Abstract
Several bacteria have evolved specialized secretion systems to deliver bacterial effector proteins into eukaryotic cells with the capacity to modulate cellular pathways to promote bacterial survival and replication. The spatial and temporal context in which effectors exert their biochemical activities is critical for their function. Understanding the mechanisms that lead to their precise subcellular localization following delivery into host cells is essential for understanding effector function in the context of infection. Recent studies have shown that bacterial effectors exploit host cellular machinery to accurately target their biochemical activities within the host cell.
Several bacterial species, which infect eukaryotic hosts, have evolved specialized protein secretion systems that mediate the transport of bacterial effector proteins into the cytosol of host cells. These effectors have the ability to modulate a variety of host cellular functions to guarantee the survival and replication of the bacteria that encode them. The most studied of these protein secretion systems are the so called Type III (T3SS) and Type IV (T4SS) secretions systems, which are present in a wide variety of bacteria that are symbiotic or pathogenic for mammals, plants or even insects1, 2 (Box1). These systems often deliver multiple proteins with a variety of biochemical activities that in a coordinated fashion, modulate complex cell processes including cytoskeletal dynamics, membrane trafficking, transcription, cell cycle progression, signal transduction and protein ubiquitination3–5. While there has been significant progress in understanding the biochemical activity of several bacterial effectors, how these diverse activities are ultimately coordinated once the effectors have been delivered into the eukaryotic host cells is less well understood. A growing number of studies have revealed that to exert their function in a spatially coordinated manner, effector proteins must be accurately targeted to their place of action. Precise targeting of bacterial effectors to specific subcellular compartments presumably has many benefits. Since bacterial effectors are delivered in relatively low concentrations, targeting to precise subcellular locations can increase their effective concentration. Perhaps more importantly, the subcellular targeting of effectors ensures the engagement of the correct targets for their various biochemical activities.
Box 1: Type III and Type IV secretion systems.
Many bacteria pathogenic or symbiotic for animals, plants or insects have evolved complex multiprotein machines known as Type III and Type IV secretion systems to deliver bacterial proteins into target eukaryotic cells1, 2. Proteins delivered by these machines posses a variety of biochemical activities capable of modulating eukaryotic cell functions. Although these machines share a common function, they clearly evolved independently presumably from flagella, in the case of type III systems, or the bacterial conjugation systems, in the case of type IV secretion systems. It is believed that these machines must have evolved out of the need to deliver multiple proteins to the same target cell, presumably to modulate complex cell biological processes in a manner in which a single biochemical activity may not be able to do.
Eukaryotic cells are organized into discrete compartments that are most often membrane bound. Therefore, eukaryotic proteins destined for these locations have specific sorting signals or targeting motifs encoded within their amino acid sequence that determine their final destination. Those signals are often recognized by multi-protein machines, which directly mediate protein transport or further modify the target proteins so that they can become transport competent. The unique compartmentalized organization of eukaryotic cells also presents a major challenge to the precise subcellular targeting of bacterial effectors, which are usually delivered directly into the cytosol of the target cell. However, bacterial effectors have evolved various strategies to reach their final destination. These mechanisms often involve the exploitation of eukaryotic-host cell machinery, some which require signal sequences embedded within the amino acid sequence of the effector while others necessitate that an effector must first undergo post-translational modification before being targeted to its final destination. Here, we will discuss different examples of both effector targeting mechanisms (Fig. 1 and Table 1). Instead of a comprehensive review of the literature, we will focus on a few of the most well characterized examples that illustrate the main themes that have been revealed as a result of studying the subcellular targeting of bacterial effectors.
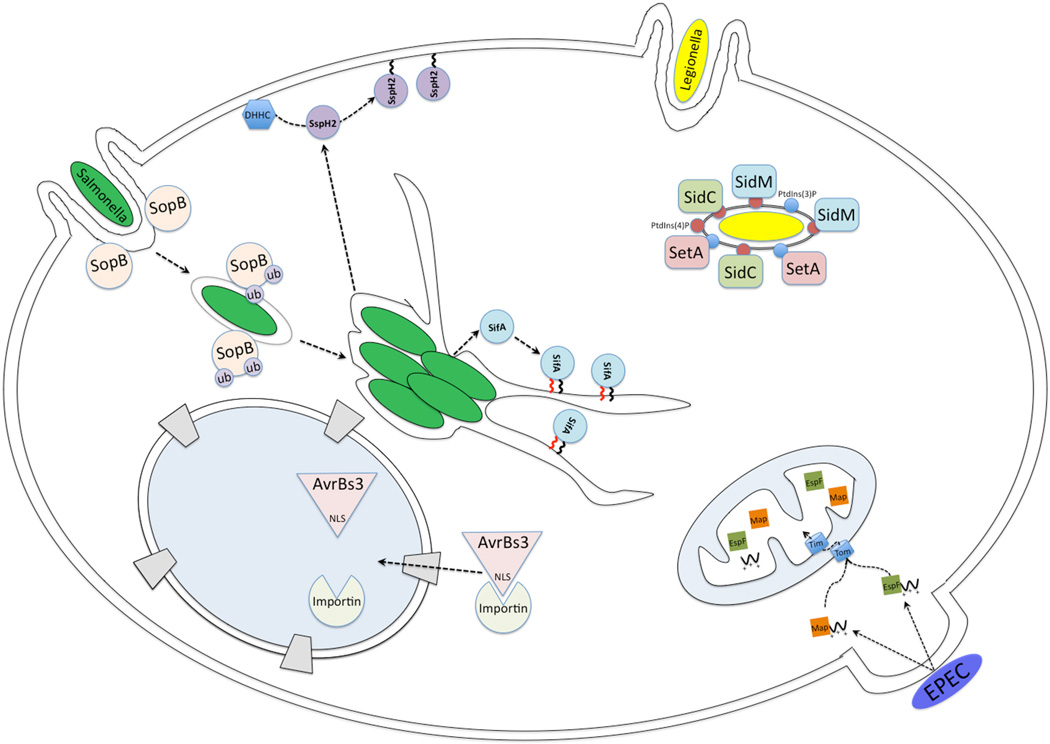
Fig. 1.
