Abstract
Alveolar rhabdomyosarcoma (ARMS) comprises a rare highly malignant tumor presumed to be associated with skeletal muscle lineage in children. The hallmark of the majority of ARMS is a chromosomal translocation that generates the PAX3-FOXO1 fusion protein, which is an oncogenic transcription factor responsible for the development of the malignant phenotype of this tumor. ARMS cells are dependent to the oncogenic activity of PAX3-FOXO1 and its expression status in ARMS tumors correlates with worst patient outcome, suggesting that blocking this activity of PAX3-FOXO1 may be an attractive therapeutic strategy against this fusion-positive disease. In this study, we screened small-molecule chemical libraries for inhibitors of PAX3-FOXO1 transcriptional activity using a cell-based readout system. We identified the Sarco/Endoplasmic Reticulum Ca2+-ATPases (SERCA) inhibitor thapsigargin as an effective inhibitor of PAX3-FOXO1. Subsequent experiments in ARMS cells demonstrated that activation of AKT by thapsigargin inhibited PAX3-FOXO1 activity via phosphorylation. Moreover, this AKT activation appears to be associated with the effects of thapsigargin on intracellular calcium levels. Furthermore, thapsigargin inhibited the binding of PAX3-FOXO1 to target genes and subsequently promoted its proteosomal degradation. In addition, thapsigargin treatment decreases the growth and invasive capacity of ARMS cells while inducing apoptosis in vitro. Finally, thapsigargin can suppress the growth of an ARMS xenograft tumor in vivo. These data reveal that thapsigargin-induced activation of AKT is an effective mechanism to inhibit PAX3-FOXO1 and a potential agent for targeted therapy against ARMS.
Keywords: Alveolar rhabdomyosarcoma, PAX3-FOXO1, small molecule screening, thapsigargin, AKT activation
Introduction
Alveolar rhabdomyosarcoma (ARMS) is an aggressive form of soft tissue pediatric sarcoma and exhibits a poor prognosis (1). The overall 5-year survival rate for ARMS patients is ~ 50% (2). Despite modern multimodal treatment approaches, the outcome for ARMS patients remains poor. This highlights the need for new therapeutic strategies to improve ARMS patients’ outcome.
The majority of ARMS is characterized by a specific chromosomal translocation t(2;13) that leads to the expression of the novel chimeric transcription factor PAX3-FOXO1. The PAX3-FOXO1 protein contains the DNA binding portions of PAX3 fused to the transactivation domain of FOXO1 (3) and acts as a potent transcriptional activator (4). Its transcriptional activity influences the expression of genes that affect cell growth, motility and apoptosis (5), and forced expression of PAX3-FOXO1 alone can induce oncogenic transformation (6). However, PAX3-FOXO1 needs additional genetic lesions to give rise to ARMS (7–9), which is also supported by mouse models of ARMS (10). The oncogenic role of PAX3-FOXO1, which is believed to be attributed by its transcriptional activity (5, 11), is necessary for the maintenance of the transformed phenotype (12–14). Besides the oncogenic role of PAX3-FOXO1 in ARMS pathogenesis, its prognostic significance has also been acknowledged in patients with this fusion-positive disease where PAX3-FOXO1-positive tumors are associated with an aggressive phenotype and significantly worse prognosis (15–17). Therefore, these evidences suggest that the inhibition of PAX3-FOXO1 activity may provide an effective treatment strategy against fusion-positive ARMS. Indeed, a variety of approaches have been taken to inhibit PAX3-FOXO1 (12, 18–20), however, thus far no pharmaceuticals have been developed against this fusion oncoprotein.
This study described the development of a screening approach to identify small-molecule inhibitors of PAX3-FOXO1 transcriptional activity. To this end, a PAX3-FOXO1-responsive luciferase reporter was used as a primary readout followed by several secondary assays in evaluating PAX3-FOXO1 activity in ARMS cells to evaluate 3,280 pharmacological compounds. The most active candidate of PAX3-FOXO1 inhibitor identified in this screen was thapsigargin (TG) that acts as a potent inhibitor of Sarco/Endoplasmic Reticulum Ca2+-ATPases (SERCA) (21). Further characterization of this compound revealed that activation of AKT by TG inhibited the transcriptional activity of PAX3-FOXO1 via phosphorylation and subsequently promoted proteosomal degradation. This TG-induced AKT activation was apparently due to the increased intracellular Ca2+ levels. Moreover, TG inhibits the malignant characteristics of ARMS cells in vitro and blocks ARMS tumor growth in vivo. These findings offer a novel preclinical rationale for a TG-based approach to target PAX3-FOXO1 activity as a therapeutic strategy against ARMS.
Materials and Methods
Cells and cell culture
Mouse ARMS cell lines U20497T and U20325 were previously described (22, 23). Human ARMS cell lines Rh7, Rh28, Rh30 and Rh41 were provided by Dr. Peter. J Houghton (Nationwide Children’s Hospital, OH). C2C12, RD, 293A and Phoenix-Ampho cell lines were authenticated as described previously (23, 24). U20497T-PRSLuc and U20325-PRSLuc cells expressing the PAX3-FOXO1-responsive reporter luciferase (6XPRS-Luc) generated through lentivirus transduction were described previously (23). A similar strategy was applied to generate Rh30-PRSLuc and RD-PRSLuc reporter cells and CMV-driven luciferase reporter U20325-CMVLuc cells. The ARMS cells were authenticated by PAX3-FOXO1 expression, a hallmark of these cells. Cell lines used in this study were not propagated more than 2 months and/or 10 passages. All these cells were grown in D-MEM with 10% FBS except C2C12 cells (20% FBS). Additional details, including reagents, inhibitors, plasmids, antibodies, and virusproduction and transduction, are described in Supplementary Materials and Methods.
Screening of small molecule libraries
To screen for inhibitors of the PAX3-FOXO1 transcriptional activity, a total of 3,280 compounds were used. U20325-PRSLuc cells were used as a screening readout to identify compounds that decreased reporter luciferase activity. Briefly, the above cells were incubated with library compounds for 24 hours followed by luciferase activity assay. The criterion used to define hits in the primary screen was the decreased luciferase activity greater than 70% of solvent DMSO control. Compounds fulfilling the above criterion were subsequently subjected to a secondary screen to evaluate compounds that decreased luciferase activity in a dose-response manner but had no effect on cell viability. Nine compounds were selected based on dose-dependent decreased luciferase activity, while maintaining greater than 80% cell viability. Details of small molecule libraries and screening of primary and secondary assays are provided in Supplementary Materials and Methods.
Luciferase assay, immunoblot and RT-PCR analysis
Luciferase assay and analysis of immunoblot and RT-PCR were done as described in Supplementary Materials and Methods.
Measurement of intracellular calcium
Intracellular free calcium in ARMS cells was determined using Fluo-4 Direct Calcium Assay kit (Invitrogen) according to the Manufacturer’s protocol. In brief, 2×104 cells per well in 96-well plate were incubated with Fluo-4 Direct calcium reagent loading solution and incubated for 30 minutes at 37°C followed by 30 minutes at room temperature. The cells were then stimulated by the addition of TG and cytosolic calcium was measured by monitoring fluorescence intensity at 516 nm, after excitation at 494 nm.
ChIP, cell proliferation, clonogenic and soft-agar colony formation assays
These assays were performed as described previously (23, 24). Additional details weredescribed in Supplementary Material and Methods.
