases the de novo synthesis of all mitochondrially encoded polypeptides. Conversely, Ppr5 overexpression decreases the de novo synthesis of mitochondrial proteins. No obvious effect can be seen on mitochondrial RNA. It has thus been proposed that Ppr5 is a negative regulator of mitochondrial translation.18 Interestingly, Ppr5 appears to be more expressed under fermentation, as shown by analysis of a Ppr5-YFP fusion (C.J.H., unpublished data).
This type of negative regulator has not been reported in S. cerevisiae, but a PPR protein, PTCD1, which is a negative regulator has been identified in humans,75 although at present there is no reason to believe that Ppr5 and PTCD1 have the same mechanism of action. PTCD1 has been proposed to lower the level of both Leu-mt-tRNAs, which are limiting, thus providing a very efficient downregulation of translation in human mitochondria.75 The fact that human and S. pombe cells have both recruited a PPR protein as a negative regulator is consistent with the hypothesis that these two organisms show a similar regulation of mitochondrial RNA metabolism, which can be correlated with the fact that the mt-mRNAs of both organisms have short, or absent 5′ UTRs. Thus, the specific translational activators targeting the 5′ UTR of each of the mt-mRNAs in S. cerevisiae might be replaced at least in part by general translation inhibitors like PTCD1 and Ppr5.
So far, the target of Ppr5 is unknown, and the S. pombe mitochondrial Leu-mt-tRNA level did not seem to be affected by the absence of Ppr5.18 Thus, it will be very interesting to find the molecular target of Ppr5 to understand the mechanism of its action. Ppr5 is the S. pombe PPR protein with the highest number of PPR motifs, so it could be expected to recognize the longest target sequence or to have more than one.
Four yeast PPR proteins have still an unknown function: YER077c, Msc6, Ppr10, Rmd9L
The functions of YER077c, Msc6, Rmd9L and Ppr10 are unknown; however, all are mitochondrial proteins. The msc6 mutant is defective in directing nuclear meiotic recombination,76 but the protein was found in mitochondria in high-throughput and proteomic studies77-79 and msc6Δ strains display reduced respiratory growth and mtDNA stability (P.G. laboratory, unpublished data). Deletion of RMD9L in combination with ∆rmd9 further destabilizes the mtDNA as noted above (§ 3.c). Recent work in our laboratories has shown that a ∆yer077c mutant is respiratory deficient and tends to lose mitochondrial DNA, and that a ∆ppr10 mutant is viable but exhibits a pleiotropic phenotype, with a spectra devoid of cytochrome b (complex III) and cytochrome aa3 (complex IV) and a sensitivity to antimycin A on glucose, reflecting an ATP synthase defect (complex V). This suggests that most of these genes probably fulfill a general function, or have a specific target with a general function.
Interactions, Genetic and Physical Partners of Yeast PPR Proteins
Many of the S. cerevisiae PPR proteins have been implicated in genetic or high-copy suppressor relationships, in two-hybrid interactions and in direct physical interactions. Most of these data, some of which have been cited in the text above, are listed in Table 2. In this section we will describe the existence of numerous indications that a complex set of interactions between regulatory factors acting at a general or specific level allows a tight coupling and spatial localization of the different steps of mitochondrial gene expression up to protein insertion and complex assembly. Several PPR proteins play key roles in this system.
In S. cerevisiae, Pet111 and Pet309, which are respectively translation factors for COX1 and COX2, have been shown by 2-hybrid and/or co-immunoprecipitation to interact with translation factors for the COX3 mRNA (Table 280). This suggests that translational activators of all the mitochondrially encoded complex IV subunits are organized as a unit at the surface of the inner membrane. As translational activators are present at limiting levels in mitochondria, this implies that there are a restricted number of foci where there is an integrated production of mitochondrially encoded complex IV subunits.
Pet111 and Pet309 also interact with the Nam1 protein, a stability factor for some mt-mRNAs that has been shown to interact with the mitochondrial RNA polymerase.81 As proposed above for Rmd9, Nam1 could stabilize the newly synthesized mRNAs until they are presented to the translation apparatus. Thus, the mitochondrial transcription machinery could be coupled to an integrated membrane bound translation system, specific for each complex, where PPR proteins play central roles.
It is also well known that translation and membrane insertion are coupled in mitochondria. For example, reducing the translation of COX2 by halving the level of Pet111 compensates for the heterozygous deletion of COX18, which encodes the Cox2 C-tail translocation factor (Table 2). Conversely, overexpression of PET111 increases the production of Cox2 but this overproduced subunit is unstable and causes imbalance in the production of other cytochrome oxidase subunits.65
Taken together, these examples show that mitochondrial gene expression is delicately balanced and highly integrated in S. cerevisiae (and probably in all organisms), and that this is achieved by the coordinated expression, activity and interaction of many factors, including several key PPR proteins that control the stability, processing and translation of one or several mt-mRNAs.
