Abstract
A small subset of the large pentatricopeptide repeat (PPR) protein family in higher plants contain a C-terminal small MutS-related (SMR) domain. Although few in number, they figure prominently in the chloroplast biogenesis and retrograde signaling literature due to their striking mutant phenotypes. In this review, we summarize current knowledge of PPR-SMR proteins focusing on Arabidopsis and maize proteomic and mutant studies. We also examine their occurrence in other organisms and have determined by phylogenetic analysis that, while they are limited to species that contain chloroplasts, their presence in algae and early branching land plant lineages indicates that the coupling of PPR motifs and an SMR domain into a single protein occurred early in the evolution of the Viridiplantae clade. In addition, we discuss their possible function and have examined conservation between SMR domains from Arabidopsis PPR proteins with those from other species that have been shown to possess endonucleolytic activity.
Keywords: pentatricopeptide repeat protein, small MutS-related domain, chloroplast, plastid, mitochondria, endonuclease, genomes uncoupled, Arabidopsis thaliana, Zea mays
Introduction
The pentatricopeptide repeat (PPR) protein family was serendipitously discovered as a result of computational analysis of the then incomplete Arabidopsis thaliana genome sequence for gene products likely to be targeted to plastids and mitochondria.1 While subsequent analysis revealed that these proteins are ubiquitous in eukaryotes, they were found to be particularly prevalent in terrestrial plants (e.g., 450 members in Arabidopsis).2–4 Since their discovery, a plethora of genetic, molecular, and biochemical evidence suggests that PPR proteins bind RNA in a highly specific manner and facilitate events such as cleavage, editing, splicing, turnover, and translation of their target organellar transcript(s).3,5,6
PPR proteins are defined by the presence of tandem repeats of degenerate 31–36 amino acid motifs and can be classified based on motif structure and the presence of additional C-terminal domains.6 The P subfamily consists of PPR proteins with orthodox 35 amino acid PPR (P) motifs, while the PLS subfamily includes PPR proteins with additional long (L) or short (S) motif variants and derive their name from their characteristic tandem arrays of P-L-S motif triplets. PLS PPR proteins are further classified, based on their C-terminal domain(s), into the E, E+, and DYW subgroups. In addition, while not yet formally recognized as subgroups, P-class PPR proteins can also be categorized by the presence of additional domains, such as the small MutS-related (SMR) domain.5
Searching the Arabidopsis genome reveals that eight proteins contain both PPR motifs and an SMR domain (Fig. 1). Despite the relatively small size of this subgroup, there has been sustained interest in this type of PPR protein since the revelation that genomes uncoupled 1 (GUN1) encodes a PPR protein with a C-terminal SMR domain.7 GUN1 is a central regulator of plastid retrograde signaling, where the developmental and/or functional state of the plastid exerts control on the expression of nuclear genes encoding plastid-localized proteins, such as photosynthesis-associated nuclear genes (PhANGs). Despite this important role, we still do not understand the precise molecular mechanisms of GUN1 and other proteins with similar domain architecture and what specific role, if any, the SMR domain plays in their function. This review will focus on this small but important group of PPR proteins that contain an SMR domain by summarizing our current knowledge from studies performed in higher plants, examining their presence in other organisms and discussing the possible role of the SMR domain.
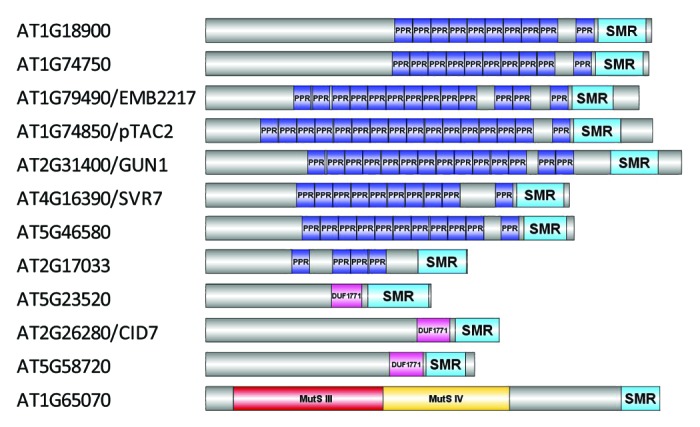
Figure 1. Proteins containing an SMR domain in the model plant Arabidopsis thaliana. A non-redundant set of 12 proteins was identified by searching the Universal Protein knowledgebase47 (UniProt; www.uniprot.org) for Arabidopsis proteins that contain the InterPro48 domain IPR002625 (Smr protein/MutS2 C-terminal domain). Proteins are denoted by their corresponding Arabidopsis Genome Identifier (AGI; ATXGXXXXX) and, if applicable, followed by their common name (e.g., GUN1). Protein domain structure is shown alongside each AGI to demonstrate presence and location of the pentatricopeptide repeat (PPR), small MutS-related (SMR), domain of unknown function (DUF) 1771, and MutS domains. The schematics of protein domain structure were created by combining TPRpred49 to predict PPR domains (those with P > 0.01 were excluded), InterProScan50 to identify other domains and DOG 1.051 for visualization of their respective positions.
The SMR Domain—What Is It?
MutS proteins are key enzymes involved in repair of mismatched DNA bases produced during biological processes such as DNA replication.8 The SMR domain was originally identified in the C-terminal region of the MutS2 protein from the cyanobacterium Synechocystis.9 MutS2 proteins suppress homologous recombination by endonucleolytic digestion of branched DNA structures formed early in this process and the nuclease activity of these proteins has been specifically attributed to the SMR domain.10 Moreover, while not all organisms have MutS2 orthologs, proteins containing SMR domains are widespread in bacterial and eukaryotic species.9,11 In a recent review, Fukui and Kuramitsu11 introduced a classification system for proteins containing SMR domains. Subfamily 1 consists of MutS2 orthologs and is restricted to proteins from bacterial and plant species, subfamily 2 includes proteins with domains in addition to the SMR domain and are usually found only in eukaryotes, while subfamily 3 comprises “stand-alone” SMR domains and comprises proteins from both prokaryotes and eukaryotes.11
In Arabidopsis, 12 proteins are found to contain an SMR domain (Fig. 1). As can be seen from the domain structure shown, AT1G65070 belongs to subfamily 1 (MutS2-like) while the remaining 11 proteins are classified as subfamily 2 SMR proteins. Of the 11 subfamily 2 SMR proteins, eight contain PPR motifs. Consistent with nomenclature used recently,12 we will refer to proteins with this domain architecture as PPR-SMR proteins (PPR-SMRs). When performing BLAST searches using PPR-SMRs, other plant PPRs are identified with C-terminal domains that potentially represent a highly degenerate SMR domain (e.g., AT3G18110/EMB1270). However, their relationship to bona fide PPR-SMRs remains to be clarified and these will not be discussed further.
