Abstract
Knowledge of the role of components of the RNA polymerase I transcription machinery is paramount to understanding regulation of rDNA expression. We describe key findings for the roles of essential transcription factor SL1 and activator upstream binding factor (UBF). We demonstrate that human SL1 can direct accurate Pol I transcription in the absence of UBF and can interact with the rDNA promoter independently and stably, consistent with studies of rodent SL1 but contrary to previous reports of human SL1. UBF itself does not bind stably to rDNA but rapidly associates and dissociates. We show that SL1 significantly reduces the rate of dissociation of UBF from the rDNA promoter. Our findings challenge the idea that UBF activates transcription through recruitment of SL1 at the rDNA promoter and suggest that the rate of pre-initiation complex (PIC) formation is primarily determined by the rate of association of SL1, rather than UBF, with the promoter. Therefore, we propose that SL1 directs PIC formation, functioning in core promoter binding, RNA polymerase I recruitment, and UBF stabilization and that SL1-promoter complex formation is a necessary prerequisite to the assembly of functional and stable PICs that include the UBF activator in mammalian cells.
RNA polymerase (Pol)1 enzymes themselves have no intrinsic ability to recognize and bind specifically to promoter DNA sequences, so pre-initiation complex formation in transcription calls for the recruitment of the Pol enzymes to the promoter via transcription factors. Basal transcription factors and (co-)activators of transcription cooperate in this function in eukaryotes where three classes of highly related enzymes, Pol I, Pol II, and Pol III, catalyze the transcription of specific sets of genes. With few exceptions, a complex of TBP and TBP-associated factor (TAF) proteins is required for the accurate initiation of transcription by all three polymerases (1). The particular complement of TAFs in each complex, although variable in Pol II transcription, is specific to each class of genes. There is evidence that binding of certain TAF subunits to TBP precludes the binding of TAFs from a different class (2). The precise roles of the TBP-TAF complexes in mediating a specific interaction between the polymerases and their respective promoters are distinct.
In mammalian Pol II transcription, the TBP-TAF complex TFIID can bind at the promoter by virtue of the specific interaction of TBP with TATA boxes and the specific interactions of the TAF proteins with other promoter sequences, whereupon Pol II and other factors are recruited to form the pre-initiation complex (PIC) (3). The initial phases of mammalian Pol III transcription from the different types of Pol III promoters converge on the recruitment of the TBP-TAF complex TFIIIB and the Pol III enzyme in formation of the PIC (4, 5). Loading of promoter type-specific TFIIIB at the promoter DNA occurs with the assistance of the specific DNA binding capabilities of TFIIIA and TFIIIC or TFIIIC alone or the multisubunit complex PBP/PTF/SNAPC, depending on the Pol III promoter type.
In mammalian Pol I transcription (reviewed in Ref. 6) the TBP-TAF complex selectivity factor SL1 (murine TIF-IB), which is composed of TBP and at least three TAFIs, is essential for Pol I transcription (2, 7–9). The species promoter selectivity of SL1, in which SL1 from humans and mice are not interchangeable between these systems, in contrast to Pol I and the activator UBF, suggests a specific interaction between SL1 and its cognate rDNA core promoter (reviewed in Ref. 10). Indeed, TIF-IB (mouse SL1) and rat SL1 can bind their cognate rDNA promoters independently of UBF (11, 12). However, this had not been demonstrated for the human system (13–15).
SL1 interacts with the hRRN3 protein (murine TIF-IA), a component of initiation-competent Pol Iβ, and this interaction is essential for the recruitment of Pol I by SL1 to the rDNA promoter (16). There is a cooperative interaction between SL1 and UBF at the human rDNA promoter, which can occur prior to the recruitment of Pol I (14, 15, 17). UBF binds as a dimer to the rDNA promoter at the upstream control element (UCE) and core element via interaction of its high mobility group (HMG) boxes 1–3 with the DNA minor groove. UBF binding induces bending of the DNA, a single dimer organizing ~140 bp in a loop of 360°, comprising an enhancesome, and the formation of tandem enhancesomes is proposed to juxtapose the UCE and core (18, 19).
It has been proposed that UBF directs PIC formation, binding at both the UCE and the core regions of the promoter and facilitating the recruitment and binding of SL1, which interacts with the bound UBF at both sites, with the recruitment of Pol I culminating in initiation of transcription (reviewed in Refs. 20, 21). This model holds that UBF stabilizes the interaction of SL1 with the rDNA promoter (13, 14) and was largely born from the finding that human SL1 alone does not produce a DNase I footprint on the rDNA promoter whereas UBF does produce a footprint, which is both extended and altered in the presence of SL1 (13, 14, 17, 22). However, a specific role for UBF in nucleating PIC formation at the promoter is inconsistent with the finding that UBF binds rather indiscriminately (23, 24) and extensively across the rDNA in vivo (25).
Here we have shown that human SL1 can bind the human rDNA promoter independently and direct the initiation of transcription specifically from the rDNA promoter in the absence of UBF. UBF interacts dynamically with the rDNA, and we have demonstrated that SL1 stabilizes UBF binding at the rDNA promoter. Our data, together with data from rodent systems (11, 12, 26), imply that SL1 drives or nucleates PIC formation at the rDNA promoter, challenging the model that UBF executes this role.
EXPERIMENTAL PROCEDURES
SL1, Pol I, and UBF Purification
Human SL1 was purified as outlined in supplemental Fig. 1. Nuclear extracts from HeLa cells were prepared essentially as described (27), except that NaCl was substituted for KCl. The resulting extract was precipitated by 60% saturation with ammonium sulfate and then centrifuged at 35,000 × g for 20 min at 4 °C. The pellet was then resuspended in TM20 buffer (50 mm Tris·HCl, pH 7.9, 0.05 m KCl, 12.5 mm MgCl2, 1 mm EDTA, 20% glycerol, 0.015% Nonidet P-40, 1 mm sodium metabisulfite, 1 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin) and dialyzed against TM10/0.1 (TM buffer with 10% glycerol and 0.1 m KCl). Soluble protein from the nuclear extract at ~40 mg/ml was then applied to a Heparin-Sepharose fast flow column (column volume (CV) of 60 ml; Amersham Biosciences) equilibrated in TM10/0.1 and eluted in three steps at 0.2, 0.4 and 0.7 m KCl, as described previously (7). A 200-ml G25 Sephadex column was washed with 1.2 CVs of TM10/0.2, and the 0.7 m Heparin-Sepharose fraction was loaded at 5 ml/min. The protein peak was eluted at a flow rate of 5 ml/min and a single fraction collected as judged by absorbance (A280) reading exceeding 100 milliabsorbance units. The desalted SL1 fraction was then loaded onto a Mono S HR 5/5 column (CV 0.98 ml; Amersham Biosciences) equilibrated in TM10/0.2. A linear salt gradient of 0.2–1 m KCl was then applied over 25 CVs, and 1-ml fractions were collected. Fractions were assayed for SL1 activity by in vitro transcription with Pol I and UBF. The SL1 activity eluted at 0.27–0.46 m KCl and was pooled and diluted to 0.2 m KCl with TM10/0. A POROS Heparin 20 column (CV 1.66 ml; Perspective) was equilibrated in TM10/0., and the pooled Mono S fractions were loaded. SL1 was eluted with a linear salt gradient of 0.2–1.0 m KCl over 30 CVs. 1-ml fractions were collected, and fractions were assayed for SL1 activity. SL1 peak fractions were reapplied to the POROS Heparin 20 columns in a second round of purification, but this time SL1 was eluted in a single step at 1.0 m KCl, resulting in ~6-fold concentration. The SL1 sample was further concentrated by a NanoSep 10-kDa molecular mass cutoff concentrator (Flowgen, UK) to a 200-μl volume. A Superose 6 HR 10/30 column (CV 23.6 ml; Amersham Biosciences) was equilibrated with TM10/0.35 at 0.1 ml/min, and these conditions were maintained following loading of the SL1 sample. The column was developed with 1.2 CV of TM10/0.35. Fractions of 0.6 ml were collected and assayed for SL1 activity. Pol I (free of UBF) was purified as described (16).
