Abstract
Nicotinamide adenine dinucleotide (NAD) is a central metabolic cofactor by virtue of its redox capacity, and as such regulates a wealth of metabolic transformations. However, the identification of the longevity protein Sir2, the founding member of the sirtuin protein family, as being NAD+-dependent reignited interest in this metabolite. The sirtuins (SIRT1-7 in mammals) utilize NAD+ to deacetylate proteins in different subcellular compartments with a variety of functions, but with a strong convergence on optimizing mitochondrial function. Since cellular NAD+ levels are limiting for sirtuin activity, boosting its levels is a powerful means to activate sirtuins as a potential therapy for mitochondrial, often age-related, diseases. Indeed, supplying excess precursors, or blocking its utilization by PARP enzymes or CD38/CD157, boosts NAD+ levels, activates sirtuins and promotes healthy aging. Here, we discuss the current state of knowledge of NAD+ metabolism, primarily in relation to sirtuin function. We highlight how NAD+ levels change in diverse physiological conditions, and how this can be employed as a pharmacological strategy.
Keywords: Aging, Metabolism, Mitochondria, PARPs, Sirtuins
1. Introduction
Nicotinamide adenine dinucleotide (NAD) is a metabolic cofactor that is present in cells either in its oxidized (NAD+) or reduced (NADH) form. Its function as a cofactor for a multitude of enzymatic reactions has been appreciated since the early 1900’s, when NAD+ was described as a “cozymase” in fermentation and its characteristics were elucidated, not in the last place by several Nobel prize winners (Berger et al., 2004). In its function as an oxidoreductase cofactor, NAD+ is critical for a wide range of enzymatic reactions, including for instance GAPDH in glycolysis. NAD redox balance is tightly regulated (we refer the reader for more information of this aspect to (Houtkooper et al., 2010a)). After a period of relative anonymity, NAD+ became again in the spotlight because it was identified as a substrate for a major class of deacetylase proteins, the sirtuins, named after the founding member of the family yeast Sir2p (Ivy et al., 1986; Rine and Herskowitz, 1987). Sirtuins have pleiotropic metabolic effects, and since NAD+ levels reflect the energy state of the cell, it was hypothesized that sirtuins could function as metabolic sensors that use NAD+ as a messenger and cosubstrate, translating this signal to a cellular adaptation (Canto and Auwerx, 2011). Due to this development, it has become apparent that pathways involved in synthesis or consumption of NAD+ are attractive targets for the management of conditions with dysfunctional metabolism, including not only obesity and diabetes, but also cancer and neurodegenerative diseases (Houtkooper and Auwerx, 2012).
In this review, we will describe the pathways contributing to NAD+ homeostasis, and will discuss their potential benefits in the management of metabolic disease.
2. NAD+ metabolism
NAD+ metabolism is a careful balance between biosynthesis on one hand and its breakdown on the other. Importantly, both sides of the balance are composed of several pathways.
NAD+ biosynthesis and salvage
NAD+ can be synthesized from various precursors (figure 1). De novo biosynthesis, which starts from the amino acid tryptophan, occurs primarily in the liver and kidney but is considered a minor contributor to the total pool of NAD+ (reviewed in detail in (Houtkooper et al., 2010a)). On the other hand, biosynthesis from nicotinic acid (NA) or nicotinamide (NAM)—both present in our diet as vitamin B3—is the primary source of NAD+. These pathways, also known as the salvage or Preiss-Handler pathway, are important for NAD+ homeostasis. This is illustrated by the human disease pellagra, which is caused by NAD+ deficiency subsequent to poor dietary intake of precursors. Pellagra is clinically characterized by the 4 “D’s”, i.e. diarrhea, dermatitis, dementia, and if untreated ultimately death. Pellagra, in dogs prevalent as black tongue disease, is caused by deficiency of NAD+ (precursors) and can be easily treated by providing the vitamin in the diet (Elvehjem et al., 1937). Synthesis of NAD+ from NA or NAM involves phosphoribosyl transfer followed by adenylyl transfer. In the case of NA, the resulting product requires a final ATP-dependent amidation step by NAD synthase to complete the synthesis of NAD+ (for more detail on NAD+ enzymology we refer the reader to (Houtkooper et al., 2010a; Magni et al., 2004).
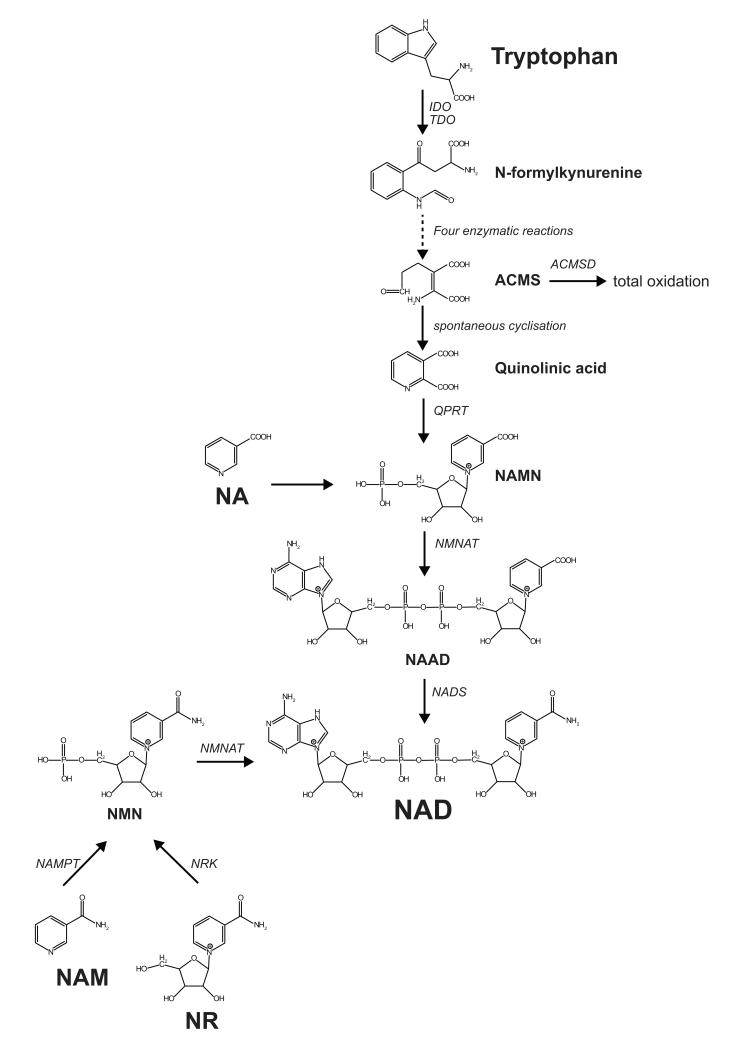
Figure 1. De novo biosynthesis and salvage pathway of NAD+.
The first step of the NAD+ de novo biosynthesis is the conversion of tryptophan into N-formylkynurenine through an enzymatic reaction catalyzed by either indoleamine 2,3-dioxygenase (IDO) or tryptophan 2,3-dioxygenase (TDO). N-Formylkynurenine is then converted in four successive enzymatic reactions into α-Amino-β-carboxymuconate-ε-semialdehyde (ACMS), which can undergo either enzymatic conversion directed to total oxidation or spontaneous cyclization to quinolinic acid. The following step is the formation of nicotinic acid mononucleotide (NAMN) through the quinolinate phosphoribosyltransferase (QPRT) activity. NAMN is then transformed to nicotinic acid adenine dinucleotide (NAAD) by the nicotinamide mononucleotide adenylyltransferase (NMNAT) enzymes. The final step in the biosynthesis of NAD+ is the amidation of NAAD by the NAD synthase enzyme. NAD+ is also synthesized through the NAD+ salvage pathway from its precursors NA, NAM, or NR. From NA, the first step in NAD+ synthesis is catalyzed by nicotinic acid phosphoribosyltransferase (NAPT) and leads to the formation of NAMN. Similarly, NAM is converted by nicotinamide phosphoribosyltransferase (NAMPT), forming NMN, which is also the product of phosphorylation of NR by nicotinamide riboside kinase (NRK). Both NAMN and NMN are then converted by NMNAT, after which the NAMN-derived NAAD requires the final amidation through NAD synthase.