PPR-SMRs—What Do We Know So Far?
Characterization of PPR-SMRs has focused on higher plant models, such as Arabidopsis and maize. Data collected thus far is derived from a combination of proteomic and mutant analyses and is summarized in Table 1 for the Arabidopsis PPR-SMRs and their maize orthologs.
Table 1. Summary of current knowledge of PPR-SMR proteins in the dicot and monocot plant models, Arabidopsis and maize.
| Arabidopsis | Maize | ||||
|---|---|---|---|---|---|
|
Locus ID (name) |
Subcellular localization |
Mutant phenotype |
Locus ID (name) |
Subcellular localization |
Mutant phenotype |
| AT2G31400 (GUN1) |
Plastid YFP7 |
Normal gross phenotype in dark or light growth conditions but shows defective de-etiolation response27,28; unable to repress PhANGs upon treatment with NF27 or lincomycin7,29 |
GRMZM2G432850 (N/A) |
Possibly plastid
57
|
? |
| AT1G74850 (pTAC2) |
Plastid MS: chloroplast,21,58,59 stromal megadalton complex,16 nucleoids,17 TAC19 |
Seedling lethal, requires exogenous carbon source for further growth, cannot produce seeds, PEP promoter usage affected19 |
GRMZM2G122116 (Zm-pTAC2) |
Plastid MS: proplastid,18 nucleoids18 |
Very pale yellow green (PML: Zm-ptac2–1) Ivory (PML: Zm-ptac2–2,-3) |
| AT4G16390 (SVR7) |
Plastid GFP6; MS: chloroplast,21,59 stromal megadalton complex,16 nucleoids17 |
Slower growth with reduced chlorophyll concentration,25,26 plastid rRNA processing defects,25 reduced translation of plastid ATP synthase subunits26 |
GRMZM2G128665 (ATP4) |
Plastid MS: proplastid,18 nucleoids18 |
Pale green, reduced translation of ATP synthase subunits12 and stability of dicistronic rpl16-rpl14 RNAs36 |
| AT5G46580 (N/A) |
Plastid MS: chloroplast,59 envelope,21 stromal megadalton complex,16 nucleoids17 |
? |
GRMZM2G438524 (PPR53) |
Plastid MS: proplastid,18 nucleoids18 |
Very pale yellow green-virescent (PML: ppr53–1) Ivory-virescent (PML: ppr53–2,-3) |
| AT2G17033 (N/A) |
Plastid
60
|
? |
GRMZM2G164202 (PPR-SMR4) |
Plastid MS: nucleoids18 |
WT-like (PML: ppr-smr4–1) |
| AT1G79490 (EMB2217) |
Mitochondrion GFP13 |
Embryo defective, developmental arrest occurs at globular stage15 |
GRMZM2G345667 (N/A) |
Mitochondrion
57
|
? |
| AT1G74750 (N/A) |
Mitochondrion
14
|
? |
GRMZM2G475897 (N/A) |
Mitochondrion 57 | ? |
| AT1G18900 (N/A) |
Mitochondrion 14 | ? | |||
Localization data was derived from the SUBA3 database60 for Arabidopsis proteins and by manual curation of proteomic data sets for maize proteins. Subcellular localizations in italics are based on predictions while those in bold are based on experimental evidence (GFP/YFP, GFP/YFP fusion studies; MS, identified by mass spectrometry of protein samples). In some cases proteins were identified from a sample corresponding to a specific suborganellar location as specified (e.g., stroma, envelope, nucleoids, TAC). Mutant phenotype descriptions are based on manual curation of the literature and, where available, seedling phenotype descriptions from the maize photosynthetic mutant library61 (PML; http://pml.uoregon.edu/photosyntheticml.html). For PML descriptions, note that these mutants have not yet been analyzed in detail and the effect of the mutation on the expression of the gene still needs to be determined before definitive phenotypes are assigned.
PPR-SMRs in higher plants are localized to both mitochondria and plastids
Of the eight Arabidopsis PPR-SMRs, three have either been found (AT1G7949013) or are predicted (AT1G74750 and AT1G1890014) to be localized to mitochondria. The corresponding maize orthologs are also predicted to be localized to the mitochondria. For the confirmed mitochondrial-localized PPR-SMR AT1G79490, it is also known that mutant lines have an embryo lethal phenotype (EMB2217), with developmental arrest occurring at the globular stage.15 Moreover, the corresponding gene has been reported to have transient, germination-specific expression at early stages of Arabidopsis seed germination, consistent with an important role in early plant development.13 However, this is the extent of the information available from the current literature for Arabidopsis mitochondrial PPR-SMRs.