Human UBF1 was extensively purified from Sf9 cells (4 × 108) infected with recombinant UBF baculoviruses at a multiplicity of infection of 10.2 Cells were harvested 40 h post-infection and lysed by sonication in 50 ml of TM10/0.4 buffer (50 mm Tris·HCl, pH 7.9, 12.5 mm MgCl2, 1 mm EDTA, 10% glycerol, 1 mm sodium metabisulfite, 1 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 0.4 m KCl). The lysate was cleared at 70,000 × g for 20 min at 4 °C, and ammonium sulfate was added to 60% over 1 h. Precipitated proteins were collected at 70,000 × g for 40 min at 4 °C, and the pellet was resuspended in 1 m (NH4)2SO4 in buffer A (25 mm HEPES, pH 7.2, 0.1 m KCl, 10% glycerol, 1 mm EDTA, 1 mm sodium metabisulfite, 1 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin) and stirred for 2 h on ice. Solution was cleared at 70,000 × g for 40 min at 4 °C, and the supernatant was loaded on a Phenyl-Sepharose HP column (XK26/20, 66 ml; Amersham Biosciences). The column was washed with 3 column volumes of 1 m (NH4)2SO4 in buffer A, and step-eluted with 1 volume of buffer A. The fraction was desalted on Sephadex G25 (Amersham Biosciences) in TM10/0.05, loaded on a DEAE-Sepharose FF column (XK26/20, 60 ml; Amersham Biosciences); this column was then developed with a linear gradient of 0.05–0.40 m KCl in TM10 over 20 column volumes. UBF peak fractions were pooled, and an equal volume of 2 m (NH4)2SO4 in buffer A was added and subsequently applied to a Phenyl-Sepharose HP column (7.5 ml; Amersham Biosciences), which was step-eluted with 1.5 column volume of buffer A. Following desalting on Sephadex G25 in TM10/0.2, the sample was loaded on a POROS Heparin 20 column (4.6 × 15 mm; Perceptive), which was then developed with a linear gradient of 0.2–1.0 m KCl in TM10 over 30 column volumes. UBF1 eluted as a single peak at 0.5 m KCl. The final purification step involved a POROS HQ10 column (4.6 × 15 mm; Perceptive), which was developed with a linear gradient of 0.2–1.0 m KCl in TM10 over 40 column volumes. UBF eluted as a single peak at 0.68 m KCl, and this fraction was concentrated on a 10-kDa cutoff concentrator (Filtron). SDS-PAGE and silver staining revealed a single protein species of 97 kDa in the UBF peak fraction that was active in transcription (data not shown).
Promoter DNA and Preparation of Immobilized Templates
Plasmid DNA prHu3 WT and deletion derivatives −83 and −26 (22, 28) were used. The pseudo-WT rDNA template (in Fig. 4) contains the prHu3 promoter sequence but diverges from it at +10 relative to the transcription start site into sequences unrelated to rDNA (29). Immobilized promoter rDNA template (fragment (Fr) 4 of 432 bp) is described (30). The nonspecific DNA fragment of 423 bp (unrelated to rDNA) was generated by PCR with a biotinylated primer and was a kind gift from Chris Stockdale. Protein binding studies with the immobilized promoter templates were performed as described (30). Immunoblotting was as described (30). TBP-specific monoclonal antibody SL39 was a kind gift from Dr. Nouria Hernandez.
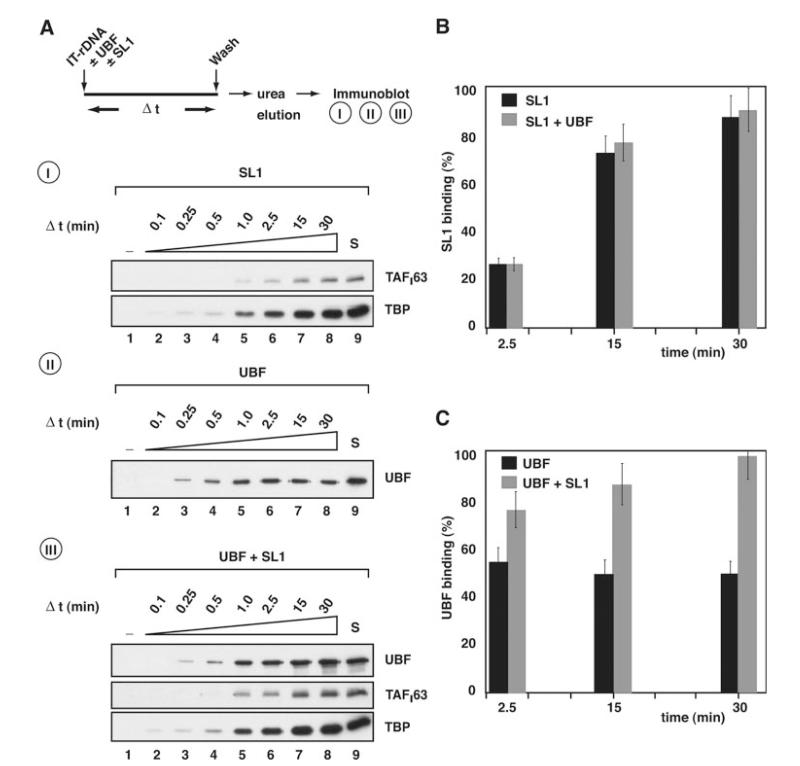
Fig. 4. Rate of association of SL1 and UBF with the rDNA promoter.