Recently, a “new” NAD+ precursor—NAM riboside (NR)—that also enhances NAD+ levels through the salvage pathways was described (Bieganowski and Brenner, 2004). Even though this pathway for NAD biosynthesis was already known in bacteria, it was only recently demonstrated that NR —which is found in milk and yeast—could also be used to synthesize NAD+ in eukaryotes (Bieganowski and Brenner, 2004). Indeed, supplementation of NR to cells or mice increases the levels of NAD+ and results in the activation of its downstream signaling cascades (Canto et al., 2012), as will be discussed in more detail below.
Sirtuins
Sirtuins are a class of metabolic regulators, of which seven orthologues exist in mammals (Blander and Guarente, 2004; Chalkiadaki and Guarente, 2012; Haigis and Sinclair, 2010; Houtkooper et al., 2012). The sirtuins differ in tissue expression, subcellular localization, enzymatic activity and targets. Sirtuins are named after their homology to yeast Sir2 (silent regulator 2) (Ivy et al., 1986; Rine and Herskowitz, 1987), which was originally described as a NAD+-dependent class III histone deacetylases (Imai et al., 2000). Sirtuins are categorized into four different classes according to the amino acid sequence-based phylogenetic analysis (Frye, 2000): Class I includes SIRT1, SIRT2, and SIRT3, Class II and Class III SIRT4 and SIRT5, respectively, and SIRT6 and SIRT7 come under Class IV. Mammalian sirtuins show a diverse subcellular localization. SIRT1, SIRT6 and SIRT7 are mainly found in the nucleus, SIRT2 is predominantly in the cytoplasm, while SIRT3, SIRT4 and SIRT5 are localized in mitochondria (Pirinen et al., 2012). It has become clear, however, that the sirtuins not only deacetylate histones, but also a wide range of other proteins (figure 2). Most of the targets are involved in stress response pathways, whether metabolic in nature, genotoxic or otherwise. In addition, some of the sirtuins were reported to ADP-ribosylate proteins rather than deacetylate (Haigis et al., 2006), and SIRT5 was shown to act as a demalonylase and desuccinylase (Du et al., 2011; Peng et al., 2011; Wang et al., 2011). Future research will have to determine whether other sirtuins also possess such activity, but it seems likely that multiple family members function as deacylases.
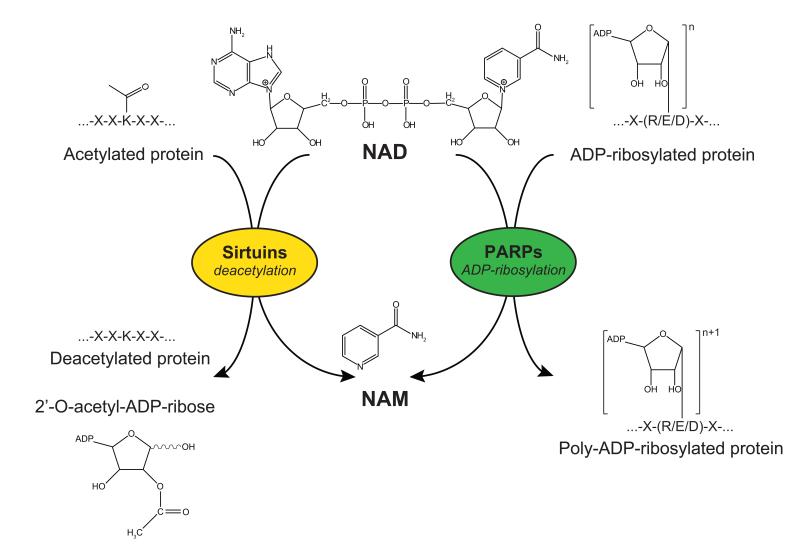
Figure 2. Sirtuins and PARPs as competing NAD+-consuming enzymes.
Sirtuins are NAD+-consuming deacetylases, using NAD+ to cleave acetyl groups from acetylated lysine residues of target proteins, in a reaction that generates NAM and 2′-O-acetyl-ADP-ribose. PARP family members are also NAD+-consuming enzymes. They catalyze a reaction in which multiple ADP-ribose groups are transferred to a mono ADP ribosylated substrate protein, forming long chains and branches of ADP-ribosyl polymers.
The nuclear sirtuin SIRT1 is the best-known member of the family especially after it was described as the target of the polyphenol, resveratrol (Howitz et al., 2003), which is found in low quantities in red wine (see section “Pharmacological control of NAD+ levels”). SIRT1 deacetylates histones, but its key activity involves the regulation of mitochondrial biogenesis and stress response through the deacetylation of PGC-1α (Rodgers et al., 2005) and FOXO1 (Brunet et al., 2004; van der Horst et al., 2004). Its role in stress response is further confirmed by the identification of p53, HIF-1α and NF-κB as SIRT1 targets (reviewed in (Canto and Auwerx, 2011; Houtkooper et al., 2012)). Less is known about the other sirtuins, but it is clear that they also impact on metabolism in various ways. The cytosolic SIRT2 was shown to deacetylate tubulin (North et al., 2003), as well as the sterol regulatory element binding protein-2 (SREBP-2) (Luthi-Carter et al., 2010), although genetic evidence for this latter association is so far lacking. Furthermore, SIRT2 deacetylates phosphoenolpyruvate carboxykinase (PEPCK) to control gluconeogenesis (Jiang et al., 2011) and was recently shown to deacetylate the receptor-interacting protein 1 (RIP1), and thereby serve as a critical component of the TNFα-mediated programmed necrosis pathway (Narayan et al., 2012). Finally, SIRT2 controls myelin formation in vivo through the atypical-PKC regulator PAR3 (Beirowski et al., 2011). The mitochondrial sirtuins—SIRT3, SIRT4 and SIRT5—deacetylate protein targets involved in oxidative phosphorylation (Ahn et al., 2008), fatty acid oxidation (Hirschey et al., 2010), ketogenesis (Shimazu et al., 2010), oxidative stress (Someya et al., 2010), glutamate metabolism (Haigis et al., 2006), urea cycle (Nakagawa et al., 2009), as well as several other mitochondrial pathways (Hebert et al., 2013), and thereby regulate multiple facets of mitochondrial metabolism (reviewed in (Houtkooper et al., 2012; Pirinen et al., 2012; Verdin et al., 2010)). Surprisingly, mice deficient in either of the mitochondrial sirtuins do not develop an overt metabolic phenotype under basal non-challenged conditions (Fernandez-Marcos et al., 2012; Haigis et al., 2006; Haigis and Sinclair, 2010; Hirschey et al., 2010; Lombard et al., 2007; Nakagawa et al., 2009). SIRT6 deacetylates both histones and DNA polymerase β, a DNA repair protein. As a result, deletion of SIRT6 in mice results in a severe premature aging phenotype associated with defects in DNA repair (Mostoslavsky et al., 2006). Additionally, Sirt6−/− mice have reduced IGF1 levels and are severely hypoglycemic (Mostoslavsky et al., 2006), possibly mediated by the HIF1α-dependent activation of glycolysis and subsequent decreases in glucose levels (Zhong et al., 2010). No in vivo molecular deacetylation targets have been described for the nucleolar SIRT7, but knockdown or overexpression of SIRT7 resulted in decreased or increased RNA polymerase I-mediated transcription, respectively (Ford et al., 2006). A thorough characterization of Sirt7−/− mice has not been performed but mice deficient for SIRT7 display hyperacetylation of p53, develop cardiomyopathy and die young (Vakhrusheva et al., 2008).