The five remaining PPR-SMRs all have experimental evidence indicating that they localize to the other endosymbiotically derived organelle, the plastid (Table 1). Extensive proteomic data are available for three of these (pTAC2, SVR7, and AT5G46580) and for the corresponding maize orthologs (Zm-pTAC2, ATP4, and PPR53). Specifically, these proteins were found in proteomic studies of Arabidopsis chloroplast stromal megadalton complexes16 as well as Arabidopsis17 and maize18 plastid nucleoids. In addition, pTAC2 and an ortholog of AT5G46580 were found in preparations of plastid transcriptionally active chromosomes (pTAC) from Arabidopsis19 and spinach,20 respectively. A minor fraction of AT5G46580 was also found in a plastid envelope-enriched sample.21 This places the AT5G46580 protein in three different compartments of the plastid: the thylakoid membranes (which nucleoids/TAC are associated with), the stroma, and the envelope. While it may be that this protein localizes to all of these plastid sub-compartments, it is possible that this protein may only be loosely associated with the nucleoids and easily removed during preparation of various sub-compartmental plastid fractions. Furthermore, while no experimental data exists for the Arabidopsis protein, the maize ortholog of AT2G17033, PPR-SMR4, has also recently been detected in nucleoid-enriched fractions.18 Finally, despite its central role in plastid retrograde signaling pathways, proteomic data for GUN1 is absent and its plastid localization is based on microscopic analysis of transiently expressed fluorescent protein fusions.7 GUN1-yellow fluorescent protein (YFP) was shown to accumulate in chloroplasts in a punctate pattern overlapping the patterns of pTAC2-cyan fluorescent protein, indicating co-localization of GUN1 and pTAC2 in actively transcribed sites of plastid nucleoids.7 However, given that more recent studies suggest that processes such as mRNA cleavage, splicing, and editing, as well as ribosome assembly, take place in association with the nucleoids,18 it is not clear whether GUN1 is specifically bound to plastid DNA and/or RNA in vivo.
Apart from the lack of GUN1 protein detected, the fact that other plastid PPR-SMRs are routinely detected in plastid proteomic studies indicates they are more abundant than other PPR proteins, which are generally considered to be low abundance proteins. For example, SVR7 was the only PPR protein (out of 450) that could be reliably detected in whole leaf protein samples,22 where its abundance was found to decrease with increasing leaf age. In addition, recent proteomic analyses have allowed relative quantitation of protein abundance to be estimated using spectral counts derived from mass spectrometry (MS) analysis.16–18,23,24 This approach is based on the observation that the number of MS/MS acquisitions of peptides coming from a protein shows a positive correlation to the relative concentration of the protein in the sample. These available data sets have allowed us to assess PPR-SMR protein abundance relative to other PPR proteins as summarized in Table 2. In general, in both Arabidopsis and maize, PPR-SMRs dominate the protein mass that can be attributed to PPR proteins, contributing 26–53% of the total PPR protein mass in samples ranging from total leaf protein extracts to purified nucleoids. While the exact reason for the high abundance of these proteins remains to be determined, we speculate that this could reflect binding to multiple targets (e.g., ATP4, see below) and/or that their targets are highly abundant (e.g., rRNA, see SVR7 below). More specifically, in maize, Zm-pTAC2 was found to be the most abundant PPR-SMR in all samples analyzed (13–34% of total PPR protein mass), with PPR53 the next most abundant PPR-SMR (6.5–21%). In Arabidopsis, pTAC2 was also the most abundant PPR-SMR in nucleoids (34%) but in other samples (total leaf and high molecular weight stromal fractions) SVR7 was consistently found to be the dominant PPR-SMR protein present (24–34%). Interestingly, the maize ortholog of SVR7, ATP4, while detected in all samples, was always found at lower levels (1–3% of total PPR protein mass) indicating different expression levels of these orthologs in the monocot and dicot lineages. It remains to be determined whether this difference underlies their reported functional divergence (see below).
Table 2. The relative abundance of PPR-SMR proteins in different Arabidopsis and maize protein samples based on normalized adjusted spectral counts as an estimate of protein abundance.
| Reference | Protein fraction description | No. proteins identified | No. PPR proteins identified | No. PPR-SMR proteins identified |
% of total protein mass attributed to PPR proteins |
% of total PPR protein mass attributed to PPR-SMR proteins |
|---|---|---|---|---|---|---|
| 24 |
Total leaf protein (Arabidopsis, Col-0, rosette leaves) |
3424 |
17 |
3 |
0.05 |
48% 32% SVR7, 14% pTAC2, 2% AT5G46580 |
| 17 |
Total leaf protein (Arabidopsis, no information) |
815 |
9 |
3 |
0.02 |
50% 34% SVR7, 14% AT5G46580, 2% pTAC2 |
| 16 |
Stromal fraction: low molecular weight (Arabidopsis, Col-0, rosette leaves, 55 d old) |
398 |
0 |
0 |
0 |
0 |
| 16 |
Stromal fraction: high molecular weight A (Arabidopsis, Col-0, rosette leaves, 55 d old) |
293 |
9 |
3 |
0.46 |
33% 24% SVR7, 4.5% pTAC2, 4.5% AT5G46580 |
| 16 |
Stromal fraction: high molecular weight B (Arabidopsis, Col-0, rosette leaves, 55 d old) |
230 |
6 |
3 |
0.47 |
53% 28% SVR7, 19% pTAC2, 6% AT5G46580 |
| 17 |
Nucleoids (Arabidopsis, Col-0, young seedlings) |
1026 |
26 |
3 |
1.04 |
47% 34% pTAC2, 12% AT5G46580, 1% SVR7 |
| 17 |
Proplastids (maize, B73, third leaf blade of 8−9 d old seedlings) |
2242 |
32 |
3 |
0.67 |
48% 24% Zm-pTAC2, 21% PPR53, 3% ATP4 |
| 18 |
Proplastids (maize, third leaf blade of 8−9 d old seedlings) |
1717 |
17 |
3 |
0.41 |
53% 34% Zm-pTAC2, 17% PPR53, 2% ATP4 |
| 23 |
Chloroplasts (maize, WT-T43, third leaf blade of 12−14 d old seedlings) |
1428 |
5 |
0 |
0.002 |
0 |
| 18 |
Nucleoids - average from base-tip-young samples (maize) |
1092 |
63 |
4 |
4.65 |
29% 16% Zm-pTAC2, 11% PPR53, 1.5% ATP4, 0.5% PPR-SMR4 |
| 18 |
Nucleoids, leaf base (maize, third leaf blade of 8−9 d old seedlings) |
678 |
46 |
4 |
4.89 |
27% 13% Zm-pTAC2, 12% PPR53, 1% ATP4, 1% PPR-SMR4 |
| 18 |
Nucleoids, leaf tip (maize, third leaf blade of 8−9 d old seedlings) |
710 |
35 |
3 |
2.68 |
26% 18% Zm-pTAC2, 6.5% PPR53, 1.5% ATP4 |
| 18 | Nucleoids, young leaves (maize, leaf blades of 7- to 8-d-old seedlings) |
827 | 55 | 4 | 6.38 |
32% 18% Zm-pTAC2, 12% PPR53, 1.5% ATP4, 0.5% PPR-SMR4 |
For quantitation of protein mass, each protein accession is scored for total MS/MS spectral counts (SPC), unique SPC (uniquely matching to an accession), and adjusted SPC (adjSPC). AdjSPC is the sum of unique SPCs and SPCs from shared peptides across accessions with SPC distributed in proportion to their unique SPC. The normalized adjSPC (NadjSPC) for each protein is calculated through division of adjSPC by the sum of all adjSPC values for the proteins from the sample (e.g., per gel lane or protein extract). Thus, NadjSPC provides a relative protein abundance measure by mass. For example, a protein with NadjSPC = 0.01 contributes approximately 1% of the protein mass of the analyzed sample. NadjSPC values were obtained from the publications indicated and used to calculate the relative abundance of PPR and PPR-SMR proteins.