A, I, following the procedure outlined, 0.7 μg of SL1 was incubated with 10 pmol immobilized ribosomal promoter template (IT-rDNA). At 0, 0.1, 0.25, 0.5, 1.0, 2.5, 15, and 30 min (lanes 1-8, respectively) equal aliquots were removed and reactions stopped by washing templates with TM10/0.05. Template-associated proteins were then eluted with 5 m urea and analyzed by immunoblotting using antibodies specific for SL1 subunits TAFI63 and TBP. II, as for panel I, except that 0.8 μg of recombinant and purified UBF was incubated with the immobilized ribosomal promoter template. Immunoblotting used antibodies specific for UBF. III, as for panel I, except that both 0.8 μg of UBF and 0.7 μg of SL1 were incubated with IT-rDNA. Immunoblotting used antibodies specific for UBF, TAFI63, and TBP. Control lanes S (lanes 9) were loaded with 125 ng of SL1 and/or 100 ng of UBF, as indicated. B, the association of SL1 with the rDNA promoter in the absence (A, panel I) or presence (A, panel III) of UBF was quantified by measuring TAFI63 ECL signals, expressed as a percentage of the control signal S (A, panels I and III, 125 ng of SL1, lanes 9) and plotted against time (black column, SL1; gray column, SL1 plus UBF). C, the association of UBF with the rDNA promoter in the absence (A, panel II) or presence (A, panel III) of SL1 was quantified by measuring UBF ECL signals, expressed as a percentage of the control signal S (A, panels II and III, 100 ng of UBF, lane 9) and plotted against time (black column, SL1; gray column, SL1 plus UBF). Standard deviations were determined from three independent experiments for each single time point. A–C, note that the amounts of SL1 and UBF used in these binding studies were chosen because they support efficient basal and/or activated transcription in an in vitro assay, as determined by titration (data not shown).
Reconstituted Transcription
Protein factors used were as follows: recombinant human UBF, highly purified Pol I (16), and highly purified SL1 (described here). In vitro transcription reactions were performed as previously described (30). Supercoiled plasmid DNA or immobilized linear DNA fragments were used as templates in transcription reactions. The transcripts were analyzed by S1 nuclease protection as described (30), with 0.1 pmol 5′-end 32P-labeled S1 oligonucleotide probe (WT protected region +1 to +40 or pseudo-WT protected region +1 to +32).
RESULTS
Basal Transcription Initiation in Vitro by SL1 and Pol I
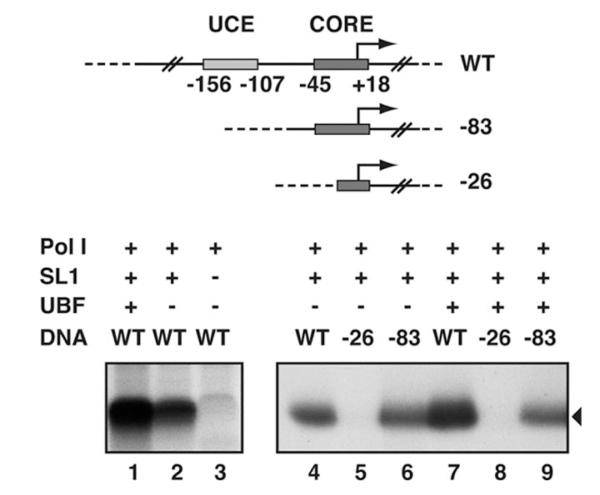
We first tested a highly purified human SL1 preparation free from UBF (see supplemental material) in combination with recombinant human UBF and extensively purified Pol I (free of UBF) for the ability to direct rRNA synthesis from the human ribosomal promoter. A high level of α-amanitin-resistant transcription initiation was achieved with UBF, SL1, and Pol I (Fig. 1, lane 1) from the rDNA promoter template (−515 to +1548). Importantly, Pol I transcription can also occur in the absence of UBF, albeit at lower levels (Fig. 1, compare lanes 1 and 2) but with an absolute requirement for SL1 (compare lanes 2 and 3). (Note that the faint band in lane 3 is transcription-unrelated; it can occur in control samples where no transcription occurs, including a negative control, for instance where no factors are added (data not shown)). Consistent with previous transcription analysis with promoter mutants in crude extracts (32, 33), “basal” levels of transcription directed by SL1 and Pol I were supported by the core promoter element (Fig. 1, lanes 4 and 6, −83 template), whereas a partial deletion of the essential core element abolished transcription (Fig. 1, lane 5, −26 template). Removal of only the UCE resulted in basal levels of transcription in the presence of UBF, SL1, and Pol I (compare lanes 7 and 9); thus, UBF activation was entirely dependent on the UCE. Collectively, the data suggest that human SL1 can interact with the core promoter element independently of UBF and that this SL1-DNA complex directs specific transcription initiation by Pol I.
Fig. 1. SL1-rDNA core promoter interactions direct basal Pol I transcription.
Human rDNA promoter plasmid construct WT (prHu3 (26)) and deletion mutants −83 (prHu3−83) and −26 (prHu3−26) (23), used in reconstituted transcription reactions, are shown schematically. The UCE and core promoter elements are labeled according to their positions relative to the transcription start site at +1. 100 ng of WT (lanes 1-4 and 7), −83 (lanes 6 and 9) or −26 (lanes 5 and 8) were incubated in the presence of recombinant hUBF (50 ng, lanes 1 and 7-9) and highly purified hSL1 (130 ng, lanes 1, 2, and 4-9) and Pol I (1 μl, lanes 1-9). Accurately initiated transcripts (black triangle) were analyzed by S1 nuclease protection and autoradiography.
SL1 Can Bind the Ribosomal Promoter Independently of UBF
So, does SL1 bind the promoter independently or does it bind only when in complex with Pol I? To address this question, an immobilized rDNA template was incubated with the highly purified fraction of SL1, which is free from UBF and Pol I. The immobilized DNA was washed to remove unbound SL1, and half the DNA templates were analyzed in a reconstituted transcription reaction to assess whether SL1 was specifically associated with the rDNA promoter elements. The other half was analyzed for bound SL1 subunits following immunoblotting (Fig. 2A). The SL1-rDNA complexes directed specific transcription initiation by Pol I in an SL1 dose-dependent manner (Fig. 2B). SL1 bound to a nonspecific DNA fragment had no detectable effect on end-to-end transcription by Pol I (data not shown). Consistent with the transcription results, the amount of TAFI110 detected on the rDNA template followed the increased amount of SL1 loaded onto the template, which demonstrated that SL1 could interact with the rDNA promoter independently (Fig. 2C, lanes 2-4). SL1 bound to a lesser extent to a similarly sized DNA fragment that is unrelated to rDNA (lanes 6-8). Similar results were obtained with TAFI63- and TBP-specific antibodies, which were more sensitive than those for TAFI110 (Fig. 2D).
