Abstract
Owing to its ability to form biofilms on implanted medical devices, the fungal pathogen Candida albicans causes frequent infections in humans. A hallmark of C. albicans biofilms is the presence of two types of cells, budding yeast cells and growing hyphae, which are bound together and embedded in extracellular matrix material. Although cell-cell adhesion is critical to biofilm formation, architecture and cohesion, we know little about the fundamental forces behind this interaction. Here, we use single-cell force spectroscopy (SCFS) to quantify the forces engaged in yeast-hyphae adhesion, focusing on the role of Als (Agglutinin-like sequence) proteins as prototypes of cell adhesion molecules. We show that adhesion between individual yeast and hyphal cells involves strong, short-range cohesive interactions (1.1 nN ± 0.2 nN; 86 ± 33 nm), and weak, long-range tether interactions (0.4 ± 0.2 nN; 234 ± 81 nm). Control experiments demonstrate that these interactions originate from cell surface proteins that are specific to C. albicans. Using mutant strains deficient for Als expression, we find that Als3 proteins, primarily expressed on the germ tube, play a key role in establishing strong cohesive adhesion. We suggest a model in which cohesive adhesion during biofilm formation originates from tight hydrophobic interactions between Als tandem repeat domains on adjacent cells. When subjected to force, the two interacting cell surfaces detach but the cell bodies remain tethered through macromolecular extensions. Our results represent the first direct, non-invasive measurement of adhesion forces between interacting fungal cells, and provide novel insights into the molecular origin of the cohesive strength of fungal biofilms.
Keywords: AFM, biofilms, cell-cell adhesion, force nanoscopy, pathogens, single-cells, Candida albicans
INTRODUCTION
The formation of biofilms on tissues and implanted devices is responsible for a wide range of microbial infections.1,2 Bacterial and fungal biofilms consist of microcolonies made of cell aggregates and enclosed in a matrix of extracellular polymeric material. Although cell-cell interactions play key roles in controlling biofilm cohesion and architecture, their molecular details are poorly understood.
A remarkable example of biofilm-forming microbe is Candida albicans, a common fungal pathogen found in hospital settings and in chronic disease patients, and which can lead to both morbidity and mortality.3,4 An important feature of C. albicans biofilms is the presence of two morphological forms of the fungus: budding yeast cells and growing hyphae.3 The development of a biofilm starts with the formation of an initial basal layer made of yeast-phase cells adhering to a substrate. Formation of microcolonies and germination of yeast cells then leads to the addition of an upper hyphal layer. The two fungal forms display major differences in their cell surface macromolecules, including the cell adhesion glycoproteins known as Als (Agglutinin-like sequence) proteins,5,6 and are believed to play distinct roles in biofilm formation. To date, how fungal morphogenesis modulates cell-cell adhesion and cohesion in C. albicans biofilms is poorly understood. Clarification of this issue is critical to our understanding of the molecular bases of biofilm formation and may contribute to the development of new antifungal therapies.
Among the eight different Als proteins produced by C. albicans, Als1 and Als3 are considered as the most important for biofilm formation.7,8 Als3 expression is induced in the initial stages of biofilm formation,7 and is primarily localized in germ tubes and hyphae.9–11 Deletion of the ALS3 gene strongly affects in vitro biofilm formation, but not under in vivo conditions, probably due to higher expression of the homologous gene ALS1 under these conditions.9,10,12 All Als proteins share three distinct functional regions that are engaged in cell adhesion. The two N-terminal immunoglobulin (Ig)-like regions show broad substrate specificity and initiate cell adhesion. These are followed by a threonine-rich region (T) containing a 7-residue sequence that strengthen cell adhesion through amyloid bonds. The central region of the protein contains a variable number of tandem repeat (TR) domains that are 36 amino acids in length, and bind to each other and to various substrates through hydrophobic interactions. Previously, we used single-molecule atomic force microscopy (AFM) to demonstrate that these three regions mediate strong recognition binding events (Ig region), amyloid-mediated clustering and interactions (T region), and strong hydrophobic interactions associated with protein unfolding (TR region).11,13–15 Yet, the extent to which these different Als-based interactions contribute to the adhesion of whole cells is unknown.
Traditional methods used in microbiology provide averaged information obtained on large populations of cells. By contrast, the emerging field of single-cell microbiology uses new tools to analyze individual cells in complex, heterogeneous populations, thereby enabling us to reveal a diversity of behaviors and rare events that would otherwise be hidden.16,17 Among these technologies, atomic force microscopy (AFM) has been instrumental in unravelling the structure, properties and interactions of living cells at the single-cell and single-molecule levels.18,19 In the cell adhesion context, AFM-based single-cell force spectroscopy (SCFS) has proven very useful for measuring the fundamental forces driving cell-substrate and cell-cell adhesion.20–23 The general principle is to immobilize a single living cell on an AFM cantilever and to measure the forces between this cell probe and a substrate or another cell. Here we use SCFS for quantifying the forces engaged in yeast-hyphae adhesion in C. albicans. We demonstrate that the adhesion between individual yeast and hyphal cells is mediated primarily by Als3 adhesins expressed on germ tubes, and involves strong, short-range cohesive interactions and weak, long-range tether interactions. The method presented here is a powerful tool in single-cell microbiology for quantifying the adhesion forces between different fungal morphotypes (yeast vs hyphae), thereby contributing to increase our understanding of the mechanisms of biofilm formation.