Apoptosis-DNA ladder formation Assay
Rh28 and Rh30 cells (5×106) were seeded in 10 cm2 dishes and incubated overnight. Cells were then treated with TG or vehicle control. After 4 days treatment, both attached and floated cells were collected, washed and lysed by cell lysis buffer (25). Cell extracts were clarified by centrifuge and SDS was added to the extracts (1% final). The extracts were then treated with RNase A (5μg/μl) for 2 hours at 56°C followed by proteinase K (2.5 μg/μl) for 2 hours at 37°C. Subsequently, DNA was extracted by with a mixture of phenol, chloroform isoamyl alcohol followed by ethanol precipitation and resolved on a 2% agarose gel and documented.
Invasion Assay
This assay was performed according to Manufacture’s protocol (BD Bioscience). Briefly, D-MEM containing 10% FBS was added to the lower chamber in a Matrigel chamber containing 24-well plate. Rh30 and U20325 cells (2×104 cells per well) treated with TG or vehicle control were suspended in D-MEM containing 1% FBS and added to the top of each insert well followed by incubation at 37°C for 24 hours. Following incubation, the non-invading cells were removed by scrubbing from the upper surface of the insert. The cells on the lower surface of the insert were stained with Hoechst dye, visualized and analyzed.
Xenograft experiments
All animal studies were conducted in accordance with the Guidance for the Care and Use of Laboratory Animals (National Institutes of Health) and approved by Institutional Animal Care and Used Committee. Details of the xenograft experiments were described in Supplementary Materials and Methods. Briefly, 5×106 Rh28 cells were injected into flank of 8–10 weeks old NOG mice. Once tumor volume reached a minimum 200 mm3, mice were treated via intravenous injection (single treatment) with TG or PBS control. Tumor volume was measured every 3 days and determined by the formula (W2×L)/2 where W = width and L=length.
Immunohistochemistry analysis
Briefly, formalin fixed dissected tumors were embedded in paraffin for immunohistochemistry analysis. Paraffin sections were the stained with hematoxylin-eosin (H&E) or subjected to immunohistochemical staining for Ki-67 and activated caspase 3. Stained slides were counterstained with hematoxylin, mounted, and images were captured. Additional details are described in Supplementary Materials and Methods.
Statistical analysis
Unpaired two-tailed student’s t test analyses were performed to ascertain statistical significance and a P value less than 0.05 was considered statistically significant.
Results
Screening of small-molecule libraries identify thapsigargin as an inhibitor of PAX3-FOXO1
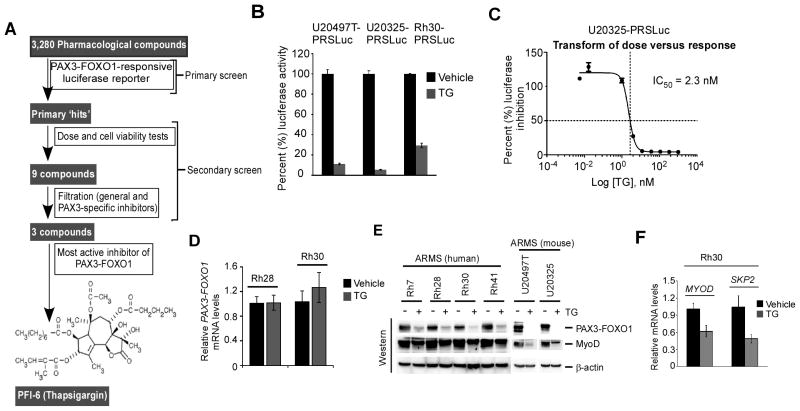
This study was aimed to identify compounds that inhibit PAX3-FOXO1 activity in ARMS cells. A screening was conducted of small-molecule libraries that consisted of 3,280 compounds to identify those that suppress PAX3-FOXO1 transcriptional activity. Fig. 1A depicted the outline of screening design that identified PFI-6 (Thapsigargin) as an inhibitor of PAX3-FOXO1 activity. In the primary screen, the library compounds were evaluated using PAX3-FOXO1-resonsive luciferase reporter ARMS-325-6XPRS-Luc cells (23), described hereafter as U20325-PRSLuc, to monitor PAX3-FOXO1 activity. The increased luciferase activity observed in these cells was abolished by PAX3-FOXO1 shRNA (23), which also depleted the PAX3-FOXO1 protein (Supplementary Fig. S1A). However, such effects were not observed with scrambled control shRNA, validating PAX3-FOXO1-mediated increase in luciferase activity in U20325-PRSLuc cells. Most PAX3-FOXO1-positive human ARMS cells, including Rh28 and Rh30 cells, express a low level of wild-type PAX3 (26) (Supplementary Fig. S1B). Since PAX3-FOXO1 contains the DNA binding regions of PAX3 (5), its presence in human ARMS cells would interfere in the identification of inhibitors of PAX3-FOXO1 activity. In contrast, mouse U20325 ARMS cells (27) express a typical fusion PAX3-FOXO1, myogenic MyoD proteins, no wild-type PAX3 (Supplementary Fig. S1C and D). Therefore, the basis of using U20325-PRSLuc cells was that wild-type PAX3 would not interfere in identifying compounds that inhibit PAX3-FOXO1-mediated reporter luciferase activity in these cells. In the primary screen, ‘hit’ compounds were selected upon treatment of U20325-PRSLuc cells with library compounds based on >70% inhibition of luciferase activity. Hit compounds were validated with secondary screening, which included a dose-response assay and parallel cell viability measurement in reporter cells. The secondary screen yielded 9 compounds (cataloged as PFI 1 to 9) that exhibit dose-dependent inhibition of luciferase activity with insignificant cell cytotoxicity (Supplementary Fig. S2A). These compounds were then subjected to additional filters to eliminate compounds that either inhibitors of general transcription or wild-type PAX3. General transcriptional inhibition was tested using U20325 cells expressing a constitutively active CMV-driven reporter luciferase. Inhibition of PAX3 was evaluated in PAX3-FOXO1 negative RD-PRSLuc cells, which are ERMS subtype RD cells that express endogenous PAX3 and the 6XPRS-luc reporter gene (Supplementary Fig. S2B). After filtering, 4 compounds remained, which were further characterized using two additional Rh30-PRSLuc and U20497T-PRSLuc (23) reporter cells. Of the 4 compounds, PFI-6 showed the strongest inhibitory effect on luciferase activity (Supplementary Fig. S2C). This compound was identified as SERCA inhibitor thapsigargin (TG). Its inhibitory effect on PAX3-FOXO1-responsive luciferase activity was re-evaluated in parallel both in human (Rh30-PRSLuc) and mouse (U20325-PRSLuc and U20497T-PRSLuc) cells. As shown in Fig. 1B, TG severely inhibits luciferase activity in these cells. The IC50 value of TG inhibited luciferase activity in U20325-PRSLuc cells was determined as 2.3 nM (Fig. 1C). Such IC50 value of TG determined in Rh30PRSLuc cells was 8.0 nM (data not shown). Because TG inhibits PAX3-FOXO1-driven reporter transcription, the effect of TG on the expression status of PAX3-FOXO1 was evaluated using Rh28 and Rh30 cells. As shown in Fig. 1D, TG did not suppress PAX3-FOXO1 mRNA levels. Therefore, the effect of TG on PAX3-FOXO1 protein levels was determined in a panel of PAX3-FOXO1 expressing ARMS cell lines (23, 28). The data demonstrated that TG decreases PAX3-FOXO1 protein levels in these cells (Fig. 1E). Moreover, TG caused a decrease in the expression of MyoD, which is a PAX3-FOXO1downstream target gene (29). In addition, the quantitative RT-PCR analysis confirmed the decreased expression of both MYOD and SKP2, another downstream target of PAX3-FOXO1 (30), in TG-treated Rh30 cells (Figure 1F). Thus, TG exerts its effects on PAX3-FOXO1-dependent gene transcription in ARMS cells in part by declining PAX3-FOXO1 protein but not mRNA levels.