The Yeast PPR Proteins—Divergence and Evolution
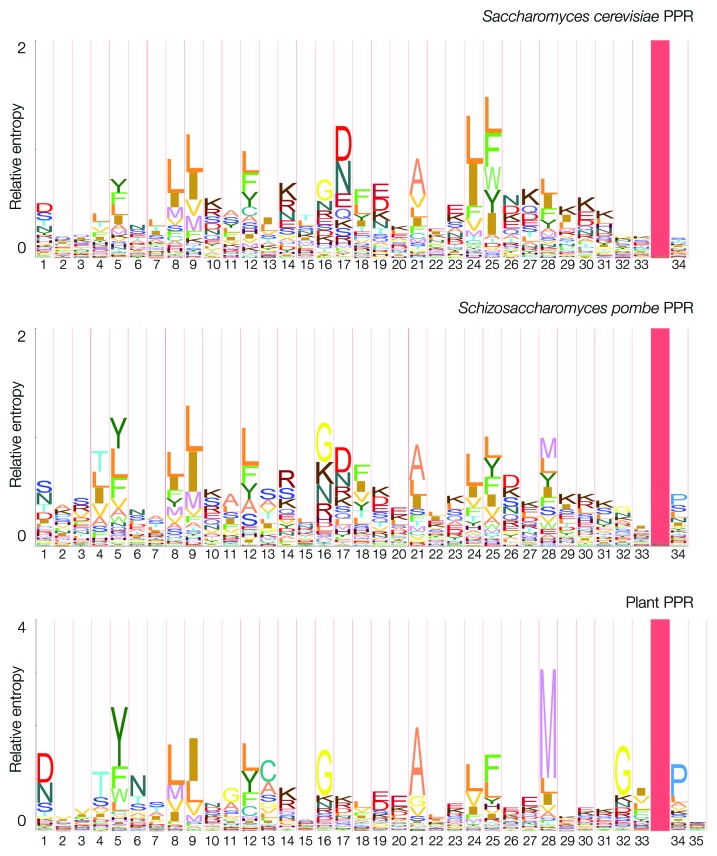
In order to gain a deeper understanding of the multifaceted roles of PPR proteins in the functioning of the yeast mitochondrial genetic system, and put them in the context of what is known about this protein family in other organisms, it is necessary to look into the comparative and evolutionary aspects of this protein family. Sequence logos are a useful tool for comparing PPR motifs and identifying conserved features. Figure 3 shows the HMM profile logos of PPR motifs from the yeasts S. cerevisiae and S. pombe, and, for comparison, of typical plant PPR motifs. The key residues that stand out in the plant PPR logo are partially conserved in yeasts, with the notable exception of the M at position 28, which is absent from the S. cerevisiae logo and only weakly conserved in S. pombe. It is, however, clear that the yeast motifs are significantly more divergent, and the information content of their HMM profiles is lower, as shown by the relative entropy and the emission probabilities at each position, confirming the observations made by Lipinski et al.4 Similarly, the PPR motifs in Trypanosoma,82 and humans83 also show significant intragenomic divergence, and their sequences differ from the plant-based general motif consensus.
Figure 3. Logos of pHMM profiles of the PPR motifs from S. cerevisiae (profile built from 85 motifs), S. pombe (profile built from 66 motifs) and of representative PPR motifs from Viridiplantae (built from 1,222 motifs originating from the Pfam database8). In the construction of the yeast profiles, only motifs detected as statistically significant using pHMM profiles4) were included. The S. cerevisiae and S. pombe motifs had to be shifted by 2 and 1 residues, respectively, in order to align them with the canonical plant signature. Logos were built using LogoMat-M,90 the height of the stack at each position represents the entropy of the position relative to the background distribution, whereas the relative size of a letter corresponds to its emission probability in the HMM model. The shaded vertical blocks correspond to insertion sites.
Recent work on the structural basis of substrate binding by plant PPR proteins84 identified amino acid positions 1 and 6 of the motif as critical for the sequence-specific recognition of the binding site in RNA, by the means of a combinatorial code, wherein one base of the RNA target is recognized by two adjacent PPR motifs, therefore if a protein has N successive PPR motifs, we would expect it to recognize N-1 bases in the RNA sequence. There is evidently some similarity between the yeast motifs and the ones observed in plants but there is clearly more variability and less information contained in the yeast profiles. Given the evolutionary distance between yeasts and plants, there is no reason to assume that substrate recognition uses the same residues in the PPR motifs. Significantly, mtDNAs of Saccharomycotina are usually characterized by a very low GC content (17% in S. cerevisiae), one may therefore speculate that a simpler substrate recognition mechanism, based on purine/pyrimidine distinction could offer sufficient specificity, but if this is the case RNA target sequences might need to be longer in yeasts. However, pending the structural elucidation of a PPR protein-RNA interaction in yeast, any discussion regarding its mechanism will remain purely speculative.