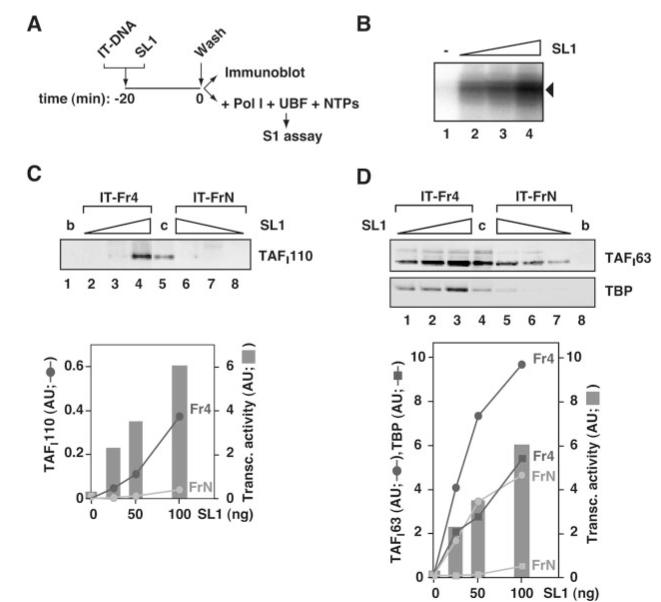
Fig. 2. SL1 binds an immobilized rDNA promoter template independently of UBF.
A, as outlined, 50, 100, and 200 ng of purified SL1 were incubated for 20 min on ice with 2 pmol immobilized rDNA promoter fragment (IT-rDNA, Fr4 of 432 bp) or an equivalent amount of nonspecific DNA (IT-FrN of 423 bp). The complexes were washed with TM10/0.2. Half the immobilized templates were used for immunoblotting and the other half for transcription reactions with Pol I, UBF, and ribonucleotides (NTPs). B, the presence of promoter rDNA-bound SL1 (0, 25, 50, and 100 ng, lanes 1-4) was monitored using reconstituted transcription reactions. Transcripts were analyzed by S1 nuclease protection. C, SL1 (25, 50, and 100 ng) associated with the rDNA promoter fragment (lanes 2-4), with the nonspecific DNA fragment (lanes 6-8), and with beads without any DNA (b, lane 1, 200 ng of SL1 input) were eluted with 5 m urea and analyzed by immunoblotting using antibodies specific for SL1 subunit TAFI110. A control of 60 ng of SL1 was loaded directly onto the gel (c, lane 5). The ECL signals (in arbitrary units) of TAFI110 associated with promoter rDNA (Fr4) and nonspecific DNA (FrN) were quantified and plotted against the amounts of SL1 used. In the same graph, transcript levels (in B) were quantified with the aid of phosphorimaging and plotted (in arbitrary units) against the amounts of SL1 used. D, SL1 associated with the immobilized rDNA promoter fragment (lanes 1-3), with the nonspecific DNA fragment (lanes 5-7), and with beads without any DNA (b, lane 8) were analyzed by immunoblotting (as in panel C). Because the antibodies for TAFI63 and TBP were more sensitive than those for TAFI110, one-fifth of the bound SL1 was analyzed. As a control SL1 was loaded directly onto the gel (c, lane 4; 7 ng). Transcript levels (from panel B) were plotted as in panel C. ECL signals were plotted against the amounts of SL1 incubated with the DNA templates (Fr4 or FrN). Non-linearity of the immunoblotting-ECL assay might account for the apparent differences in the relative amounts of SL1 bound to nonspecific and promoter DNA detected by the different antibodies (C and D).
Importantly, there is a correlation between the level of transcription and the amount of SL1 on the rDNA template, suggesting that, although some SL1 is bound to the DNA template non-specifically and indeed SL1 displays a relaxed sequence-specificity, a proportion of the rDNA-bound SL1 is accurately positioned to function in transcription (Fig. 2, C and D, graphs). Taken together, these results demonstrate the autonomous binding of human SL1 to the rDNA promoter and support the concept of UBF-independent basal transcription initiation in vitro.
SL1 Commits an rDNA Promoter Template for Transcription by Pol I
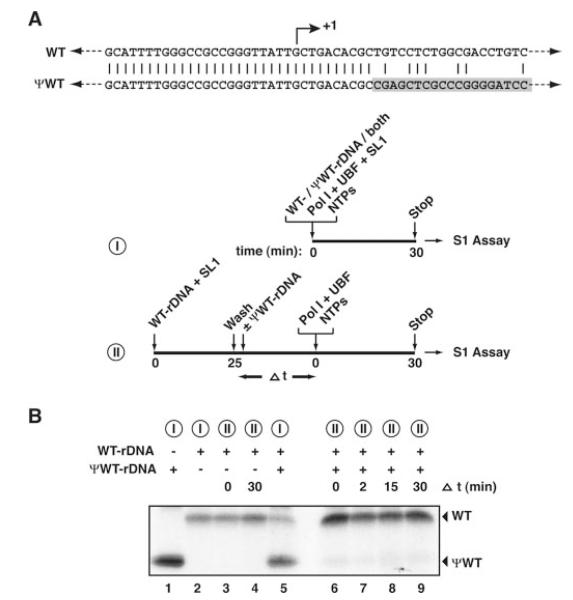
Immunoblotting of an immobilized rDNA promoter detects all SL1 bound, specifically and non-specifically, whereas the transcription assay detects only the promoter-bound fraction of SL1. Therefore, to study the stability of promoter-bound SL1, we performed template commitment assays (Fig. 3). In the assays, we used two promoter templates (Fig. 3A), transcription from which was distinguishable due to divergence of the template sequences at +10 and to the use of different length specific probes in the S1 nuclease protection assay. Transcription initiated from WT and pseudo-WT rDNA templates, in the presence of Pol I, SL1, UBF, and nucleotides, when incubated separately (Fig. 3B, lanes 2 and 1, respectively) or together (lane 5). However, when SL1 was pre-bound to the WT template and then this protein-DNA complex was incubated with the pseudo-WT template, little or no transcription was detected from the pseudo-WT template following the addition of Pol I, UBF, and nucleotides even after incubation with the pseudo-WT template for up to 30 min (Fig. 3B, lanes 6-9). The weak band observed with the pseudo-WT template (lanes 6-9), which does not increase with increasing time of incubation with the pseudo-WT template, might represent low levels of transcription supported either by a small amount of unstably bound SL1 or unbound SL1 not removed in the wash step. We conclude that SL1 binds stably to an rDNA promoter template and commits the template for transcription by Pol I.
Fig. 3. SL1 commits the rDNA promoter template for transcription.