A number of intracellular bacteria have evolved specialized secretion systems to deliver bacterial effector proteins into eukaryotic cells. These effector proteins modulate host cell pathways to promote bacterial survival and replication. Once translocated into the host cell cytosol, many bacterial effector proteins (some of them depicted in this diagram) exploit eukaryotic-host cell machinery to reach their final subcellular localization. L. pneumophila utilizes a T4SS to deliver phosphoinositide-binding effectors, including SidC, SidM, and SetA, which recruit host cell membrane to the Legionella-containing vacuole (LCV) to help establish an intracellular replication niche. The Enteropathogenic Escherichia coli (EPEC) T3SS effectors, Map and EspF encode putative N-terminal mitochondrial targeting signals, and when transiently expressed in eukaryotic cells, localize to mitochondria by interacting with components of the classical mitochondrial import machinery. The plant pathogen, Xanthomonas translocates a family of effectors, including AvrBs3, which localize to the plant nucleus where they alter host gene expression during infection. These effectors encode a classic nuclear localization signal (NLS) that is recognized by the host cell nuclear import pathway leading to their nuclear targeting. The Salmonella T3SS effector, SopB, requires host-mediated monoubiquitination to localizes to the salmonella-containing vacuole (SCV) late during infection. Salmonella also utilizes host-mediated lipidation to target effectors to their site of action. Salmonella effector, SspH2 is S-palmitoylated by host enzymes resulting in its plasma membrane localization. Another Salmonella effector, SifA, is both S-palmitoylated and prenylated resulting in localization to the SCV.
Table 1.
Examples of the Subcellular Localization of Bacterial Effectors
| Bacteria | Effector | Subcellular Localization | Targeting Motif/s | Refs |
|---|---|---|---|---|
| Agrobacterium tumefaciens ssp | VirE2 | Nuclear | Bipartite NLS | 100, 105 |
| VirD2 | Nuclear | Bipartite NLS | 102, 103 | |
| Escherichia coli ssp | Map | Plasma Membrane, Mitochondria | Protein-Protein Interaction, Mitochondrial Presequence | 71, 73, 77 |
| EspF | Mitochondria, Nucleolus, plasma membrane | Mitochondrial Presequence, ?, ? | 72, 79, 106 | |
| Tir | Plasma Membrane | ? | 82, 83, 107 | |
| Legionella ssp | AnkB | Legionella-containing vacuole | Prenylation | 30 |
| Lpl28061 | Legionella-containing vacuole | Prenylation | 29 | |
| SidC | Legionella-containing vacuole | PtdIns(4)P binding domain | 58, 59 | |
| SidM | Legionella-containing vacuole | PtdIns(4)P binding domain | 60, 61, 108 | |
| LidA | Legionella-containing vacuole | PtdIns(4)P binding domain | 60, 61 | |
| SetA | Legionella-containing vacuole | PtdIns(3)P binding domain | 62 | |
| Pseudomonas ssp | AvrB | Plasma Membrane | N-Myristoylation/S-Palmiotylation | 17 |
| AvrRpm1 | Plasma Membrane | N-Myristoylation/S-Palmiotylation | 17 | |
| HopF2 | Plasma Membrane | N-Myristoylation/S-Palmiotylation | 19 | |
| HopZ | Plasma Membrane | N-Myristoylation/S-Palmiotylation | 18 | |
| AvrPphB | Plasma Membrane | N-Myristoylation/S-Palmiotylation | 22 | |
| ORF4 | Plasma Membrane | N-Myristoylation/S-Palmiotylation | 22 | |
| NopT | Plasma Membrane | N-Myristoylation/S-Palmiotylation | 22 | |
| AvrPto | Plasma Membrane | N-Myristoylation/S-Palmiotylation | ||
| ExoU | Plasma Membrane | PtdIns(4,5)P2 binding domain | 63, 64 | |
| Multiple Effector Family2 | Mitochondria/Chloroplast | Mitochondrial /Chlotoplsdt Presequence | 70 | |
| HopG1 | Mitochondria | ? | 109 | |
| Salmonella ssp | SspH2 | Plasma Membrane | S-Palmitoylation | 10 |
| SseI | Plasma Membrane | S-Palmitoylation | 10 | |
| SifA | Salmonella containing vacuole and Salmonella induced Filaments | Prenylation | 32–34 | |
| SipB | Mitochondria | ? | 110 | |
| SopA | Mitochondria | ? | 111 | |
| SopB | Salmonella containing vacuole | Ubiquitination | 49, 50 | |
| Xanthomonas ssp | AvrBs3 | Nuclear | Monopartite NLS | 96, 98, 99 |
| AvrXa5 | Nuclear | Monopartite NLS | 98, 112 | |
| AvrXa7 | Nuclear | Monopartite NLS | 113 | |
| AvrXa10 | Nuclear | Monopartite NLS | 98 | |
| PthA | Nuclear | Monopartite NLS | 98 | |
| Shigella ssp | IpaH1 | Plasma Membrane | S-Palmitoylation | 10 |
| IpaH4.5 | Plasma Membrane | S-Palmitoylation | 10 | |
Ivanov et al showed eight different Legionella effectors require the CAAX motif for membrane localization.
Subcellular localization of multiple Pseudomonas effectors including Hop, Avr, Hol, Hrp family members was predicted by the program TargetP (http://www.cbs.dtu.dk/services/TargetP-1.0/).
Membrane-Targeting by Host-mediated Lipidation
An increasing number of bacterial effectors are covalently modified by the attachment of a variety of lipid groups upon their translocation into the cytoplasm of eukaryotic cells. The addition of lipid groups increases the hydrophobicity of proteins, which can facilitate their tethering to intracellular membranes and, as a result, affects protein localization and function. While some proteins are exclusively modified by a single lipid group, others are sequentially modified with different lipids allowing for multiple layers of targeting information. Lipidation of proteins not only promotes their membrane association but can also direct their association with liquid-order domains of the membrane, such as lipid rafts and caveolae. The major forms of protein lipidation in eukaryotes are S-palmitoylation, N-myristoylation, and prenylation.
S-palmitoylation
S-palmitoylation has an important role in regulating protein stability, protein-protein interactions and protein activity6–8. This modification is unique among lipid modifications in that it is generally reversible, and can therefore impact protein function by dynamically controlling membrane association and/or protein-protein interactions. S-palmitoylation is characterized by the addition of a saturated 16-carbon palmitic acid to a specific cysteine residue in a protein through a thioester bond8. Unlike other lipid modifications, S-palmitoylation has no clear amino acid sequence requirement other than the presence of a cysteine residue. In eukaryotic cells palmitoylation is catalyzed by a family of 23 Asp-His-His-Cys (DHHC) motif-containing proteins that have palmitoyltransferase activity9. DHHC proteins are polytopic membrane proteins found on a variety of cellular membranes with a conserved cytoplasmicfacing cysteine-rich domain that includes the DHHC catalytic motif. S-palmitoylation has also been shown to play an important role in regulating protein stability, protein-protein interactions and protein activity6–8. This modification is unique among lipid modification in that it is generally reversible, and therefore can impact protein function by dynamically controlling membrane association and/or protein-protein interactions.