Other NAD+ consumers: PARPs and CD38/CD157
NAD+ is not only consumed by sirtuins, but also by the members of the poly(ADP-ribose) polymerase (PARPs) family and the NAD glycohydrolases CD38 and CD157 (figure 2). The nuclear PARP1 accounts for most of the PARP activity in vivo and is the best studied family member, but critical functions for other PARPs are emerging (Luo and Kraus, 2012; Schreiber et al., 2006). PARPs are best characterized for their role in DNA damage pathways, but more generally PARPs regulate adaptive stress responses, including inflammatory, oxidative, proteotoxic, and genotoxic stresses (Luo and Kraus, 2012). For example, when protein translation is stalled, PARP13 localizes to cytosolic stress granules and regulates microRNA expression and activity, alleviating the protein stress (Leung et al., 2011). More recently, however, the role of the different PARPs role in metabolism has become more apparent, as Parp1 and Parp2 knockout mice are protected against high-fat diet induced obesity (Bai et al., 2011a; Bai et al., 2011b; Bai et al., 2007). Based on the functional links between PARPs and sirtuins, it was tempting to speculate that the levels of NAD+, the joint co-substrate, could in fact dictate these functions. The in vivo characterization of mutant mice for PARP1—the major PARP isoform—further corroborated this hypothesis (Bai et al., 2011b). Parp1−/− mice were protected from high-fat diet induced obesity and showed overall improved fitness compared to control littermates. The effects of Parp1 deletion were due to elevation of NAD+ levels and subsequent SIRT1-dependent activation of mitochondrial metabolism in brown adipose tissue and muscle (Bai et al., 2011b). Importantly, this genetic evidence was confirmed by pharmacological studies using PARP inhibitors (Bai et al., 2011b), as will be further discussed in section “Pharmacological control of NAD+ levels”. Interestingly, Parp2−/− mice were also protected against diet-induced obesity, but this effect was not mediated through changes in NAD+ levels and activation of SIRT1 as the case in Parp1−/− mice, but rather through the induction of muscle SIRT1 expression (Bai et al., 2011a). While PARPs are stress response proteins, the NAD+-consuming CD38 is an ubiquitous, but still quite enigmatic enzyme, involved in maintaining calcium homeostasis (Lee, 2012). Although CD38 is often referred to as an ectoenzyme, it may also have intracellular activity (Lee, 2012), although its full potential as an NAD+ consumer is yet to be discovered. Still, even if most CD38 activity occurs outside the cell, the resultant metabolites can be transported inside, most likely in the form of NR (Nikiforov et al., 2011). Similar to PARP deficient mice, CD38−/− mice display highly elevated NAD+ levels that are accompanied by SIRT1 activation and, at the organismal level, increased energy expenditure (Barbosa et al., 2007). The role of CD157, also known as Bst1, is not characterized in the context of metabolic disease. It is interesting to note, however, that a recent study demonstrated a role for CD157 in the response of intestinal Paneth cells to CR (Yilmaz et al., 2012). When Paneth cells are exposed to caloric restriction (CR), CD157 is activated to produce cyclic ADP-ribose, which in turn signals to intestinal stem cells to switch on maintenance programs rather than differentiation (Yilmaz et al., 2012). Whether or not CD157 is involved in metabolic regulation in other tissues as well remains to be investigated.
3. Modulation of NAD levels by physiological processes
Fasting and exercise
SIRT1 activity is generally increased during restrictive metabolic conditions, and decreased in situations of caloric excess. Complying with the fact that SIRT1 activity is regulated by NAD+, these observations shed light on the potential role of NAD+ as a metabolic sensor in stress conditions, where the levels of NAD+ are generally affected. During fasting and exercise, the level of NAD+ increases (Canto et al., 2009; Canto et al., 2010). Interestingly, this increase in NAD+ levels is linked with sirtuin activation. In a similar way, CR in mouse models leads to an increase in the level of NAD+ in different tissues, such as muscle, liver and white adipose tissue (Canto et al., 2010; Chen et al., 2008). Conversely, caloric excess by means of a high-fat diet (Kim et al., 2011), but also aging (Braidy et al., 2011; Yoshino et al., 2011), lead to reduced NAD+ levels. Several studies have revealed through the prism of the sirtuin family the potential involvement NAD+ in longevity modulation during CR, and in a larger extent in the physiological aging mechanism.
Caloric restriction, NAD+ and aging
Aging is characterized by a progressive accumulation of molecular, cellular and organ damage, leading to dys- or malfunction of many metabolic processes and a generalized physiological decline. If this decline is uncompensated, it can result in the development of age-associated diseases, like neurodegenerative diseases, such as Alzheimer’s and Parkinson’s disease, metabolic disorders, such as type 2 diabetes and atherosclerosis, or cancer. Despite the complexity of the aging process, many studies have demonstrated over the last two decades that aging is subject to regulation by common signaling pathways, transcription factors and their co-regulators. Among the different proposed mechanisms that impact on and modulate longevity, CR is by far the most consistent and reproducible intervention that increases lifespan and protects against the decline of biological functions with age in many different species.
CR is defined as a moderate limitation of food intake below the ad libitum level, without malnutrition. It was already known for ages that moderation and composition of diet can influence the aging process (Schafer, 2005), but the modern day founder was Clive McCay, who put its benefits in the scientific spotlight in 1935 (McCay et al., 1989). CR remains the most effective and reproducible intervention to extend lifespan and delay the development of age-associated diseases in divergent species, from yeast to monkeys (Houtkooper et al., 2010b; Koubova and Guarente, 2003). The concept that enhanced mitochondrial function upon CR contributes to its beneficial effects on lifespan was extended to humans, in which a general improvement in metabolic health occurs during CR ((Civitarese et al., 2007) and reviewed in (Holloszy and Fontana, 2007)). The implication of NAD+ in aging is closely intertwined with the major role proposed for the NAD+-dependent sirtuin enzymes in CR.
The role of NAD+ in CR emerged from studies in yeast, where pioneering work revealed that longevity mediated by CR requires the NAD+ biosynthesis pathway and the activity of the yeast sirtuin homolog Sir2 (Lin et al., 2000). It was proposed that increased Sir2 activity leads to the repression of recombination events at the homologous repeats present in the ribosomal DNA, preventing as such the formation of extra-chromosomal ribosomal DNA circles, which is one of the causes of replicative aging in yeast. Thus, the increased dosage of SIR2 in yeast prevents the formation of extra-chromosomal ribosomal DNA circles and prolongs lifespan, whereas its inhibition has the opposite effect, reducing the replicative life span by 50% (Kaeberlein et al., 1999). Interestingly, mimicking CR by reducing glucose concentration of the growth medium from 2% to 0.5% is sufficient to extend lifespan to a similar level as by overexpressing Sir2, and these effects were dependent on the Sir2 gene or the nicotinate phosphoribosyltransferase 1 (NPT1) gene, which is involved in the biosynthesis of NAD (Lin et al., 2000).
The worm genome comprises four genes sharing homology with Sir2, with sir-2.1 being the closest homolog of Sir2 (Frye, 2000), and sir-2.1 is required for the lifespan extension in response to the CR-mutation eat-2 (Wang and Tissenbaum, 2006). In the fruitfly Drosophila melanogaster, which expresses five homologs of Sir2 (Frye, 2000), CR extends lifespan and increases dSir2 mRNA expression, but was unable to mediate lifespan extension in flies where dSir2 had been deleted (Rogina and Helfand, 2004). As discussed before, the mammalian genome encodes seven sirtuins and within this family of proteins, SIRT1 is the most extensively studied in the context of the lifespan regulation. Several in vivo studies assign a role for SIRT1 to explain the longer life in mice under CR. In fact, in Sirt1−/− mice the beneficial effects on metabolism and longevity induced by a CR diet are attenuated, although it should be noted that these mice are very sick to start with (Boily et al., 2008; Chen et al., 2005). Conversely, transgenic mice, constitutively overexpressing the Sirt1 gene, exhibit a range of features that are reminiscent of the phenotypes seen in CR mice, as they are lighter and metabolically more active, show improved glucose homeostasis, and develop less cancer (Banks et al., 2008; Bordone et al., 2007; Herranz et al., 2010; Pfluger et al., 2008).