RESULTS AND DISCUSSION
Probing cell-cell adhesion
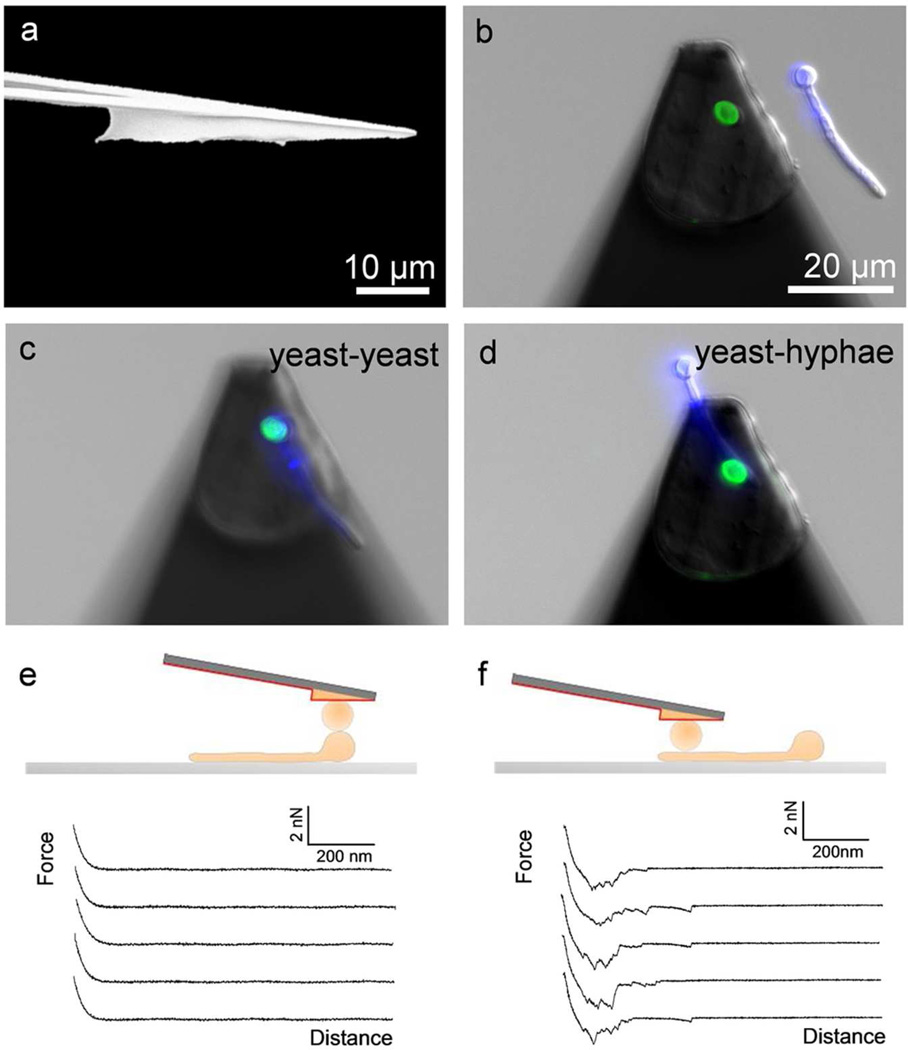
Cell probes were prepared by attaching single fungal cells on tip-less cantilevers using polydopamine.24,25 An important issue in SCFS is the standard tilt (~10°) of the cantilever, which can cause non-uniform, tangential load on the cell, and, in turn, cell sliding and rolling.26 To circumvent this problem, cells were attached on wedged cantilevers prepared using the protocol of Stewart et al. (Figure 1a).26 Wedged-cantilevers were coated with a thin film of polydopamine.24,25 Using an integrated AFM-inverted optical microscope, the polydopamine probes were then approached toward a single yeast cell deposited on a glass petri dish in buffer, kept in contact for 10 s, and then withdrawn (Figure 1b).
Figure 1.
Force spectroscopy of cell-cell adhesion using wedged cantilevers. (a) Scanning electron microscopy image of a wedged cantilever prepared using UV-curable glue. (b) Non-destructive method for studying yeast-hyphae adhesion: single yeast cells from Candida albicans (labeled with Con A-FITC, green) were attached on a polydopamine-coated wedged cantilever and approached towards a C. albicans hyphae (Calcofluor White, blue) immobilized on a hydrophobic substrate. (c-f) To measure yeast-hyphae adhesion forces, the yeast cell probe was positioned on top of the yeast region (c, e) or the germ tube region (d, f) of the hyphae, and multiple force-distance curves were recorded (e, f).
To probe the adhesion between single yeast and hyphal cells, a yeast cell probe was approached towards an hyphal cell immobilized on a hydrophobic substrate.11 This immobilization method may select for a certain subpopulation of cells, but this was the only approach available to firmly attached hyphal cells without using chemicals that could alter the cell surface. As shown in Figure 1c,d, the set-up is better visualized by labelling the cells with green ConA-FITC (yeast cells) and blue Calcofluor White (hyphae). Note that to avoid interference with the labelling molecules, the cells were not labelled for SCFS measurements. The use of an integrated AFM-inverted optical microscope enabled us to accurately position the yeast probe over the yeast (Figure 1c) or the germ tube (Figure 1d) regions of the hyphae cell. The viability of the cells was checked using a LIVE/DEAD yeast viability staining protocol.27 Fluorescence images showed that both yeast and germinating cells were alive before and after the SCFS experiments (Supporting Information Figure 1, red color), thus confirming that cell probe preparation and SCFS measurements were minimally invasive.
Representative force-distance curves recorded between individual yeast and hyphal cells are shown in Figure 1e,f. While the curves obtained on the yeast region of the hyphae showed no adhesion (Figure 1e), those recorded on the germ tube featured large adhesion forces with multiple peaks and extended rupture lengths (Figure 1f). These force profiles indicate that the molecular bonds formed between the two cells break sequentially until they have completely separated from each other.20,22
Quantifying the adhesion forces between yeasts and hyphae
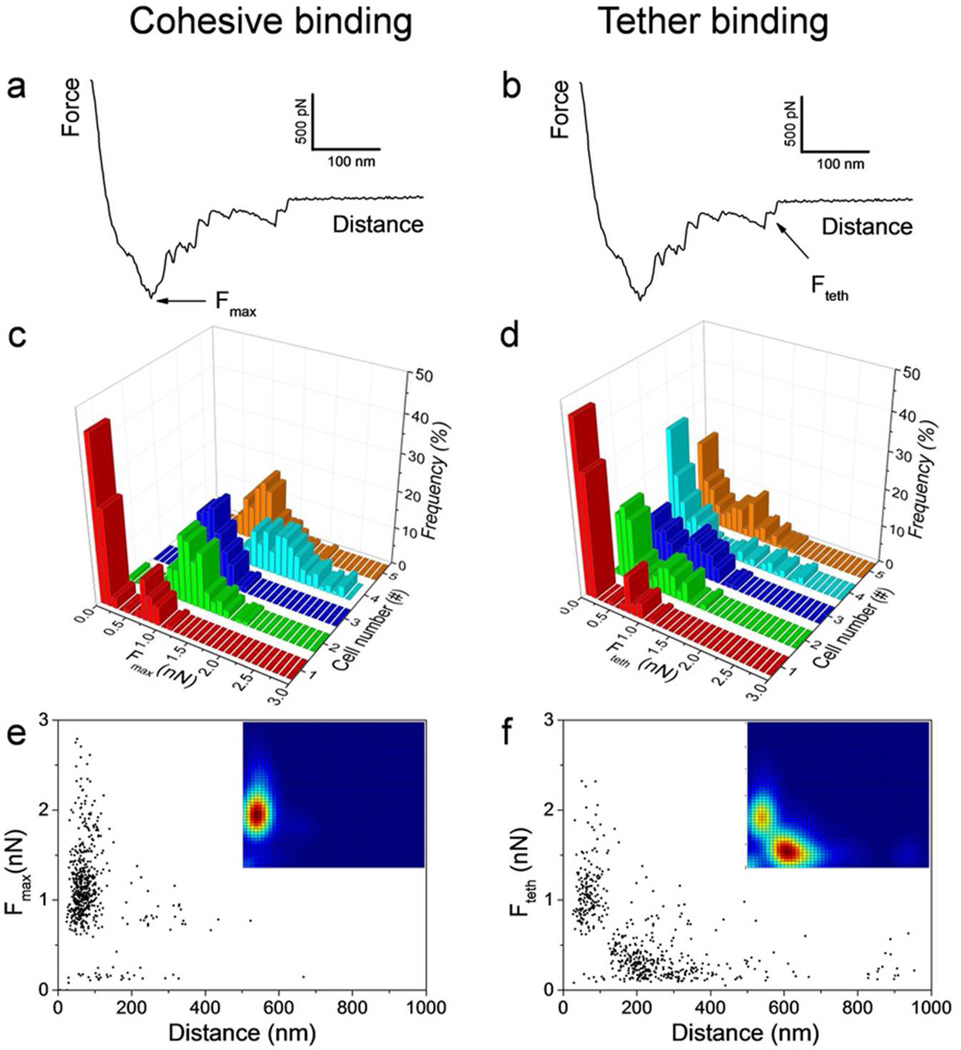