Salmonella enterica is the cause of gastroenteritis and typhoid fever, a life-threatening systemic disease of humans. A recent study has shown that host-mediated S-palmitoylation of an N-terminal cysteine residue of the Salmonella enterica T3SS effector SspH2 results in its targeting to the host-cell plasma membrane10. Based on amino acid sequence similarity, several other potentially palmytoilated Salmonella T3SS effectors were identified, including SseI. Both SspH2 and SseI are targeted to the plasma membrane in a palmitoylation-dependent manner. A screen of each of the DHHC family members identified a subset of enzymes able to catalyze the palmitolylation of SspH2 and SseI10. Two of these enzymes, DHHC-3 and -7, were able to directly palmitoylate SspH2 and SseI in vitro. DHHC-3 and -7 are phylogenetically related and have been shown to be active toward other N-terminally palmitoylated substrates. Although both SspH2 and SseI are stably palmitoylated, they are targeted to different domains of the plasma membrane of polarized cells. This finding suggests that while essential for plasma membrane localization, palmitoylation is not sufficient to target these effectors to their final destination. SspH2 belongs to a family of bacterial effectors that possess a C-terminal NEL E3 ligase domain, which have been implicated in immune response modulation during infection11, 12. The C-terminal domain of SseI shares amino acid sequence similarity with a family of bacterial deamidases13 and has been reported to regulate cell migration13, 14, a function that requires its palmitoylation and plasma membrane targeting10. Thus, Salmonella has evolved to exploit host-mediated palmitoylation to target at least two distinct biochemical activities to their sites of action at the host-cell plasma membrane.
N-myristoylation
N-myristoylation results in the covalent attachment of myristic acid via an amid bond primarily to the alpha-amino group of an N-terminal glycine15. This modification is catalyzed by an N-myristoyltransferase following exposure of a GXXX(S/T/C) N-terminal consensus sequence. Although N-myristoylation has been historically described as a co-translational modification that occurs following the removal of the initiator methionine residue of a protein, there are known examples of post-translational myristoylation15, 16. In many instances, protein N-myristoylation is necessary but not sufficient to promote stable and permanent membrane association. Consequently, this modification often occurs together with S-palmitoylation of proximal cysteine residues or a polybasic amino acid domain next to the N-terminus of the protein.
Pseudomonas syringae, which causes a variety of diseases in plants, encodes a large number of T3SS effectors that promote virulence in susceptible hosts. A subset of these effectors, including AvrB, AvrRpm1, HopF2, and multiple HopZ family members, encode N-terminal eukaryotic consensus sites for myristolyation17–19. When expressed in susceptible plant cells, AvrB and AvrRmp1 are targeted to the plasma membrane17. Substitution of the critical N-terminal glycine residue with an alanine (G2A) in either protein resulted in their mislocalization to the cytosol. In addition, both effectors are also S-palmitoylated on a cysteine immediately following the glycine, and this palmitoylation increases membrane association17. Therefore, myristoylation and S-palmitoylation of these Avr effectors are required for correct plasma membrane localization. Both AvrB and AvrRmp1 have been shown to interact and induce phosphoylation of RIN4, a negative regulator of basal plant defense, which is localized to the plasma membrane of plant cells20,21. Consistent with the importance of this modification for effector function, myristoylation, and to a lesser extent palmitoylation, were shown to be required for the hypersensitive response triggered by AvrB and AvrRmp1 in resistant hosts and the virulence phenotype of AvrRmp1 in susceptible hosts17.
While AvrB and AvrRmp1 possess conventional myristoylation sites found at the very N-terminus of eukaryotic proteins, another subset of Avr type III effectors, including AvrPphB, ORF4, and NopT, possess internal myristoylation sites within their full-length protein sequence22. Interestingly, following translocation into host cells, these effectors undergo an autoproteolytic-processing event that exposes a new N-terminus possessing a myristoylation consensus sequence. This process is analogous to the myristoylation of certain eukaryotic proteins following cleavage by caspases, which reveals cryptic myristoylation consensus sites23. These processed effectors are then myristoylated, as well as S-palmitoylated, by the host and targeted to the plasma membrane22. AvrPphB specifically cleaves the Arabidopsis plasma membrane-localized protein kinase, PBS1, to initiate the RPS5-dependent hypersensitive response24, 25. Not only is lipidation required for plasma membrane localization, but dual lipidation is required for the activity of AvrPphB in resistant plants22. Therefore, P. syringae encodes proteins bearing both conventional and cryptic eukaryotic myristoylation motifs that can functionally target bacterial effectors to the plasma membrane of the host cell during infection.
Prenylation
Prenylation involves the covalent addition of a 15-carbon farnesyl or a 20-carbon geranylgeranyl isoprenoid group to a cysteine residue within a conserved C-terminal tetrapeptide CaaX (Cys-aliphatic-aliphatic-X) motif26. The residue located at position ‘X’ determines whether the protein will be farnesylated or geranylgeranylated. When “X” is a serine, methionine, or glutamine, the protein is recognized by FTase whereas a leucine at this position results in modification by GGTase-I26. Prenyltransferases catalyze the transfer of isoprenoid group to the cysteine residue of the CaaX motif and are only found in eukaryotic cells. Following prenylation, the terminal ‘aaX’ sequence is cleaved off by a Ras-converting enzyme-1 located in the ER membrane27. Then the C-terminal prenylated cysteine residue is then methylated by isoprenyl-cysteine carboxyl methyltransferase28. Prenylation is a permanent posttranslational modification required for protein stability that results in the localization of proteins to intracellular membranes through attachment of the modified C-terminus.
Legionella pneumophila, the cause of Legionnaires disease, encodes several T4SS effectors with eukaryotic CaaX motifs within their sequences, which are required for subcellular localization29–31. For example, once translocated, the L. pneumophila effector AnkB is farnesylated on the cysteine residue within a conserved CaaX motif by the host cell protein farnesyltransferase (PFT), and as a result becomes associated with the cytosolic face of the Legionella containing vacuole (LCV)30. Mutation of the critical cysteine residue of the CaaX motif to an alanine or inhibition of host PFT resulted in loss of farnesylation and of targeting to the LCV. The LCV membrane is derived from the ER, which contains the eukaryotic enzymes required for processing farnesylated proteins. Therefore, it has been proposed that immediately following translocation into the LCV, AnkB is locally prenylated, anchoring it into the LCV membrane. AnkB also contains an eukaryotic F-box domain that is thought to recruit polyubiquitinated proteins to the LCV. AnkB is required for rapid acquisition of polyubiquitinated proteins and is essential for intracellular proliferation. Legionella strains expressing prenylation-deficient AnkB are unable to recruit polyubiquitinated proteins to the LCV and are attenuated in a mouse model of Legionnaires’ disease30, 31.