PPR-SMR mutant analysis reveals diverse phenotypes and putative targets
Genetic approaches show that, despite their similarity in protein architecture, the gross and molecular mutant phenotypes for plastid PPR-SMRs differ dramatically. For example, at the level of plant vitality and growth, Arabidopsis mutant phenotypes range from seedling lethal (ptac219) to moderately slower growth and paler leaves (svr725,26) to a normal, wild-type-like phenotype (gun17) under normal growth conditions. Similarly, this is the case for maize PPR-SMR protein orthologs with seedling phenotypes also ranging from wild-type-like (ppr-smr-4) to very pale yellow-green (Zm-ptac2 and ppr53; Table 1).
“Genomes uncoupled” (GUN) refers to the mutant phenotype where nuclear and plastid gene expression is uncoupled. Twenty years ago, gun mutants were identified from a mutagenized collection of plants containing the GUS reporter gene driven by the promoter of a gene encoding a light harvesting complex protein, LHCB1.2.27 Mutants impaired in plastid-to-nuclear signaling were identified by screening seedlings in the presence of the carotenoid biosynthesis inhibitor, norflurazon (NF).27 The initial publication from this screen identified three gun mutants (gun1, gun2, gun3), in which LHCB1.2 expression was not repressed after NF treatment, compared with the control line. Since then, these and other gun mutants have been characterized, but it was not until 2007 that GUN1 was found to be a plastid-localized PPR-SMR protein.7 As well as the classical “genomes uncoupled” phenotype, characterized by the inability to repress PhANG gene expression when plastid function is inhibited, gun1 mutants are also retarded in their ability to de-etiolate, indicating that GUN1 plays a role in the transition from heterotrophic to photoautotrophic growth.28 Moreover, gun1 is unique among the gun mutants in that impaired repression of PhANGs occurs when the seedlings are subjected to treatment with either NF or plastid translation inhibitors,7,29 such as lincomycin. This indicates that GUN1 is required for a retrograde signaling pathway involving plastid gene expression as well as another pathway involving carotenoid biosynthesis. For detailed information and further discussions on GUN1 and plastid retrograde signaling, we direct the reader to recent reviews in this area.30–33
PTAC2 was identified as one of 18 novel components of plastid transcriptionally active chromosomes (pTACs).19 The ptac2 mutant is only viable when an exogenous carbon source is available and, when this is provided, it develops yellow cotyledons and pale green primary leaves, but is unable to proceed to reproductive growth. Examination of the ultrastructure of the plastids in the ptac2 mutant indicates that plastid development is severely impaired. Analysis of transcript abundance of plastome-encoded genes suggests an involvement of pTAC2 in plastid-encoded-polymerase (PEP)-dependent transcription and processing of chloroplast RNAs as the ptac2 mutant plants showed a strongly reduced accumulation of transcripts generated by PEP.19,34
The svr7 mutant was identified during a screen for suppressors of var2 variegation.25 VAR2 encodes a plastid protease (FtsH), and in its absence, leaves develop a characteristic variegated pattern, including white sectors where chloroplasts fail to develop.35 However, the svr7/var2 double mutant lacks these white sectors. Processing of 23S, 16S, and 4.5S rRNA is perturbed in svr7.25 In addition, a specific reduction in the accumulation of the ATP synthase subunits A, B, E, and F and reduced ribosome association of atpB/E and rbcL mRNAs in the svr7 mutant has also been observed, indicating that SVR7 is involved in translational activation of these transcripts.26 Given its similarity to GUN1, the authors also investigated if the svr7 mutant displays a “gun” phenotype by testing PhANG responses upon treatment with NF. These experiments indicated that the svr7 mutant is, like wild-type, able to repress PhANG expression upon inhibition of chloroplast function and, thus, does not display a “gun” phenotype.26
ATP4, the maize ortholog of SVR7, has also been characterized.12 RNA co-immunoprecipitation assays identified the dicistronic plastid atpB/E mRNA as a ligand for ATP4 in vivo. As for the svr7 mutant, polysome analysis indicates that translation of the atpB/E transcript is perturbed in the atp4 mutant. However, atp4 also shows reduced translation of the atpA transcript and exhibits a more extreme phenotype compared with svr7 with apparent loss of the plastid ATP synthase complex. Also, in contrast to svr7, the accumulation of processed atpF and psaJ transcripts12 and the stabilization of dicistronic rpl16-rpl14 RNAs36 is affected in the atp4 mutant. Thus, the phenotypes of atp4 and svr7 mutants suggest that the functions of these orthologs are not strictly conserved. Furthermore, while over-accumulation of plastid rRNA precursors is observed in the atp4 mutant, as seen for svr7, the authors note that these differences are likely to be secondary as they are not specific to atp4 and are also observed in other mutants impaired in plastid gene expression and/or ATP synthase activity.12
PPR-SMRs—Which Organisms Have Them?