To gain insight into the molecular origin of the measured adhesion forces, force profiles obtained for multiple cells were examined in detail, and the relevant parameters compiled into histograms and scatter plots (Figure 2). As shown in Figure 2a,b, the curves recorded at a loading rate (LR) of 120.000 pN/s between yeasts and hyphae showed two characteristic features, i.e. a large adhesion force peak (Fmax = 1.1 nN ± 0.2 nN; mean ± standard deviation; n = 500 curves from 5 independent cell combinations; 92 % of the curves;) at short distance (86 ± 33 nm), that was often followed by a second peak of weaker adhesion (Fteth = 0.4 ± 0.2 nN; 61 % of the curves) at longer distance (234 ± 81 nm). The histograms in Figure 2c,d demonstrate that the general features of the curves did not substantially change when recording consecutive force curves or when comparing different cells. To correlate the force and distance values for each Fmax and Fteth peaks, both parameters were plotted in scatter plots (Figure 2e,f), and in bivariate diagrams (insets) in which the probability density is displayed as a color map (low and high densities are shown in blue and red, respectively). From these plots, it can clearly be seen that the strong adhesion events (Fmax) occur at short distances, while most weak adhesion events (Fteth) are of much longer range. These observations lead us to conclude that strong adhesion events represent the largest cell- cell binding strength at close contact, referred to as “cohesive interactions”, while weaker events correspond to extended macromolecular bonds, referred to as “tether interactions”, occurring after the two cell bodies have detached from each other.21,22
Figure 2.
Force spectroscopy of yeast-hyphae adhesion. (a, b) Representative force curves obtained for the yeast-hyphae interaction showed two characteristic features, i.e. strong, short-range adhesion force peaks (cohesive adhesion, Fmax), followed by weak, long-range force peaks (tether adhesion, Frupt). (c, d) Adhesion force histograms of the Fmax and Frupt peaks obtained for 5 different yeast/hyphae combinations (n = 100 for each combination). (e, f) Scatter plots of the adhesion force versus distance obtained for Fmax and Frupt peaks selected randomly from 5 different experiments (n = 500 data points). The insets show the corresponding bivariate color-coded maps of probability function, in which blue and red represent low and high frequency events.
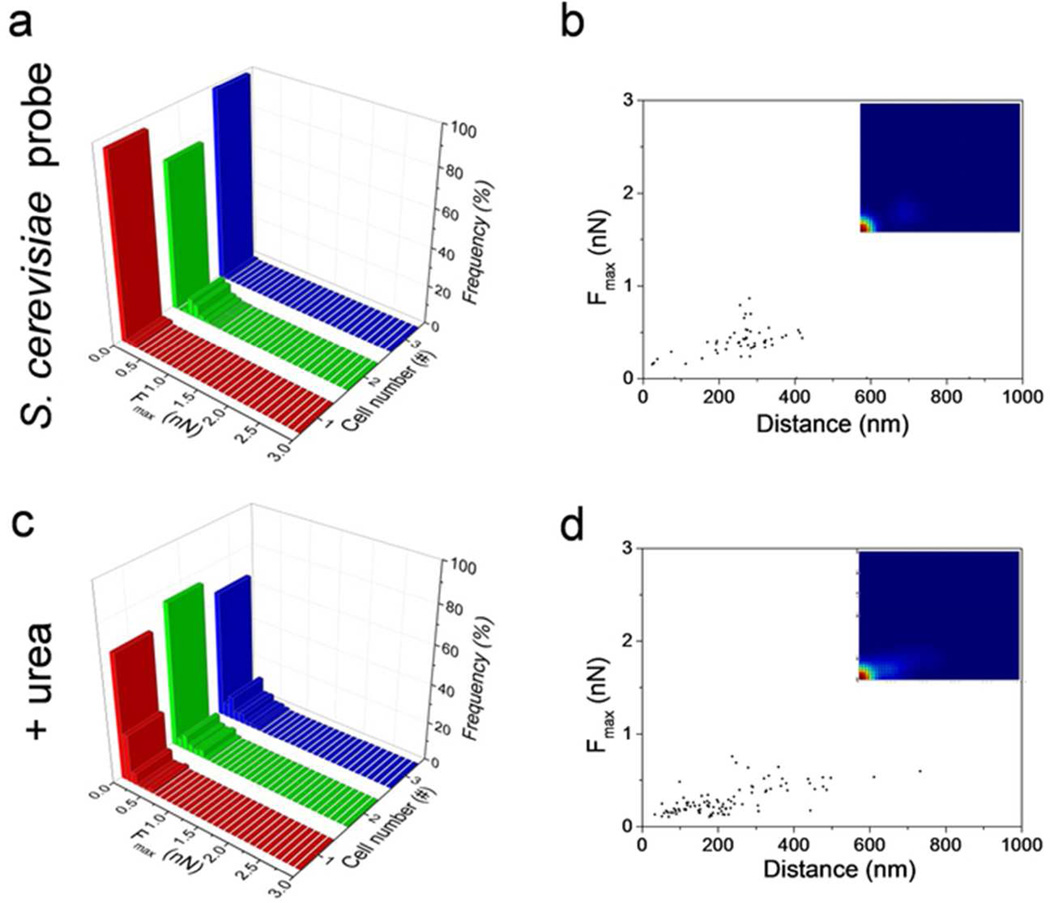
Control experiments were performed to determine whether cohesive interactions originate from cell surface proteins that are specific to C. albicans. Force curves recorded between non-pathogenic yeast Saccharomyces cerevisiae and C. albicans hyphae yielded a very low adhesion frequency (8 %, Figure 3a,b; see red color in the lower left corner of the probability density map), and the adhesion profiles showed only weak tether interactions (Fmax = Fteth = 0.4 ± 0.1 nN; 257 ± 86 nN; LR = 120.000 pN/s). In addition, treatment of C. albicans hyphae with urea (4 M) led to similar results (Figure 3c,d; Fmax = Fteth = 0.3 ± 0.2 nN; 222 ± 132 nN ), thus indicating that disruption of hydrogen bonds in cell surface proteins abolish cell-cell adhesion. These observations lead us to conclude that the measured yeast-hyphae interactions require C. albicans surface proteins.