A striking feature of the PPR proteins in yeasts is their very rapid evolutionary divergence. In genome-wide pair wise comparisons of orthologous sequences in selected members of Saccharomycetales, the PPR proteins were always among the fastest evolving, with average sequence identity about 2-fold lower than the genomic average.4 This is a unique feature of the PPR proteins, as neither the structurally related TPR proteins, nor the functionally related mitochondrial ribosomal proteins exhibit this extraordinary evolutionary divergence. In comparisons between more distant species (such as S. cerevisiae and S. pombe) it makes the assignment of individual PPR proteins to orthologous groups on the basis of sequence similarity very challenging.
A plausible scenario for the origin of the PPR proteins, is that they evolved in Eukaryotes to replace the intrinsic organellar gene expression mechanisms lost in the course of the degenerative evolution of the endosymbiont, and to aid in the integration of the organellar and nuclear gene expression, in a process sometimes referred to as the “domestication” of the endosymbiont.85
The marked divergence of yeast PPR family members may reflect the rapid evolution of their target RNA sequences, encoded in the quickly changing mitochondrial genomes. Compatibility between the mitochondrial RNAs and the nuclear-encoded proteins involved in their expression plays a very important role in the evolution of yeasts. Nucleo-mitochondrial incompatibility between closely related Saccharomyces species was demonstrated to act as a variant of the Dobzhansky-Muller interspecies reproductive barrier,86-89 and considering the high evolutionary variability of yeast mitochondrial genomes, could be one of the factors driving speciation in this group. At least one PPR protein, Aep2, is among the factors playing a key role in speciation through the cyto-nuclear incompatibility of two closely related Saccharomyces species: S. cerevisiae and S. bayanus.87,89 In hybrid cells, S. bayanus Aep2p fails to recognize the 5′ UTR of the S. cerevisiae mitochondrial ATP9 mRNA, resulting in respiratory deficiency. This functional incompatibility cannot be attributed to single amino acid changes, but involves multiple critical residues in the protein sequence,89 suggesting that the co-evolution of mitochondrial transcript sequences and the proteins that recognize them play a role in driving the rapid evolution of PPR proteins in yeasts.
Conclusion
At present, we would expect the future of research on PPR proteins of yeasts to follow two main directions, which might be described as the structural and physiological pathways: (1) improving the identification of new PPR proteins and deciphering the PPR protein-RNA binding code, and (2) precisely defining the diverse roles that the individual PPR proteins play in mitochondrial gene expression.
Improving PPR protein identification and deciphering the PPR-RNA binding code will require refining and complementing the algorithms that are used by combining the phylogenetically informed sequence profiles with predictions of secondary and tertiary structure, taking into account such biophysical hallmarks of the PPR proteins, as the presence of a positively charged RNA-binding surface (see above §1). In S. cerevisiae and S. pombe, PPR proteins have been found that are involved in the regulation of at least one component of all the respiratory complexes; however, these yeasts lack complex I. Therefore, it is highly probably that when other complex I containing yeasts are examined, new PPR proteins will be found that regulate some of the mitochondrially encoded subunits of this complex.
In terms of deciphering the PPR-RNA code, a first step will be the precise identification of at least one RNA target sequence, there are several examples where a small RNA region has been delimited, but in all cases these are too long to be the actual target sequence. In this context, Pet111 and Dmr1 from S. cerevisiae and Ppr1 from S. pombe are good candidates for further study; Dmr1 is particularly interesting as the protein can be purified and this purified protein interacts with RNA in vitro.31 The ultimate objective will be to solve the structure of a PPR protein crystallized with its RNA target.
Defining the precise role of individual PPR proteins has been hampered by several factors, the absence of an in vitro mitochondrial translation system, the extremely low levels of most of the PPR proteins found in mitochondria (see for example ref. 62) and the highly integrated nature of mitochondrial gene expression. All these factors combine to make the biochemical analysis of the function of individual PPR proteins technically very challenging. But if we are to understand the function of this fascinating family of proteins, it is a challenge that we must try and meet.
At the present time there is much excitement and activity surrounding the deciphering of the PPR-RNA code, this is indeed an important objective, but to our mind, it is not the ultimate objective. We know that PPR proteins bind to RNA, deciphering the code will help us to determine more easily where they bind, but it will not necessarily tell us what they do. In some ways, that tacit assumption that deciphering the code will answer all our questions is a little reminiscent of widely held pre-genomic belief that when we had the sequence, “we would understand the way a cell worked.” So while the code is important, we should not allow it to tempt us to abandon the more difficult physiological path, or like Robert Frost, we might come to regret the “Road not taken.”
Acknowledgments
C.J.H. and N.B. are supported by the Association Française contre les Myopathies grant 15680, the Institut Fédératif de Recherche 115 and the CNRS. P.G. was supported by the EU Operational Programme Innovative Economy via the Foundation for Polish Science grant TEAM/2010-6/6, and by the Ministry of Science and Higher Education of Poland grant N N303 550639. We are grateful to Malgorzata Komor for her help in the analysis of S. pombe PPR motifs in silico and to Cyrielle Sophie, Katja Hansen, Laurent Dujeancourt, Karolina Labedzka-Dmoch, Karolina Wiecek and Bartosz Zapisek for sharing unpublished results.