Figure 1.
Small-molecule libraries screen for the identification of PAX3-FOXO1 inhibitors. (A) Diagram depicting the schematic screen procedures that used to identify thapsigargin as an inhibitor of PAX3-FOXO1 starting from 3,280 compounds. (B) Luciferase reporter assay. Indicated PAX3-FOXO1-responsive reporter ARMS cells were incubated with 250 nM TG or vehicle control for 24 hours and the luciferase activity was determined after the addition of luciferin agent (Using Luciferases Assay Kit, Promega). Luciferase activity was normalized to the vehicle control and values were expressed after protein normalization. Error bars, ± SEM (n=3) (C) Effect of different concentrations of TG (indicated as log value) on luciferase activity in U20325-PRSLuc cells incubated with TG or vehicle control for 24 hours. Values were expressed as percent (%) inhibition compared to control. IC50 value of TG on reporter gene transcription in these cells determined from the indicated dose versus response curve as shown. (D) Quantitative RT-PCR analysis of PAX3-FOXO1 performed in indicated cells incubated with 8 nM TG or vehicle control for 24 hours. CT values were normalized to ACTB. Error bars, ± SEM (n=3). (E) Immunoblot of extracts from a panel of indicated ARMS cells treated with TG (8.0 nM and 2.3 nM for human and mouse ARMS cells respectively) or vehicle control for 24 hours. As indicated, PAX3-FOXO1 and MyoD were detected with FOXO1 and MyoD antibodies respectively, and β-actin for loading control. (F) Quantitative RT-PCR analysis of MYOD and SKP2 performed in Rh30 cells incubated with 8.0 nM TG or vehicle control for 16 hours. CT values were normalized to ACTB. Error bars, ± SEM (n=3).
Thapsigargin-induced AKT activity causes transcriptional inactivation of PAX3-FOXO1 via phosphorylation
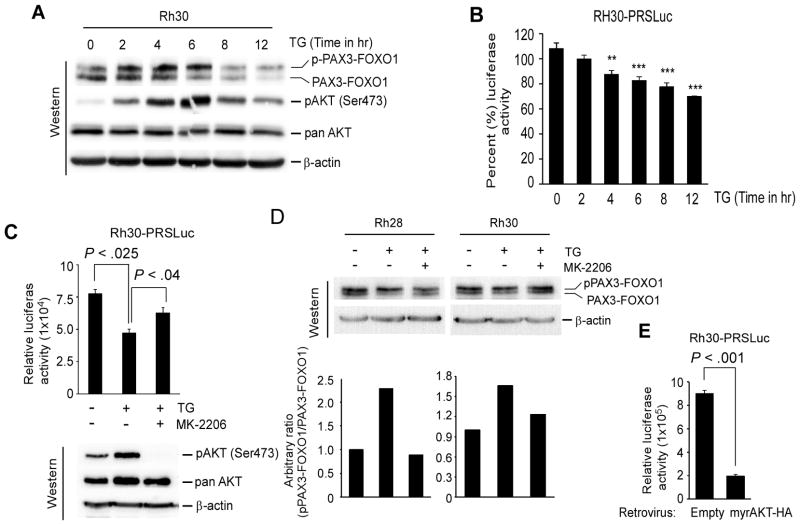
Phosphorylation modulates PAX3-FOXO1 transcriptional activity (18, 23, 31), specifically, acute AKT activation inhibits PAX3-FOXO1-dependent gene transcription by inducing its phosphorylation (23). Since TG stimulates activation of AKT (32), phosphorylation status of AKT at Ser437 (as a measurement of AKT activation) and PAX3-FOXO1 was evaluated in Rh30 cells treated with TG for 2, 4, 6, 8 and 12 hours. Increased AKT activation was detected in TG-treated cells as early as 2 hours, but decreased at the latter two time points (Fig. 2A). Additionally, an increased level of phosphorylated PAX3-FOXO1 was predominant in TG-treated cells at 2, 4 and 6 hours, but the level of it declined with decreased levels of activated AKT at latter time points. The TG-induced AKT activation coupled with increased levels of phosphorylated PAX3-FOXO1 was correlated with a gradual decrease in luciferase activity in Rh30-PRSLuc cells treated with TG for same time points (Fig. 2B). The data further showed that the decreased levels of PAX3-FOXO1 at latter time points was associated with decrease in luciferase activity in Rh30PRSLuc cells treated with TG. The role of TG-induced AKT activation in modulating PAX3-FOXO1 activity was further characterized in Rh30-PRSLuc cells incubated with TG in the presence or absence of the AKT kinase inhibitor MK-2206. The data showed that while TG alone decreases PAX3-FOXO1-mediated reporter luciferase activity, MK-2206 abolishes this TG effect (Fig. 2C). Immunoblot confirmed that TG-induced AKT activation in these cells was blocked by MK-2206. Next, the effect of TG-induced AKT activation on PAX3-FOXO1 phosphorylation status was examined by repeating the above experiment in Rh28 and Rh30 cells. The data showed that the levels of phosphorylated PAX3-FOXO1 induced by TG decreased with MK-2206 co-incubation (Fig. 2D). Finally, to verify that the inhibition of PAX3-FOXO1 activity by TG was due to AKT activation, luciferase activity in Rh30-PRSLuc cells was determined following the expression of a constitutive active form of AKT (myrAKT-HA). Indeed, constitutive active AKT retrovirus but not control retrovirus decreases luciferase activity (Fig. 2E). Together, the results suggest that TG inhibited the transcriptional activity of PAX3-FOXO1 via phosphorylation due to an increase in the levels of activated AKT.
Figure 2.
Thapsigargin mediates AKT activation can increase PAX3-FOXO1 phosphorylation and inhibit its transcriptional activity in ARMS cells. (A) Immunoblot of extracts from Rh30 cells incubated with 8.0 nM TG for indicated time points, probed for PAX3-FOXO1, activated pAKT (Ser473), total AKT, and β-actin for loading control. (B) Luciferase activity in Rh30-PRSLuc reporter cells treated with 8.0 nM TG for indicated time points. Values were normalized to protein. Error bars, ± SEM, (n=3). P-value ** = 0.001 to 0.01, *** = 0.0001 to 0.001. (C) Luciferase activity in Rh30-PRSLuc cells treated with vehicle control or 8.0 nM TG along with or without AKT inhibitor MK-2206 (1.0 μM) for 4 hours. Values were expressed after normalization with protein. Error bars, ± SEM, (n=3). (top). Immunoblot of cell extracts probed for activated pAKT (Ser473), total AKT, and β-actin for loading control (bottom). (D) Immunoblot of extracts from indicated cells treated with TG along with or without MK-2206 as described in (C), probed with FOXO1 and β-actin antibodies (top). Relative level of phosphorylated PAX3-FOXO1 (upper) is represented as a bar graph (bottom). (E) Luciferase activity in Rh30-PRSLuc cells transduced with retrovirus expressing a constitutively active AKT (myrAKT-HA) or control retrovirus (empty) grown for 2 days. Values were expressed after protein normalization. Error bars, ± SEM, (n=3).