Figure 3.
Yeast-hyphae adhesion involves cell surface proteins that are specific to C. albicans. (a, c) Adhesion force histograms and (b, d) scatter plots (n = 300 data points; insets: probability density maps) of the strongest adhesion force peaks (Fmax), obtained for the interaction between S. cerevisiae cell probes and C. albicans hyphae (a, b) and between native C. albicans cell probes and C. albicans hyphae treated with 4 M urea (c, d).
Als proteins on the germ tube largely contribute to cell-cell adhesion
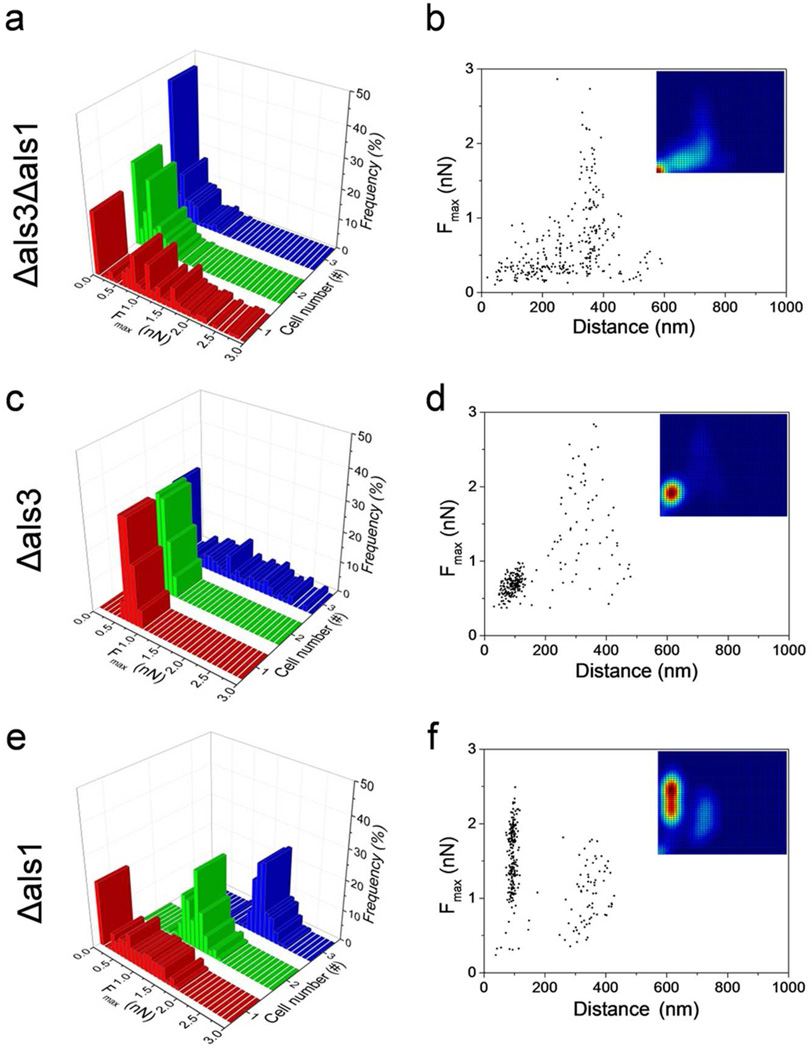
Als glycoproteins play key roles in mediating adhesion and biofilm formation in C. albicans. Although some Als, like Als3, are known to be primarily expressed on germ tubes, it is unclear whether changes in Als expression during fungal morphogenesis modulate cell-cell adhesion. To address this issue, we measured the forces between WT yeast cells and germ tubes from the double mutant als3Δ/als3Δ als1Δ/als1Δ deficient for Als3 and Als1 expression (Supporting Information Figure 2a; Figure 4a,b; LR = 120.000 pN/s). Force signatures of the double mutant were very different from those of the WT. First, the adhesion probability (i.e. fraction of force curves showing adhesion events) dropped from 92 % to 66 % (red color in the lower left corner of the probability density map). Second, most adhesion curves showed only weak tether events (Fmax = Fteth = 0.6 ± 0.5 nN; n = 300 curves) with extended rupture distances (279 ± 117 nm), while strong/short cohesive events were essentially missing. These observations demonstrate that Als3 and (or) Als1 plays a crucial role in establishing strong cohesive adhesion between yeast and hyphal cells.
Figure 4.
Role of Als proteins in mediating yeast-hyphae adhesion. (a, c, e) Adhesion force histograms and (b, d, f) scatter plots (n = 300 data points; insets: probability density maps) of the strongest adhesion force peaks (Fmax) obtained for the interaction between C. albicans cell probes and C. albicans hyphae from the double mutant als3Δ/als3Δ als1Δ/als1Δ (Δals3Δals1) (a, b), and the simple mutants als3Δ/als3Δ (Δals3) (c, d) and als1Δ/als1Δ (Δals1) (e, f). For each mutant, data from 3 different yeast/hyphae combinations are shown.