The Salmonella enterica T3SS effector SifA contains a C-terminal CaaX motif, (331CLCCFL) and is geranylgeranylated on cysteine333 by a host geranylgeranyltransferase32, 33. SifA localizes to the Salmonella containing vacuole (SCV) as well as tubular membrane structures known as Salmonella-induced filametns (SIFs) that extend from the SCV during infection34. Inhibition of SifA prenylation leads to the mislocalization of SifA to the cytosol32. While prenylation alone can facilitate interaction with cellular membrane, some prenylated proteins require either a stretch of basic amino acids or S-palmitoylation upstream for their stable association to membranes. It was shown that cysteine331 of SifA is palmitoylated, which presumably strengthens its association with membranes32. SifA is required for Salmonella pathogenesis and is thought to regulate membrane fusion to help provide sufficient membrane as the SCV expands34. While deletion of SifA results in a replication defect in macrophages and reduced colonization in mouse studies, prenylation- and palmitoylation-deficient SifA strains did not show this phenotype32. Therefore, while targeting of SifA to the SCV and tubular extensions requires prenylation, the contribution of this localization for SifA function remains unclear.
Membrane-targeting by Host Mediated Ubiquitination
Ubiquitinylation results in the covalent attachment of ubiquitin to a lysine residue on a target protein and constitutes a common and important post-translational modification in eukaryotic cells35. Following the initial conjugation, a substrate can remain monoubiquitinylated at a single lysine residue, or additional ubiquitin molecules can either be ligated to one of the seven lysines in each of the first ubiquitin molecule, resulting in polyubiquitinylation or can be ligated to multiple lysine residues, resulting in multiubiquitinylation. The type of ubiquitinylation and the topology of the ubiquitin chains that are formed direct the substrate’s fate36.
Protein ubiquitination can signal for proteasome-dependent degradation but can also modulate protein function by, for example, changing protein localization or allowing the formation of multi-protein complexes37–41. Many bacterial effector proteins have evolved to directly modulate the ubiquitination pathway by acting as deubiquitinases or E3 ligases, thus altering the function or stability of host target proteins12, 42, 43. Other effector proteins, however, utilize ubiquitination to modulate their own function. For example, it has been shown that the half-life of some effectors is controlled by ubiquitin-mediated degradation44, 45. In other cases, ubiquitination can be critical to targeting the effector protein to its final destination. For example, SopB is a Salmonella T3SS effector protein that is involved in at least three distinct phenotypes, all of which are strictly dependent on its phosphoinositide phosphatase activity. By stimulating the RhoG exchange factor SGEF, SopB is capable of mediating actin-dependent Salmonella-internalization into non-phagocytic cells46. In addition, SopB is required for the activation of the protein kinase Akt, by still poorly understood mechanisms, which promotes intracellular survival47. Lastly, SopB can modulate the phosphoinositide composition of the SCV, thus modulating the membrane traffic of this compartment48. This functional diversification correlates with the temporal regulation of SopB localization. Early during infection, SopB localizes to the plasma membrane where it activates RhoG and Akt. Later, it localizes to the SCV, where it modulates the phosphoinositide composition of the SCV membrane. The differential localization of SopB is strictly dependent on its monoubiquitination since a SopB mutant that cannot be ubiquitinated, remains at the plasma membrane49, 50. Although this mutant can mediate RhoG-dependent entry and Akt activation, it is unable to localize to the SCV resulting in a defect in intracellular growth. The ubiquitin-mediated relocalization of SopB is reminiscent of the ubiquitin-mediated relocalization of the epidermal growth factor receptor from the plasma membrane to an endocytic compartment51, 52. However, although both proteins are re-localized in an ubiquitin-dependent manner, the cellular machinery necessary for this relocalization appears to be different.
Membrane-targeting by Phosphoinositide Binding
Phosphoinositides play an essential role in regulating a variety of cellular processes ranging from membrane traffic to actin dynamics53–55. The reversible phosphorylation of the inositol group of phosphoinositides at different positions and in different combinations can generate seven different species, which concentrate in different intracellular membranes. This differential distribution is highly regulated and allows for effective subcellular targeting of host membrane-associated signaling events. Phosphoinositides serve as anchor moieties for a variety of proteins with binding domains that are selective for different phosphoinositide species. Several bacterial effector proteins have been shown to use phosphoinositides to accurately target their biochemical activities56, 57. For example, several Legionella pneumophila T4SS effectors proteins are targeted to the Legionella-containing vacuole by binding different phosphoinositides: SidC and SidM are targeted by binding PtdIns(4)P58–61, SetA by interacting with PtdIns(3)P62, and LidA by binding both phosphoinositides60, 61. Together these effector influence the remodeling and maturation of LCV by promoting the interaction of the LCV with host vesicles and organelles. Interestingly, the discrete domains of these effectors that bind PtdIns(4)P or PtdIns(3)P do not exhibit any detectable sequence or structural similarity to equivalent domains in eukaryotic proteins, suggesting that these domains may have been the result of convergent evolution, a common theme in the evolution of effector proteins. The Pseudomonas aeruginosa T3SS effector protein ExoU is another example of an effector that is targeted to its final destination by phosphoinositide binding. ExoU is a phospholipase that is targeted to the plasma membrane by a discrete α-helical bundle domain at its C-terminus that has high affinity for PtdIns(4,5)P2, which is abundant at the cytoplasmic side of the plasma membrane63, 64. Targeting of the phospholipase activity of ExoU to the plasma membrane via its C-terminal PI binding domain leads to rapid necrotic host cell death my disrupting host cell membranes, which promotes Pseudomonas pathogenesis64–66.
Mitochondria/Chloroplast Targeting
Eukaryotic cells use similar mechanisms to target proteins to mitochondria and chloroplasts. Most eukaryotic mitochondria/chloroplast proteins are synthesized in the cytosol as preproteins that encode either an N-terminal presequence or an internal targeting signal within the precursor protein67, 68. These presequences form an amphiphilic α-helix with positively charged residues on one side of the helix and hydrophobic residues aligned on the other side. During synthesis, many mitochondria/chloroplast preproteins associate with molecular chaperones that maintain them in a partially unfolded conformation. These preproteins are then targeted to the mitochondria/chloroplast and can be imported into the organelle where they can undergo proteolyic cleavage. The targeting mechanisms for mitochondrial proteins have been examined in detail. The targeting signals are recognized by receptor subunits of the TOM and TIM complexes present in mitochondrial membranes. Following binding of the presequence to the Tom/Tim complex, preproteins are imported across the mitochondrial membranes in an unfolded state. Once translocated, the presequence is usually cleaved off, and chaperones in the mitochondrial matrix mediate protein folding69. Mitochondrial import is generally described as a posttranslational process, implying that proteins are completely synthesized before import starts67.