Thapsigargin-induced Ca2+ levels correlate with AKT activation inhibited PAX3-FOXO1 transcriptional activity followed by its proteasomal degradation
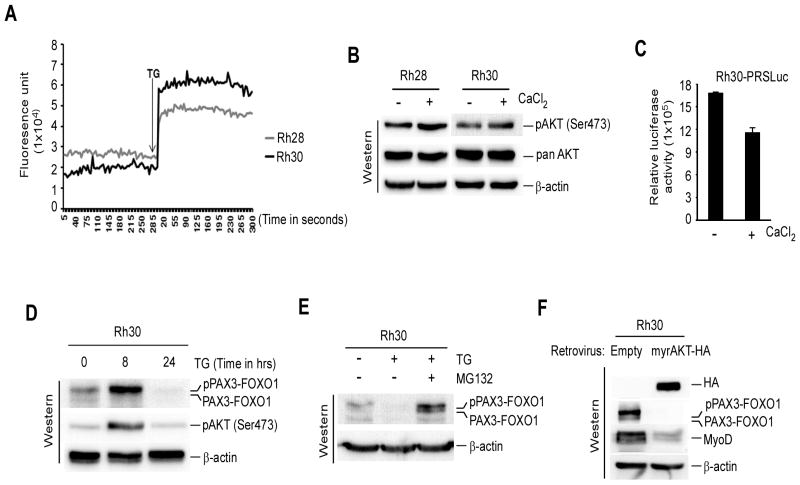
Thapsigargin disrupts the endoplasmic reticulum (ER) Ca2+ store and elevates cytosolic Ca2+ levels, which affects downstream signaling pathways involved in multiple cellular responses (21, 32, 33). Since TG activated AKT inhibits PAX3-FOXO1 transcriptional activity in ARMS cells (Fig. 2), it was possible that the activation of AKT and subsequent inhibition of PAX3-FOXO1 activity was caused by TG-induced rise of cytosolic Ca2+ levels in these cells. Indeed, TG treatment resulted in an elevation of cytosolic Ca2+ levels in Rh28 and RH30 cells (Fig. 3A). Furthermore, CaCl2 treatment caused an increase in AKT activation in above ARMS cells (Fig. 3B) and a decrease in PAX3-FOXO1-responsive reporter luciferase activity in Rh30-PRSLuc cells (Fig. 3C). These results suggest that the effect of TG on AKT activation and PAX3-FOXO1 transcriptional activity is at least partially due to its effect on intracellular calcium levels.
Figure 3.
Thapsigargin-induced intracellular calcium levels link to AKT activation coupled with suppression of PAX3-FOXO1 activity and phosphorylated PAX3-FOXO1 undergoes proteasomal degradation (A) Intracellular calcium concentration in indicated ARMS cells treated with 100 nM TG. The arrow indicates the time when TG was added. (B) Immunoblot of extracts from cells in (A) treated with or without 1.0 mM CaCl2 for 24 hours probed for activated pAKT(Ser473), total AKT and β-actin. (C) Luciferase activity in Rh30-PRSLuc cells treated with or without CaCl2 as described in (B) and values was expressed after protein normalization. Error bars, ± SEM, (n=3). (D) Immunoblot of extracts from Rh30 cells treated with 8.0 nM TG for indicated time points, probed to detect the levels of PAX3-FOXO1, activated pAKT (Ser473), and β-actin for loading control. (E) Immunoblot of extracts from Rh30 cells treated with 8.0 nM TG alongside with or without 100 nM MG132 or vehicle control for 24 hours probed for PAX3-FOXO1 and β-actin. (F) Immunoblot of extracts from Rh30 cells expressing with or without myrAKT-HA through retroviral transduction confirming reduced levels of PAX3-FOXO1 and its target MyoD. β-actin served as a loading control.
Although TG-induced AKT activation correlated with increased levels of phosphorylated PAX3-FOXO1 in ARMS cells (Fig. 2A), TG also decreased PAX3-FOXO1 protein levels in these cells (Fig. 1E). It was postulated that the AKT activation mediated increase in phosphorylated PAX3-FOXO1 will subsequently be declined in ARMS cells treated with TG. Indeed, an increased levels of phosphorylated PAX3-FOXO1 followed by decreased protein levels was observed in TG-treated RH30 cells (Fig. 3D). Increased level of phosphorylated PAX3-FOXO1 was further correlated with AKT activation in TG-treated cells. Therefore, the mechanism that decreases PAX3-FOXO1 protein levels in TG-treated ARMS cells was examined. Studies demonstrated that TG induces the ER-stress response pathways leading to proteasome-dependent degradation of proteins (34). To evaluate whether ER-stress signals were related to the decreased PAX3-FOXO1 protein levels, Rh28 and Rh30 cells were treated with DTT, which causes ER-stress by interfering with disulfide bond stability (35, 36), and analyzed by immunoblot analysis. A reduced level of PAX3-FOXO1 protein was observed in cells treated with TG as anticipated, but not with DTT (Supplementary Fig. S3), suggesting that the decreased PAX3-FOXO1 protein levels may be caused by proteosomal degradation independent of ER-stress signals. Therefore, PAX3-FOXO1 protein levels was evaluated in Rh30 cells by incubating with TG alone or co-incubating with proteosomal inhibitor MG132. The data showed that the decreased PAX3-FOXO1 protein levels in TG-treated cells was blocked by MG132 (Fig. 3E), demonstrating its proteosomal-dependent degradation. The data presented thus far suggest that activation of AKT by TG promotes PAX3-FOXO1 phosphorylation which leads to its proteosomal degradation. Therefore, the effect of AKT activation on PAX3-FOXO1 protein levels was examined in Rh30 cells that have transduced with retrovirus expressing myrAKT-HA. As shown in Fig. 3F, PAX3-FOXO1 protein was practically absent in cells that expressed HA-tagged myrAKT. Moreover, the absence of PAX3-FOXO1 protein was associated with decreased expression of its target MyoD in these cells. Together, these results suggest that TG elevates AKT-stimulated phosphorylation of PAX3-FOXO1 and subsequently leads to its proteasomal degradation.