To clarify the respective roles of Als1 and Als3 in yeast-hyphae adhesion, we analyzed germ tubes from the als1Δ/als1Δ (Figure 4c,d) and als3Δ/als3Δ (Figure 4e,f) single mutants (Supporting Information Figure 2b,c). We found that knockout of Als1 alone (als1Δ/als1Δ mutant) leads to almost full recovery of the cohesive interactions. Most curves (93 %) showed a strong adhesion peak (Fmax = 1.5 ± 0.4 nN; n=300 from 3 independent yeast/hyphae) at short distance (94 ± 13 nm), generally followed (65 %) by a second peak of weaker adhesion (Fteth = 0.7 ± 0.5 nN) at longer distance (190 ± 75 nm), a behavior very similar to that of the WT. Notably, expression of Als1 alone (als3Δ/als3Δ mutant) also led to the reappearance of some cohesive adhesion peaks (Fmax = 0.7 ± 0.1 nN; 93 ± 23 nm) but these were less frequently observed (68 %) and of lower magnitude. These results show that the localized accumulation of Als3 proteins - and to a lesser extent Als1 proteins - on hyphae is the main driving force of the yeast-hyphae adhesion.
Als3 proteins provide remarkable cohesive strength to cell-cell adhesion bonds
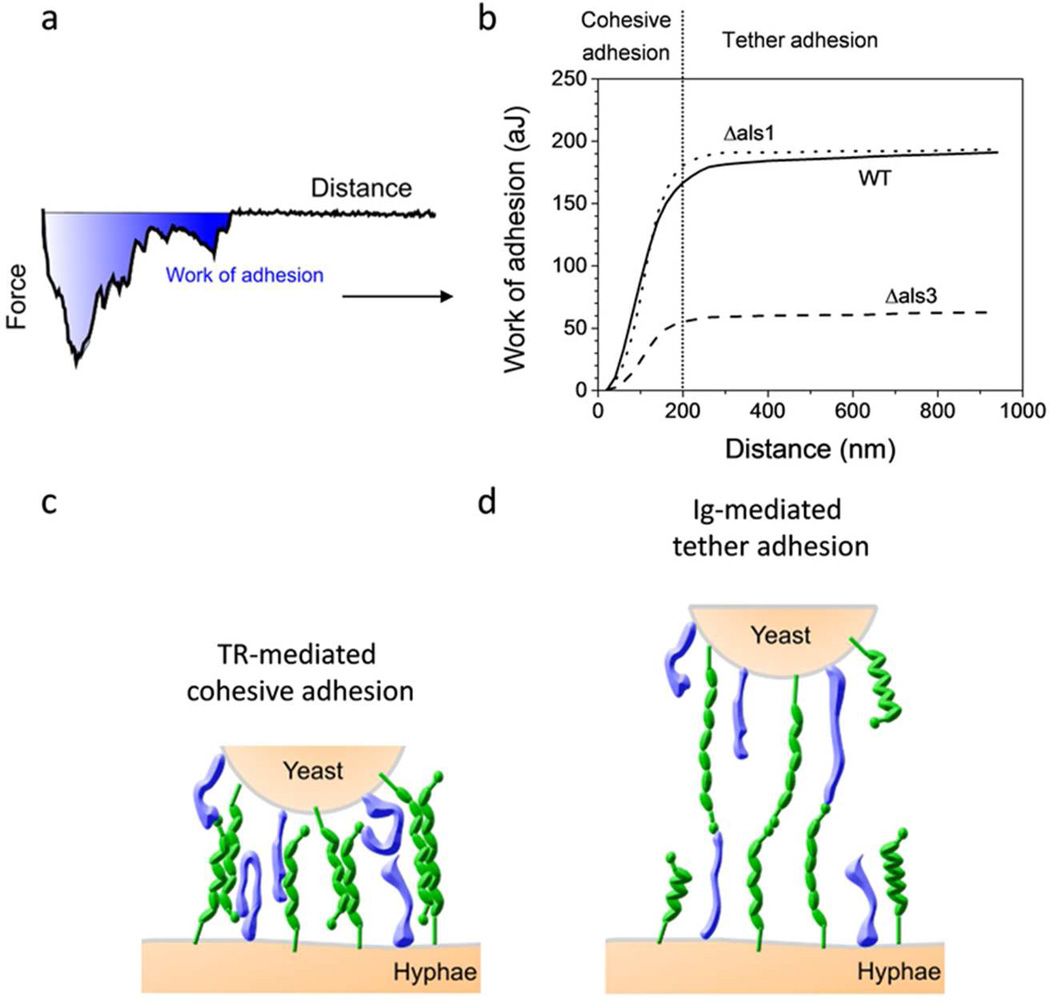
To better understand the molecular nature of Als3-mediated cell-cell adhesion, we calculated the work of adhesion (or detachment) from the area under the retraction curves.20,28 Figure 5 shows the variation of the cumulative work of adhesion as a function of the distance, calculated for the WT and for the single mutants (averaged from 20 force curves recorded on 3 different cells). For the WT, it can clearly be seen that strongly adhesive interactions contribute to most of the work of adhesion (up to ~180 10−18 J) at short distances (0–200 nm), while weak tether interactions only have a marginal contribution (~25 10−18 J) at longer distances. While the als1Δ/als1Δ mutant shows the same behaviour as the WT, the als3Δ/als3Δ mutant displays a dramatic decrease of work of adhesion (~50 10−18 J).
Figure 5.
Mechanism of Als-mediated yeast-hyphal adhesion: interplay of cohesive and tether interactions. (a) To get a molecular view of Als3-mediated cell-cell adhesion, the work of adhesion was calculated from the area under the retraction curves. (b) Variation of the cumulative work of adhesion as a function of the distance, calculated for the WT and for the als1Δ/als1Δ (Δals1) and als3Δ/als3Δ (Δals3) single mutants. (c, d) Proposed adhesion mechanism in which strong, short-range cohesive interactions are mediated by tight hydrophobic bonds between Als TR domains on adjacent yeast and hyphae (c), while weak, long-range tether interactions involve binding of terminal Ig domains on fully stretched Als to specific ligands (d). Als proteins are shown in green, non-Als proteins such as Hwp1 and Eap1 are shown in blue. The size and surface density of the molecules are not on scale.