The beneficial effects mediated by SIRT1 under CR conditions have been proposed to be due to an improvement of the mitochondrial function and biogenesis (Guarente and Picard, 2005), but also to a global increase in stress resistance and maintenance of the cellular and mitochondrial homeostasis (figure 3). Recent studies in worms added a new layer of complexity in this dynamic mechanism, by showing that an early burst of ROS is required for the induction of the ROS defense pathway and for the lifespan extension under CR conditions (Mouchiroud et al., 2011; Schulz et al., 2007). Moreover, compelling new evidence suggest that other pro-longevity pathways, induced by CR and/or stress conditions, could also potentially involve SIRT1, such as mitophagy, mitochondrial dynamics (fission/fusion) and mitochondrial unfolded response (Durieux et al., 2011; Egan et al., 2011; Yang et al., 2011). Further studies are needed to decipher the exact role of NAD+ and SIRT1 in lifespan regulation through these mechanisms.
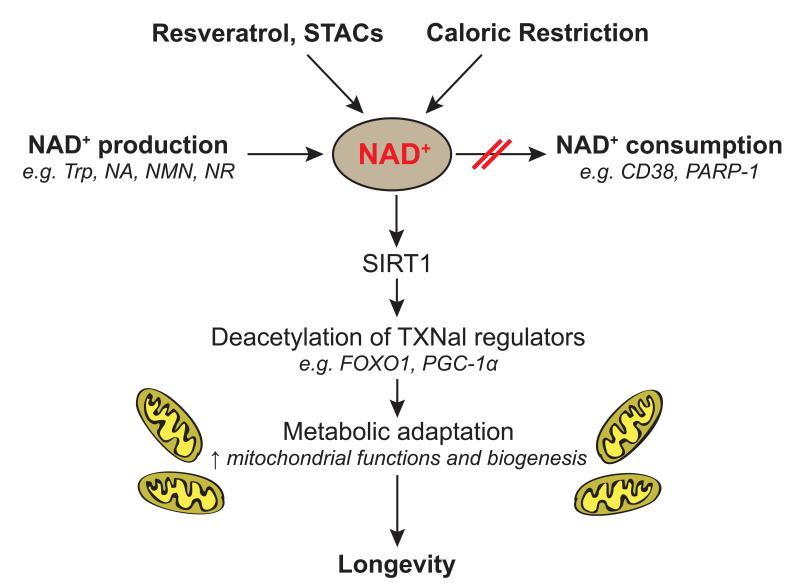
Figure 3. NAD+ as a keystone for mitochondrial regulation.
NAD+ is a rate-limiting metabolite for the SIRT1 enzymatic activity. SIRT1 activity can be increased by different types of physiological or experimental interventions that increase NAD+ levels, such as caloric restriction, treatment with STACs/resveratrol, enhancement of NAD+ biosynthesis through supplementation with precursors (NA, NR, NMN), or through inhibition of NAD+-consuming activities, such as the PARPs or CD38. This stimulation of the SIRT1 deacetylation activity leads to an improvement of the mitochondrial adaptation and ultimately to beneficial effects on health and lifespan.
However, the requirement of sirtuin proteins in longevity modulation is not without controversy and still the object of an intense debate. Initial work reported that increased expression of the yeast protein Sir2 and of related sirtuin proteins in Caenorhabditis elegans and Drosophila melanogaster extends lifespan (Rogina and Helfand, 2004; Tissenbaum and Guarente, 2001). These observations were recently challenged by showing that the effect of overexpression of worm sir-2.1 and fly SIR2 on lifespan is, at best, limited or even absent (Burnett et al., 2011; Viswanathan and Guarente, 2011). In view of the predominant role of SIRT1 in metabolic homeostasis in mammals, we speculate that SIRT1 is rather a major keystone in health maintenance and stress response, instead of being crucial for the determination of lifespan per se. As such, NAD+ serves as a central metabolite that communicates the metabolic state under such stressful conditions and activates the sirtuins to trigger adaptive and protective responses. Further studies are needed to elucidate the role of the others sirtuin family members in longevity regulation.
4. Pharmacological control of NAD+ levels
Resveratrol and STACs
As briefly mentioned above, various compounds can modulate the levels of NAD+ and thereby activate sirtuin enzymes. One of the best described is the polyphenol 3,5,4′-trihydroxystillbene, which was originally isolated in 1939 from the roots of the plant white hellebore (Veratrum grandiflorum O. Loes) (Takaoka, 1939). This fact is reflected in the common name, of the compound, i.e., resveratrol, a combination of res (from the fact that it is a resorcinol or dihydroxy benzene), veratr (Veratrum) and ol (for the alcoholic groups) (figure 4). A few decades later, in 1963, resveratrol was extracted from the roots of another plant, Japanese knotweed (Reynoutria japonica), commonly used in traditional Chinese and Japanese medicine (reviewed in (Baur and Sinclair, 2006)). In the wild, resveratrol is found in many edible fruits, such as grapes, blueberries, cranberries or peanuts (Baur and Sinclair, 2006). It is also present in red wine at concentration ranging from 0.1 to 14.3 milligrams per liter (Soleas et al., 1997). Interestingly, higher levels of resveratrol are made by plants in response to infection or nutrient stress, thereby qualifying it as a phytoalexin. Resveratrol has attracted the attention of the scientific community when it was demonstrated that this molecule could be the origin cardioprotective effects specific to red wine which is commonly referred to as «the French paradox » (Kopp, 1998; Pace-Asciak et al., 1995). Since then resveratrol has been shown to be effective in preventing and delaying the progression of various diseases such as cancer (Jang et al., 1997), cardiovascular disease and glucose intolerance (Timmers et al., 2011) and ischemic stroke (Sinha et al., 2002; Wang et al., 2002). In 2003, an in vitro high throughput screening of small chemical compounds identified resveratrol as the most potent activator of SIRT1, able to extend the yeast lifespan (Howitz et al., 2003). As in yeast, treatment with resveratrol increases lifespan of worms and fly in a SIRT1 (sir-2.1 or Sir2p, respectively) dependent manner (Howitz et al., 2003; Wood et al., 2004), although this is controversial (Bass et al., 2007; Kaeberlein et al., 2005). In mammals, resveratrol supplementation in mice fed with a high fat diet (HFD) improved physiological parameters, as these mice showed a decrease in HFD-induced weight gain, an improved glucose metabolism, and less damage to the pancreas and heart, all features associated with increased activity of AMPK and PGC-1α, culminating in increased mitochondrial number and function (Baur et al., 2006; Lagouge et al., 2006). Ultimately, this beneficial metabolic profile leads to a longer life expectancy (Pearson et al., 2008). This effect on longevity is, however, only observed when mice are fed with a HFD. Under chow diet, treatment with resveratrol does not extend mice lifespan, although it seems to improve their overall health (Barger et al., 2008; Pearson et al., 2008). In chow fed mice, resveratrol significantly attenuated several hallmarks of aging, such as reduced inflammatory and apoptotic events in vascular endothelium, limiting the formation of cataracts, preservation of bone density and conservation of motor activity with age (Barger et al., 2008; Baur et al., 2006; Lagouge et al., 2006; Pearson et al., 2008). These resveratrol treated mice also exhibit a transcriptional profile in heart, liver and muscle that is similar to that seen in animals under CR, supports the idea that resveratrol mimics the effects of food limitation in ad libitum fed individuals. The fact that the effects of the CR mimetic, resveratrol, also depend on the diet contributed to the controversy that CR could work only in animals maintained under “regular” laboratory conditions. Indeed, it has been questioned whether the beneficial effects observed under CR were not due to a simple “rescue” of the deleterious effects brought by the state of overnutrition specific of the artificial diets prepared in laboratories (Harper et al., 2006; Longo and Finch, 2003; Martin et al., 2010; Prentice, 2005). These observations could explain the conflicting results obtained with CR in non-human primates, where two independent studies—both of which using a different control diet and feeding regimen—showed a different extent of CR health benefits (Colman et al., 2009; Mattison et al., 2012). Since it was known that resveratrol acts as an inhibitor of the ATP synthase complex in the oxidative phosphorylation (Zheng and Ramirez, 2000), it was hypothesized that the effects of resveratrol could be mediated by activation of the AMP-activated protein kinase (AMPK), rather than direct SIRT1 activation (Beher et al., 2009; Canto et al., 2010; Pacholec et al., 2010; Um et al., 2010). Indeed, AMPK is activated upon resveratrol treatment and resveratrol’s effects are lost in cells or tissues devoid of AMPK (Canto et al., 2010; Um et al., 2010) (figure 5). Following AMPK activation, expression of NAMPT increased, leading to increased NAD+ levels and activation of SIRT1 (Canto et al., 2009; Canto et al., 2010; Fulco et al., 2008). It should be noted that the mode of action of resveratrol is still subject of debate. Recent reports suggest that resveratrol may act through phosphodiesterase 4 inhibition, thereby mobilizing calcium stores and activating AMPK (Park et al., 2012), or through allosteric activation of SIRT1 that is dependent on structural hydrophobic motifs in SIRT1 substrates (Hubbard et al., 2013), but further work is needed to clarify whether this also plays a physiological role.