Thapsigargin inhibits PAX3-FOXO1 binding on the regulatory elements of its target genes
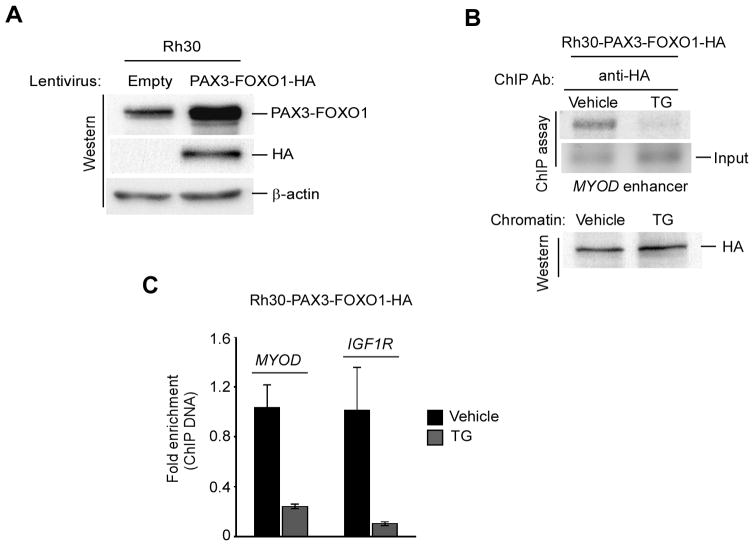
Because TG inhibits PAX3-FOXO1-driven gene transcription in ARMS cells (Fig. 2B), it may be resulted from TG-induced inhibition of PAX3-FOXO1 binding to the regulatory regions of its target genes. Hence, the effect of TG on such PAX3-FOXO1 binding to its targets was decided to perform in human ARMS cells by ChIP assay. Genome-wide binding study demonstrated that PAX3-FOXO1 binding sites are enriched with PAX3 motifs and strongly associated with induced expression of genes in human ARMS cells (29). Although human ARMS cells express low levels of wild-type PAX3 (26) (Supplementary Fig. S1B), which constitutes the N-terminal DNA binding side of PAX3-FOXO1, the use of PAX3-specific antibodies to examine TG-induced effects specifically on PAX3-FOXO1 binding by ChIP assay was avoided in these cells. The rational was that it may produce a wild-type PAX3-specific background noise for such PAX3-FOXO1 binding by ChIP in human ARMS cells. Moreover, the use of FOXO1-specific antibodies is avoided for the above ChIP assay because PAX3-FOXO1 binding to its target genes is not contributed by the DNA binding domain of wild-type FOXO1 (37). However, PAX3-FOXO1-specific antibodies are not yet commercially available. Therefore, PAX3-FOXO1 binding study was performed in human ARMS Rh30 cells following the overexpression of HA-epitope tagged PAX3-FOXO1 and using HA antibodies for ChIP. As shown in Fig. 4A, an increased level of PAX3-FOXO1 along with the expression of HA protein was observed in RH30 cells transduced with lentivirus expressing PAX3-FOXO1-HA. These were labeled as Rh30-PAX3-FOXO1-HA cells and subsequently used for ChIP to evaluate the effect of TG on PAX3-FOXO1 binding to the core enhancer regulatory region of its known target MYOD (29). A reduction in PAX3-FOXO1 binding to the enhancer region of MYOD was observed in cells incubated with TG (Fig. 4B). Moreover, the analysis of chromatin used for ChIP showed that the expression of PAX3-FOXO1-HA was not affected by TG, suggesting that the deficiency of PAX3-FOXO1 chromatin occupancy on MYOD was not due to the decreased levels of PAX3-FOXO1 protein in TG-treated cells. The quantitative PCR analysis of ChIP DNA also showed that PAX3-FOXO1 chromatin occupancy on MYOD and second intron of IGF1R, which is a target bound and directly regulated by PAX3-FOXO1 (29), was severely impaired in TG-treated these cells (Fig. 4C). Together, the data imply that TG eradicates PAX3-FOXO1 binding to its targets resulting in the suppression of PAX3-FOXO1-mediated gene transcription in ARMS cells.
Figure 4.
Thapsigargin prevents PAX3-FOXO1 recruitment to chromatin of downstream target genes. (A) Immunoblot of cell extracts shows the expression of HA-epitope and the increased levels of PAX3-FOXO1 in Rh30 cells that have transduced with lentivirus expressing PAX3-FOXO1-HA but not empty virus. β-actin served as a loading control. (B) ChIP assay using anti-HA antibody on the MYOD enhancer in Rh30-PAX3-FOXO1 cells (as shown in A) treated with TG or vehicle control for 4 hours (top). Immunoblot of chromatins used for ChIP shows equivalent levels PAX3-FOXO1-HA present either TG or vehicle treated cells (bottom). (C) Quantitative ChIP analyses using anti-HA antibody on the MYOD and IGF1R enhancer regions in chromatins used as in (B). Error bars, ± SEM, (n=3).
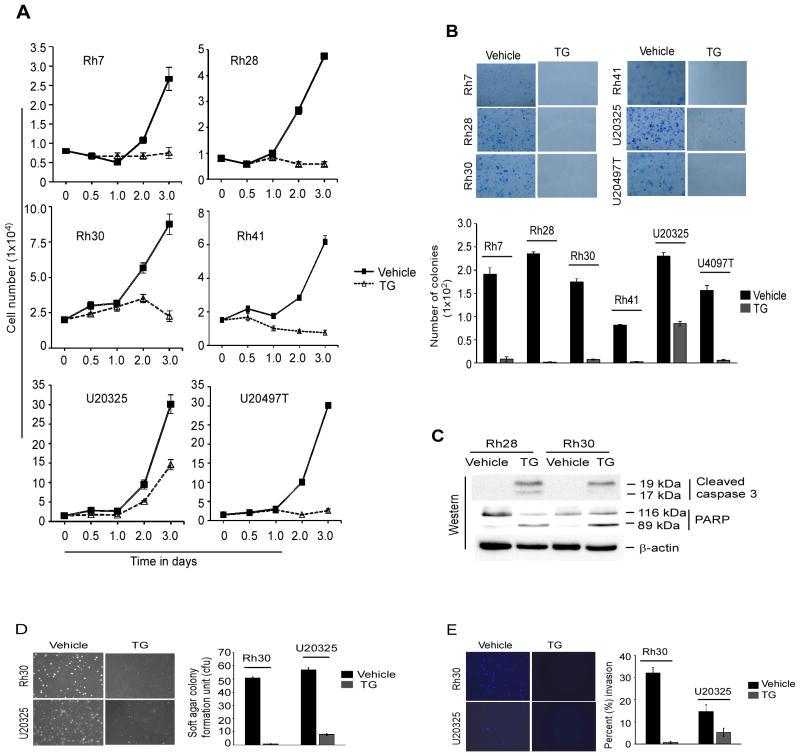
Thapsigargin inhibits ARMS cell tumorigenic potential and induces apoptosis in vitro
Studies have indicated that the inhibition of either PAX3-FOXO1 activity or expression blocks the growth and induces apoptosis of ARMS cells (14, 18, 20, 31). Since TG suppresses PAX3-FOXO1 activity and subsequently its abolition in ARMS cells (Fig. 2 and 3), the effect of TG on ARMS cell growth was examined. The results showed that TG severely impaired the growth of a panel of ARMS cell lines (Fig. 5A). The effectiveness of TG on the survival ability of these ARMS cells to grow into colonies was also examined and the data showed a severe decrease in number of colonies in TG-treated cells (Fig. 5B). These results provoked the exploration of apoptosis in TG-treated ARMS cells by examining the levels of cleaved caspase 3 and PARP, which are biomarkers of apoptosis. Indeed, an increased level of these apoptotic markers were observed in Rh28 and Rh30 cells treated with TG (Fig. 5C). Moreover, TG-induced apoptosis in these cells was evident by detecting apoptotic DNA fragmentation, a key biochemical hallmark of apoptosis (Supplementary Fig. S4).
Figure 5.
Thapsigargin inhibits malignant phenotypes and induces apoptosis in ARMS cells in vitro. (A) Growth of a panel of indicated ARMS cell lines treated with TG (concentration used as described in Fig. 1E) or vehicle control for indicated times points. Error bars, ± SEM, (n=3). (B) Clonogenic survival assay performed in TG and vehicle control treated ARMS cells as in (A). Viable colonies stained with methylene blue (top) and quantified by counting ten randomly selected fields. Error bars, ± SEM (bottom). (C) Immunoblot of extracts from indicated ARMS cells treated with 8.0 nM TG or vehicle control for 4 days probed for cleaved caspase 3 and PARP. β-actin served as a loading control. (D) Soft-agar colony formation assay performed in Rh30 and U20325 cells treated with 8.0 nM and 2.3 nM TG respectively or vehicle control for 20 day. Colonies were stained and counted in three randomly selected fields in triplicates; quantification represented as bar graph. Error bars, ± SEM. (E) Matrigel invasion assay performed in indicated ARMS cells treated with TG (as in D) or vehicle control for 24 hours. Invaded cells were stained, visualized and counted in 10 randomly selected fields; quantification is represented. Error bars, ± SEM
Studies have also implicated that PAX3-FOXO1 expression in ARMS confers an aggressive phenotype in vivo such as tumorigenic and metastatic potential (5, 38). The anchorage-independent growth of tumor cells in vitro is generally assumed to be closely related to the above in vivo events. Therefore, the effect of TG on the ability of ARMS cells to exhibit anchorage-independent cell growth was evaluated in Rh30 and U20325 cells by examining colony-forming capacity in semi-solid soft agar media. The results showed that TG inhibited the growth of these cells as evidenced by the decreased number of colonies (Fig. 5D). Additionally, the effect of TG was evaluated on invasive behavior of ARMS cells, one of the hallmarks of the metastatic potential. This was performed by treating Rh30 and U20325 cells with TG and measuring the invasiveness with a Matrigel invasion assay. The data showed that TG also inhibited these cells invasion through Matrigel (Fig. 5E). Together, these in vitro results demonstrate that TG is able to block ARMS cell growth, survival, metastatic ability and induce apoptosis.