Can we relate the work of adhesion to a surface density of interacting Als molecules? The work of adhesion of a single Als protein can be estimated from earlier single-molecule measurements13,14 to be ~20 aJ (estimated from single-molecule experiments performed at similar temperature and with a loading rate of 100.000 pN/s), meaning that the ~200 aJ value corresponds to the detachment of ~10 molecules. Note that this calculation assumes parallel loading of all bonds and linearly-additive adhesion forces, which may not be strictly correct for such single-live cell experiments. Nevertheless, the obtained values may be converted into protein surface densities, considering the cell-cell contact area. As a rough approximation, the contact zone of two deformable spheres pressed on each other may be estimated by the following Hertz model equation: in which A is the radius of the contact area, R the effective radius (1/R= 1/R1+1/R2), F the applied load, and E* the effective young modulus (1/E*=((1-v12)/E1)+((1-v22)/E2) in which E1, E2 are the elastic moduli and v1, v2 the Poisson’s ratios associated with each body). Considering a radius of 1.5 µm, a young modulus of 0.6 MPa (determined from indentation measurements) and a Poisson’s ratio of 0.3 for the yeast cell and a radius of 0.6 µm, a young modulus of 7 MPa and a Poisson’s ratio of 0.3 for the germ tube, we found an area radius of ~100 nm, thus yielding a contact area of ~0.03 µm2. The number of molecules in interaction in this area gives a surface density of ~320 Als/µm2, a value in the range of the average cell surface concentration expected for Candida adhesins. Altogether, these observations demonstrate that Als3 provides strongly cohesive adhesion energy between yeast and hyphae cells, which is likely to play an important role in biofilm formation.
We suggest that yeast-hyphae cohesive adhesion originates from tight hydrophobic interactions between Als TR domains on adjacent cells, while tether adhesion would involve the terminal Ig domains on fully stretched Als proteins. Supporting this view, we observed that addition of Ig-ligand peptide "KLRSMAYKIPTHRR" to WT hyphae had a moderate effect, i.e. slight decrease of adhesion frequency and of the maximum adhesion force, suggesting that Ig domains are important for proper extension of the TR regions (Supporting Information Figure 3a); by contrast, addition of the peptide to the double mutant als3Δ/als3Δ als1Δ/als1Δ completely abolished all adhesion signatures, including tether interactions, confirming that these events are mediated by the Ig region (Supporting Information Figure 3b).
CONCLUSIONS
We have shown that the non-destructive attachment of single yeast cells to AFM cantilevers using polydopamine is a valuable method for measuring cell-cell adhesion forces in fungal pathogens. The use of wedged cantilevers makes it possible to avoid tangential load on the cell, thus cell sliding and rolling problems. Using this single-cell technology, we have provided the first direct measurement of adhesive forces between interacting fungal cells. We demonstrated that yeast-hyphae adhesion involves both strong (short-range) cohesive interactions and weak (long-range) tether interactions that are essentially mediated by Als3 proteins expressed on the germ tube. Our experiments provide direct evidence that hyphal formation results in stronger cell-cell interactions, thus that hyphal transition is important for strong biofilm formation. This finding explains why mutants altered in the yeast-to-hyphae transition are strongly affected in their ability to produce thick biofilms.29
Our results favor a model in which strong cohesive adhesion between yeast and hyphae originates from tight hydrophobic interactions between Als TR domains on adjacent cells, i.e. Als3 domains on hyphae and other domains like Als1 or Als5 domains on yeasts (Figure 5c). This mechanism is consistent with the notion that germ tubes are highly hydrophobic,11, 30 and that cell surface hydrophobicity is important for C. albicans adhesion.30–32 Upon cell-cell separation, force-induced unfolding of the TR domains may enhance hydrophobic interactions through increased exposure of hydrophobic residues, up to distances of ~200 nm corresponding to fully stretched TR domains. Further separation would lead to detachment of the interacting TR domains, while the cells would remain connected through weak tether interactions, a phenomenon which increases the lifetime of the adhesive bond (Figure 5d).22 As tether bonds were rarely observed in control conditions (germinating yeast cell, germ tube treated with urea or probed with a S. cerevisiae), we believe they involve binding of the terminal Ig domains of fully stretched Als3 to their cognate ligands, i.e. peptide sequences containing the “τϕ+” motif. In natural biofilms, we expect that Als3 Ig domains will bind to other Als proteins but also to Hwp1 and related cell surface proteins found on C. albicans hyphae. It has indeed been shown that, whereas als1Δ/als1Δ als3Δ/als3Δ or hwp1Δ/hwp1Δ mutants were not able to form biofilms, mixing of such mutant cells resulted in full biofilm formation, suggesting that interaction between the Als and Hwp1 proteins is important for cell-cell interactions and biofilm formation.22
In our cell-cell experiments, one cell (the yeast cell attached to the cantilever) had a sparse surface expression of adhesins, while the other (the germ tube) produced much more adhesins.11, 33 Under these conditions, we predict that a substantial fraction of the probed interactions are between Als adhesins on the hyphae and non-Als proteins on the yeast cell (Figure 5). Indeed the decreased interactions between yeast cells and als mutant hyphae supports this interpretation. Although we do not know the exact identities of the interacting molecules on the yeast cells, hydrophobic cell wall mannoproteins are surely involved.30 One candidate would be Eap1, a hydrophobic mannoprotein expressed on C. albicans yeast cells. The mature form is composed of about 600 amino acid residues in tandem repeats and it mediates binding to polystyrene.34 Eap1 also displays multiple copies of the “τϕ+” motif that binds to Als Ig-like regions, including a pair in tandem near the N-terminus.35 Therefore, this protein would participate in the bulk hydrophobic interactions involved in cohesive adhesion (Figure 5c). The N-terminal position “τϕ+” sequences would allow maximal extension before bond rupture (Figure 5d). We anticipate that the interactions between pairs of highly adhesive cells, e.g. two hyphal cells, would be even stronger than what we measured here. Such strength of adhesion explains the high resistance of C. albicans biofilms to disruption by shear.36,37
METHODS
Microorganisms and cultures
C. albicans SC5314 cells were cultivated on YPD medium (1% yeast extract, 2% Bacto-peptone, 2% D-glucose, supplemented with 2% agar) at 37°C.38 Few colonies were inoculated in YPD liquid medium and agitated overnight (30°C, 200 rpm). Non-germinated cells were harvested by centrifugation, washed 3 times with sodium acetate buffer and resuspended in 10 mL buffer to a concentration of ~106 cells/mL. For hyphae formation, germination was induced by inoculating 250 µL of cell suspension in 8 mL of RPMI 1640 medium buffered with MOPS (Sigma) at pH 7, and agitated at 37°C, 200 rpm, for 90 min unless otherwise stated. We used mutant strains with deletion of both alleles of ALS genes (kindly provided by Aaron Mitchell, Carnegie Mellon University, Pittsburgh, PA).8 These mutants were derived from CAI-438 and included the single mutants als1Δ/als1Δ (Δals1)30 and als3Δ/als3Δ (Δals3)12 and the double mutant als3Δ/als3Δ als1Δ/als1Δ (Δals3Δals1).