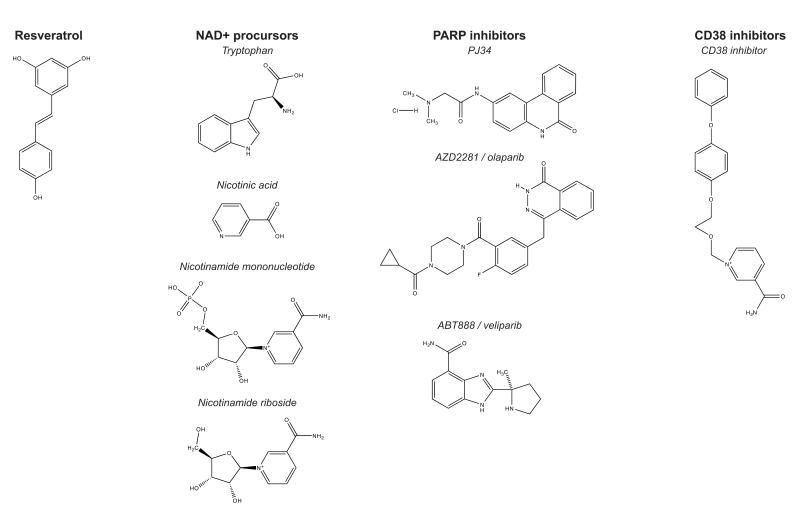
Figure 4. Compounds increasing NAD+ levels.
Chemical structures of compounds that increase NAD+ levels. We distinguish four types of NAD+ boosters, including resveratrol, the primary NAD+ precursors NA, NMN and NR, the PARP inhibitors PJ34, olaparib and veliparib, and the CD38 inhibitor 1-{[2-(4-phenoxyphenoxy)ethoxy]methyl}-3-(aminocarbonyl)-pyridinium chloride (Dong et al., 2011).
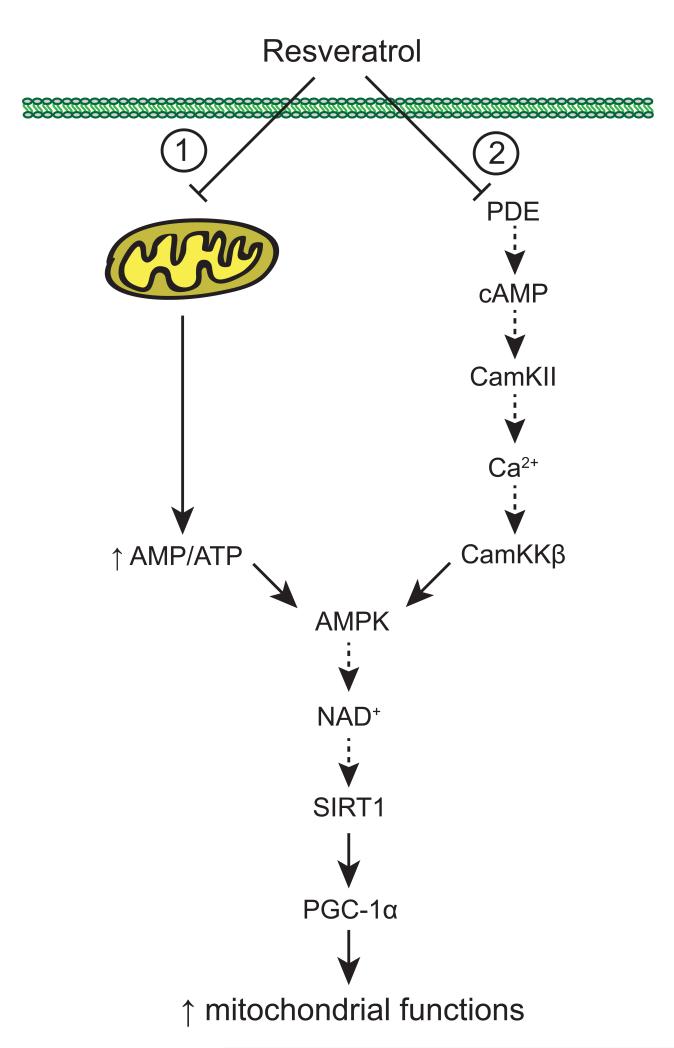
Figure 5. Mechanisms of action of resveratrol.
Resveratrol promotes mitochondrial biogenesis and functions through indirect AMPK and SIRT1 activation. First, compelling evidence suggests that the metabolic actions of resveratrol are based on its ability to act as a mitochondrial poison by inhibiting ATP synthase activity (1). The resulting energy stress will in turn activate AMPK, leading to the stimulation of SIRT1 by increasing NAD+ levels. SIRT1 will then activate downstream targets through deacetylation, ultimately leading to an improvement of mitochondrial function. Another potential explanation of how resveratrol acts is based on a recent report showing that resveratrol inhibits phosphodiesterase (PDE) 4 activity and induces cAMP signaling resulting in Ca2+ release and, ultimately the activation of the CamKKb-AMPK pathway (2).
Regardless of these issues, resveratrol treatment in mice induces mitochondrial biogenesis and energy expenditure (Baur et al., 2006; Lagouge et al., 2006). The required dose (200-400 mg/kg/day), however, was in a range that is normally incompatible with human consumption (15-30 g per day for a 75 kg person). Importantly and reassuring from a clinical point of view, resveratrol supplementation in obese humans reached beneficial effects at a far lower dose (150 mg per day) (Timmers et al., 2011). It should be noted, however, that resveratrol failed to exert beneficial effects in non-obese female subjects (Yoshino et al., 2012), in line with mouse data where resveratrol is particularly effective in high-fat diet fed mice (Baur et al., 2006; Lagouge et al., 2006).