Amino acid sequence analysis revealed that the N-terminal T3SS signal of most Pseudomonas syringae effectors share a similar amino acid composition to chloroplast and mitochondrial presequences70. As a result, most P. syringae effectors are predicted to localize to chloroplasts or mitochondria70. However, this similarity may merely imply an evolutionarily conserved mechanism used by these organelles and T3SS to recognize and target substrates. Therefore, caution should be exercised before assigning mitochondrial/chloroplast localization to bacterial T3SS effectors based on bioinformatic analysis alone.
In addition to P. syringae, the enteropathogenic Escherichia coli (EPEC) T3SS effectors Map and EspF encode putative N-terminal mitochondrial presequences with cleavage signals71, 72. When transiently expressed, the N-terminus of Map can be cleaved and Map localizes to host mitochondria, indicating that host cell mitochondrial targeting machinery can recognize Map as a substrate71. In vitro assays showed that Map mitochondrial import involves components of the Mitochondrial import pathway (Tom22, Tom40 and mtHsp70), which is consistent with Map import into the mitochondria matrix via the classical import mechanism73. In addition, it has been reported that Map disrupts the mitochondrial membrane potential leading to mitochondrial fragmentation71,73. More recent structural and functional studies, however, have shown that EPEC Map is a guanine nucleotide exchange factor (GEF)74, 75. In addition to a GEF domain, Map possesses a canonical C-terminal PSD-95/Disc Large/ZO-1 (PDZ)-binding motif that interacts with the PDZ domains of the Ezrin-binding protein, EBP50/NHERF176, 77. Following translocation, Map localizes to the plasma membrane at the bacterial/host interface77. This localization results in CDC42 activation, leading to the localized formation of actin-rich membrane protrusions77. Although this particular activity is at odds with the reported mitochondrial localization and disruption, it has been observed that at later time points, Map does appear to colocalize with the mitochondria. It is therefore possible that Map may have a dual function, working as a GEF for CDC42 required for actin rearrangements at the plasma membrane promoting bacterial attachment early during infection and for disrupting mitochondrial function later in infection
Some studies have suggested that the EPEC T3SS effector EspF is targeted to host mitochondria72, 76, 78–80. However, other studies have also observed that EspF can be targeted to the nucleus and/or plasma membrane78, 79. Some of these disparate subcellular localizations could be a result of epitope tagging or in some instances overexpression, which can alter the physiological localization of proteins. Some of these disparate subcellular localizations could be a result of epitope tagging or in some instances overexpression, which can alter the physiological localization of proteins. In fact, the EPEC effector Tir is inserted into the host plasma membrane following translocation81,82,83, however when Tir is transiently expressed with a C-terminal GFP tag, Tir is mistargeted to the mitochondria84. EspF has been attributed a number of functions, including disruption of tight junctions, inhibiting phagocytosis, plasma membrane remodeling, cytoskeletal rearrangements, disruption of the nucleolus, mitochondrial dysfunction, and apoptosis85, 86. However, mitochondrial targeting of EspF is not required for the majority of these functions. Therefore, it is possible that mitochondrially targeted EspF does not represent the functional pool of EspF during infection. Finally, it should be observed that when some bacterial proteins are ectopically expressed within mammalian cells they can localize to the host mitochondria despite clear evidence that they are not otherwise targeted to the mitochondria87, suggesting that cryptic mitochondrial targeting signals may be common in bacterial proteins and therefore their targeting to mitochondria should be interpreted with caution.
Nuclear Localization
Given the central importance of nuclear functions in cellular physiology, it is not surprising that many effector proteins have evolved to carry out their function within the nucleus of infected cells. Most proteins that function in the nucleus are selectively imported from the cytosol through nuclear pore complexes, large structures that form channels that allow proteins smaller than the diameter of the channel to passively diffuse into the nucleus. Globular proteins larger than 50kD are unable to passively diffuse through the nuclear pore complex and therefore require active import, which is dependent on nuclear localization signals (NLS).
Several pathways for nuclear import have been described. The classical NLS for nuclear protein import consists of either one (monopartite) or two (bipartite) short amino acid sequences rich in the positively charge amino acids, lysine and arginine. NLSs may be present in multiple copies and located throughout the protein, usually forming a loop on the surface of the fully folded protein. Signals are recognized by soluble carriers of the importin α and importin β superfamilies88. In classical nuclear import, importin α recognizes and binds in the cytoplasm both the nuclear cargo and importin β, which then mediates the interaction of the complex with the nuclear pore as it translocates into the nucleus89–91. Once the complex reaches the nucleus, RanGTP binds to importin β causing a conformational change that results in the release of the importin α-cargo complex into the nucleus92.
In most instances, the transport of bacterial effector proteins to the nucleus is carried out by the cellular nuclear import machinery. For example, members of the the plant pathogen genus Xanthomonas encode a large family of transcription activator-like (TAL) T3SS effectors, including AvrBs3, AvrXa5, AvrXa7, AvrXa10, and PthA, which localize to the plant nucleus and regulate plant gene expression during infection93. TAL effectors are characterized by a central DNA binding region consisting of nearly identical tandem 34 amino acid repeats followed by classic monopartite NLS, and an acidic transcriptional activation domain (AAD)93, 94. In susceptible host species, the TAL effector AvrBs3 of X. campestris pv. Vesicatoria modulates gene expression inducing hypertrophy of plant host cells95. In contrast, in resistant species AvrBs3 triggers the hypersensitive response leading to localized host cell death. Both responses require the NLS and AAD of AverBs3 supporting a functional role for AvrBs3 in the host nucleus95–97. Sequence analysis revealed three putative NLSs encoded within the C-terminal domain of AvrBs3, which are conserved in other members of this gene family98. NLS2 and NLS of AvrBs3 were shown to be required for AvrBs3 to be fully active while NLS1 is insufficient for AvrBs3 activity96, 99. AvrBs3 was shown by both, yeast two-hybrid and in vitro binding assays, to interact with two members of the classic nuclear import pathway in pepper Bs3 plants, CalMPα1 and CalMPα299. CalMPα1 and CalMPα2 are importin α proteins, which mediate nuclear import by binding the NLS of substrate proteins in the cytosol. The interaction of AvrBs3 with CalMPα1 and CalMPα2 was shown to be dependent on the NLSs of AvrBs399. Therefore, following translocation into the host cell cytosol, AvrBs3 is targeted to the nucleus by the classical eukaryotic nuclear import pathway where it alters host gene expression.