Fluorescence microscopy
Following germination for 90 min in RPMI, cells were washed twice by centrifugation in buffer. Cells were resuspended in 20 µL/mL were incubated in a 25 µM solution of Calcofluor White M2R for 30 min at 37°C. Non-germinated yeast cells were washed twice by centrifugation in buffer. Cells were resuspended in 20 µL/mL in acetate buffer supplemented with 1mM Ca2+ and 1mM Mn2+ and incubated in a 30 µg/mL solution of fluorescein isothiocyanate (FITC)-conjugated Concanavalin A for 30 min at 37°C. Images were acquired with a Zeiss Axio Observer Z1 equipped with a Hamamatsu camera C10600.
Viability tests
The viability of yeast and hyphal cells was tested using a LIVE/DEAD yeast viability kit (Molecular Probes). Prior to attachment, a cell suspension (106 cell/mL in 2% glucose Hepes solution) was mixed with FUN1 cell stain (5 µM), mixed thoroughly and incubated for 30 min in the dark at 30°C. Labelled cells were then attached to polydopamine probes or solid substrates, and their viability checked by fluorescence microscopy.
Atomic force microscopy
AFM measurements were performed at room temperature (20°C) in sodium acetate buffer using a Bioscope Catalyst from Bruker corporation (Santa Barbara, CA). Germinating cells were immobilized through hydrophobic attachment on solid substrata. To this end, glass coverslips coated with a thin layer of gold were immersed overnight in a 1 mM solution of 1-dodecanethiol (Sigma), rinsed with ethanol and dry under N2. After induction of germ tube formation in RPMI, the cells were harvested and rinsed three times in acetate buffer. Drops (200 µL) of the concentrated suspension were deposited on the hydrophobic substrates and let to stand for 3 h. After rinsing the unattached cells with buffer, the forces between yeast cell probes (see below) and immobilized hyphae were measured. Force curves were recorded with a maximum applied force upon approach of 1 nN, using a constant approach and retraction speed of 1000 nm.s−1 and a surface delay of 1 s. For blocking experiments, 0.2 mg/mL of "KLRSMAYKIPTHRR" peptides (Eurogentec) were injected in the solution 30 min before the experiment.
Cell probes
We used tipless, oxide sharpened microfabricated cantilevers with a nominal spring constant of ~0.12 N/m (Microlevers, Veeco Metrology Group). The spring constants of the cantilevers were measured prior to the experiments using the thermal noise method. For cell probe preparation, a tipless cantilever was immersed for 1 h in a 10 mM Tris buffer solution (pH 8.5) containing 4 mg/mL dopamine hydrochloride (99%, Sigma-Aldrich). The cantilever was then washed and dried under N2, brought into contact with an isolated non-germinated cell for 10 s, and the obtained cell probe was then transferred without dewetting over a germinated cell for further force measurements.
Supplementary Material
ACKNOWLEDGEMENTS
Work at the Université catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS), the Universite catholique de Louvain (Fonds Spéciaux de Recherche), the Région Wallonne, the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communaute francaise de Belgique (Concerted Research Action). Work at VIB, KU Leuven was supported by Flemish Science Foundation (FWO), the KU Leuven and by the Interuniversity Attraction Poles Programme, initiated by the Belgian Science Policy Office (IAP; P7/28). Work at Brooklyn College was supported by NIH grant R01 GM 098616. Y.F.D. and D.A. are Research Director and Postdoctoral Researcher of the FRS-FNRS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest. The authors declare no competing financial interest.
Supporting Information Available: Additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Costerton JW, Stewart PS, Greenberg EP. Bacterial Biofilms: A Common Cause of Persistent Infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Kolter R, Greenberg EP. Microbial Sciences: The Superficial Life of Microbes. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 3.Douglas LJ. Candida Biofilms and Their Role in Infection. Trends Microbiol. 2003;11:30–36. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 4.Finkel JS, Mitchell AP. Genetic Control of Candida albicans Biofilm Development. Nat. Rev. Microbiol. 2011;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoyer LL. The Als Gene Family of Candida albicans. Trends Microbiol. 2001;9:176–180. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- 6.Lipke PN, Garcia MC, Alsteens D, Ramsook CB, Klotz SA, Dufrene YF. Strengthening Relationships: Amyloids Create Adhesion Nanodomains in Yeasts. Trends Microbiol. 2012;20:59–65. doi: 10.1016/j.tim.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nailis H, Vandenbroucke R, Tilleman K, Deforce D, Nelis H, Coenye T. Monitoring Als1 and Als3 Gene Expression During in Vitro Candida albicans Biofilm Formation under Continuous Flow Conditions. Mycopathologia. 2009;167:9–17. doi: 10.1007/s11046-008-9148-6. [DOI] [PubMed] [Google Scholar]
- 8.Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. Complementary Adhesin Function in C. albicans Biofilm Formation. Curr. Biol. 2008;18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Filler SG. Candida Albicans Als3, a Multifunctional Adhesin and Invasin. Eukaryotic Cell. 2011;10:168–173. doi: 10.1128/EC.00279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman DA, Oh SH, Zhao X, Zhao H, Hutchins JT, Vernachio JH, Patti JM, Hoyer LL. Monoclonal Antibodies Specific for Candida albicans Als3 That Immunolabel Fungal Cells in vitro and in vivo and Block Adhesion to Host Surfaces. J. Microbiol. Methods. 2009;78:71–78. doi: 10.1016/j.mimet.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beaussart A, Alsteens D, El-Kirat-Chatel S, Lipke PN, Kucharíková S, Van Dijck P, Dufrêne YF. Single-Molecule Imaging and Functional Analysis of Als Adhesins and Mannans During Candida albicans Morphogenesis. ACS Nano. 2012;6:10950–10964. doi: 10.1021/nn304505s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nobile CJ, Andes DR, Nett JE, Smith Jr FJ, Yue F, Phan QT, Edwards Jr JE, Filler SG, Mitchell AP. Critical Role of Bcr1-Dependent Adhesins in C. Albicans Biofilm Formation in Vitro and in Vivo. PLoS Pathogens. 