All of the studies undertaken to date that have identified PPR-SMR proteins or investigated their function have done so using higher plant species and have focused on single Arabidopsis or maize proteins, and there is little information on the evolutionary relationships between PPR-SMR proteins within or across species. What has not been previously examined is the extent to which this protein architecture, where PPR and SMR domains are coupled into a single protein, is present in other organisms. As whole genome sequences become accessible through the recent increases in sequencing data available for organisms representing diverse lineages, this provides an opportunity to examine the presence of PPR-SMR proteins in a wide range of organisms to determine their origins and diversification.
PPR-SMR protein sequences were collated in two ways—by searching for proteins containing both PPR and SMR domains in Uniprot using the InterPro identifiers IPR002625 and IPR002885 and by BLAST using the SMR domains of the Arabidopsis members of this PPR subgroup. Sequences obtained were manually curated so major clades were represented by organisms for which complete genome sequences were available, where possible, and truncated and redundant sequences were removed. It is already known that proteins containing SMR domains are found in both prokaryotic and eukaryotic organisms9 while PPR motifs are confined to eukaryotes.3 Thus, it is not surprising that PPR-SMR proteins are essentially only found in eukaryotic organisms and, interestingly, largely confined to the Viridiplantae clade. One major exception to this is sequences found for PPR-SMR proteins in two strains of Legionella longbeachae.37 However, given the paucity of PPR proteins encoded in other bacterial species it is likely that these sequences are remnants of a horizontal gene transfer event, as has been previously suggested to explain PPR genes identified in an isolated number of bacterial species.3
PPR-SMR proteins were also found in heterokont species (brown algae, diatoms). Until recently, heterokont chloroplasts were thought to be derived from the secondary endosymbiosis of an ancestral red algae by a eukaryotic host—the “chromalveolate hypothesis.”38 However, we were unable to find PPR-SMR sequences in red algal genomes (Chondrus crispus and Cyanidioschyzon merolae). This suggests that the PPR-SMR proteins found in heterokonts may be derived from an ancestral endosymbiont from the green algal lineage, supporting the more recent hypothesis that endosymbiosis of a green algae into the ancestral host cell preceded the engulfment of a red algae.39,40
Our Bayesian phylogenetic analysis (Fig. 2) also reveals that orthologs of all Arabidopsis proteins are present in species representing the major angiosperm clades, including both dicots and monocots. However, the putative mitochondrial PPR-SMRs AT1G18900 and AT1G74750 are represented by a single ortholog in most other flowering plants, with Arabidopsis lyrata the only exception, indicating that a recent gene duplication event accounts for the extra protein present in Arabidopsis species. Five PPR-SMRs were identified in the lycophyte and bryophyte models, Selaginella moellendorfii and Physcomitrella patens, respectively. Homologs of GUN1, pTAC2, SVR7/AT5G46580, EMB2217, and AT1G18900/AT1G74750 were found in Selaginella while homologs of GUN1, pTAC2, AT2G17033, and AT1G18900/AT1G74750 were found in Physcomitrella. This suggests that the SVR7/AT5G46580 and EMB2217 clades arose when tracheophytes evolved. However, the discovery of PPR-SMR proteins in chlorophytes (Micromonas, Chlorella, and Ostreococcus) suggests that this type of PPR protein emerged early in the evolution of the PPR protein family in chloroplast-containing lineages.
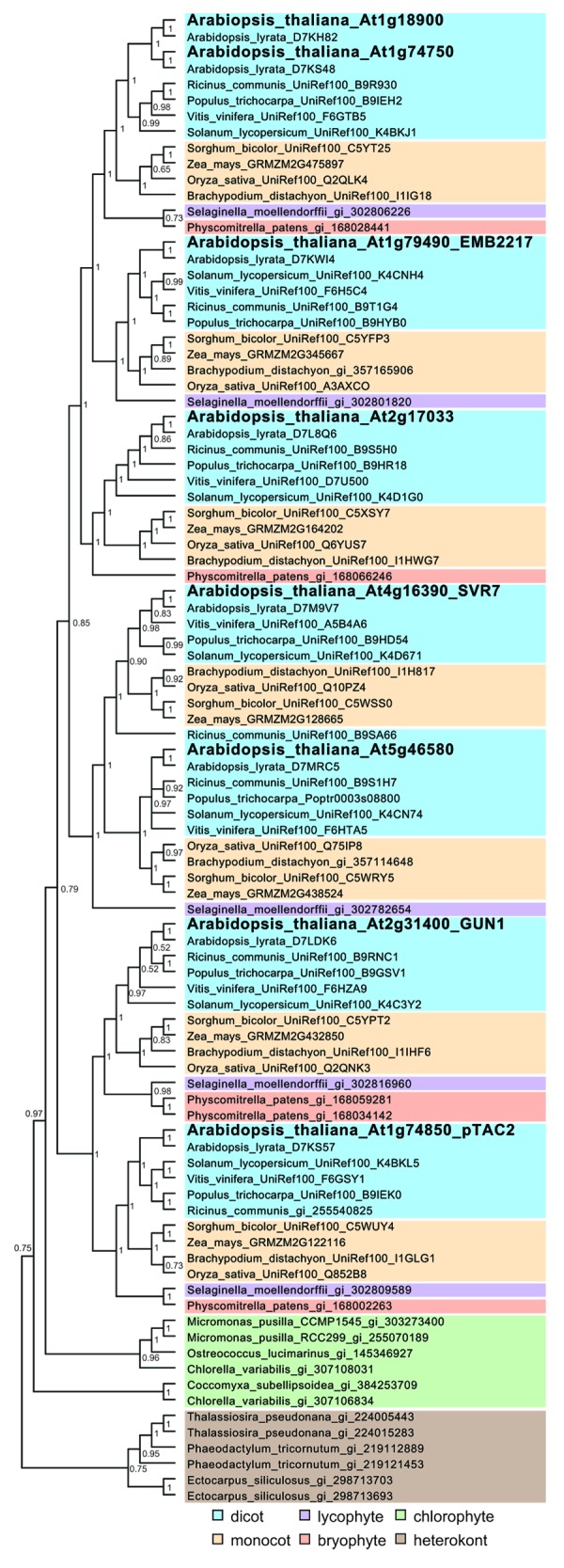
Figure 2. Bayesian phylogenetic tree of PPR-SMR protein sequences from a range of different species. Sequences of PPR-SMR proteins were obtained from BLAST searches and InterPro domain searches (IPR002625 and IPR002885) and aligned using MUSCLE.52 A phylogenetic tree was constructed using MrBayes version 3.2.153 which employs Markov Chain Monte Carlo (MCMC) sampling to approximate the posterior probabilities of phylogenies54 (shown above the branches). MrBayes 3.2.1 was run in parallel on the Fornax supercomputer (located at iVEC@UWA) utilizing the BEAGLE library55 with a mixed model of molecular evolution (determined using jModelTest56), utilizing 12 chains for 50 million generations and trees sampled every 1000 generations. All runs reached a plateau in likelihood score, which was indicated by the standard deviation of split frequencies (0.0015), and the potential scale reduction factor was close to one, indicating the MCMC chains converged. Sequences are color shaded based on their lineage as indicated.