Glossary
Abbreviations:
- mtDNA
mitochondrial DNA
- mRNA
messenger RNA
- mt
mitochondrial
- mt-mRNA
mitochondrial mRNA
- mtRNA
mitochondrial RNA
- NA
not applicable
- ND
not determined
- ORF
open reading frame
- (p)HMM
(profile) Hidden Markov Model(s)
- PPR
penta-tricopeptide repeat(s)
- rRNA
ribosomal RNA
- tRNA
transfert RNA
- TPR
tetra-tricopeptide repeat(s)
- UTR
untranslated region
Nomenclature Convention
For S. cerevisiae, wild-type genes and genes with dominant mutations are written in uppercase letters and genes with recessive mutations in lowercase letters. For S. pombe, both wild-type genes and genes with mutation, either dominant or recessive, are written in lowercase letters.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/25392
References
- 1.Small ID, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–7. doi: 10.1016/S0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura T, Yagi Y, Kobayashi K. Mechanistic insight into pentatricopeptide repeat proteins as sequence-specific RNA-binding proteins for organellar RNAs in plants. Plant Cell Physiol. 2012;53:1171–9. doi: 10.1093/pcp/pcs069. [DOI] [PubMed] [Google Scholar]
- 3.Castandet B, Araya A. The nucleocytoplasmic conflict, a driving force for the emergence of plant organellar RNA editing. IUBMB Life. 2012;64:120–5. doi: 10.1002/iub.581. [DOI] [PubMed] [Google Scholar]
- 4.Lipinski KA, Puchta O, Surendranath V, Kudla M, Golik P. Revisiting the yeast PPR proteins--application of an Iterative Hidden Markov Model algorithm reveals new members of the rapidly evolving family. Mol Biol Evol. 2011;28:2935–48. doi: 10.1093/molbev/msr120. [DOI] [PubMed] [Google Scholar]
- 5.Main ER, Lowe AR, Mochrie SG, Jackson SE, Regan L. A recurring theme in protein engineering: the design, stability and folding of repeat proteins. Curr Opin Struct Biol. 2005;15:464–71. doi: 10.1016/j.sbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Kobe B, Kajava AV. When protein folding is simplified to protein coiling: the continuum of solenoid protein structures. Trends Biochem Sci. 2000;25:509–15. doi: 10.1016/S0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
- 7.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–63. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 8.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–22. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karpenahalli MR, Lupas AN, Söding J. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics. 2007;8:2. doi: 10.1186/1471-2105-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–6. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foury F, Roganti T, Lecrenier N, Purnelle B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998;440:325–31. doi: 10.1016/S0014-5793(98)01467-7. [DOI] [PubMed] [Google Scholar]
- 12.Schäfer B. Genetic conservation versus variability in mitochondria: the architecture of the mitochondrial genome in the petite-negative yeast Schizosaccharomyces pombe. Curr Genet. 2003;43:311–26. doi: 10.1007/s00294-003-0404-5. [DOI] [PubMed] [Google Scholar]
- 13.Schäfer B. RNA maturation in mitochondria of S. cerevisiae and S. pombe. Gene. 2005;354:80–5. doi: 10.1016/j.gene.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann B, Nickel J, Speer F, Schäfer B. The 3′ ends of mature transcripts are generated by a processosome complex in fission yeast mitochondria. J Mol Biol. 2008;377:1024–37. doi: 10.1016/j.jmb.2008.01.038. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J Biol Chem. 2007;282:12610–8. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markov DA, Savkina M, Anikin M, Del Campo M, Ecker K, Lambowitz AM, et al. Identification of proteins associated with the yeast mitochondrial RNA polymerase by tandem affinity purification. Yeast. 2009;26:423–40. doi: 10.1002/yea.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haffter P, Fox TD. Nuclear mutations in the petite-negative yeast Schizosaccharomyces pombe allow growth of cells lacking mitochondrial DNA. Genetics. 1992;131:255–60. doi: 10.1093/genetics/131.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kühl I, Dujeancourt L, Gaisne M, Herbert CJ, Bonnefoy N. A genome wide study in fission yeast reveals nine PPR proteins that regulate mitochondrial gene expression. Nucleic Acids Res. 2011;39:8029–41. doi: 10.1093/nar/gkr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nouet C, Bourens M, Hlavacek O, Marsy S, Lemaire C, Dujardin G. Rmd9p controls the processing/stability of mitochondrial mRNAs and its overexpression compensates for a partial deficiency of oxa1p in Saccharomyces cerevisiae. Genetics. 2007;175:1105–15. doi: 10.1534/genetics.106.063883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steele DF, Butler CA, Fox TD. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc Natl Acad Sci USA. 1996;93:5253–7. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonnefoy N, Remacle C, Fox TD. Genetic transformation of Saccharomyces cerevisiae and Chlamydomonas reinhardtii mitochondria. Methods Cell Biol. 2007;80:525–48. doi: 10.1016/S0091-679X(06)80026-9. [DOI] [PubMed] [Google Scholar]