2006;2:0636–0649. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alsteens D, Garcia MC, Lipke PN, Dufrene YF. Force-Induced Formation and Propagation of Adhesion Nanodomains in Living Fungal Cells. Proc. Natl. Acad. Sci. USA. 2010;107:20744–20749. doi: 10.1073/pnas.1013893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alsteens D, Dupres V, Klotz SA, Gaur NK, Lipke PN, Dufrêne YF. Unfolding Individual Als5p Adhesion Proteins on Live Cells. ACS Nano. 2009;3:1677–1682. doi: 10.1021/nn900078p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alsteens D, Ramsook CB, Lipke PN, Dufrêne YF. Unzipping a Functional Microbial Amyloid. ACS Nano. 2012;6:7703–7711. doi: 10.1021/nn3025699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lidstrom ME, Konopka MC. The Role of Physiological Heterogeneity in Microbial Population Behavior. Nat. Chem. Biol. 2010;6:705–712. doi: 10.1038/nchembio.436. [DOI] [PubMed] [Google Scholar]
- 17.Brehm-Stecher B, Johnson EA. Single-Cell Microbiology : Tools, Technologies, and Applications. Micriobiol. Mol. Biol. Rev. 2004;68:538–559. doi: 10.1128/MMBR.68.3.538-559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller DJ, Dufrêne YF. Atomic Force Microscopy as a Multifunctional Molecular Toolbox in Nanobiotechnology. Nat. Nanotech. 2008;3:261–269. doi: 10.1038/nnano.2008.100. [DOI] [PubMed] [Google Scholar]
- 19.Müller DJ, Helenius J, Alsteens D, Dufrêne YF. Force Probing Surfaces of Living Cells to Molecular Resolution. Nat. Chem. Biol. 2009;5:383–390. doi: 10.1038/nchembio.181. [DOI] [PubMed] [Google Scholar]
- 20.Benoit M, Gabriel D, Gerisch G, Gaub HE. Discrete Interactions in Cell Adhesion Measured by Single-Molecule Force Spectroscopy. Nat. Cell Biol. 2000;2:313–317. doi: 10.1038/35014000. [DOI] [PubMed] [Google Scholar]
- 21.Krieg M, Arboleda-Estudillo Y, Puech PH, Kafer J, Graner F, Muller DJ, Heisenberg CP. Tensile Forces Govern Germ-Layer Organization in Zebrafish. Nat. Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 22.Helenius J, Heisenberg CP, Gaub HE, Muller DJ. Single-Cell Force Spectroscopy. J. Cell Sci. 2008;121:1785–1791. doi: 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- 23.Friedrichs J, Helenius J, Müller DJ. Quantifying Cellular Adhesion to Extracellular Matrix Components by Single-Cell Force Spectroscopy. Nat. Protoc. 2010;5:1353–1361. doi: 10.1038/nprot.2010.89. [DOI] [PubMed] [Google Scholar]
- 24.Lee H, Dellatore SM, Miller WM, Messersmith PB. Mussel-Inspired Surface Chemistry for Multifunctional Coatings. Science. 2007;318:426–430. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang S, Elimelech M. Bioinspired Single Bacterial Cell Force Spectroscopy. Langmuir. 2009;25:9656–9659. doi: 10.1021/la902247w. [DOI] [PubMed] [Google Scholar]
- 26.Stewart MP, Hodel AW, Spielhofer A, Cattin CJ, Muller DJ, Helenius J. Wedged Afm-Cantilevers for Parallel Plate Cell Mechanics. Methods. 2013;2:186–194. doi: 10.1016/j.ymeth.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 27.Millard PJ, Roth BL, Thi HPT, Yue ST, Haugland RP. Development of the Fun-1 Family of Fluorescent Probes for Vacuole Labeling and Viability Testing of Yeasts. Appl. Environ. Microbiol. 1997;63:2897–2905. doi: 10.1128/aem.63.7.2897-2905.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sariisik E, Docheva D, Padula D, Popov C, Opfer J, Schieker M, Clausen-Schaumann H, Benoit M. Probing the Interaction Forces of Prostate Cancer Cells with Collagen I and Bone Marrow Derived Stem Cells on the Single Cell Level. PLoS ONE. 2013;8:e57706. doi: 10.1371/journal.pone.0057706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramage G, VandeWalle K, López-Ribot JL, Wickes BL. The Filamentation Pathway Controlled by the Efg1 Regulator Protein Is Required for Normal Biofilm Formation and Development in Candida albicans. FEMS Microbiol. Lett. 2002;214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- 30.Masuoka J, Hazen KC. Cell Wall Protein Mannosylation Determines Candida albicans Cell Surface Hydrophobicity. Microbiology. 1997;143:3015–3021. doi: 10.1099/00221287-143-9-3015. [DOI] [PubMed] [Google Scholar]
- 31.Yoshijima Y, Murakami K, Kayama S, Liu D, Hirota K, Ichikawa T, Miyake Y. Effect of Substrate Surface Hydrophobicity on the Adherence of Yeast and Hyphal Candida: Original Article. Mycoses. 2010;53:221–226. doi: 10.1111/j.1439-0507.2009.01694.x. [DOI] [PubMed] [Google Scholar]
- 32.Ener B, Douglas LJ. Correlation between Cell-Surface Hydrophobicity of Candida albicans and Adhesion to Buccal Epithelial Cells. FEMS Microbiol. Lett. 1992;99:37–42. doi: 10.1016/0378-1097(92)90284-u. [DOI] [PubMed] [Google Scholar]
- 33.Hoyer LL, Payne TL, Bell M, Myers AM, Scherer S. Candida albicans Als3 and Insights into the Nature of the Als Gene Family. Curr. Genet. 1998;33:451–459. doi: 10.1007/s002940050359. [DOI] [PubMed] [Google Scholar]
- 34.Li F, Palecek SP. Distinct Domains of the Candida albicans Adhesin Eap1p Mediate Cell-Cell and Cell-Substrate Interactions. Microbiology. 2008;154:1193–1203. doi: 10.1099/mic.0.2007/013789-0. [DOI] [PubMed] [Google Scholar]
- 35.Klotz SA, Gaur NK, Lake DF, Chan V, Rauceo J, Lipke PN. Degenerate Peptide Recognition by Candida albicans Adhesins Als5p and Als1p. Infect. Immun. 2004;72:2029–2034. doi: 10.1128/IAI.72.4.2029-2034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Kohler JR, Kadosh D, Lopez-Ribot JL. Dispersion as an Important Step in the Candida albicans Biofilm Developmental Cycle. PLoS Pathog. 2010;6:e1000828. doi: 10.1371/journal.ppat.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, et al. Portrait of Candida albicans Adherence Regulators. PLoS Pathog. 2012;8:e1002525. doi: 10.1371/journal.ppat.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gillum AM, Tsay EYH, Kirsch DR. Isolation of the Candida albicans Gene for Orotidine-5′-Phosphate Decarboxylase by Complementation of S. cerevisiae Ura3 and E. Coli Pyrf Mutations. Molecular and General Genetics MGG. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.