The SMR Domain of PPR-SMRs—What Is Its Function?
The function of the SMR domain in PPR-SMR proteins has not yet been comprehensively explored. Currently, the only examination of the specific role of the SMR domain that has been published has come from the characterization of GUN1, which reported DNA-binding activity of the SMR domain, using a non-specific substrate (calf thymus DNA).7 However, studies of SMR domain-containing proteins in other organisms have focused on its specific role as a nuclease and evidence now exists for endonucleolytic activity in members representing all three subfamilies of SMR-domain-containing proteins.10,11,41–43
Functional characterization of the C-terminal domain of the human BCL3 binding protein (a subfamily 2 SMR domain) provided the first evidence for endonuclease activity of the SMR domain.43 The recombinant domain was found to non-specifically incise a supercoiled plasmid DNA to generate an open circular form of the plasmid, demonstrating the nicking endonuclease activity of the protein. A specific role for the SMR domain of this protein in binding DNA was later demonstrated.44 Nuclease and DNA-binding activity was also confirmed for the subfamily 1 SMR domain of the MutS2 protein from Thermus thermophilus10 and for a subfamily 3 “stand-alone” SMR domain-containing protein, YdaL, from Escherichia coli.42 Interestingly, the Leishmania donovani mRNA cycling sequence-binding protein (LdCSBP) containing a CCCH Zn-finger RNA-binding domain and a subfamily 2 SMR domain has been reported to possess RNA endonuclease activity.41 The SMR domain of LdCSBP alone exhibits both DNA and RNA endonuclease activity, but the full-length protein shows only sequence-specific RNA cleavage activity.41
Given these reported activities of SMR domain-containing proteins, it is tempting to speculate on possible functions of PPR-SMR proteins. One possibility would be that PPR-SMRs are factors with a dual function in both DNA and RNA metabolism, whereby the PPR motifs confer RNA binding activity while the SMR domain confers DNA binding activity. This would be consistent with a role in transcription (e.g., pTAC2). Alternatively, by analogy to LdCSBP, the observation that a protein containing RNA binding and SMR domains can act as an RNA endonuclease raises the question of whether PPR-SMR proteins can act as sequence-specific RNA endonucleases, where the PPR motifs confer RNA sequence specificity and the SMR domain confers endonuclease activity. This possibility would be consistent with a role in mRNA processing (e.g., SVR7 and ATP4).
While these proposed functions require rigorous experimental verification, given that endonucleolytic activity has been reported for four SMR domain-containing proteins, a comparison of the SMR domains from these proteins with those from Arabidopsis PPR-SMRs was undertaken to determine if conserved residues are present (Fig. 3). From an examination of different SMR domains belonging to the different SMR subfamilies conserved motifs specific to each subfamily have been identified.11 Subfamilies 1 and 3 have a characteristic HGXG centrally within the SMR domain. In contrast, subfamily 2 contains a TGXG motif at the same position. These motifs are perfectly conserved in those proteins known to have endonuclease activity (Fig. 3). For the Arabidopsis proteins five of the eight SMR domains linked to PPR motifs have a TGXG motif at this position. The three SMR domains that diverge from this motif are in pTAC2, EMB2217, and AT2G17033. Subfamily 2 SMR domains are also characterized by an LDXH motif toward the N terminus of the SMR domain. This is conserved in the subfamily 2 SMR domains already verified to confer DNA and RNA endonuclease activity (human B3BP protein and the Leishmania CSBP protein). For the Arabidopsis proteins, only two of the eight SMR domains linked to PPR motifs have LDXH at this position, namely GUN1 and AT2G17033. Thus, the only Arabidopsis PPR-SMR containing both conserved motifs is GUN1.
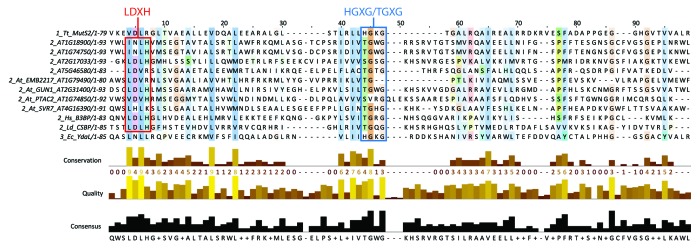
Figure 3. SMR domain alignment to assess amino acid sequence conservation. The SMR domains of the eight Arabidopsis PPR-SMR proteins were aligned with SMR domains from proteins that have been experimentally demonstrated to have endonucleolytic activity.10,41–43 The sequences are denoted by the SMR subfamily type (1_, 2_, or 3_) followed by the AGI (for Arabidopsis proteins) or alternative identifier (Tt_MutS2 – Thermus thermophilus MutS2 protein; Hs_B3BP – Homo sapiens BCL3 binding protein; Ld_CSBP – Leishmania donovani cycling sequence binding protein; Ec_YdaL – Escherichia coli YdaL protein), and the length of the SMR domain (e.g., /1–93). Alignment was performed using MUSCLE52 and visualized using Jalview (www.jalview.org) with ClustalX coloring by conservation. The positions of previously described conserved regions11 are indicated on the alignment: the LDXH motif present in subfamily 2 SMR domains and the centrally located HGXG/TGXG (subfamilies 1 and 3/subfamily 2) are bounded by the red and blue boxes, respectively.