- 22.Deshpande AP, Patel SS. Mechanism of transcription initiation by the yeast mitochondrial RNA polymerase. Biochim Biophys Acta. 2012;1819:930–8. doi: 10.1016/j.bbagrm.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–73. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 24.Howard MJ, Lim WH, Fierke CA, Koutmos M. Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5′ processing. Proc Natl Acad Sci USA. 2012;109:16149–54. doi: 10.1073/pnas.1209062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–57. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 26.Stribinskis V, Gao GJ, Sulo P, Ellis SR, Martin NC. Rpm2p: separate domains promote tRNA and Rpm1r maturation in Saccharomyces cerevisiae mitochondria. Nucleic Acids Res. 2001;29:3631–7. doi: 10.1093/nar/29.17.3631. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kassenbrock CK, Gao GJ, Groom KR, Sulo P, Douglas MG, Martin NC. RPM2, independently of its mitochondrial RNase P function, suppresses an ISP42 mutant defective in mitochondrial import and is essential for normal growth. Mol Cell Biol. 1995;15:4763–70. doi: 10.1128/mcb.15.9.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stribinskis V, Heyman HC, Ellis SR, Steffen MC, Martin NC. Rpm2p, a component of yeast mitochondrial RNase P, acts as a transcriptional activator in the nucleus. Mol Cell Biol. 2005;25:6546–58. doi: 10.1128/MCB.25.15.6546-6558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stribinskis V, Ramos KS. Rpm2p, a protein subunit of mitochondrial RNase P, physically and genetically interacts with cytoplasmic processing bodies. Nucleic Acids Res. 2007;35:1301–11. doi: 10.1093/nar/gkm023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams EH, Butler CA, Bonnefoy N, Fox TD. Translation initiation in Saccharomyces cerevisiae mitochondria: functional interactions among mitochondrial ribosomal protein Rsm28p, initiation factor 2, methionyl-tRNA-formyltransferase and novel protein Rmd9p. Genetics. 2007;175:1117–26. doi: 10.1534/genetics.106.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puchta O, Lubas M, Lipinski KA, Piatkowski J, Malecki M, Golik P. DMR1 (CCM1/YGR150C) of Saccharomyces cerevisiae encodes an RNA-binding protein from the pentatricopeptide repeat family required for the maintenance of the mitochondrial 15S ribosomal RNA. Genetics. 2010;184:959–73. doi: 10.1534/genetics.110.113969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno JI, Buie KS, Price RE, Piva MA. Ccm1p/Ygr150cp, a pentatricopeptide repeat protein, is essential to remove the fourth intron of both COB and COX1 pre-mRNAs in Saccharomyces cerevisiae. Curr Genet. 2009;55:475–84. doi: 10.1007/s00294-009-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreno JI, Patlolla B, Belton KR, Jenkins BC, Radchenkova PV, Piva MA. Two independent activities define Ccm1p as a moonlighting protein in Saccharomyces cerevisiae. Biosci Rep. 2012;32:549–57. doi: 10.1042/BSR20120066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labouesse M, Netter P, Schroeder R. Molecular basis of the ‘box effect’, A maturase deficiency leading to the absence of splicing of two introns located in two split genes of yeast mitochondrial DNA. Eur J Biochem. 1984;144:85–93. doi: 10.1111/j.1432-1033.1984.tb08434.x. [DOI] [PubMed] [Google Scholar]
- 35.Davies SM, Rackham O, Shearwood AM, Hamilton KL, Narsai R, Whelan J, et al. Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett. 2009;583:1853–8. doi: 10.1016/j.febslet.2009.04.048. [DOI] [PubMed] [Google Scholar]
- 36.Haque ME, Koc H, Cimen H, Koc EC, Spremulli LL. Contacts between mammalian mitochondrial translational initiation factor 3 and ribosomal proteins in the small subunit. Biochim Biophys Acta. 2011;1814:1779–84. doi: 10.1016/j.bbapap.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci USA. 2011;108:415–20. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchirico ME. Understanding mitochondrial biogenesis through gene relocation. Amherst, MA: University of massachusetts, 1998. [Google Scholar]
- 39.Sanchirico M, Tzellas A, Fox TD, Conrad-Webb H, Periman PS, Mason TL. Relocation of the unusual VAR1 gene from the mitochondrion to the nucleus. Biochem Cell Biol. 1995;73:987–95. doi: 10.1139/o95-106. [DOI] [PubMed] [Google Scholar]
- 40.Ellis TP, Helfenbein KG, Tzagoloff A, Dieckmann CL. Aep3p stabilizes the mitochondrial bicistronic mRNA encoding subunits 6 and 8 of the H+-translocating ATP synthase of Saccharomyces cerevisiae. J Biol Chem. 2004;279:15728–33. doi: 10.1074/jbc.M314162200. [DOI] [PubMed] [Google Scholar]