Agrobacterium tumefaciens also encodes a T4SS bacterial effector, VirD2 that has been shown to interact with the eukaryotic nuclear import machinery. Agrobacterium pathogenesis requires the transfer of the bacterial tumor inducing (Ti) plasmid to the host plant cell. The transferred DNA (T-DNA) is imported into the cell nucleus and integrates into the plant cell genome. Expression of T-DNA leads to tumor formation in the host plant. The nuclear import of T-DNA requires the interaction with two Agrobacterium effectors, VirD2 and VirE2. VirD2 associates with the 5’ end of single-stranded T-DNA, while VirE2 coats the rest of the ssDNA molecule100, 101. Both VirD2 and VirE2 possess functional NLSs. VirD2 encodes two NLSs, and the C-terminal-proximal NLS has been shown to target reporter proteins to the nucleus of plant, animal and yeast cells102, 103. VirD2 was shown to bind the Arabidopsis importin α protein AtKAPα104. Binding of VirD2 to AtKAPα is dependent on the presence of the NLS sequence in VirD2, consistent with AtKAPα acting as an import receptor for the classical nuclear import pathway.
Concluding Remarks
Some bacteria have evolved simple mechanisms to deliver exotoxins, which encompass a limited number of biochemical activities, into eukaryotic cells by encoding all the required information for the transport from the bacteria into eukaryotic cells within a single polypeptide. However, the need to deliver multiple (in some instances more than 100) biochemical activities (effector proteins) to the same cell in a temporally and spatially restricted manner have demanded the evolution of much more complex delivery machines such as the T3SS or T4SS. The diversity of the biochemical activities of these effectors and the coordination necessary for them to exert their appropriate function demand that they must be targeted precisely to the appropriate subcellular location. It is becoming increasingly clear that the precise targeting of these effectors requires host cellular machinery that is usurped by these effectors to reach their final destination. The challenge for the future will be to visualize the fate of the translocated effectors in live cells during bacterial infection. This is difficult because in addition to the intrinsic limitations presented by the protein delivery systems themselves, effector proteins are delivered at very low concentrations. Therefore, more sensitive live imaging techniques will be required to address this important issue. It is clear that understanding of the contribution of the different effectors to bacterial pathogenesis demands a better understanding of their subcellular localization once translocated. It is also possible that information gained by the study of the targeting mechanisms utilized by different effectors may lead to the development of novel therapeutic strategies to combat infections by these pathogens.
Acknowledgements
Work on the subjects discussed in this article has been supported by NIH grants AI055472 and AI079022 (to J.E.G.). S. W. H. was supported by an NRSA fellowship from the NIH.
References
- 1.Christie P, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 3.Galán J. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubber A, Roy C. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 5.Desveaux D, Singer A, Dangl J. Type III effector proteins: doppelgangers of bacterial virulence. Curr Opin Plant Biol. 2006;9:376–382. doi: 10.1016/j.pbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 7.Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci STKE. 2006;2006:re14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- 8.Smotrys JE, Linder ME. Palmitoylation of intracellular signaling proteins: regulation and function. Annu Rev Biochem. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- 9.Greaves J, Chamberlain LH. Palmitoylation-dependent protein sorting. J Cell Biol. 2007;176:249–254. doi: 10.1083/jcb.200610151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicks SW, Charron G, Hang HC, Galan JE. Subcellular targeting of Salmonella virulence proteins by host-mediated S-palmitoylation. Cell Host Microbe. 2011;10:9–20. doi: 10.1016/j.chom.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quezada CM, Hicks SW, Galan JE, Stebbins CE. A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proc Natl Acad Sci U S A. 2009;106:4864–4869. doi: 10.1073/pnas.0811058106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hicks SW, Galan JE. Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr Opin Microbiol. 2010;13:41–46. doi: 10.1016/j.mib.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaughlin LM, et al. The Salmonella SPI2 effector SseI mediates long-term systemic infection by modulating host cell migration. PLoS Pathog. 2009;5:e1000671. doi: 10.1371/journal.ppat.1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worley MJ, Nieman GS, Geddes K, Heffron F. Salmonella typhimurium disseminates within its host by manipulating the motility of infected cells. Proc Natl Acad Sci U S A. 2006;103:17915–17920. doi: 10.1073/pnas.0604054103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- 16.Towler DA, Eubanks SR, Towery DS, Adams SP, Glaser L. Amino-terminal processing of proteins by N-myristoylation. Substrate specificity of N-myristoyl transferase. J Biol Chem. 1987;262:1030–1036. [PubMed] [Google Scholar]
- 17.Nimchuk Z, et al. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell. 2000;101:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- 18.Lewis JD, Abada W, Ma W, Guttman DS, Desveaux D. The HopZ family of Pseudomonas syringae type III effectors require myristoylation for virulence and avirulence functions in Arabidopsis thaliana. J Bacteriol. 2008;190:2880–2891. doi: 10.1128/JB.01702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert-Seilaniantz A, Shan L, Zhou JM, Tang X. The Pseudomonas syringae pv. tomato DC3000 type III effector HopF2 has a putative myristoylation site required for its avirulence and virulence functions. Mol Plant Microbe Interact. 2006;19:130–138. doi: 10.1094/MPMI-19-0130. [DOI] [PubMed] [Google Scholar]
- 20.Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 21.Marathe R, Dinesh-Kumar SP. Plant defense: one post, multiple guards?! Mol Cell. 2003;11:284–286. doi: 10.1016/s1097-2765(03)00072-8. [DOI] [PubMed] [Google Scholar]
- 22.Dowen RH, Engel JL, Shao F, Ecker JR, Dixon JE. A family of bacterial cysteine protease type III effectors utilizes acylation-dependent and -independent strategies to localize to plasma membranes. J Biol Chem. 2009;284:15867–15879. doi: 10.1074/jbc.M900519200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin DD, Beauchamp E, Berthiaume LG. Post-translational myristoylation: Fat matters in cellular life and death. Biochimie. 2011;93:18–31. doi: 10.1016/j.biochi.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 24.Shao F, et al. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 25.Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109:575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- 26.Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 27.Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 28.Dai Q, et al. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J Biol Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- 29.Ivanov SS, Charron G, Hang HC, Roy CR. Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J Biol Chem. 2010;285:34686–34698. doi: 10.1074/jbc.M110.170746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price CT, Al-Quadan T, Santic M, Jones SC, Abu Kwaik Y. Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J Exp Med. 2010;207:1713–1726. doi: 10.1084/jem.20100771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price CT, Jones SC, Amundson KE, Kwaik YA. Host-mediated post-translational prenylation of novel dot/icm-translocated effectors of legionella pneumophila. Front Microbiol. 2010;1:131. doi: 10.3389/fmicb.2010.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinicke AT, et al. A Salmonella typhimurium effector protein SifA is modified by host cell prenylation and S-acylation machinery. J Biol Chem. 2005;280:14620–14627. doi: 10.1074/jbc.M500076200. [DOI] [PubMed] [Google Scholar]