A, the WT-rDNA and pseudo(ψ)-WT-rDNA templates are identical in the promoter region, diverging at +10, just downstream of the transcription start site at +1. Outline of experimental procedures: I, samples 1, 2 and 5, II, samples 3, 4 and 6–9. B, I, pseudo-WT-rDNA (200 ng, lane 1), WT-rDNA immobilized on beads (Fr 4, 50 ng, lane 2), or WT-rDNA plus pseudo-WT-rDNA (lane 5) were incubated with Pol I, SL1, UBF, and NTPs for 30 min at 30 °C. Transcripts were detected following S1 nuclease protection assay with oligonucleotides specific for the WT or pseudo-WT templates, yielding protected fragments of 40 and 32 nucleotides, respectively. II, SL1 (5 μl of highly purified Superose 6 SL1) was incubated with WT-rDNA (50 ng, lanes 3, 4, and 6-9) at 4 °C for 25 min. The SL1-rDNA complexes produced were incubated in the absence (lanes 3 and 4) or presence (lanes 6-9) of 200 ng of pseudo-WT-rDNA for 0 (lanes 3 and 6), 2 (lane 7), 15 (lane 8), or 30 (lanes 4 and 9) min. Transcription was initiated with the addition of Pol I, UBF, and NTPs. Transcripts were detected as in I.
Dynamic Interactions of the Pol I Transcription Factors at the rDNA Promoter
We next considered the possibility that UBF could activate transcription by affecting the rate of formation or the stability of SL1-rDNA complexes during ordered PIC assembly at the promoter. SL1 and/or UBF were incubated with an excess of immobilized template rDNA, and the amount of each factor bound to the rDNA was determined at various time points (Fig. 4). Progressively more SL1 was bound to the rDNA over time, and it appeared that most if not all SL1 input had bound after 30 min (Fig. 4A, panel I, and 4B), consistent with stable binding of SL1 to the template DNA. UBF binding to the rDNA reached equilibrium after 1–2.5 min at 50% of the input UBF (Fig. 4A, panel II, and 4C), suggesting a high association and dissociation rate for UBF alone (Fig. 4A, compare panels I and II). Strikingly, UBF failed to increase the rate at which SL1 binds the rDNA (Fig. 4A, compare panels I and III, and 4B). Significantly, however, the dynamic equilibrium of UBF-DNA binding is shifted by SL1, as a result of which ~100% of the UBF input was associated with the rDNA template after 30 min (Fig. 4A, compare panels II and III, and 4C). Collectively, these data suggest that SL1 stabilizes UBF at the rDNA promoter.
SL1 Reduces the Dissociation of UBF from DNA
Stabilization by SL1 of UBF at the rDNA promoter may result from the ability of SL1 to decrease the rate of dissociation of UBF from the rDNA. To test this, we analyzed the dissociation of UBF and/or SL1 over time from preassembled rDNA promoter-protein complexes in the presence of an excess of nonspecific DNA, which serves as a “sink” for dissociated factors (Fig. 5). To reaffirm the correlation between the level of transcription and the amount of SL1 and/or UBF on the rDNA template, and thereby demonstrate that a fraction of the SL1 and/or UBF bound was specifically associated with the rDNA promoter, we performed binding studies and transcription assays using rDNA promoter templates with binding sites for both proteins.
Fig. 5. SL1 stabilizes the association of UBF with the rDNA promoter.
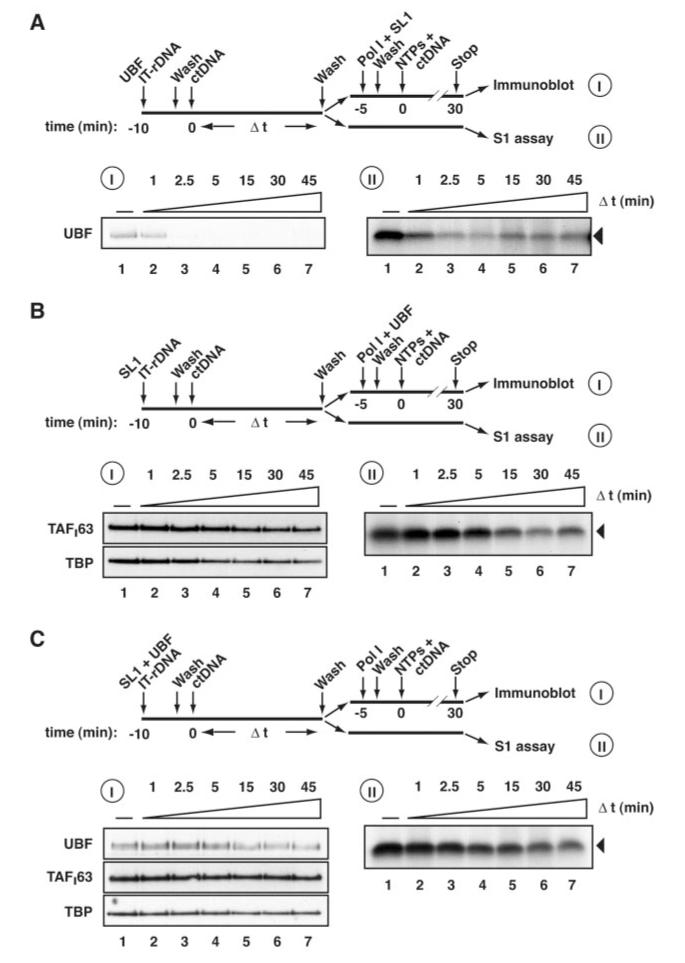
A, following the procedure outlined, 0.4 μg of UBF was first incubated with 6 pmol immobilized human rDNA (IT-rDNA) for 10 min at 4 °C. The templates were then washed with TM10/0.05. 0.5 μg of sheared calf thymus (ct) DNA was added, equal aliquots were removed at 0, 1, 2.5, and 5 min, and the templates were washed with TM10/0.05. Template-associated proteins were analyzed by immunoblotting (I, lanes 1-4) following elution with 5 m urea and using antibodies specific for UBF or the SL1 subunits TAFI63 and TBP. Template-associated proteins were also analyzed by reconstituted transcription assays (I, lanes 1-4) in which purified Pol I, SL1 were incubated together with the protein-IT-rDNA complexes for 5 min. Following a wash with TM10/0.05, transcription was initiated by the addition of NTPs, limited to a single round by the addition of 0.5 μg of ctDNA, and stopped after 30 min. Transcripts were analyzed by S1 nuclease protection. B, as for panel A, except that 0.6 μg of SL1 was first incubated with IT-rDNA and that in the reconstituted transcription assays, purified Pol I and UBF were incubated together with the protein-IT-rDNA complexes. C, as for panel A except that both 0.4 μg of UBF and 0.6 μg of SL1 were first incubated with IT-rDNA and that, in the reconstituted transcription assays, purified Pol I was incubated together with the protein-IT-rDNA complexes.