- 41.Lee C, Tibbetts AS, Kramer G, Appling DR. Yeast AEP3p is an accessory factor in initiation of mitochondrial translation. J Biol Chem. 2009;284:34116–25. doi: 10.1074/jbc.M109.055350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Payne MJ, Schweizer E, Lukins HB. Properties of two nuclear pet mutants affecting expression of the mitochondrial oli1 gene of Saccharomyces cerevisiae. Curr Genet. 1991;19:343–51. doi: 10.1007/BF00309594. [DOI] [PubMed] [Google Scholar]
- 43.Ackerman SH, Gatti DL, Gellefors P, Douglas MG, Tzagoloff A. ATP13, a nuclear gene of Saccharomyces cerevisiae essential for the expression of subunit 9 of the mitochondrial ATPase. FEBS Lett. 1991;278:234–8. doi: 10.1016/0014-5793(91)80124-L. [DOI] [PubMed] [Google Scholar]
- 44.Ellis TP, Lukins HB, Nagley P, Corner BE. Suppression of a nuclear aep2 mutation in Saccharomyces cerevisiae by a base substitution in the 5′-untranslated region of the mitochondrial oli1 gene encoding subunit 9 of ATP synthase. Genetics. 1999;151:1353–63. doi: 10.1093/genetics/151.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sparks KA, Dieckmann CL. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 1998;26:4676–87. doi: 10.1093/nar/26.20.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staples RR, Dieckmann CL. Suppressor analyses of temperature-sensitive cbp1 strains of Saccharomyces cerevisiae: the product of the nuclear gene SOC1 affects mitochondrial cytochrome b mRNA post-transcriptionally. Genetics. 1994;138:565–75. doi: 10.1093/genetics/138.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Islas-Osuna MA, Ellis TP, Marnell LL, Mittelmeier TM, Dieckmann CL. Cbp1 is required for translation of the mitochondrial cytochrome b mRNA of Saccharomyces cerevisiae. J Biol Chem. 2002;277:37987–90. doi: 10.1074/jbc.M206132200. [DOI] [PubMed] [Google Scholar]
- 48.Chen W, Dieckmann CL. Genetic evidence for interaction between Cbp1 and specific nucleotides in the 5′ untranslated region of mitochondrial cytochrome b mRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6203–11. doi: 10.1128/mcb.17.11.6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Islas-Osuna MA, Ellis TP, Mittelmeier TM, Dieckmann CL. Suppressor mutations define two regions in the Cbp1 protein important for mitochondrial cytochrome b mRNA stability in Saccharomyces cerevisiae. Curr Genet. 2003;43:327–36. doi: 10.1007/s00294-003-0405-4. [DOI] [PubMed] [Google Scholar]
- 50.Krause K, Lopes de Souza R, Roberts DG, Dieckmann CL. The mitochondrial message-specific mRNA protectors Cbp1 and Pet309 are associated in a high-molecular weight complex. Mol Biol Cell. 2004;15:2674–83. doi: 10.1091/mbc.E04-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayer SA, Dieckmann CL. The yeast CBP1 gene produces two differentially regulated transcripts by alternative 3′-end formation. Mol Cell Biol. 1989;9:4161–9. doi: 10.1128/mcb.9.10.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carignani G, Groudinsky O, Frezza D, Schiavon E, Bergantino E, Slonimski PP. An mRNA maturase is encoded by the first intron of the mitochondrial gene for the subunit I of cytochrome oxidase in S. cerevisiae. Cell. 1983;35:733–42. doi: 10.1016/0092-8674(83)90106-X. [DOI] [PubMed] [Google Scholar]
- 53.Manthey GM, McEwen JE. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–43. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tavares-Carreón F, Camacho-Villasana Y, Zamudio-Ochoa A, Shingú-Vázquez M, Torres-Larios A, Pérez-Martínez X. The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J Biol Chem. 2008;283:1472–9. doi: 10.1074/jbc.M708437200. [DOI] [PubMed] [Google Scholar]
- 55.Coffin JW, Dhillon R, Ritzel RG, Nargang FE. The Neurospora crassa cya-5 nuclear gene encodes a protein with a region of homology to the Saccharomyces cerevisiae PET309 protein and is required in a post-transcriptional step for the expression of the mitochondrially encoded COXI protein. Curr Genet. 1997;32:273–80. doi: 10.1007/s002940050277. [DOI] [PubMed] [Google Scholar]
- 56.Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci USA. 2003;100:605–10. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem J. 2004;382:331–6. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruzzenente B, Metodiev MD, Wredenberg A, Bratic A, Park CB, Cámara Y, et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–56. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chujo T, Ohira T, Sakaguchi Y, Goshima N, Nomura N, Nagao A, et al. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 2012;40:8033–47. doi: 10.1093/nar/gks506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poutre CG, Fox TD. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987;115:637–47. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Costanzo MC, Bonnefoy N, Williams EH, Clark-Walker GD, Fox TD. Highly diverged homologs of Saccharomyces cerevisiae mitochondrial mRNA-specific translational activators have orthologous functions in other budding yeasts. Genetics. 2000;154:999–1012. doi: 10.1093/genetics/154.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Green-Willms NS, Butler CA, Dunstan HM, Fox TD. Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J Biol Chem. 2001;276:6392–7. doi: 10.1074/jbc.M009856200. [DOI] [PubMed] [Google Scholar]