Conclusions and Perspectives
PPR proteins that contain a C-terminal SMR domain represent a small but enigmatic subset of the PPR protein family whose members in higher plants show diverse protein abundance and varied putative functions in organellar RNA metabolism. Phylogenetic analysis indicates that PPR-SMRs are confined to green plants and algae but that they are ancient proteins that have modestly diversified during angiosperm evolution. Despite their ancient origins and important roles in extant plant species, we still lack knowledge of their specific roles in organelle gene expression—we don’t know their exact RNA-binding sites nor their mechanism of action, and whether already reported effects on RNA processing, accumulation, and translation are direct or indirect. While the exact identity of their target RNAs remains to be confirmed, recent breakthroughs in predicting binding sites45 and identifying binding site “footprints”46 should enable more rapid progress in this area. Also, given that PPR proteins have been suggested to be specific factors for effector proteins and several have been identified in protein complexes, identifying interaction partners may also shed light on their specific functions.
The role of the SMR domain, which makes these proteins unique, remains elusive. While we have shown that amino acids in the SMR domain are conserved in some Arabidopsis PPR-SMR proteins compared with SMR domains with known endonucleolytic activity, future experiments that test the endonuclease activity of an SMR domain derived from a PPR protein as well as targeting specific amino acids to identify catalytic residues would be invaluable. Finally, if RNA endonuclease activity can be confirmed for an SMR domain, coupling this with PPR motifs that have been designed to target a specific transcript will enable “engineering” of sequence-specific RNA endonucleases. However, furthering our basic understanding of this small but unique subset of the PPR protein family is essential for these potentially exciting applications to come to fruition.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors’ research in this area is supported by the Australian Research Council: Centre of Excellence in Plant Energy Biology (CE0561495) to IS and Discovery Early Career Researcher Award (DE120101117) to KAH. SL was a recipient of a joint PhD Scholarship (International Research Fees for China) from the China Scholarship Council (CSC) and the University of Western Australia. The authors thank Dr Christopher Harris for facilitating the implementation of MrBayes on the Fornax supercomputer located at iVEC@UWA.
Glossary
Abbreviations:
- DUF
domain of unknown function
- EMB
embryo defective
- GFP
green fluorescent protein
- GUN
genomes uncoupled
- LHC
light harvesting complex
- MS
mass spectrometry
- NF
norflurazon
- PEP
plastid-encoded RNA polymerase
- PhANG
photosynthesis-associated nuclear gene
- PPR
pentatricopeptide repeat
- pTAC
plastid transcriptionally active chromosome
- rRNA
ribosomal RNA
- SMR
small MutS-related
- SVR
suppressor of variegation
- YFP
yellow fluorescent protein
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26172
References
- 1.Small ID, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–7. doi: 10.1016/S0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 2.O’Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol. 2008;25:1120–8. doi: 10.1093/molbev/msn057. [DOI] [PubMed] [Google Scholar]
- 3.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–70. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Fujii S, Small I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011;191:37–47. doi: 10.1111/j.1469-8137.2011.03746.x. [DOI] [PubMed] [Google Scholar]
- 5.Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem Soc Trans. 2007;35:1643–7. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- 6.Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–9. doi: 10.1126/science.1140516. [DOI] [PubMed] [Google Scholar]
- 8.Sachadyn P. Conservation and diversity of MutS proteins. Mutat Res. 2010;694:20–30. doi: 10.1016/j.mrfmmm.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Moreira D, Philippe H. Smr: a bacterial and eukaryotic homologue of the C-terminal region of the MutS2 family. Trends Biochem Sci. 1999;24:298–300. doi: 10.1016/S0968-0004(99)01419-X. [DOI] [PubMed] [Google Scholar]
- 10.Fukui K, Kosaka H, Kuramitsu S, Masui R. Nuclease activity of the MutS homologue MutS2 from Thermus thermophilus is confined to the Smr domain. Nucleic Acids Res. 2007;35:850–60. doi: 10.1093/nar/gkl735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukui K, Kuramitsu S. Structure and Function of the Small MutS-Related Domain. Mol Biol Int. 2011;2011:691735. doi: 10.4061/2011/691735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zoschke R, Kroeger T, Belcher S, Schöttler MA, Barkan A, Schmitz-Linneweber C. The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J. 2012;72:547–58. doi: 10.1111/j.1365-313X.2012.05081.x. [DOI] [PubMed] [Google Scholar]
- 13.Narsai R, Law SR, Carrie C, Xu L, Whelan J. In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol. 2011;157:1342–62. doi: 10.1104/pp.111.183129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Law SR, Narsai R, Taylor NL, Delannoy E, Carrie C, Giraud E, Millar AH, Small I, Whelan J. Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol. 2012;158:1610–27. doi: 10.1104/pp.111.192351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muralla R, Lloyd J, Meinke D. Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS One. 2011;6:e28398. doi: 10.1371/journal.pone.0028398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olinares PD, Ponnala L, van Wijk KJ. Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol Cell Proteomics. 2010;9:1594–615. doi: 10.1074/mcp.M000038-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang M, Friso G, Nishimura K, Qu X, Olinares PD, Majeran W, Sun Q, van Wijk KJ. Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships; the concept of orthoproteomics. J Proteome Res. 2013;12:491–504. doi: 10.1021/pr300952g. [DOI] [PubMed] [Google Scholar]