UBF dissociated rapidly from the rDNA when incubated with the rDNA template prior to the addition of SL1 and Pol I (Fig. 5A, panel I). The dissociation of UBF from the rDNA, as reflected in the reduced signals for UBF following immunoblotting of the protein-DNA complexes, correlated with decreased levels of transcription achieved in reconstituted transcription assays (Fig. 5A, panel II), suggesting that the UBF-promoter DNA complexes observed were primarily functional. Interestingly, the time taken for UBF to completely dissociate from the rDNA (<2.5 min) is similar to that required to reach equilibrium between the association and dissociation of UBF with the rDNA template (Figs. 5A, panel I, 4A, panel II, and 4C). This reaffirms a dynamic interaction of UBF with the rDNA in the absence of other factors, where rates of promoter association and dissociation appear similar, resulting in rapid exchange of UBF on the promoter.
In contrast to the transient association of UBF with the promoter, SL1-rDNA complexes appear less dynamic. The SL1 complex remains relatively stably bound to its target DNA (Fig. 5B, panel I), again reflected in the levels of transcription achieved in reconstituted transcription assays (panel II). SL1 starts to dissociate detectably from the template following 5 min of exposure to an excess of calf thymus DNA (Fig. 5B, lanes 4-7).
Incubation of SL1 and UBF together with the rDNA template significantly reduces the dissociation rate of UBF (Fig. 5, compare UBF binding in A, panel I, and C, panel I). Because SL1 itself is relatively stably bound to DNA, only a relatively small effect of UBF on SL1-rDNA complex stability was detectable (Fig. 5, compare B, panels I and II, with C, panels I and II).
Taken together, these data confirm previous findings that SL1 and UBF interact in a cooperative manner at the promoter. Importantly, SL1 reduces the dissociation of UBF from the rDNA promoter. Therefore, SL1-promoter complex formation appears to be a necessary prerequisite to the assembly of functional and stable PICs that include the UBF activator.
DISCUSSION
The TBP-TAF complex SL1 and UBF are required for efficient Pol I transcription initiation, and these proteins exhibit a cooperative interaction at the rDNA promoter. DNase I footprinting experiments had led to the proposal that UBF functions to recruit human SL1 to the promoter during PIC assembly (13, 14, 34). However, UBF binding to DNA is neither sequence-specific nor restricted to the rDNA promoter region in vivo (25); therefore it is unclear how SL1 would be targeted specifically to the promoter by UBF. Furthermore, the promoter selectivity and species specificity of Pol I transcription has been attributed to the SL1/TIF-IB complex (22), suggesting a specific interaction between SL1 and its cognate promoter. Therefore, we re-examined the model that PIC formation is directed by UBF binding at the rDNA promoter. Here we present data that implies that it is the SL1 complex, rather than UBF, that nucleates PIC formation at the rDNA promoter. We have demonstrated that human SL1 can direct Pol I to initiate specific transcription from the rDNA promoter in the absence of UBF, indicating that there is no requirement for UBF, either in the recruitment of SL1 for PIC formation or in the accurate initiation of Pol I transcription. This basal transcription requires the core promoter region but not the UCE. Moreover, we have shown that SL1 can bind stably to the rDNA promoter independently of UBF. The human Pol I transcriptional system therefore resembles the rodent system (11, 12), inasmuch as TIF-IB/SL1 interacts with the rDNA promoter independently and UBF has a stimulatory, rather than an indispensable, role in transcription.
UBF activates transcription above the levels seen with SL1 alone. Here, our binding and transcription studies indicate that SL1 is stably bound to and can commit the rDNA promoter, whereas the association and dissociation kinetics for UBF at the rDNA promoter demonstrate a relatively unstable and dynamic interaction of UBF with rDNA. Activation of transcription by UBF cannot be explained by the relatively subtle influence of UBF on the rate of dissociation and overall stability of SL1 at the rDNA promoter. On the contrary, here we have shown that SL1 stabilizes the interaction of UBF on the rDNA template by reducing its rate of dissociation from the promoter and that this correlates with the stabilization of productive PICs engaged in activated and accurate transcription.
The previously reported extension and enhancement of the DNase I footprint with UBF and SL1, compared with UBF alone (13, 14, 17, 22), could be due to altered interactions of UBF with the DNA or to SL1 directly contacting these regions or to a combination of both. UBF binds DNA with relaxed specificity, and it is thought that UBF may have affinity for sequences that are flexible in the conformations they can adopt, rather than for a specific sequence (23, 24, 35, 36). SL1 binding might influence the rDNA promoter structure and thereby increase the affinity of UBF for such DNA. It has been proposed that a dimer of UBF functions as an architectural protein that bends the DNA 360°, organizing it into a structure termed an enhancesome, and that formation of tandem enhancesomes juxtaposes the core and UCE sequences (18, 19, 37, 38). SL1 could potentially be involved in maintaining the bent structure of the DNA caused by UBF, fixing the UBF, SL1, and DNA in a stable structure or perhaps influencing the extent of the distortion. Finally, UBF has a highly acidic tail, and although the exact function of this carboxyl-terminal domain is unknown it is also found in the related proteins HMG1 and HMG2. This acidic tail generally lowers the affinity of HMG boxes for most DNA substrates in vitro (reviewed in Ref. 39), and in the case of UBF an influence of the tail on HMG box 1 DNA binding has been reported (38). Given the reported interaction of SL1 with the tail of UBF (15, 40, 41), we think it is possible that SL1 suppresses the effect of the tail on HMG box-DNA interactions and so stabilizes UBF at the rDNA promoter.
We have shown that SL1 itself can form a stable interaction with the rDNA template. Paradoxically, fluorescence recovery after photobleaching studies in vivo had suggested that SL1 exchanges rapidly (42). However, in that study it was not possible to distinguish between TAFI subunits incorporated into the SL1 complex and free TAFI subunits or between promoter DNA-bound, non-promoter DNA-bound, and unbound SL1. Nonetheless, in vivo there may well be factors present that can assist in the association and dissociation of SL1 from the DNA.