Thapsigargin inhibits the growth of human ARMS xenografts in vivo
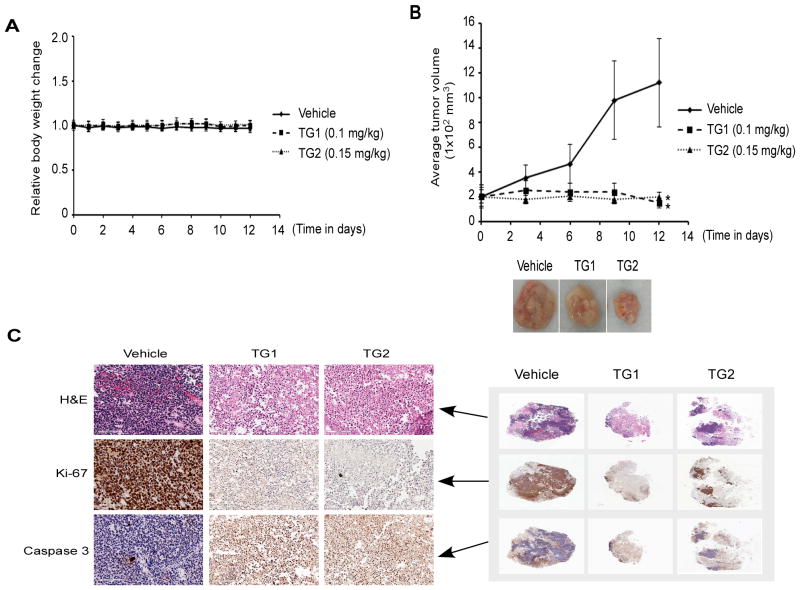
Finally, the in vivo effect of TG on tumor growth was evaluated using an Rh28 ARMS xenograft mouse model. Initial dose-finding experiment in wild-type mice demonstrated the maximum tolerable single intravenous dose of TG, which did not produce mortality, was 0.2 mg/kg body weight. Subsequently, Rh28 xenografts were treated with TG (single administration) at two different doses (0.1 mg/kg and 0.15 mg/kg); control mice received a one-time PBS treatment and tumor growth was measured. As anticipated, neither of the above one-time dosing regimens of TG produced any significant changes in body weight from treatment to the time of euthanization (Fig. 6A). However, the mice that were treated with TG either 0.1 or 0.15 mg/kg showed a significant reduced tumor growth when measuring the tumor volume (Fig. 6B). To further characterize the effect of TG on tumor growth in vivo, the resected tumors from both TG-treated and control mice were sectioned and stained with H&E or used for immunohistochemical analysis. As shown in Fig. 6C, H&E staining of tumor sections showed less viable round cell morphology in TG-treated mice (Fig. 6C). Moreover, tumors sections stained with antibody against proliferation marker Ki-67 and apoptosis-inducing activated caspase 3 evidently showedthe decreased Ki-67 but increased activated caspase 3 positive-cells in TG-treated mice. Together, the results display in vivo inhibition of tumor-cell proliferation and concomitant increased apoptosis in ARMS tumor model following TG treatment.
Figure 6.
Thapsigargin inhibits ARMS xenograft tumor growth in vivo. Mice bearing Rh28 xenograft tumors were treated with either vehicle (PBS control) or indicated two different doses of TG and monitored for 15 days. (A) Comparison of body weight of mice bearing tumors for TG treated versus control. (B) Growth of tumors (*P <0.05) for TG-treated versus control (top). Photographic represents the largest tumor size harvested on day 15 from TG-treated versus control mice (bottom). (C) Immunohistological staining of H&E (top), Ki67 (middle) and activated caspase 3 (bottom) under 20X magnification of tumor sections (left) from TG-treated versus control mice on day 15 (right).
Discussion
In this study, a cell-based screening system was used to identify small-molecule inhibitors of PAX3-FOXO1, a unique fusion oncogenic transcription factor present only in childhood cancer ARMS (5, 17). The system constituted a PAX3-FOXO1-responsive reporter gene in PAX3-deficient ARMS cells expressing endogenous PAX3-FOXO1 protein to screen a library of 3,280 compounds for inhibitors of PAX3-FOXO1 transcriptional activity. This system was an important simplification on measuring transcriptional activity solely mediated by PAX3-FOXO1, which successfully identified 3 lead compounds that inhibited PAX3-FOXO1 activity. The most promising compound identified was the sarco-endoplasmic reticulum Ca2+-ATPases (SERCA) inhibitor thapsigargin (TG). This compound was then evaluated for its potential as an inhibitor of PAX3-FOXO1. The results showed that TG elevates AKT-coupled phosphorylation of PAX3-FOXO1, resulting in the suppression of its activity and subsequently proteosomal degradation in ARMS cells. Moreover, TG inhibited the ARMS cells growth and invasiveness and induced the apoptosis of these cells in vitro. Most importantly, TG suppressed the growth of ARMS xenografts in vivo. These results underscore the robustness of this cell-based system in identifying TG as a first-in-class inhibitor of PAX3-FOXO1 suppressing ARMS cell growth and malignant phenotypes. Moreover, this system can be expanded in evaluating additional small-molecule libraries to identify potential inhibitors of PAX3-FOXO1.
The screening-identified TG was originally isolated from plant Thapsia garganica L. (Linnaeus) (39). Although TG is most widely used inhibitor of SERCA to study intracellular Ca2+ signaling (21), its action is also associated with the activation of apoptosis in cells (33). This apoptotic aspect of TG has recently exploited to develop a TG-derived prodrug for cancer therapy (40). First and foremost, this study uncovered that TG inhibits PAX3-FOXO1 transcriptional activity as demonstrated by suppression of its driven reporter gene transcription and downstream target MyoD and SKP2 expression in ARMS cells. Studies have reported TG-induced activation of multiple kinase signaling pathways (41), including activation of AKT (32). In ARMS cells, AKT activation also results in PAX3-FOXO1 phosphorylation-coupled decreased transcriptional activity (23). The data in this study reveal that TG inhibited PAX3-FOXO1 transcriptional activity is concurrent with AKT activation leading to PAX3-FOXO1 phosphorylation in ARMS cells. In support of the above view is that AKT inhibitor MK-2206 overcomes TG inhibited PAX3-FOXO1-responsive gene transcription. Although the increase of intracellular calcium levels by TG accounts for its action, most stunning in this esteem is that CaCl2 alone increased AKT activation alongside with decreased PAX3-FOXO1-responsive gene transcription. Since TG-induced AKT activation is downstream of calcium release (32), the findings of CaCl2 suggest that TG-induced suppression of PAX3-FOXO1 activity results from activation of AKT via Ca2+-induced signaling pathways. Currently, it is unclear how TG/Ca2+ activates AKT in ARMS cells and experiments are aimed at elucidating the mechanisms of this activation.