Increasing SIRT1 activity through the use of synthetic SIRT activating compounds or STACs, such as SRT1720, also prevents diet-induced obesity and delays the onset of associated metabolic abnormalities in mice models (Feige et al., 2008; Milne et al., 2007). Similar to resveratrol, the small molecule SIRT1 activator SRT1720, which is 1,000-fold more potent than resveratrol, also extends both mean and maximum mouse lifespan in mice fed with HFD (Minor et al., 2011). This effect is potentially explained by the improved metabolic homeostasis, as typified by insulin sensitization and increased mitochondrial and locomotor activity (Feige et al., 2008; Milne et al., 2007). It is important to mention that there is, however, still some debate whether SRT1720, as well as other SIRT1-activators, are targeting SIRT1 in a specific manner (Beher et al., 2009; Dai et al., 2010; Pacholec et al., 2010), although recent evidence suggests that this may be dictated by specific hydrophobic residues in SIRT1 substrates (Hubbard et al., 2013). Human efficacy studies with such synthetic SIRT1 activators should be reported in the near future.
NAD+ boosters
A more specific approach to modify NAD+ levels involves the supplementation of NAD+ precursors or NAD+-consumption inhibitors (figure 3). The precursors NA, NMN, and NR, but also PARP or CD38 inhibitors increase NAD levels in various cell types and tissues of mice (Bai et al., 2011b; Barbosa et al., 2007; Canto et al., 2012; Yoshino et al., 2011).
NA, also called niacin, has been used to treat dietary tryptophan deficits (pellagra) and hyperlipidemia (Elvehjem et al., 1937; Karpe and Frayn, 2004; Sauve, 2008). Along another line, reduced NAMPT expression in Nampt+/− mice, which decreased plasma NMN levels and lowered NAD+ levels in brown adipose tissue, at least in female mice, was shown to impair glucose-stimulated insulin secretion (Revollo et al., 2007). This effect can be rescued by NMN supplementation, which indicates that the maintenance of NAD+ levels is crucial for pancreatic function (Revollo et al., 2007). A recent study has confirmed this observation by demonstrating that NAMPT activity is compromised by HFD and aging, and could contribute to the pathogenesis of type 2 diabetes (Yoshino et al., 2011). Enhancing NAD+ biosynthesis by intraperitoneal injection of NMN indeed improved glucose homeostasis in obese mice (Yoshino et al., 2011). NR is another potent naturally occurring NAD+ precursor. Dietary NR supplementation increases NAD+ levels in brown adipose tissue, muscle and liver, but not in brain and white adipose tissue (Canto et al., 2012). In responsive tissues, NR activates both SIRT1 and SIRT3 activity, improves mitochondrial function and thereby alleviates metabolic dysfunction associated with HFD-induced obesity (Canto et al., 2012). These observations indicate that NR could also be used to prevent and/or treat the decline in mitochondrial function observed upon age-associated diseases.
Another attractive angle to modulate NAD+ levels consists in targeting the activity of other (non-sirtuin) NAD+-consuming enzymes, such as PARPs and CD38 (figure 3). Following the hypothesis that NAD+ is the rate-limiting factor for the activation of SIRT1, SIRT1 activity is reduced when PARP1 is activated. Conversely, genetic or pharmacological inactivation of PARP1 (Bai et al., 2011b) or CD38 (Barbosa et al., 2007; Dong et al., 2011) function increases NAD+ levels resulting in SIRT1 activation and the induction of a gene expression program that stimulates mitochondrial metabolism. In line with this premise, Parp1 or Cd38 knockout mice show improved metabolic function and are protected against diet-induced obesity (Bai et al., 2011b; Barbosa et al., 2007). It is important to note that in Parp1−/− mice only SIRT1 is activated (Bai et al., 2011b), which is in contrast to the dual SIRT1 and SIRT3 activation observed with the NAD+ precursor NR. This is, however, consistent with the nuclear localization and activity of PARP1, raising NAD+ levels in this compartment, while NR can be converted to NAD+ in mitochondria as well (Canto et al., 2012). While PARP inhibitors have not yet been tested for metabolic effects, their clinical development for cancer therapy could provide interesting opportunities in this direction (Audeh et al., 2010; Gelmon et al., 2011). In fact, we expect that boosting oxidative metabolism through modulating NAD+ levels could in itself prove to be a powerful anti-cancer regimen and actually inhibit the “Warburg effect”. A potential issue could be the pancreatic dysfunction observed in Parp2−/− mice (Bai et al., 2011a) that may be replicated when mice are treated with pan-PARP inhibitors. Small molecule CD38 inhibitors (or CD157 inhibitors if identified) may circumvent this problem, but further insights in the NAD+ accumulation and trafficking are required (Sauve et al., 1998; Sauve and Schramm, 2002). Additionally, prolonged studies are required to exclude potential long-term adverse effects of both the PARP and CD38 inhibitors.
5. Conclusions and future perspectives
Following the description of the sirtuin enzyme family, both allosteric sirtuin activating compounds and small molecules that modulate NAD+ levels took center stage as a potential ways to therapeutically target sirtuin signaling for the treatment of diseases linked with mitochondrial dysfunction. Research on NAD+ modulators led to the identification of various physiological and pharmaceutical interventions that resulted in an increase in NAD+ levels and an activation of SIRT1. These include direct biosynthesis precursors, e.g. NR and NMN, inhibitors of its utilization, e.g. PARP and CD38/CD157 inhibitors, and indirect effectors, e.g. resveratrol. Improving on this relatively rich pharmacology, we recently identified a compound (NR) that activates both SIRT1 and SIRT3, since it is metabolized throughout the cell, not only in the nucleus. It is furthermore likely that novel pharmacophores, which affect the activity of other enzymes in the NAD biosynthesis pathway, will emerge as potential tools to modulate NAD+ levels (e.g. targeting NR kinase). This highlights the importance of understanding the basic biochemistry underlying NAD+ homeostasis. Despite these specificities, all compounds that increase NAD+ levels improve metabolic homeostasis and protect against metabolic diseases, although to various degrees. Future work on the differences between these distinct classes of compounds that modulate NAD+ levels and affect sirtuin signaling—for instance defining their molecular targets and/or tissue-specific effects—will elucidate how the compounds can be optimally employed clinically to treat both common polygenic forms of metabolic diseases, such as type 2 diabetes, as well as rare monogenic diseases that cause metabolic dysfunction, such as inherited mitochondrial diseases.
Importantly, the potential therapeutic impact of changing NAD+ levels is not limited to the metabolic realm discussed in this manuscript, but can be extended to other age-associated diseases such as neurodegenerative disorders and cancer. Indeed, CR permits to maintain neuronal plasticity with age (Adams et al., 2008; Halagappa et al., 2007; Patel et al., 2005) and reduces cancer risk (Kalaany and Sabatini, 2009), suggesting that increasing NAD+ levels could also be interesting within this context (Speakman and Mitchell, 2011). In line with these beneficial effects of CR on age-associated pathologies, transgenic mice with SIRT1 overexpression are not only protected against HFD-induced metabolic pathology, but also show protection against tumor development (Herranz et al., 2010) and progression of neurodegenerative disease (Donmez et al., 2010; Jeong et al., 2012; Wareski et al., 2009). Importantly, some of the NAD+ boosting compounds may display adverse effects that may preclude their use for relatively mild metabolic disturbances that can also be treated by changes in life style, while this may be acceptable for the severely debilitating inherited conditions (Houtkooper and Auwerx, 2012). Finally, although it is clear that increasing NAD+ levels improves stress response and prevents development of metabolic disease, for instance upon HFD, it remains to be seen whether or not increasing NAD+ levels may lead to increased lifespan in higher species.
Acknowledgements
The authors thank the members of the Auwerx lab for discussions.