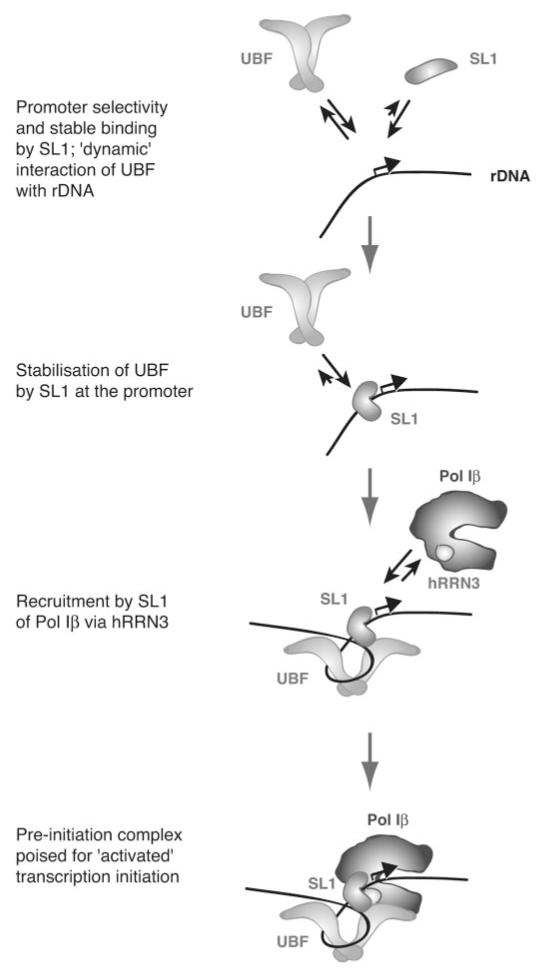
The experiments described here imply that human SL1 can interact with DNA with some sequence specificity, because at least a proportion of the SL1 bound to the rDNA template is correctly positioned at the promoter, judging from the transcriptional readout. UBF does not increase the specificity of SL1 binding in vitro.2 The affinity of SL1 for promoter DNA may be a consequence of specific interactions of its subunits either with the promoter sequences (43, 44) or with the sequence-directed structures that arise from DNA curvature, duplex stability, and twist angle variations that are conserved between rDNA promoters from different species (45). Our Surface Plasma Resonance data (not shown) indicate that the association rate of SL1 with promoter DNA is higher than that with nonspecific DNA sequences, whereas the dissociation rates are comparable. The specificity of binding of human SL1 is nonetheless insufficient to produce a DNase I footprint (13, 14, 17, 22), and indeed SL1 binds nonspecific DNA. The TBP subunit of SL1 is a minor groove DNA-binding protein and, as such, interacts rather non-specifically with DNA. TBP can form a stable interaction with both TATA-containing and TATA-less DNA and has been shown to interact with the rDNA promoter in Acanthamoeba (46). It has been proposed that TBP straddles and slides along the DNA (47). Together with our findings, this raises the interesting possibility that human SL1 initially interacts with DNA non-specifically and then tracks along the DNA until it encounters the rDNA promoter binding site, whereupon it docks, perhaps undergoing a conformational change that “locks” it onto the DNA (see Fig. 6). There is evidence that most DNA-binding proteins first bind DNA non-specifically and then translocate to their specific sites; the intramolecular processes involved in such a switch from non-specific binding to specific binding to base pairs in the target DNA sequence have recently been elucidated for the lactose repressor paradigm (48).
Fig. 6. An integrated model for the role of SL1 in directing productive pre-initiation complex formation at the rRNA gene promoter in mammalian Pol I transcription.
See “Discussion” for details.
In vivo, nucleolar compartmentalization and the chromatin scaffold might additionally contribute to Pol I transcription specificity. The specificity could be increased further by the interaction of SL1 with the Pol I enzyme subunits or other factors; perhaps significantly with respect to the latter, we have observed an increased rate of PIC assembly from nuclear extracts compared with that from purified factors (30). We have found no evidence to suggest that UBF increases the specificity of the transcription reaction, in that SL1 itself is sufficient to recruit and position Pol I for initiation of transcription, UBF does not significantly influence the interaction of SL1 with the rDNA template, and, crucially, in a separate study we have determined that UBF does not detectably alter the affinity of Pol I for the promoter.2 Indeed, we have shown in that study that UBF functions in a rate-limiting step occurring after PIC formation. It may thus be significant that SL1 increases the “lifetime” of the activator UBF-DNA complex. The paradigm for transcriptional activator function is stimulation of transcription by recruitment of the TBP-TAF complex, other basal transcription factors, and/or the RNA polymerase (49). The Pol I activator UBF does not fit this paradigm in that neither does it recruit TBP-TAF complex SL1, as shown here, nor is it essential for the recruitment of Pol I to the rDNA promoter, as shown elsewhere.2 It is possible that, in vivo, UBF interactions increase the local concentrations of the Pol I transcription factors and the Pol I enzyme at rDNA and thereby indirectly influence PIC formation (50).
Data presented here for the human Pol I transcription system, together with data from the rodent systems, suggest an integrated model for PIC formation in mammalian Pol I transcription in which SL1, rather than UBF, nucleates PIC formation. UBF dynamically “samples” rDNA sequences, including the promoter, but binds stably at the promoter only in the presence of SL1. The rate of PIC formation therefore depends on the rate of SL1-DNA association and not on that of UBF-DNA. We propose that SL1 instigates the initial protein-DNA and protein-protein interactions that culminate in targeting of Pol Iβ, the initiation-competent form of the enzyme, via hRRN3 (16), to previously inactive rDNA promoters (Fig. 6). SL1 and UBF remain promoter bound throughout single and multiple rounds of transcription in vitro (30). Even during mitosis when rRNA synthesis is down-regulated, SL1 and UBF remain associated with the chromosomal rDNA repeats (51), unlike Pol I (52). Together with the apparent low dissociation rate for SL1 on the immobilized rDNA promoter in vitro, this leads us to propose that SL1 may serve as a molecular scaffold on the rDNA, stabilizing the assembly and binding of other factors such as Pol I and UBF and committing the rDNA genes to successive rounds of transcription reinitiation. SL1 is a potential target for signal transduction pathways that regulate the levels of rDNA transcription (31) and thereby affect cellular growth and proliferation. It is anticipated that SL1 activity will be regulated not only at the level of its binding to DNA but also at the levels of Pol I recruitment and UBF stabilization.
Supplementary Material
Acknowledgments
We thank Nouria Hernandez for TBP-specific and Brian McStay for UBF-specific antibodies. We thank the National Cell Culture Center (Minneapolis) for growing HeLa cells. We thank colleagues in the Zomerdijk laboratory and Angus Lamond and Tom Owen-Hughes for advice and critical reading of the manuscript.
Footnotes
The on-line version of this article (available at http://www.jbc.org) contains one supplemental figure.
The abbreviations used are: Pol I, RNA Polymerase I; HMG, high mobility group; IT-rDNA, immobilized rDNA promoter template; PIC, pre-initiation complex; SL1, selectivity factor 1; TBP, TATA box-binding protein; TAF, TBP-associated factor; UBF, upstream binding factor; UCE, upstream control element; WT, wild type; CV, column volume; Fr, fragment.