In ARMS cells, PAX3-FOXO1 phosphorylation either by PKC or GSK3 positively regulate its transcriptional activity (18, 31) and the blockade of phosphorylation on PAX3 domain of PAX3-FOXO1 results in its binding incompetence to targets, thereby transcriptional inactivation (18). In TG-treated ARMS cells, however, the data show that AKT activation mediated inhibition of PAX3-FOXO1 activity via phosphorylation impairs its binding to the target genes. Because PAX3-FOXO1 contains two consensus AKT phosphorylation site in the FOXO1 domain (42), TG-inhibited binding of PAX3-FOXO1 on its target genes may be resulted from the phosphorylation of these AKT sites. In that case, it may be concluded that phosphorylation of AKT sites in PAX3-FOXO1 impedes its binding to target gene transcription. This inhibition of PAX3-FOXO1 binding by TG may arise as a consequence of AKT activation mediating a blockade of downstream effector GSK3, which has been implicated in PAX3-FOXO1 phosphorylation and subsequent transactivation (31). Experiments are now underway in assessing these scenarios in TG-treated ARMS cells.
It has been shown that the inhibition of PAX3-FOXO1 phosphorylation blocks its transcriptional activity alongside with increased protein levels in ARMS cells (18). However, the data presented here show that TG-induced transcriptional inactive phosphorylated PAX3-FOXO1 protein subsequently declines in these cells. It seems that there is a phosphorylation-dependent transcriptionally-inactive PAX3-FOXO1 degradation in TG-treated ARMS cells. The ER-stress inducer TG triggers proteasome-dependent degradation of cellular proteins (43). Although pharmacological blockade of proteosomal activity inhibits the decline of PAX3-FOXO1 protein in TG-treated ARMS cells, it can be argued against TG-induced proteosomal activity in this scenario. Supporting evidence is the inability of DTT, a known ER-stress inducer, to decrease PAX3-FOXO1 protein levels in ARMS cells. It was hypothesized that TG-induced AKT activation might trigger proteasome-dependent degradation of phosphorylated PAX3-FOXO1 in these cells. This is supported by the results that show deceased levels of PAX3-FOXO1 protein in ARMS cells expressing a constitutively active form of AKT. Taken together, the data in this study offer that the regulation of PAX3-FOXO1 activity and expression by AKT such that phosphorylation blocks activity and promotes degradation of PAX3-FOXO1. An emphasis to this view is the therapeutic significance of TG in the inhibition of PAX3-FOXO1 activity and expression in ARMS cells. This was predicted based on studies that have shown the persistent dependence of ARMS cells on the presence of PAX3-FOXO1, as its down-regulation reduces growth, motility and induces apoptosis (9, 12–14, 44). Indeed, the data presented here show that TG is effective in inhibiting cell growth and invasion, and inducing apoptosis of ARMS cells in vitro. Most importantly, TG is effective in reducing tumor growth in an Rh28 ARMS xenograft in vivo.
In summary, this study identifies TG as an inhibitor of PAX3-FOXO1 transcriptional activity and expression in ARMS cells. Moreover, the findings highlight the significance of TG-induced AKT activation in the alteration of PAX3-FOXO1 function resulted inhibition of ARMS. Central to this perspective is the finding that TG-induced Ca2+-dependent AKT activation, via unknown mechanisms, leads to PAX3-FOXO1 phosphorylation and inhibition of its binding to suppress target gene expression and subsequent proteasomal degradation. Such TG actions on PAX3-FOXO1 eventually facilitate the block of ARMS. On the basis of these data, we conclude that TG could be developed as a potential therapeutic agent for such and possibly related translocation-positive childhood ARMS.
Supplementary Material
Acknowledgments
Financial Supports: This work was supported by Public Health Service grant AR051502 from National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) and Roswell Park Alliance Foundation grant to A.K.M.
The authors thank the Small Molecule Screening Core, Pathology Resource Network and Animal Core (Roswell Park Cancer Institute), Jean Veith for intravenous injection into mice. The authors also thank to Dr. Norman J. Karin and David W. Wolff for critical suggestions and editorial support.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exits.
References
- 1.Charytonowicz E, Cordon-Cardo C, Matushansky I, Ziman M. Alveolar rhabdomyosarcoma: is the cell of origin a mesenchymal stem cell? Cancer letters. 2009;279:126–36. doi: 10.1016/j.canlet.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 2.Ognjanovic S, Linabery AM, Charbonneau B, Ross JA. Trends in childhood rhabdomyosarcoma incidence and survival in the United States, 1975–2005. Cancer. 2009;115:4218–26. doi: 10.1002/cncr.24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr FG, Nauta LE, Davis RJ, Schafer BW, Nycum LM, Biegel JA. In vivo amplification of the PAX3-FKHR and PAX7-FKHR fusion genes in alveolar rhabdomyosarcoma. Human molecular genetics. 1996;5:15–21. doi: 10.1093/hmg/5.1.15. [DOI] [PubMed] [Google Scholar]
- 4.Bennicelli JL, Edwards RH, Barr FG. Mechanism for transcriptional gain of function resulting from chromosomal translocation in alveolar rhabdomyosarcoma. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:5455–9. doi: 10.1073/pnas.93.11.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linardic CM. PAX3-FOXO1 fusion gene in rhabdomyosarcoma. Cancer letters. 2008;270:10–8. doi: 10.1016/j.canlet.2008.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheidler S, Fredericks WJ, Rauscher FJ, 3rd, Barr FG, Vogt PK. The hybrid PAX3-FKHR fusion protein of alveolar rhabdomyosarcoma transforms fibroblasts in culture. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9805–9. doi: 10.1073/pnas.93.18.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linardic CM, Downie DL, Qualman S, Bentley RC, Counter CM. Genetic modeling of human rhabdomyosarcoma. Cancer research. 2005;65:4490–5. doi: 10.1158/0008-5472.CAN-04-3194. [DOI] [PubMed] [Google Scholar]
- 8.Naini S, Etheridge KT, Adam SJ, Qualman SJ, Bentley RC, Counter CM, et al. Defining the cooperative genetic changes that temporally drive alveolar rhabdomyosarcoma. Cancer research. 2008;68:9583–8. doi: 10.1158/0008-5472.CAN-07-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia SJ, Holder DD, Pawel BR, Zhang C, Barr FG. High expression of the PAX3-FKHR oncoprotein is required to promote tumorigenesis of human myoblasts. The American journal of pathology. 2009;175:2600–8. doi: 10.2353/ajpath.2009.090192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes & development. 2004;18:2614–26. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Begum S, Emami N, Cheung A, Wilkins O, Der S, Hamel PA. Cell-type-specific regulation of distinct sets of gene targets by Pax3 and Pax3/FKHR. Oncogene. 2005;24:1860–72. doi: 10.1038/sj.onc.1208315. [DOI] [PubMed] [Google Scholar]
- 12.Bernasconi M, Remppis A, Fredericks WJ, Rauscher FJ, 3rd, Schafer BW. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13164–9. doi: 10.1073/pnas.93.23.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebauer M, Wachtel M, Niggli FK, Schafer BW. Comparative expression profiling identifies an in vivo target gene signature with TFAP2B as a mediator of the survival function of PAX3/FKHR. Oncogene. 2007;26:7267–81. doi: 10.1038/sj.onc.1210525. [DOI] [PubMed] [Google Scholar]