- 33.Boucrot E, Beuzon CR, Holden DW, Gorvel JP, Meresse S. Salmonella typhimurium SifA effector protein requires its membrane-anchoring C-terminal hexapeptide for its biological function. J Biol Chem. 2003;278:14196–14202. doi: 10.1074/jbc.M207901200. [DOI] [PubMed] [Google Scholar]
- 34.Stein MA, Leung KY, Zwick M, Garcia-del Portillo F, Finlay BB. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol Microbiol. 1996;20:151–164. doi: 10.1111/j.1365-2958.1996.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 35.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 36.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 39.Huang J, Xu LG, Liu T, Zhai Z, Shu HB. The p53-inducible E3 ubiquitin ligase p53RFP induces p53-dependent apoptosis. FEBS Lett. 2006;580:940–947. doi: 10.1016/j.febslet.2005.09.105. [DOI] [PubMed] [Google Scholar]
- 40.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubori T, Nagai H. Bacterial effector-involved temporal and spatial regulation by hijack of the host ubiquitin pathway. Front Microbiol. 2011;2:145. doi: 10.3389/fmicb.2011.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins CA, Brown EJ. Cytosol as battleground: ubiquitin as a weapon for both host and pathogen. Trends Cell Biol. 2010;20:205–213. doi: 10.1016/j.tcb.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Kubori T, Galan JE. Temporal regulation of salmonella virulence effector function by proteasome-dependent protein degradation. Cell. 2003;115:333–342. doi: 10.1016/s0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- 45.Kubori T, Shinzawa N, Kanuka H, Nagai H. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 2010;6:e1001216. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel JC, Galan JE. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J Cell Biol. 2006;175:453–463. doi: 10.1083/jcb.200605144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steele-Mortimer O, et al. Activation of Akt/protein kinase B in epithelial cells by the Salmonella typhimurium effector sigD. J Biol Chem. 2000;275:37718–37724. doi: 10.1074/jbc.M008187200. [DOI] [PubMed] [Google Scholar]
- 48.Hernandez LD, Hueffer K, Wenk MR, Galan JE. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science. 2004;304:1805–1807. doi: 10.1126/science.1098188. [DOI] [PubMed] [Google Scholar]
- 49.Knodler LA, Winfree S, Drecktrah D, Ireland R, Steele-Mortimer O. Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cell Microbiol. 2009;11:1652–1670. doi: 10.1111/j.1462-5822.2009.01356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel JC, Hueffer K, Lam TT, Galan JE. Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell. 2009;137:283–294. doi: 10.1016/j.cell.2009.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315:683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 52.von Zastrow M, Sorkin A. Signaling on the endocytic pathway. Curr Opin Cell Biol. 2007;19:436–445. doi: 10.1016/j.ceb.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Matteis MA, Godi A. PI-loting membrane traffic. Nat Cell Biol. 2004;6:487–492. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- 54.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 55.Michell RH. Inositol derivatives: evolution and functions. Nat Rev Mol Cell Biol. 2008;9:151–161. doi: 10.1038/nrm2334. [DOI] [PubMed] [Google Scholar]
- 56.Pizarro-Cerda J, Cossart P. Subversion of phosphoinositide metabolism by intracellular bacterial pathogens. Nat Cell Biol. 2004;6:1026–1033. doi: 10.1038/ncb1104-1026. [DOI] [PubMed] [Google Scholar]
- 57.Weber SS, Ragaz C, Hilbi H. Pathogen trafficking pathways and host phosphoinositide metabolism. Mol Microbiol. 2009;71:1341–1352. doi: 10.1111/j.1365-2958.2009.06608.x. [DOI] [PubMed] [Google Scholar]
- 58.Weber SS, Ragaz C, Reus K, Nyfeler Y, Hilbi H. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2006;2:e46. doi: 10.1371/journal.ppat.0020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ragaz C, et al. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol. 2008;10:2416–2433. doi: 10.1111/j.1462-5822.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- 60.Brombacher E, et al. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem. 2009;284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 62.Jank T, et al. Domain organization of Legionella effector SetA. Cell Microbiol. 2012;14:852–868. doi: 10.1111/j.1462-5822.2012.01761.x. [DOI] [PubMed] [Google Scholar]
- 63.Rabin SD, Veesenmeyer JL, Bieging KT, Hauser AR. A C-terminal domain targets the Pseudomonas aeruginosa cytotoxin ExoU to the plasma membrane of host cells. Infect Immun. 2006;74:2552–2561. doi: 10.1128/IAI.74.5.2552-2561.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gendrin C, et al. Structural basis of cytotoxicity mediated by the type III secretion toxin ExoU from Pseudomonas aeruginosa. PLoS Pathog. 2012;8:e1002637. doi: 10.1371/journal.ppat.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finck-Barbancon V, et al. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 66.Allewelt M, Coleman FT, Grout M, Priebe GP, Pier GB. Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect Immun. 2000;68:3998–4004. doi: 10.1128/iai.68.7.3998-4004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 68.Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 69.Wiedemann N, Frazier AE, Pfanner N. The protein import machinery of mitochondria. J Biol Chem. 2004;279:14473–14476. doi: 10.1074/jbc.R400003200. [DOI] [PubMed] [Google Scholar]
- 70.Guttman DS, et al. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science. 2002;295:1722–1726. doi: 10.1126/science.295.5560.1722. [DOI] [PubMed] [Google Scholar]
- 71.Kenny B, Jepson M. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell Microbiol. 2000;2:579–590. doi: 10.1046/j.1462-5822.2000.00082.x. [DOI] [PubMed] [Google Scholar]
- 72.Nougayrede JP, Donnenberg MS. Enteropathogenic Escherichia coli EspF is targeted to mitochondria and is required to initiate the mitochondrial death pathway. Cell Microbiol. 2004;6:1097–1111. doi: 10.1111/j.1462-5822.2004.00421.x. [DOI] [PubMed] [Google Scholar]
- 73.Papatheodorou P, et al. The enteropathogenic Escherichia coli (EPEC) Map effector is imported into the mitochondrial matrix by the TOM/Hsp70 system and alters organelle morphology. Cell Microbiol. 2006;8:677–689. doi: 10.1111/j.1462-5822.2005.00660.x. [DOI] [PubMed] [Google Scholar]