K. I. Panov, J. K. Friedrich, J. Russell, and J. C. B. M. Zomerdijk, submitted for publication.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Hernandez N. Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 2.Comai L, Zomerdijk JC, Beckmann H, Zhou S, Admon A, Tjian R. Science. 1994;266:1966–1972. doi: 10.1126/science.7801123. [DOI] [PubMed] [Google Scholar]
- 3.Verrijzer CP, Tjian R. Trends Biochem. Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 4.Geiduschek EP, Kassavetis GA. J. Mol. Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 5.Schramm L, Hernandez N. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 6.Russell J, Zomerdijk JCBM. Trends Biochem. Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comai L, Tanese N, Tjian R. Cell. 1992;68:965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- 8.Zomerdijk JCBM, Beckmann H, Comai L, Tjian R. Science. 1994;266:2015–2018. doi: 10.1126/science.7801130. [DOI] [PubMed] [Google Scholar]
- 9.Zomerdijk JCBM, Tjian R. In: Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Paule MR, editor. Springer Verlag; New York: 1998. pp. 67–73. [Google Scholar]
- 10.Heix J, Grummt I. Curr. Opin. Genet. Dev. 1995;5:652–656. doi: 10.1016/0959-437x(95)80035-2. [DOI] [PubMed] [Google Scholar]
- 11.Schnapp A, Grummt I. J. Biol. Chem. 1991;266:24588–24595. [PubMed] [Google Scholar]
- 12.Smith SD, O’Mahony DJ, Kinsella BT, Rothblum LI. Gene Expr. 1993;3:229–236. [PMC free article] [PubMed] [Google Scholar]
- 13.Learned RM, Learned TK, Haltiner MM, Tjian RT. Cell. 1986;45:847–857. doi: 10.1016/0092-8674(86)90559-3. [DOI] [PubMed] [Google Scholar]
- 14.Bell SP, Learned RM, Jantzen HM, Tjian R. Science. 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- 15.Jantzen HM, Chow AM, King DS, Tjian R. Genes Dev. 1992;6:1950–1963. doi: 10.1101/gad.6.10.1950. [DOI] [PubMed] [Google Scholar]
- 16.Miller G, Panov KI, Friedrich JK, Trinkle-Mulcahy L, Lamond AI, Zomerdijk JCBM. EMBO J. 2001;20:1373–1382. doi: 10.1093/emboj/20.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jantzen HM, Admon A, Bell SP, Tjian R. Nature. 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- 18.Bazett Jones DP, Leblanc B, Herfort M, Moss T. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 19.Stefanovsky VY, Pelletier G, Bazett-Jones DP, Crane-Robinson C, Moss T. Nucleic Acids Res. 2001;29:3241–3247. doi: 10.1093/nar/29.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grummt I. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 21.Moss T, Stefanovsky VY. Cell. 2002;109:545–548. doi: 10.1016/s0092-8674(02)00761-4. [DOI] [PubMed] [Google Scholar]
- 22.Learned RM, Cordes S, Tjian R. Mol. Cell. Biol. 1985;5:1358–1369. doi: 10.1128/mcb.5.6.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu CH, McStay B, Jeong SW, Reeder RH. Mol. Cell. Biol. 1994;14:2871–2882. doi: 10.1128/mcb.14.5.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Copenhaver GP, Putnam CD, Denton ML, Pikaard CS. Nucleic Acids Res. 1994;22:2651–2657. doi: 10.1093/nar/22.13.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Sullivan AC, Sullivan GJ, McStay B. Mol. Cell. Biol. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith SD, Oriahi E, Lowe D, Yang-Yen HF, O’Mahony D, Rose K, Chen K, Rothblum LI. Mol. Cell. Biol. 1990;10:3105–3116. doi: 10.1128/mcb.10.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dignam JD, Martin PL, Shastry BS, Roeder RG. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 28.Learned RM, Tjian R. J. Mol. Appl. Genet. 1982;1:575–584. [PubMed] [Google Scholar]
- 29.Smale ST, Tjian R. Mol. Cell. Biol. 1985;5:352–362. doi: 10.1128/mcb.5.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panov KI, Friedrich JK, Zomerdijk JC. Mol. Cell. Biol. 2001;21:2641–2649. doi: 10.1128/MCB.21.8.2641-2649.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James MJ, Zomerdijk JCBM. J. Biol. Chem. 2004;279:8911–8918. doi: 10.1074/jbc.M307735200. [DOI] [PubMed] [Google Scholar]
- 32.Haltiner MM, Smale ST, Tjian R. Mol. Cell. Biol. 1986;6:227–235. doi: 10.1128/mcb.6.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Learned RM, Smale ST, Haltiner MM, Tjian R. Proc. Natl. Acad. Sci. U. S. A. 1983;80:3558–3562. doi: 10.1073/pnas.80.12.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bell SP, Jantzen HM, Tjian R. Genes Dev. 1990;4:943–954. doi: 10.1101/gad.4.6.943. [DOI] [PubMed] [Google Scholar]
- 35.Putnam CD, Copenhaver GP, Denton ML, Pikaard CS. Mol. Cell. Biol. 1994;14:6476–6488. doi: 10.1128/mcb.14.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pikaard CS, McStay B, Schultz MC, Bell SP, Reeder RH. Genes Dev. 1989;3:1779–1788. doi: 10.1101/gad.3.11.1779. [DOI] [PubMed] [Google Scholar]
- 37.Moss T, Stefanovsky VY. Prog. Nucleic Acids Res. Mol. Biol. 1995;50:25–66. doi: 10.1016/s0079-6603(08)60810-7. [DOI] [PubMed] [Google Scholar]
- 38.Leblanc B, Read C, Moss T. EMBO J. 1993;12:513–525. doi: 10.1002/j.1460-2075.1993.tb05683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas JO, Travers AA. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 40.Tuan JC, Zhai W, Comai L. Mol. Cell. Biol. 1999;19:2872–2879. doi: 10.1128/mcb.19.4.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kihm AJ, Hershey JC, Haystead TA, Madsen CS, Owens GK. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14816–14820. doi: 10.1073/pnas.95.25.14816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- 43.Rudloff U, Eberhard D, Tora L, Stunnenberg H, Grummt I. EMBO J. 1994;13:2611–2616. doi: 10.1002/j.1460-2075.1994.tb06551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beckmann H, Chen JL, O’Brien T, Tjian R. Science. 1995;270:1506–1509. doi: 10.1126/science.270.5241.1506. [DOI] [PubMed] [Google Scholar]
- 45.Marilley M, Pasero P. Nucleic Acids Res. 1996;24:2204–2211. doi: 10.1093/nar/24.12.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bric A, Radebaugh CA, Paule MR. J. Biol. Chem. 2004;279:31259–31267. doi: 10.1074/jbc.M311828200. [DOI] [PubMed] [Google Scholar]
- 47.Coleman RA, Pugh BF. J. Biol. Chem. 1995;270:13850–13859. doi: 10.1074/jbc.270.23.13850. [DOI] [PubMed] [Google Scholar]
- 48.Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 49.Ptashne M, Gann A. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 50.Mais C, Wright JE, Prieto JL, Raggett SL, McStay B. Genes Dev. 2005;19:50–64. doi: 10.1101/gad.310705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jordan P, Mannervik M, Tora L, Carmo Fonseca M. J. Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leung AK, Gerlich D, Miller G, Lyon C, Lam YW, Lleres D, Daigle N, Zomerdijk J, Ellenberg J, Lamond AI. J. Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.