LM is supported by a “Fondation pour la Recherche Médicale” fellowship, and RHH is supported by an AMC Postdoc fellowship and a ZonMw-VENI grant (number 91613050). The work in the LISP is supported by grants of the Ecole Polytechnique Fédérale de Lausanne, the EU Ideas programme (Sirtuins; ERC-2008-AdG-23118), The Velux Stiftung, NIH (1R01HL 106511-01A1), and the Swiss National Science Foundation (SNF 31003A-124713 and CRSII3-136201). JA is the Nestle Chair in Energy Metabolism.
Footnotes
Declaration of interest The authors declare no conflict of interest with respect to this publication.
References
- Adams MM, Shi L, Linville MC, Forbes ME, Long AB, Bennett C, Newton IG, Carter CS, Sonntag WE, Riddle DR, et al. Caloric restriction and age affect synaptic proteins in hippocampal CA3 and spatial learning ability. Experimental neurology. 2008;211:141–149. doi: 10.1016/j.expneurol.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci USA. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN, Oaknin A, Loman N, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C, Brunyanszki A, Huber A, Szanto M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H, et al. PARP-2 Regulates SIRT1 Expression and Whole-Body Energy Expenditure. Cell Metab. 2011a;13:450–460. doi: 10.1016/j.cmet.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, et al. PARP-1 Inhibition Increases Mitochondrial Metabolism through SIRT1 Activation. Cell Metab. 2011b;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 [corrected] controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma [corrected] heterodimer. J Biol Chem. 2007;282:37738–37746. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa MT, Soares SM, Novak CM, Sinclair D, Levine JA, Aksoy P, Chini EN. The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. Faseb J. 2007;21:3629–3639. doi: 10.1096/fj.07-8290com. [DOI] [PubMed] [Google Scholar]
- Barger JL, Kayo T, Vann JM, Arias EB, Wang J, Hacker TA, Wang Y, Raederstorff D, Morrow JD, Leeuwenburgh C, et al. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS One. 2008;3:e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass TM, Weinkove D, Houthoofd K, Gems D, Partridge L. Effects of resveratrol on lifespan in Drosophila melanogaster and Caenorhabditis elegans. Mech Ageing Dev. 2007;128:546–552. doi: 10.1016/j.mad.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- Beher D, Wu J, Cumine S, Kim KW, Lu SC, Atangan L, Wang M. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem Biol Drug Des. 2009;74:619–624. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- Beirowski B, Gustin J, Armour SM, Yamamoto H, Viader A, North BJ, Michan S, Baloh RH, Golden JP, Schmidt RE, et al. Sir-two-homolog 2 (Sirt2) modulates peripheral myelination through polarity protein Par-3/atypical protein kinase C (aPKC) signaling. Proc Natl Acad Sci USA. 2011;108:E952–961. doi: 10.1073/pnas.1104969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Ramirez-Hernandez MH, Ziegler M. The new life of a centenarian: signalling functions of NAD(P) Trends Biochem Sci. 2004;29:111–118. doi: 10.1016/j.tibs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Bieganowski P, Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PloS one. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- Braidy N, Guillemin GJ, Mansour H, Chan-Ling T, Poljak A, Grant R. Age related changes in NAD+ metabolism oxidative stress and sirt1 activity in wistar rats. PLoS ONE. 2011;6:e19194. doi: 10.1371/journal.pone.0019194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. Targeting Sirtuin 1 to Improve Metabolism: All You Need Is NAD+? Pharmacol Rev. 2011 doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, et al. The NAD(+) Precursor Nicotinamide Riboside Enhances Oxidative Metabolism and Protects against High-Fat Diet-Induced Obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. Interdependence of AMPK and SIRT1 for Metabolic Adaptation to Fasting and Exercise in Skeletal Muscle. Cell Metab. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkiadaki A, Guarente L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat Rev Endocrinol. 2012;8:287–296. doi: 10.1038/nrendo.2011.225. [DOI] [PubMed] [Google Scholar]
- Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Kustigian L, Carney D, Case A, Considine T, Hubbard BP, Perni RB, Riera TV, Szczepankiewicz B, Vlasuk GP, et al. SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J Biol Chem. 2010;285:32695–32703. doi: 10.1074/jbc.M110.133892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Si YQ, Sun SY, Pu XP, Yang ZJ, Zhang LR, Zhang LH, Leung FP, Lam CM, Kwong AK, et al. Design, synthesis and biological characterization of novel inhibitors of CD38. Org Biomol Chem. 2011;9:3246–3257. doi: 10.1039/c0ob00768d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvehjem C, Madden R, Strong F, Woolley D. Relation of nicotinic acid and nicotinic acid amide to canine black tongue. J Am Chem Soc. 1937;59:1767–1768. [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Fernandez-Marcos PJ, Jeninga EH, Canto C, Harach T, de Boer VC, Andreux P, Moullan N, Pirinen E, Yamamoto H, Houten SM, et al. Muscle or liver-specific Sirt3 deficiency induces hyperacetylation of mitochondrial proteins without affecting global metabolic homeostasis. Sci Rep. 2012;2:425. doi: 10.1038/srep00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, Hirte H, Huntsman D, Clemons M, Gilks B, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- Guarente L, Picard F. Calorie restriction--the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halagappa VK, Guo Z, Pearson M, Matsuoka Y, Cutler RG, Laferla FM, Mattson MP. Intermittent fasting and caloric restriction ameliorate age-related behavioral deficits in the triple-transgenic mouse model of Alzheimer’s disease. Neurobiology of disease. 2007;26:212–220. doi: 10.1016/j.nbd.2006.12.019. [DOI] [PubMed] [Google Scholar]
- Harper JM, Leathers CW, Austad SN. Does caloric restriction extend life in wild mice? Aging Cell. 2006;5:441–449. doi: 10.1111/j.1474-9726.2006.00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, et al. Calorie Restriction and SIRT3 Trigger Global Reprogramming of the Mitochondrial Protein Acetylome. Mol Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Auwerx J. Exploring the therapeutic space around NAD+ J Cell Biol. 2012;199:205–209. doi: 10.1083/jcb.201207019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010a;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Williams RW, Auwerx J. Metabolic networks of longevity. Cell. 2010b;142:9–14. doi: 10.1016/j.cell.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Hubbard BP, Gomes AP, Dai H, Li J, Case AW, Considine T, Riera TV, Lee JE, E SY, Lamming DW, et al. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Ivy JM, Klar AJ, Hicks JB. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta RG, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- Jeong H, Cohen DE, Cui L, Supinski A, Savas JN, Mazzulli JR, Yates JR, 3rd, Bordone L, Guarente L, Krainc D. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat Med. 2012;18:159–165. doi: 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaany NY, Sabatini DM. Tumours with PI3K activation are resistant to dietary restriction. Nature. 2009;458:725–731. doi: 10.1038/nature07782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe F, Frayn KN. The nicotinic acid receptor--a new mechanism for an old drug. Lancet. 2004;363:1892–1894. doi: 10.1016/S0140-6736(04)16359-9. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim JH, Noh S, Hur HJ, Sung MJ, Hwang JT, Park JH, Yang HJ, Kim MS, Kwon DY, et al. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J Proteome Res. 2011;10:722–731. doi: 10.1021/pr100892r. [DOI] [PubMed] [Google Scholar]
- Kopp P. Resveratrol, a phytoestrogen found in red wine. A possible explanation for the conundrum of the ‘French paradox’? Eur J Endocrinol. 1998;138:619–620. doi: 10.1530/eje.0.1380619. [DOI] [PubMed] [Google Scholar]