- 14.Kikuchi K, Tsuchiya K, Otabe O, Gotoh T, Tamura S, Katsumi Y, et al. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochemical and biophysical research communications. 2008;365:568–74. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Barr FG. Chromosomal translocations involving paired box transcription factors in human cancer. The international journal of biochemistry & cell biology. 1997;29:1449–61. doi: 10.1016/s1357-2725(97)00095-2. [DOI] [PubMed] [Google Scholar]
- 16.Missiaglia E, Williamson D, Chisholm J, Wirapati P, Pierron G, Petel F, et al. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:1670–7. doi: 10.1200/JCO.2011.38.5591. [DOI] [PubMed] [Google Scholar]
- 17.Stegmaier S, Poremba C, Schaefer KL, Leuschner I, Kazanowska B, Bekassy AN, et al. Prognostic value of PAX-FKHR fusion status in alveolar rhabdomyosarcoma: a report from the cooperative soft tissue sarcoma study group (CWS) Pediatric blood & cancer. 2011;57:406–14. doi: 10.1002/pbc.22958. [DOI] [PubMed] [Google Scholar]
- 18.Amstutz R, Wachtel M, Troxler H, Kleinert P, Ebauer M, Haneke T, et al. Phosphorylation regulates transcriptional activity of PAX3/FKHR and reveals novel therapeutic possibilities. Cancer research. 2008;68:3767–76. doi: 10.1158/0008-5472.CAN-07-2447. [DOI] [PubMed] [Google Scholar]
- 19.Rodeberg DA, Nuss RA, Heppelmann CJ, Celis E. Lack of effective T-lymphocyte response to the PAX3/FKHR translocation area in alveolar rhabdomyosarcoma. Cancer immunology, immunotherapy : CII. 2005;54:526–34. doi: 10.1007/s00262-004-0625-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng FY, Cui J, Liu L, Chen T. PAX3-FKHR sensitizes human alveolar rhabdomyosarcoma cells to camptothecin-mediated growth inhibition and apoptosis. Cancer letters. 2009;284:157–64. doi: 10.1016/j.canlet.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treiman M, Caspersen C, Christensen SB. A tool coming of age: thapsigargin as an inhibitor of sarco-endoplasmic reticulum Ca(2+)-ATPases. Trends in pharmacological sciences. 1998;19:131–5. doi: 10.1016/s0165-6147(98)01184-5. [DOI] [PubMed] [Google Scholar]
- 22.Taniguchi E, Nishijo K, McCleish AT, Michalek JE, Grayson MH, Infante AJ, et al. PDGFR-A is a therapeutic target in alveolar rhabdomyosarcoma. Oncogene. 2008;27:6550–60. doi: 10.1038/onc.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jothi M, Nishijo K, Keller C, Mal AK. AKT and PAX3-FKHR cooperation enforces myogenic differentiation blockade in alveolar rhabdomyosarcoma cell. Cell Cycle. 2012:11. doi: 10.4161/cc.11.5.19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee MH, Jothi M, Gudkov AV, Mal AK. Histone methyltransferase KMT1A restrains entry of alveolar rhabdomyosarcoma cells into a myogenic differentiated state. Cancer research. 2011;71:3921–31. doi: 10.1158/0008-5472.CAN-10-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. The EMBO journal. 2001;20:1739–53. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis RJ, Barr FG. Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8047–51. doi: 10.1073/pnas.94.15.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Giovanni C, Landuzzi L, Nicoletti G, Lollini PL, Nanni P. Molecular and cellular biology of rhabdomyosarcoma. Future Oncol. 2009;5:1449–75. doi: 10.2217/fon.09.97. [DOI] [PubMed] [Google Scholar]
- 28.Mendes RE, Jones RN, Deshpande LM, Ross JE, Sader HS. Daptomycin activity tested against linezolid-nonsusceptible gram-positive clinical isolates. Microb Drug Resist. 2009;15:245–9. doi: 10.1089/mdr.2009.0045. [DOI] [PubMed] [Google Scholar]
- 29.Cao L, Yu Y, Bilke S, Walker RL, Mayeenuddin LH, Azorsa DO, et al. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer research. 2010;70:6497–508. doi: 10.1158/0008-5472.CAN-10-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishijo K, Chen QR, Zhang L, McCleish AT, Rodriguez A, Cho MJ, et al. Credentialing a preclinical mouse model of alveolar rhabdomyosarcoma. Cancer research. 2009;69:2902–11. doi: 10.1158/0008-5472.CAN-08-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng FY, Dong H, Cui J, Liu L, Chen T. Glycogen synthase kinase 3 regulates PAX3-FKHR-mediated cell proliferation in human alveolar rhabdomyosarcoma cells. Biochemical and biophysical research communications. 2010;391:1049–55. doi: 10.1016/j.bbrc.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber M, Hughes MR, Krystal G. Thapsigargin-induced degranulation of mast cells is dependent on transient activation of phosphatidylinositol-3 kinase. J Immunol. 2000;165:124–33. doi: 10.4049/jimmunol.165.1.124. [DOI] [PubMed] [Google Scholar]
- 33.Denmeade SR, Isaacs JT. The SERCA pump as a therapeutic target: making a “smart bomb” for prostate cancer. Cancer biology & therapy. 2005;4:14–22. doi: 10.4161/cbt.4.1.1505. [DOI] [PubMed] [Google Scholar]
- 34.Hampton RY. ER-associated degradation in protein quality control and cellular regulation. Current opinion in cell biology. 2002;14:476–82. doi: 10.1016/s0955-0674(02)00358-7. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Korennykh AV, Behrman SL, Walter P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16113–8. doi: 10.1073/pnas.1010580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puthalakath H, O’Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–49. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 37.Barr FG. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene. 2001;20:5736–46. doi: 10.1038/sj.onc.1204599. [DOI] [PubMed] [Google Scholar]
- 38.Wang C. Childhood rhabdomyosarcoma: recent advances and prospective views. Journal of dental research. 2012;91:341–50. doi: 10.1177/0022034511421490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasmussen U, Broogger Christensen S, Sandberg F. Thapsigargine and thapsigargicine, two new histamine liberators from Thapsia garganica L. Acta pharmaceutica Suecica. 1978;15:133–40. [PubMed] [Google Scholar]
- 40.Denmeade SR, Mhaka AM, Rosen DM, Brennen WN, Dalrymple S, Dach I, et al. Engineering a prostate-specific membrane antigen-activated tumor endothelial cell prodrug for cancer therapy. Science translational medicine. 2012;4:140ra86. doi: 10.1126/scitranslmed.3003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chao TS, Abe M, Hershenson MB, Gomes I, Rosner MR. Src tyrosine kinase mediates stimulation of Raf-1 and mitogen-activated protein kinase by the tumor promoter thapsigargin. Cancer research. 1997;57:3168–73. [PubMed] [Google Scholar]
- 42.del Peso L, Gonzalez VM, Hernandez R, Barr FG, Nunez G. Regulation of the forkhead transcription factor FKHR, but not the PAX3-FKHR fusion protein, by the serine/threonine kinase Akt. Oncogene. 1999;18:7328–33. doi: 10.1038/sj.onc.1203159. [DOI] [PubMed] [Google Scholar]
- 43.Hong M, Li M, Mao C, Lee AS. Endoplasmic reticulum stress triggers an acute proteasome-dependent degradation of ATF6. Journal of cellular biochemistry. 2004;92:723–32. doi: 10.1002/jcb.20118. [DOI] [PubMed] [Google Scholar]
- 44.Liu L, Wang YD, Wu J, Cui J, Chen T. Carnitine palmitoyltransferase 1A (CPT1A): a transcriptional target of PAX3-FKHR and mediates PAX3-FKHR-dependent motility in alveolar rhabdomyosarcoma cells. BMC cancer. 2012;12:154. doi: 10.1186/1471-2407-12-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.