- 18.Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 2012;158:156–89. doi: 10.1104/pp.111.188474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006;18:176–97. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melonek J, Matros A, Trösch M, Mock HP, Krupinska K. The core of chloroplast nucleoids contains architectural SWIB domain proteins. Plant Cell. 2012;24:3060–73. doi: 10.1105/tpc.112.099721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics. 2010;9:1063–84. doi: 10.1074/mcp.M900325-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baerenfaller K, Massonnet C, Walsh S, Baginsky S, Bühlmann P, Hennig L, Hirsch-Hoffmann M, Howell KA, Kahlau S, Radziejwoski A, et al. Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Mol Syst Biol. 2012;8:606. doi: 10.1038/msb.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friso G, Majeran W, Huang M, Sun Q, van Wijk KJ. Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol. 2010;152:1219–50. doi: 10.1104/pp.109.152694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Rudella A, Ramirez Rodriguez V, Zybailov B, Olinares PD, van Wijk KJ. Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell. 2009;21:1669–92. doi: 10.1105/tpc.108.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Yu F, Rodermel S. An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol. 2010;154:1588–601. doi: 10.1104/pp.110.164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zoschke R, Qu Y, Zubo YO, Börner T, Schmitz-Linneweber C. Mutation of the pentatricopeptide repeat-SMR protein SVR7 impairs accumulation and translation of chloroplast ATP synthase subunits in Arabidopsis thaliana. J Plant Res. 2013;126:403–14. doi: 10.1007/s10265-012-0527-1. [DOI] [PubMed] [Google Scholar]
- 27.Susek RE, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–99. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- 28.Mochizuki N, Susek R, Chory J. An intracellular signal transduction pathway between the chloroplast and nucleus is involved in de-etiolation. Plant Physiol. 1996;112:1465–9. doi: 10.1104/pp.112.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCormac AC, Terry MJ. The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for the GUN1 and GUN5 plastid-signalling pathways during de-etiolation. Plant J. 2004;40:672–85. doi: 10.1111/j.1365-313X.2004.02243.x. [DOI] [PubMed] [Google Scholar]
- 30.Cottage A, Gray JC. Timing the switch to phototrophic growth: a possible role of GUN1. Plant Signal Behav. 2011;6:578–82. doi: 10.4161/psb.6.4.14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leister D. Retrograde signaling in plants: from simple to complex scenarios. Front Plant Sci. 2012;3:135. doi: 10.3389/fpls.2012.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfannschmidt T. Plastidial retrograde signalling--a true “plastid factor” or just metabolite signatures? Trends Plant Sci. 2010;15:427–35. doi: 10.1016/j.tplants.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Terry MJ, Smith AG. A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci. 2013;4:14. doi: 10.3389/fpls.2013.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, de la Luz Gutiérrez-Nava M, Cantero A, Delannoy E, Jiménez LF, Lurin C, Small I, et al. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 2008;56:590–602. doi: 10.1111/j.1365-313X.2008.03634.x. [DOI] [PubMed] [Google Scholar]
- 35.Chen M, Choi Y, Voytas DF, Rodermel S. Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J. 2000;22:303–13. doi: 10.1046/j.1365-313x.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- 36.Zoschke R, Watkins KP, Barkan A. A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell. 2013;25:2265–75. doi: 10.1105/tpc.113.111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cazalet C, Gomez-Valero L, Rusniok C, Lomma M, Dervins-Ravault D, Newton HJ, Sansom FM, Jarraud S, Zidane N, Ma L, et al. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet. 2010;6:e1000851. doi: 10.1371/journal.pgen.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46:347–66. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- 39.Dorrell RG, Smith AG. Do red and green make brown?: perspectives on plastid acquisitions within chromalveolates. Eukaryot Cell. 2011;10:856–68. doi: 10.1128/EC.00326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science. 2009;324:1724–6. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- 41.Bhandari D, Guha K, Bhaduri N, Saha P. Ubiquitination of mRNA cycling sequence binding protein from Leishmania donovani (LdCSBP) modulates the RNA endonuclease activity of its Smr domain. FEBS Lett. 2011;585:809–13. doi: 10.1016/j.febslet.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Gui WJ, Qu QH, Chen YY, Wang M, Zhang XE, Bi LJ, Jiang T. Crystal structure of YdaL, a stand-alone small MutS-related protein from Escherichia coli. J Struct Biol. 2011;174:282–9. doi: 10.1016/j.jsb.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe N, Wachi S, Fujita T. Identification and characterization of BCL-3-binding protein: implications for transcription and DNA repair or recombination. J Biol Chem. 2003;278:26102–10. doi: 10.1074/jbc.M303518200. [DOI] [PubMed] [Google Scholar]
- 44.Diercks T, Ab E, Daniels MA, de Jong RN, Besseling R, Kaptein R, Folkers GE. Solution structure and characterization of the DNA-binding activity of the B3BP-Smr domain. J Mol Biol. 2008;383:1156–70. doi: 10.1016/j.jmb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruwe H, Schmitz-Linneweber C. Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins? Nucleic Acids Res. 2012;40:3106–16. doi: 10.1093/nar/gkr1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32(Database issue):D115–9. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40(Database issue):D306–12. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karpenahalli MR, Lupas AN, Söding J. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics. 2007;8:2. doi: 10.1186/1471-2105-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33(Web Server issue):W116-20. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009;19:271–3. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
- 52.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green PJ. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika. 1995;82:711–32. doi: 10.1093/biomet/82.4.711. [DOI] [Google Scholar]
- 55.Ayres DL, Darling A, Zwickl DJ, Beerli P, Holder MT, Lewis PO, Huelsenbeck JP, Ronquist F, Swofford DL, Cummings MP, et al. BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Syst Biol. 2012;61:170–3. doi: 10.1093/sysbio/syr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–6. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 57.Small I, Peeters N, Legeai F, Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–90. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 58.Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjölander K, Gruissem W, Baginsky S. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol. 2004;14:354–62. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- 59.Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One. 2008;3:e1994. doi: 10.1371/journal.pone.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA. SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res. 2013;41(Database issue):D1185–91. doi: 10.1093/nar/gks1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stern DB, Hanson MR, Barkan A. Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends Plant Sci. 2004;9:293–301. doi: 10.1016/j.tplants.2004.04.001. [DOI] [PubMed] [Google Scholar]