- Koubova J, Guarente L. How does calorie restriction work? Genes Dev. 2003;17:313–321. doi: 10.1101/gad.1052903. [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Lee HC. Cyclic ADP-ribose and nicotinic acid adenine dinucleotide phosphate (NAADP) as messengers for calcium mobilization. J Biol Chem. 2012;287:31633–31640. doi: 10.1074/jbc.R112.349464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Finch CE. Evolutionary medicine: from dwarf model systems to healthy centenarians? Science. 2003;299:1342–1346. doi: 10.1126/science.1077991. [DOI] [PubMed] [Google Scholar]
- Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi-Carter R, Taylor DM, Pallos J, Lambert E, Amore A, Parker A, Moffitt H, Smith DL, Runne H, Gokce O, et al. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc Natl Acad Sci U S A. 2010;107:7927–7932. doi: 10.1073/pnas.1002924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S. Enzymology of NAD+ homeostasis in man. Cell Mol Life Sci. 2004;61:19–34. doi: 10.1007/s00018-003-3161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B, Ji S, Maudsley S, Mattson MP. “Control” laboratory rodents are metabolically morbid: why it matters. Proc Natl Acad Sci U S A. 2010;107:6127–6133. doi: 10.1073/pnas.0912955107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, Allison DB, Young JE, Bryant M, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–321. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. 1935. Nutrition. 1989;5:155–171. discussion 172. [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor RK, Baur JA, Gomes AP, Ward TM, Csiszar A, Mercken EM, Abdelmohsen K, Shin YK, Canto C, Scheibye-Knudsen M, et al. SRT1720 improves survival and healthspan of obese mice. Sci Rep. 2011;1 doi: 10.1038/srep00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Mouchiroud L, Molin L, Kasturi P, Triba MN, Dumas ME, Wilson MC, Halestrap AP, Roussel D, Masse I, Dalliere N, et al. Pyruvate imbalance mediates metabolic reprogramming and mimics lifespan extension by dietary restriction in Caenorhabditis elegans. Aging Cell. 2011;10:39–54. doi: 10.1111/j.1474-9726.2010.00640.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan N, Lee IH, Borenstein R, Sun J, Wong R, Tong G, Fergusson MM, Liu J, Rovira II, Cheng HL, et al. The NAD-dependent deacetylase SIRT2 is required for programmed necrosis. Nature. 2012;492:199–204. doi: 10.1038/nature11700. [DOI] [PubMed] [Google Scholar]
- Nikiforov A, Dolle C, Niere M, Ziegler M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J Biol Chem. 2011;286:21767–21778. doi: 10.1074/jbc.M110.213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Pace-Asciak CR, Hahn S, Diamandis EP, Soleas G, Goldberg DM. The red wine phenolics trans-resveratrol and quercetin block human platelet aggregation and eicosanoid synthesis: implications for protection against coronary heart disease. Clin Chim Acta. 1995;235:207–219. doi: 10.1016/0009-8981(95)06045-1. [DOI] [PubMed] [Google Scholar]
- Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, Rehmann H, Taussig R, Brown AL, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NV, Gordon MN, Connor KE, Good RA, Engelman RW, Mason J, Morgan DG, Morgan TE, Finch CE. Caloric restriction attenuates Abeta-deposition in Alzheimer transgenic models. Neurobiol Aging. 2005;26:995–1000. doi: 10.1016/j.neurobiolaging.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, et al. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8:157–168. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.012658. M111 012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirinen E, Lo Sasso G, Auwerx J. Mitochondrial sirtuins and metabolic homeostasis. Best Pract Res Clin Endocrinol Metab. 2012;26:759–770. doi: 10.1016/j.beem.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice AM. Starvation in humans: evolutionary background and contemporary implications. Mech Ageing Dev. 2005;126:976–981. doi: 10.1016/j.mad.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Revollo JR, Korner A, Mills KF, Satoh A, Wang T, Garten A, Dasgupta B, Sasaki Y, Wolberger C, Townsend RR, et al. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6:363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA. NAD+ and vitamin B3: from metabolism to therapies. J Pharmacol Exp Ther. 2008;324:883–893. doi: 10.1124/jpet.107.120758. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Munshi C, Lee HC, Schramm VL. The reaction mechanism for CD38. A single intermediate is responsible for cyclization, hydrolysis, and base-exchange chemistries. Biochemistry. 1998;37:13239–13249. doi: 10.1021/bi981248s. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Schramm VL. Mechanism-based inhibitors of CD38: a mammalian cyclic ADP-ribose synthetase. Biochemistry. 2002;41:8455–8463. doi: 10.1021/bi0258795. [DOI] [PubMed] [Google Scholar]
- Schafer D. Aging, longevity, and diet: historical remarks on calorie intake reduction. Gerontology. 2005;51:126–130. doi: 10.1159/000082198. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Fisch G, Teague B, Tamborlane WV, Banyas B, Allen K, Savoye M, Rieger V, Taksali S, Barbetta G, et al. Prevalence of impaired glucose tolerance among children and adolescents with marked obesity. N Engl J Med. 2002;346:802–810. doi: 10.1056/NEJMoa012578. [DOI] [PubMed] [Google Scholar]
- Soleas GJ, Diamandis EP, Goldberg DM. Resveratrol: a molecule whose time has come? And gone? Clin Biochem. 1997;30:91–113. doi: 10.1016/s0009-9120(96)00155-5. [DOI] [PubMed] [Google Scholar]
- Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR, Mitchell SE. Caloric restriction. Mol Aspects Med. 2011;32:159–221. doi: 10.1016/j.mam.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Takaoka MJ. Resveratrol, a new phenolic compound, from Veratrum grandiflorum. Journal of the Chemical Society of Japan. 1939;60:1090–1100. [Google Scholar]
- Timmers S, Konings E, Bilet L, Houtkooper RH, van de Weijer T, Goossens GH, Hoeks J, van der Krieken S, Ryu D, Kersten S, et al. Calorie Restriction-like Effects of 30 Days of Resveratrol Supplementation on Energy Metabolism and Metabolic Profile in Obese Humans. Cell Metab. 2011;14:612–622. doi: 10.1016/j.cmet.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Um JH, Park SJ, Kang H, Yang S, Foretz M, McBurney MW, Kim MK, Viollet B, Chung JH. AMP-activated protein kinase-deficient mice are resistant to the metabolic effects of resveratrol. Diabetes. 2010;59:554–563. doi: 10.2337/db09-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477:E1–2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- Wang Q, Xu J, Rottinghaus GE, Simonyi A, Lubahn D, Sun GY, Sun AY. Resveratrol protects against global cerebral ischemic injury in gerbils. Brain Res. 2002;958:439–447. doi: 10.1016/s0006-8993(02)03543-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mechanisms of ageing and development. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhao F, Peng J, Xu J. Protein 8-class secondary structure prediction using conditional neural fields. Proteomics. 2011;11:3786–3792. doi: 10.1002/pmic.201100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareski P, Vaarmann A, Choubey V, Safiulina D, Liiv J, Kuum M, Kaasik A. PGC-1{alpha} and PGC-1{beta} regulate mitochondrial density in neurons. J Biol Chem. 2009;284:21379–21385. doi: 10.1074/jbc.M109.018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Yang CC, Chen D, Lee SS, and Walter L. The dynamin-related protein DRP-1 and the insulin signaling pathway cooperate to modulate Caenorhabditis elegans longevity. Aging Cell. 2011;10:724–728. doi: 10.1111/j.1474-9726.2011.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J, Conte C, Fontana L, Mittendorfer B, Imai S, Schechtman KB, Gu C, Kunz I, Rossi Fanelli F, Patterson BW, et al. Resveratrol supplementation does not improve metabolic function in nonobese women with normal glucose tolerance. Cell metabolism. 2012;16:658–664. doi: 10.1016/j.cmet.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide Mononucleotide, a Key NAD(+) Intermediate, Treats the Pathophysiology of Diet- and Age-Induced Diabetes in Mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Ramirez VD. Inhibition of mitochondrial proton F0F1-ATPase/ATP synthase by polyphenolic phytochemicals. Br J Pharmacol. 2000;130:1115–1123. doi: 10.1038/sj.bjp.0703397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]