- 63.Mulero JJ, Fox TD. PET111 acts in the 5′-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics. 1993;133:509–16. doi: 10.1093/genetics/133.3.509. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bonnefoy N, Bsat N, Fox TD. Mitochondrial translation of Saccharomyces cerevisiae COX2 mRNA is controlled by the nucleotide sequence specifying the pre-Cox2p leader peptide. Mol Cell Biol. 2001;21:2359–72. doi: 10.1128/MCB.21.7.2359-2372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fiori A, Perez-Martinez X, Fox TD. Overexpression of the COX2 translational activator, Pet111p, prevents translation of COX1 mRNA and cytochrome c oxidase assembly in mitochondria of Saccharomyces cerevisiae. Mol Microbiol. 2005;56:1689–704. doi: 10.1111/j.1365-2958.2005.04658.x. [DOI] [PubMed] [Google Scholar]
- 66.Strick CA, Fox TD. Saccharomyces cerevisiae positive regulatory gene PET111 encodes a mitochondrial protein that is translated from an mRNA with a long 5′ leader. Mol Cell Biol. 1987;7:2728–34. doi: 10.1128/mcb.7.8.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dunstan HM, Green-Willms NS, Fox TD. In vivo analysis of Saccharomyces cerevisiae COX2 mRNA 5′-untranslated leader functions in mitochondrial translation initiation and translational activation. Genetics. 1997;147:87–100. doi: 10.1093/genetics/147.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Helfenbein KG, Ellis TP, Dieckmann CL, Tzagoloff A. ATP22, a nuclear gene required for expression of the F0 sector of mitochondrial ATPase in Saccharomyces cerevisiae. J Biol Chem. 2003;278:19751–6. doi: 10.1074/jbc.M301679200. [DOI] [PubMed] [Google Scholar]
- 69.Zeng X, Hourset A, Tzagoloff A. The Saccharomyces cerevisiae ATP22 gene codes for the mitochondrial ATPase subunit 6-specific translation factor. Genetics. 2007;175:55–63. doi: 10.1534/genetics.106.065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rak M, Tzagoloff A. F1-dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc Natl Acad Sci USA. 2009;106:18509–14. doi: 10.1073/pnas.0910351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soto IC, Fontanesi F, Myers RS, Hamel P, Barrientos A. A heme-sensing mechanism in the translational regulation of mitochondrial cytochrome c oxidase biogenesis. Cell Metab. 2012;16:801–13. doi: 10.1016/j.cmet.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grüschke S, Römpler K, Hildenbeutel M, Kehrein K, Kühl I, Bonnefoy N, et al. The Cbp3-Cbp6 complex coordinates cytochrome b synthesis with bc(1) complex assembly in yeast mitochondria. J Cell Biol. 2012;199:137–50. doi: 10.1083/jcb.201206040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ziaja K, Michaelis G, Lisowsky T. Nuclear control of the messenger RNA expression for mitochondrial ATPase subunit 9 in a new yeast mutant. J Mol Biol. 1993;229:909–16. doi: 10.1006/jmbi.1993.1095. [DOI] [PubMed] [Google Scholar]
- 74.Payne MJ, Finnegan PM, Smooker PM, Lukins HB. Characterization of a second nuclear gene, AEP1, required for expression of the mitochondrial OLI1 gene in Saccharomyces cerevisiae. Curr Genet. 1993;24:126–35. doi: 10.1007/BF00324676. [DOI] [PubMed] [Google Scholar]
- 75.Rackham O, Davies SM, Shearwood AM, Hamilton KL, Whelan J, Filipovska A. Pentatricopeptide repeat domain protein 1 lowers the levels of mitochondrial leucine tRNAs in cells. Nucleic Acids Res. 2009;37:5859–67. doi: 10.1093/nar/gkp627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thompson DA, Stahl FW. Genetic control of recombination partner preference in yeast meiosis. Isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics. 1999;153:621–41. doi: 10.1093/genetics/153.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 78.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA. 2003;100:13207–12. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res. 2006;5:1543–54. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- 80.Naithani S, Saracco SA, Butler CA, Fox TD. Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:324–33. doi: 10.1091/mbc.E02-08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rodeheffer MS, Boone BE, Bryan AC, Shadel GS. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J Biol Chem. 2001;276:8616–22. doi: 10.1074/jbc.M009901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pusnik M, Small I, Read LK, Fabbro T, Schneider A. Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol Cell Biol. 2007;27:6876–88. doi: 10.1128/MCB.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rackham O, Filipovska A. The role of mammalian PPR domain proteins in the regulation of mitochondrial gene expression. Biochim Biophys Acta. 2012;1819:1008–16. doi: 10.1016/j.bbagrm.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 84.Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nick P. Rise and fall of plant mitochondria. Protoplasma. 2010;241:1–2. doi: 10.1007/s00709-010-0139-3. [DOI] [PubMed] [Google Scholar]