- 74.Alto NM, et al. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 75.Bulgin R, et al. Bacterial guanine nucleotide exchange factors SopE-like and WxxxE effectors. Infect Immun. 2010;78:1417–1425. doi: 10.1128/IAI.01250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hodges K, Alto NM, Ramaswamy K, Dudeja PK, Hecht G. The enteropathogenic Escherichia coli effector protein EspF decreases sodium hydrogen exchanger 3 activity. Cell Microbiol. 2008;10:1735–1745. doi: 10.1111/j.1462-5822.2008.01163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simpson N, et al. The enteropathogenic Escherichia coli type III secretion system effector Map binds EBP50/NHERF1: implication for cell signalling and diarrhoea. Mol Microbiol. 2006;60:349–363. doi: 10.1111/j.1365-2958.2006.05109.x. [DOI] [PubMed] [Google Scholar]
- 78.Alto NM, et al. The type III effector EspF coordinates membrane trafficking by the spatiotemporal activation of two eukaryotic signaling pathways. J Cell Biol. 2007;178:1265–1278. doi: 10.1083/jcb.200705021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dean P, et al. The enteropathogenic E. coli effector EspF targets and disrupts the nucleolus by a process regulated by mitochondrial dysfunction. PLoS Pathog. 2010;6:e1000961. doi: 10.1371/journal.ppat.1000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marches O, et al. EspF of enteropathogenic Escherichia coli binds sorting nexin 9. J Bacteriol. 2006;188:3110–3115. doi: 10.1128/JB.188.8.3110-3115.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Batchelor M, et al. Structural basis for recognition of the translocated intimin receptor (Tir) by intimin from enteropathogenic Escherichia coli. EMBO J. 2000;19:2452–2464. doi: 10.1093/emboj/19.11.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu H, et al. Point mutants of EHEC intimin that diminish Tir recognition and actin pedestal formation highlight a putative Tir binding pocket. Mol Microbiol. 2002;45:1557–1573. doi: 10.1046/j.1365-2958.2002.03137.x. [DOI] [PubMed] [Google Scholar]
- 83.Luo Y, et al. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature. 2000;405:1073–1077. doi: 10.1038/35016618. [DOI] [PubMed] [Google Scholar]
- 84.Malish HR, et al. Potential role of the EPEC translocated intimin receptor (Tir) in host apoptotic events. Apoptosis. 2003;8:179–190. doi: 10.1023/a:1022974710488. [DOI] [PubMed] [Google Scholar]
- 85.Wong AR, et al. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol Microbiol. 2011;80:1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x. [DOI] [PubMed] [Google Scholar]
- 86.Holmes A, Muhlen S, Roe AJ, Dean P. The EspF effector, a bacterial pathogen's Swiss army knife. Infect Immun. 2010;78:4445–4453. doi: 10.1128/IAI.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pistor S, Chakraborty T, Niebuhr K, Domann E, Wehland J. The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton. EMBO J. 1994;13:758–763. doi: 10.1002/j.1460-2075.1994.tb06318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 89.Gorlich D, et al. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 90.Gorlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 91.Gorlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 92.Lee SJ, Matsuura Y, Liu SM, Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 93.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 94.Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 95.Marois E, Van den Ackerveken G, Bonas U. The xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol Plant Microbe Interact. 2002;15:637–646. doi: 10.1094/MPMI.2002.15.7.637. [DOI] [PubMed] [Google Scholar]
- 96.Van den Ackerveken G, Marois E, Bonas U. Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell. 1996;87:1307–1316. doi: 10.1016/s0092-8674(00)81825-5. [DOI] [PubMed] [Google Scholar]
- 97.Lahaye T, Bonas U. Molecular secrets of bacterial type III effector proteins. Trends Plant Sci. 2001;6:479–485. doi: 10.1016/s1360-1385(01)02083-0. [DOI] [PubMed] [Google Scholar]
- 98.Yang Y, Gabriel DW. Xanthomonas avirulence/pathogenicity gene family encodes functional plant nuclear targeting signals. Mol Plant Microbe Interact. 1995;8:627–631. doi: 10.1094/mpmi-8-0627. [DOI] [PubMed] [Google Scholar]
- 99.Szurek B, Marois E, Bonas U, Van den Ackerveken G. Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 2001;26:523–534. doi: 10.1046/j.0960-7412.2001.01046.x. [DOI] [PubMed] [Google Scholar]
- 100.Citovsky V, Zupan J, Warnick D, Zambryski P. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science. 1992;256:1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 101.Zupan JR, Citovsky V, Zambryski P. Agrobacterium VirE2 protein mediates nuclear uptake of single-stranded DNA in plant cells. Proc Natl Acad Sci U S A. 1996;93:2392–2397. doi: 10.1073/pnas.93.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Howard EA, Zupan JR, Citovsky V, Zambryski PC. The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell. 1992;68:109–118. doi: 10.1016/0092-8674(92)90210-4. [DOI] [PubMed] [Google Scholar]
- 103.Rossi L, Hohn B, Tinland B. The VirD2 protein of Agrobacterium tumefaciens carries nuclear localization signals important for transfer of T-DNA to plant. Mol Gen Genet. 1993;239:345–353. doi: 10.1007/BF00276932. [DOI] [PubMed] [Google Scholar]
- 104.Ballas N, Citovsky V. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc Natl Acad Sci U S A. 1997;94:10723–10728. doi: 10.1073/pnas.94.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Citovsky V, Warnick D, Zambryski P. Nuclear import of Agrobacterium VirD2 and VirE2 proteins in maize and tobacco. Proc Natl Acad Sci U S A. 1994;91:3210–3214. doi: 10.1073/pnas.91.8.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nagai T, Abe A, Sasakawa C. Targeting of enteropathogenic Escherichia coli EspF to host mitochondria is essential for bacterial pathogenesis: critical role of the 16th leucine residue in EspF. J Biol Chem. 2005;280:2998–3011. doi: 10.1074/jbc.M411550200. [DOI] [PubMed] [Google Scholar]
- 107.Hartland EL, et al. Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol Microbiol. 1999;32:151–158. doi: 10.1046/j.1365-2958.1999.01338.x. [DOI] [PubMed] [Google Scholar]
- 108.Murata T, et al. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 109.Block A, et al. The Pseudomonas syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity. Cell Microbiol. 2010;12:318–330. doi: 10.1111/j.1462-5822.2009.01396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hernandez LD, Pypaert M, Flavell RA, Galan JE. A Salmonella protein causes macrophage cell death by inducing autophagy. J Cell Biol. 2003;163:1123–1131. doi: 10.1083/jcb.200309161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Layton AN, Brown PJ, Galyov EE. The Salmonella translocated effector SopA is targeted to the mitochondria of infected cells. J Bacteriol. 2005;187:3565–3571. doi: 10.1128/JB.187.10.3565-3571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zou H, et al. Identification of an avirulence gene, avrxa5, from the rice pathogen Xanthomonas oryzae pv. oryzae. Sci China Life Sci. 2010;53:1440–1449. doi: 10.1007/s11427-010-4109-y. [DOI] [PubMed] [Google Scholar]
- 113.Yang B, Zhu W, Johnson LB, White FF. The virulence factor AvrXa7 of Xanthomonas oryzae pv. oryzae is a type III secretion pathway-dependent nuclear-localized double-stranded DNA-binding protein. Proc Natl Acad Sci U S A. 2000;97:9807–9812. doi: 10.1073/pnas.170286897. [DOI] [PMC free article] [PubMed] [Google Scholar]