- 86.Chou JY, Hung YS, Lin KH, Lee HY, Leu JY. Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS Biol. 2010;8:e1000432. doi: 10.1371/journal.pbio.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chou JY, Leu JY. Speciation through cytonuclear incompatibility: insights from yeast and implications for higher eukaryotes. Bioessays. 2010;32:401–11. doi: 10.1002/bies.200900162. [DOI] [PubMed] [Google Scholar]
- 88.Herbert CJ, Dujardin G, Labouesse M, Slonimski PP. Divergence of the mitochondrial leucyl tRNA synthetase genes in two closely related yeasts Saccharomyces cerevisiae and Saccharomyces douglasii: a paradigm of incipient evolution. Mol Gen Genet. 1988;213:297–309. doi: 10.1007/BF00339595. [DOI] [PubMed] [Google Scholar]
- 89.Lee HY, Chou JY, Cheong L, Chang NH, Yang SY, Leu JY. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell. 2008;135:1065–73. doi: 10.1016/j.cell.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 90.Schuster-Böckler B, Schultz J, Rahmann S. HMM Logos for visualization of protein families. BMC Bioinformatics. 2004;5:7. doi: 10.1186/1471-2105-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Finnegan PM, Ellis TP, Nagley P, Lukins HB. The mature AEP2 gene product of Saccharomyces cerevisiae, required for the expression of subunit 9 of ATP synthase, is a 58 kDa mitochondrial protein. FEBS Lett. 1995;368:505–8. doi: 10.1016/0014-5793(95)00727-Q. [DOI] [PubMed] [Google Scholar]
- 92.Manthey GM, Przybyla-Zawislak BD, McEwen JE. The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondrial inner membrane. Eur J Biochem. 1998;255:156–61. doi: 10.1046/j.1432-1327.1998.2550156.x. [DOI] [PubMed] [Google Scholar]
- 93.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–86. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 94.Saracco SA, Fox TD. Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol Biol Cell. 2002;13:1122–31. doi: 10.1091/mbc.01-12-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mulero JJ, Fox TD. Reduced but accurate translation from a mutant AUA initiation codon in the mitochondrial COX2 mRNA of Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:383–90. doi: 10.1007/BF00281787. [DOI] [PubMed] [Google Scholar]
- 96.Bonnefoy N, Fox TD. In vivo analysis of mutated initiation codons in the mitochondrial COX2 gene of Saccharomyces cerevisiae fused to the reporter gene ARG8m reveals lack of downstream reinitiation. Mol Gen Genet. 2000;262:1036–46. doi: 10.1007/PL00008646. [DOI] [PubMed] [Google Scholar]
- 97.Mulero JJ, Fox TD. Alteration of the Saccharomyces cerevisiae COX2 mRNA 5′-untranslated leader by mitochondrial gene replacement and functional interaction with the translational activator protein PET111. Mol Biol Cell. 1993;4:1327–35. doi: 10.1091/mbc.4.12.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khalimonchuk O, Bird A, Winge DR. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J Biol Chem. 2007;282:17442–9. doi: 10.1074/jbc.M702379200. [DOI] [PubMed] [Google Scholar]
- 99.Perez-Martinez X, Butler CA, Shingu-Vazquez M, Fox TD. Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol Biol Cell. 2009;20:4371–80. doi: 10.1091/mbc.E09-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lutz MS, Ellis SR, Martin NC. Proteasome mutants, pre4-2 and ump1-2, suppress the essential function but not the mitochondrial RNase P function of the Saccharomyces cerevisiae gene RPM2. Genetics. 2000;154:1013–23. doi: 10.1093/genetics/154.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Groom KR, Heyman HC, Steffen MC, Hawkins L, Martin NC. Kluyveromyces lactis SEF1 and its Saccharomyces cerevisiae homologue bypass the unknown essential function, but not the mitochondrial RNase P function, of the S. cerevisiae RPM2 gene. Yeast. 1998;14:77–87. doi: 10.1002/(SICI)1097-0061(19980115)14:1<77::AID-YEA201>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 102.Rogowska AT, Puchta O, Czarnecka AM, Kaniak A, Stepien PP, Golik P. Balance between transcription and RNA degradation is vital for Saccharomyces cerevisiae mitochondria: reduced transcription rescues the phenotype of deficient RNA degradation. Mol Biol Cell. 2006;17:1184–93. doi: 10.1091/mbc.E05-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bryan AC, Rodeheffer MS, Wearn CM, Shadel GS. Sls1p is a membrane-bound regulator of transcription-coupled processes involved in Saccharomyces cerevisiae mitochondrial gene expression. Genetics. 2002;160:75–82. doi: 10.1093/genetics/160.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Daoud R, Forget L, Lang BF. Yeast mitochondrial RNase P, RNase Z and the RNA degradosome are part of a stable supercomplex. Nucleic Acids Res. 2012;40:1728–36. doi: 10.1093/nar/gkr941. [DOI] [PMC free article] [PubMed] [Google Scholar]